Abstract
2D nanoslit devices, where two crystals with atomically flat surfaces are separated by only a few nanometers, have attracted considerable attention because their tunable control over the confinement allows for the discovery of unusual transport behavior of gas, water, and ions. Here, the passage of double‐stranded DNA molecules is studied through nanoslits fabricated from exfoliated 2D materials, such as graphene or hexagonal boron nitride, and the DNA polymer behavior is examined in this tight confinement. Two types of events are observed in the ionic current: long current blockades that signal DNA translocation and short spikes where DNA enters the slits but withdraws. DNA translocation events exhibit three distinct phases in their current‐blockade traces—loading, translation, and exit. Coarse‐grained molecular dynamics simulation allows the different polymer configurations of these phases to be identified. DNA molecules, including folds and knots in their polymer structure, are observed to slide through the slits with near‐uniform velocity without noticeable frictional interactions of DNA with the confining graphene surfaces. It is anticipated that this new class of 2D‐nanoslit devices will provide unique ways to study polymer physics and enable lab‐on‐a‐chip biotechnology.
Keywords: 2D nanoslits, biopolymers, DNA translocation, graphene, nanofluidics
The passage of double‐stranded DNA molecules through nanoslits fabricated from exfoliated 2D materials, such as graphene or hexagonal boron nitride, is studied, and the DNA polymer behavior in this tight confinement is examined. DNA molecules, including folds and knots, are observed to slide through the slits with near‐uniform velocity without noticeable frictional interactions with the confining graphene surfaces.
The translocation of biopolymers through nanoscale constrictions such as pores and channels has inspired a new class of lab‐on‐a‐chip sensors that can detect, sort, and process DNA, RNA, and proteins for diagnostics and sequencing applications.[ 1 , 2 , 3 , 4 ] In particular, nanopores have been used abundantly for biophysics studies and DNA sequencing,[ 5 ] while lab‐on‐a‐chip platforms allow extensive microfluidic integration of different sensors and reactions.[ 6 ] Such methods require nanoscale manipulation and an understanding of the physics governing the transport of biopolymers. Despite decades of research efforts to design and fabricate different geometric confinements[ 7 ] to probe various aspects of the transport process, the fundamentals of the biopolymer transport phenomena through artificial nanochannels have not been fully resolved. One challenge is the multitude of forces involved in the transport process on the nanoscale. Molecular transport is driven by the interplay of entropic, electroosmotic, and electrophoretic forces experienced by the biopolymers.[ 7 , 8 , 9 , 10 , 11 , 12 ] For example, nanoconfinement‐induced entropic barriers hinder the insertion of large DNA polymer coils driven by the electrophoretic force into much smaller nanopores and channels that can be as small as the length scale of natural biochannels and porins. Another challenge lies in mimicking smooth and atomically precise surfaces that would allow researchers to disentangle the intrinsic polymer behavior from surface interactions.[ 13 ] Silicon nitride/oxide‐based substrates have been extensively used for nanofluidic channels to translocate biopolymers, but they suffer from significant (few nanometers root mean square (rms)) surface roughness and inhomogeneous surfaces.[ 14 , 15 , 16 ] Attempts with carbon nanotubes (CNTs), which feature smooth inner surfaces, face challenges of device integration and leakage due to the stringent requirements of a perfect seal as well as a lack of systematic control over the channel length and diameter.[ 17 , 18 , 19 ] Until recently, it was impossible to fabricate individual artificial channels with atomic scale dimensions or even sub‐nanometric size tunable nanofluidic devices which has hindered the further development of the field.[ 20 ]
Here we report the first study of DNA transport through 2D nanoslits with precisely designed dimensions and atomically smooth surfaces.[ 21 ] The nanoslits were fabricated using exfoliated 2D materials such as graphene or hexagonal boron nitride (hBN). They were assembled in a trilayer stack (Figure 1a), where the basal planes of the two crystals (top and bottom) provide atomically flat walls, while the third crystal with a precise controlled thickness serves as a spacer layer that separates these walls. The height of the confinement was determined by the thickness of the spacer layer that can be set at any value from a monolayer (0.34 nm) to tens of nanometers. The entire crystal can then be etched down to the desired length of the slit. Entry and exit to the slits were provided through out‐of‐plane bulk access reservoirs. Previous studies have demonstrated remarkable water‐, ion‐, and gas‐transport properties[ 21 , 22 , 23 ] of such devices. Furthermore, the surface properties and chemical interactions of such nanoslits can be tuned for studying various surface interactions and sensor applications. Exploration of the DNA transport properties through nanoslits may experimentally answer questions about the strength of DNA–graphene interactions hypothesized by many previous studies[ 1 , 24 , 25 , 26 ] and allow studies of DNA in strongly confined 2D slits on scales not previously probed by conventional nanofluidic devices.
Figure 1.

DNA translocation through 2D‐nanoslit devices. a) Cross‐sectional view of a 2D‐nanoslit device in the DNA translocation setup. DNA is introduced from the cis (negatively biased) side and a positive voltage is applied to drive the DNA through the slit. b) Oblique‐view schematic of the 2D nanoslit. The device is made by sandwiching graphene spacers between top and bottom layers of graphite crystals to form an atomically smooth surface within the slit. The graphene spacer defines the height of the device to an accuracy of a single layer—0.34 nm. The entire graphene crystal was then masked and etched perpendicularly to define the length of the slit. The final device geometry was w = 110 nm, h = 6.5 nm, l = 400 nm. c) Typical I–V curve of our 2D‐nanoslit device at 1 m LiCl. The conductance is linear across this range. The measured conductance of our graphene device is 1.2 × 10−8 S.
The 2D‐nanoslit devices were fabricated following the procedure previously outlined by Keerthi et al.[ 22 ] (for details, see the Supporting Information). Schematically, the device is represented in Figure 1b. The spacer that controls the height of the nanoslit is made from few‐layer thick graphene. To smooth out any roughness coming from the supporting SiN surface, thicker stacks of graphene were used as the bottom (≈20–30 nm) and top crystals (200 nm). The length, width, and height, of the major 2D‐nanoslit device that we here report on was l = 400 nm, w = 110 nm, h = 6.5 nm, respectively (for other devices, see the Supporting Information). To facilitate the detailed detection and analysis of translocation events of individual DNA molecules, our device contained only a single nanoslit. First, using 1 m LiCl solution, we checked that the current‐voltage characteristic was linear across the +300 to −300 mV range (Figure 1c). From the slope of the I–V graph, we measured a slit conductance of 1.2 × 10−8 S—in line with the expected conductance of 1.9 × 10−8 S as estimated from the bulk ionic conductivity and geometric size. The 2D‐nanoslit devices could be filled properly and did not show intrinsic instabilities. When left at a constant voltage (300 mV), the device showed a stable open slit current for many hours (overnight). The devices were extremely stable and could be used for weeks. For the DNA translocation experiments, the initial salt solution was replaced with 4 m LiCl in order to increase the signal‐to‐noise resolution of the currents.[ 27 ]
Upon addition of DNA (5 kbp, linear dsDNA) to the entry (cis, negative‐voltage‐biased) side of the device, we observed clear temporary reductions in the ionic current through the 2D‐nanoslit device at 300 mV. As is well known from the field of nanopores, such current events indicate the presence of DNA in the slit.[ 28 ] Events are characterized by their average blockade current and the time that the DNA spends within the slit. Figure 2a plots these quantities in a scatter plot (N = 796), where each dot represents a separate single‐molecule event collected from our graphene device with the dimensions reported in the preceding paragraph (l = 400 nm, w = 110 nm, h = 6.5 nm). Two main populations appear: a first one that we attribute to the translocation of DNA through the slit (termed “DNA translocation region”), and a second region that comprises of much faster transient events (termed “spike region”). These two populations of events are separated by a line that represent a threshold value obtained by integrating the average current blockade level over the total time of the event (0.2 nA ms in this case)—similar to DNA event detection in nanopore experiments[ 29 ] where translocations are separated from failed attempts by this threshold. Conveniently, this threshold value is independent of the folding conformation of DNA during the translocation. We interpret the high‐amplitude (above threshold) dips in the current trace with a long passage of time (>1 ms) as events where the DNA molecule fully translocates through the slit. Here, DNA is electrophoretically driven across the slit, temporarily displacing ions within the slit, which leads to a drop in the measured current. This process is directly analogous to the blockade of the ionic current in solid state nanopores upon passage of DNA, but the dips are much longer here (tens of milliseconds versus ≈0.5 ms for nanopores) owing to the length of the 2D‐nanoslit which exceeds the typical nanopore channel length by about two orders of magnitude. Example current traces are shown in Figure 2b,c where we see individual dips that last from a few ms to more than 60 ms. Note that the event rate is low (less than 1 event per second), indicating that co‐translocation of two DNA are extremely unlikely. Translocation times were broadly distributed, with a median translocation time of 8.4 ms (Figure 2a insert).
Figure 2.

Current blockades produced by DNA in 2D‐nanoslit devices. a) Scatter plot of the average current blockade versus the time of DNA events. The data was collected from a nanoslit device with graphene top and bottom layers which is schematically drawn in Figure 1b with the following dimensions of l = 400 nm, w = 110 nm, h = 6.5 nm. Two regions can be seen. The first is a DNA translocation region, which is marked by deep current blockades and long passage times (red). The inset shows a histogram of the event duration, with a median at 8.4 ms. The second region is the spike region (blue) marked by small current blockades and fast dwell times. b) Representative current traces for DNA translocation events (top, red) and a spike event (bottom, blue). We interpret the latter events as DNA probing the entrance of the slit but withdrawing due to entropic force. Data were filtered with a 3 kHz low pass filter. c) Six representative current traces produced by DNA translocation. The traces have an armchair shape and vary in the length and depth of the current blockades.
Closer examination of the DNA translocation events showed that most events had a characteristic armchair‐like shape, displayed in the example events of Figure 2c, with a linear rise, plateau, and a linear decay. The rise, dwell, and fall times as well as the maximum blockade amplitude reached during the event varied vastly. The blockade current ranged between 0.5 and 1% of the open slit current, and a large spread in the total dwell time and average blockade current was observed (over 2 orders of magnitude in time and between 0.07 and 0.3 nA). We hypothesize that these variations originate from different number of DNA loops that were captured from the randomly oriented polymer blob and simultaneously translocated through the nanoslit—a scenario that we will further examine below. Remarkably, we did not see any clogging of the 2D‐nanoslit and devices remained stable for hours showing clear DNA translocations, suggesting that any interactions of the DNA with the graphene surface were transient in nature.
Turning to the second population of observed events (Figure 2b, spikes), these events had a distinctive spike‐like shape and exhibited smaller blockade current amplitudes and shorter dwell times. We interpret these blockades as transient excursions of DNA into the entrance of the nanoslit that did not result into complete translocations, but rather, the DNA withdrew back into the entry reservoir. Similar phenomena have been reported in a variety of nanofluidic experiments with microslits, nanopores, and other micro‐ or nanofluidic devices that presented an entropic barrier to polymer entry.[ 11 , 30 , 31 ] A polymer can access way less configurations within a narrow slit than in free solution, and hence the entry into the slit presents an entropic barrier. This barrier will be lower for slits with taller heights (h) and for shorter DNA.[ 32 ] Shorter DNA therefore is expected to yield more translocation events. We verified this with DNA of different lengths, 10 and 1 kbp, in nanoslits of similar dimensions (see the Supporting Information): while the 10 kbp DNA exhibited no translocation events but instead exclusively probing spike events, abundant translocation events were observed when 1 kbp DNA was used (see the Supporting Information).
In order to evaluate the DNA translocation process in microscopic detail, we simulated the DNA translocation through graphene slits using coarse‐grained molecular dynamic simulations. The device geometry and size, driving voltages, and salt conditions from the experiments were all recreated in the simulation setup (Figure 3a). Our simulations employed a custom coarse‐grained model of DNA[ 33 ] and represented the graphene nanostructure as a frictionless repulsive potential. Prior to translocation, a 5 kbp DNA molecule was equilibrated in the 160 nm × 110 nm × 500 nm entry reservoir near the entrance of the slit to generate a random starting configuration of the DNA. A 3D electrostatic potential, as determined from continuum calculations, was applied to drive the DNA through the nanoslit. Indeed, the DNA was captured into and translocated through the slit. Instantaneous DNA conformations were used to compute the blockade current by means of a steric exclusion model[ 34 ] yielding a current profile of the entire event. Note that, as is generally found,[ 35 ] the timescales of events observed in the molecular dynamics simulations are typically faster than the nominal timescales by several orders of magnitudes and thereby cannot directly be compared to the dwell times in actual experiments. Figure 3b shows three typical current traces obtained from a simulation of a DNA translocation (See the Supporting Information more examples from simulations).
Figure 3.

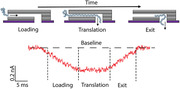
MD simulation of DNA translocation through a 2D nanoslit. a) To‐scale coarse‐grained model of the experimental 2D‐nanoslit device. b) Ionic‐current‐blockade traces obtained from the simulations of DNA translocation through the slit at 1 V. Events typically showed a ≈1–2% change of the open slit current and had an armchair shape (See the Supporting Information for other simulation runs). c) Video stills from one simulation trajectory showing the three stages of a DNA translocation event—loading, translation and exit—and the corresponding three regions in the armchair current blockade. d) Zoomed‐in view of a simulated current blockade trace at 1 V. e) Zoomed‐in view of an experimental current blockade trace that matches the armchair shape of the simulated current trace well. f) Snapshots of a simulation showing the DNA polymer exploring and probing the entrance of the slit at four different time points. g) Current‐blockade signal for a DNA probing event. Similar to experimentally obtained traces, the current blockade shows up as quick spikes with a low current‐blockade amplitude.
Current traces typically showed a ≈1–2% decrease relative to the open slit current and generally had an arm‐chair shape. Video stills captured at three different time points (Figure 3c) illustrate different stages of the DNA translocation process, which we can directly relate to the current traces obtained from the experiment. The gradual entry of DNA into the slit decreases the current (“loading”) until the leading segment exits the slit. From that point on, the amount of DNA in the slit remains approximately constant leading to a plateau in the current blockade (“translation”). Eventually, the trailing segments of the DNA enter the slit to translocate as well, which leads to a return to the open slit current (“exit”). Taken together, our simulations reproduce and explain the typical armchair shape of current blockade traces (Figure 3d) that was observed in the experiments (Figures 3e and 2c).
The simulations also allowed us to investigate the capture process of the DNA in greater detail. Upon initialization of the simulation, the DNA did not immediately get captured into the slit for translocation even under a strong applied voltage. Instead, segments of the DNA were observed to probe the entry of the slit with a partial insertion which subsequently retracted back into the reservoir, a phenomenon that occurred repeatedly. Figure 3f shows four video stills from such a process. Eventually, the DNA overcame the entropic barrier and fully translocated through the slit. Figure 3g shows the calculated current for such probing processes. Generally, much smaller blockade currents (<0.3% of the open slit current) were observed for the probing event that lasted much shorter than complete translocations. These findings match well with the results obtained from the experiments in the spike region.
The DNA transits from a 3D configuration in the polymer blob at the entrance of the slit to a significantly confined arrangement in the slit, as the slit height is much less than the persistence length of DNA (i.e., DNA persistence length ≈ 50 nm, slit height ≈ 6.5 nm), which forces the DNA into a 2D configuration. During this process, we were able to observe the capture and translocation of DNA features such as folds, loops, and knots in our simulations. For example, a series of video stills (Figure 4a) shows a DNA loop, which formed spontaneously upon the DNA capture, translocating through the slit (as marked by the dashed red box). The observation of such features is surprising as one might think that forces (i.e., the combination of electrophoretic, entropic, and frictional surface‐interaction forces) in the translocation process would pull the DNA taut and smooth out any of these features. Contrary to such expectations, loops did not unfold or get stretched out by forces experienced by the DNA during the translocation process, but instead appeared to “slide” through the slit without much internal rearrangements.
Figure 4.

DNA dynamics in 2D‐nanoslit. a) Video stills of a DNA loop translocating through the 2D nanoslit (top view). Upon capture into the slit, folds and loops along the DNA polymer chain (shown in the red dotted rectangle) are preserved throughout the entire translocation event. b) Distance versus time curves for every 100 bp segments along the same DNA molecule. The origin of the Y axis is defined to be at the entrance of the slit. Three segments are highlighted to show the general trend: green—leading segment, i.e., first segment of DNA to enter the nanoslit; cyan—middle segment; and red—trailing segment, the last one to enter the slit. Segments that were close together did not vary much in their displacement, which preserved local DNA conformations, such as loop and folds, throughout the translocation. c) Calculated velocity of DNA during the translocation. The maximum velocity increases for the trailing segments of the DNA. d) Snapshots illustrating the diversity of DNA polymer conformations in the simulations and the corresponding ionic current traces. The video stills correspond to the time point marked by the arrow in the current trace (full videos in the Supporting Information). Similar current traces were experimentally observed in the translocation measurements, confirming that complex DNA conformations can translocate through the 2D nanoslit without becoming unraveled.
To gain a more quantitative understanding of the translocation dynamics of folds in the DNA, we plotted the displacement versus time curve separately for each of the subsequent 100 bp segments comprising the DNA molecule (Figure 4b). Three such DNA segments are highlighted in particular, the leading DNA segment (the segment that first entered the slit), the middle segment (halfway during the translocation process), and the trailing segment (last segment of DNA that entered the slit). Neighboring segments are observed to have rather similar displacement curves, which indeed acts to preserve particular DNA features such as a loop or a fold throughout the translocation, as the segments slide together along the nanoslit in a frictionless manner with a near‐constant velocity. Second, while the first strand always gets pulled into the slit from a volume near the slit entrance, later segments are pulled in from further out of the reservoir (negative distances denote distances away from the slit into the entry reservoir, Figure 4b). The plot of the segment's velocities (Figure 4c) shows that nearby segments travel through the slit at nearly the same speed, suggesting that they experienced similar forces. However, there is a noticeable (≈25%) increase in the maximum velocity that is attained by the segments towards the end of the DNA event. This can be attributed to a decrease in the amount of the DNA that still is residing in the entry reservoir before the slit, as more and more DNA is unwounded and loaded into the channel, yielding a reduced drag arising from pulling the remaining polymer blob and hence an increased velocity for the DNA translocating through the slit. We note that this drag force is the only observed factor affecting the otherwise near constant velocity transport of the DNA.
Armed with these microscopic insights, we are able to explain almost all features seen in the experimental current traces. We returned to the experiments and realized that the large variation in amplitudes reached by the different current traces are likely due to the presence of a varying number of folds, loops, or knots in the DNA polymer that reside within the slit.[ 36 , 37 ] Current traces from the simulations gave us hints on how to identify such features, for example an abrupt change in the current level. We illustrate this with three event types from the simulations in Figure 4d, with corresponding experimental traces that exhibit the same profile in the current blockade (full videos are provided in the Supporting Information). First, current traces often exhibited multiple plateau levels, for example the one in Figure 4d top (denoted 2–1–2 event) where the current started with a plateau level that subsequently decreased by a factor of 2, to subsequently return to the plateau value again. These events are due to DNA that is translocating in a folded manner before straightening out into a single helix segment in the middle of the event. This in fact can be used to identify the blockade level of an individual double stranded DNA molecule, to characterize and sort the rest of the translocation events, such as the 3–1 event shown in Figure 4d where a large DNA segment made of multiple folds slides along the channel middle (see the Supporting Information for the categorization of the events). Indeed, we also observed such 3–1 events in the experimental traces, which suggests an interaction‐free surface of our 2D‐nanoslits that allows these features to slide through. We also observed the translocation of a single DNA knot (Figure 4d) which shows up as an abrupt half‐unit step in both the simulation and experiment current traces.
Taken together, these traces indicate that the dominating force during the translocation is the electrophoretic force that acts equally on the entire DNA polymer that resides within the slit. This allows loops and folds to translocate through without being pulled internally and relaxed. It is remarkable that these features persist even under the extreme slit‐like confinement (6 nm) for at least ≈10 ms, a confinement and time regime not probed by conventional nanofluidic experiments. We do not observe any evidence of graphene–DNA interactions, contrary to other graphene devices.[ 24 , 25 , 26 ] Additional experiments in hBN nanoslits show similar translocation profiles as the events obtained from graphene devices, suggesting that the translocation of the DNA is dominated by the electrophoretic driving force and slides along the atomically smooth surface (as shown by AFM characterization reported in the Supporting Information). In a liquid environment, the graphene surface may pick up a slight negative surface charge from residual OH− groups which however will be readily screened by Li+ ions in the high‐salt buffer,[ 38 ] resulting in weak DNA–graphene interactions. The discrepancy with other reported graphene–DNA interaction may arise from the conventionally followed fabrication protocols in the literature that damage the graphene sheets, allowing for defect sites to interact with dsDNA,[ 39 , 40 , 41 ] whereas our approach directly benefits from the atomically flat graphene planes of the exfoliated 2D materials which are not postprocessed.
In summary, we studied translocation of DNA molecules through ultrathin nanoslits fabricated from 2D materials. Remarkably we did not require to apply any additional coating to overcome the clogging that is commonly reported in graphene nanopore devices, and our devices remained stable for hours.[ 42 ] Clear changes in the ionic current allowed us to identify three distinct phases of DNA translocation, namely loading, translation, and exit. The entry process is dominated by the entropic cost of confining the DNA into the 2D nanoslit. Coarse‐grained simulations provided a microscopic picture that was fully consistent with the experimentally obtained ionic current traces. Neighboring segments of the DNA slid together and translocated with the same velocity in a near‐frictionless manner. Towards the end of the translocation, DNA segments faced a weaker retarding force due to the lower amount of DNA residing outside the entrance, thereby yielding higher translocation velocities. The weak forces involved in the translocation process did not stretch the DNA during translocation but allowed folds and knots to persist inside the slit. The folds and knots slid through the entire length of the nanoslit suggesting that interactions with the graphene surface are transient and weak.
Looking ahead, the 2D nanoslits provide a novel tool for probing biopolymer properties as they constitute a precisely engineered confinement with atomically smooth surfaces. We envision that the use of optically transparent 2D materials should allow future 2D nanoslit devices to be integrated with optical microscopy for fluorescence‐based nanofluidic experiments. Such devices can be used to probe the evolution of DNA knots and folds under confinement with low surface interactions. An obvious next step for future investigations will be to study the interplay of entropy and polymer configuration under ultrahigh confinement by varying the height of the slits and the length of DNA in different salt concentrations and voltages. This may pave the way for future 2D sensing devices that can, for example, be used to separate different lengths of biopolymers. Furthermore, the use of these frictionless 2D nanoslits can be expected to shed light on the basic physics of biopolymer transport.
Experimental Section
DNA‐Sensing Experiments
2D nanoslit devices were loaded in a poly(ether ether ketone) (PEEK) flow cell. Ag/AgCl electrodes and an Axopatch 200B amplifier (Molecular Devices) was used for current detection. The traces were recorded at 100 kHz and further low pass filtered at 5 kHz for with the Transanalyzer Matlab package.[ 43 ]
For the DNA sensing experiments, 5 µL of 500 ng µL−1 of the stock solution (NoLimits Individual DNA Fragments, Thermo Fisher Scientific) was pipetted into the negatively‐biased cis compartment of the flow cell containing 500 µL of 4 m LiCl solution (buffered with 40 × 10−3 m Tris‐HCl, 4 × 10−3 m EDTA, pH 8). LiCl was used to maximize the translocation time and signal‐to‐noise (SNR) ratio as is commonly done in ionic sensing measurements. Li ions can bind strongly to the negative backbone of the DNA, screening most of the charge, and hence decreasing the electrophoretic force on the DNA.[ 27 ] This diluted the DNA to a final concentration of 1 ng µL−1 in the reservoir. The DNA was electrophoretically driven through the slit with a bias voltage of 300 mV and detected by measuring changes in the current flowing through the 2D slit.
MD Simulation
All simulations of dsDNA translocation through the nanoslit were performed using the Atomic Resolution Brownian Dynamics (ARBD) package.[ 44 ] COMSOL software (COMSOL Multiphysics 5.3a) was used to obtain the electrostatic potential that was applied in ARBD. Prior to translocation simulations, a 5000‐bp DNA strand was equilibrated in a 160 nm3 volume using a multi‐resolution simulation protocol[ 33 ] to create different DNA conformation. These conformations were then used to initialize the simulations of the dsDNA translocation. The pre‐obtained electrostatic potential was then used to drive the DNA into the slit and begin the translocation process.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Supporting Information
Supplemental Video 1
Supplemental Video 2
Supplemental Video 3
Supplemental Video 4
Supplemental Video 5
Supplemental Video 6
Acknowledgements
B.R. and A.K. acknowledge Dr. Alexander Zhukov for his help with FIB milling of the Si nitride apertures. C.D. would like to acknowledge funding from ERC grant SynDiv 669598 and the Netherlands Organization for Scientific Research (NWO/OCW) as part of the Frontiers of Nanoscience and Basyc programs. B.R. acknowledges the Royal Society fellowship and enhancement award RGF\EA\181000, and funding from the European Union's H2020 Framework Programme/ERC Starting Grant agreement number 852674—AngstroCAP. A.K. acknowledges the Ramsay Memorial Fellowship and the Royal Society research grant RGS\R2\202036. A.A. and A.C. acknowledge support through National Science Foundation (USA) under grant DMR‐1827346 and the National Institutes of Health under grant P41‐GM104601. Supercomputer time was provided through Leadership Resource Allocation MCB20012 on Frontera.
Yang W., Radha B., Choudhary A., You Y., Mettela G., Geim A. K., Aksimentiev A., Keerthi A., Dekker C., Translocation of DNA through Ultrathin Nanoslits. Adv. Mater. 2021, 33, 2007682. 10.1002/adma.202007682
Contributor Information
Aleksei Aksimentiev, Email: aksiment@illinois.edu.
Ashok Keerthi, Email: ashok.keerthi@manchester.ac.uk.
Cees Dekker, Email: c.dekker@tudelft.nl.
References
- 1. Heerema S. J., Dekker C., Nat. Nanotechnol. 2016, 11, 127. [DOI] [PubMed] [Google Scholar]
- 2. Kasianowicz J. J., Bezrukov S. M., Nat. Biotechnol. 2016, 34, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Howorka S., Siwy Z., ACS Nano 2016, 10, 9768. [DOI] [PubMed] [Google Scholar]
- 4. Persson F., Tegenfeldt J. O., Chem. Soc. Rev. 2010, 39, 985. [DOI] [PubMed] [Google Scholar]
- 5. Dekker C., Nat. Nanotechnol. 2007, 2, 209. [DOI] [PubMed] [Google Scholar]
- 6. Samiei E., Tabrizian M., Hoorfar M., Lab Chip 2016, 16, 2376. [DOI] [PubMed] [Google Scholar]
- 7. Reccius C. H., Stavis S. M., Mannion J. T., Walker L. P., Craighead H. G., Biophys. J. 2008, 95, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu B., Albertorio F., Hoogerheide D. P., Golovchenko J. A., Biophys. J. 2011, 101, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Dorp S., Keyser U. F., Dekker N. H., Dekker C., Lemay S. G., Nat. Phys. 2009, 5, 347. [Google Scholar]
- 10. Plesa C., van Loo N., Ketterer P., Dietz H., Dekker C., Nano Lett. 2015, 15, 732. [DOI] [PubMed] [Google Scholar]
- 11. Turner S. W. P., Cabodi M., Craighead H. G., Phys. Rev. Lett. 2002, 88, 4. [DOI] [PubMed] [Google Scholar]
- 12. Balducci A., Mao P., Han J., Doyle P. S., Macromolecules 2006, 39, 6273. [Google Scholar]
- 13. Salieb‐Beugelaar G. B., Teapal J., van Nieuwkasteele J., Wijnperlé D., Tegenfeldt J. O., Lisdat F., van den Berg A., Eijkel J. C. T., Nano Lett. 2008, 8, 1785. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Reisner W., Nanotechnology 2015, 26, 455301. [DOI] [PubMed] [Google Scholar]
- 15. Xia D., Yan J., Hou S., Small 2012, 8, 2787. [DOI] [PubMed] [Google Scholar]
- 16. Menard L. D., Ramsey J. M., Nano Lett. 2011, 11, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu H. H., He J., Tang J., Liu H. H., Pang P., Cao D., Krstic P., Joseph S., Lindsay S., Nuckolls C., Science 2010, 327, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holt J. K., Park H. G., Wang Y., Stadermann M., Artyukhin A. B., Grigoropoulos C. P., Noy A., Bakajin O., Science 2006, 312, 1034. [DOI] [PubMed] [Google Scholar]
- 19. Geng J., Kim K., Zhang J., Escalada A., Tunuguntla R., Comolli L. R., Allen F. I., Shnyrova A. V., Cho K. R., Munoz D., Wang Y. M., Grigoropoulos C. P., Ajo‐Franklin C. M., Frolov V. A., Noy A., Nature 2014, 514, 612. [DOI] [PubMed] [Google Scholar]
- 20. Bocquet L., Nat. Mater. 2020, 19, 254. [DOI] [PubMed] [Google Scholar]
- 21. Radha B., Esfandiar A., Wang F. C., Rooney A. P., Gopinadhan K., Keerthi A., Mishchenko A., Janardanan A., Blake P., Fumagalli L., Lozada‐Hidalgo M., Garaj S., Haigh S. J., Grigorieva I. V., Wu H. A., Geim A. K., Nature 2016, 538, 222. [DOI] [PubMed] [Google Scholar]
- 22. Keerthi A., Geim A. K., Janardanan A., Rooney A. P., Esfandiar A., Hu S., Dar S. A., Grigorieva I. V., Haigh S. J., Wang F. C., Radha B., Nature 2018, 558, 420. [DOI] [PubMed] [Google Scholar]
- 23. Mouterde T., Keerthi A., Poggioli A. R., Dar S. A., Siria A., Geim A. K., Bocquet L., Radha B., Nature 2019, 567, 87. [DOI] [PubMed] [Google Scholar]
- 24. Shankla M., Aksimentiev A., Nat. Nanotechnol. 2019, 14, 858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shankla M., Aksimentiev A., Nat. Commun. 2014, 5, 5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu M., Zhao H., Chen S., Yu H., Quan X., Chem. Commun. 2012, 48, 564. [DOI] [PubMed] [Google Scholar]
- 27. Kowalczyk S. W., Wells D. B., Aksimentiev A., Dekker C., Nano Lett. 2012, 12, 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J., Stein D., McMullan C., Branton D., Aziz M. J., Golovchenko J. A., Nature 2001, 412, 166. [DOI] [PubMed] [Google Scholar]
- 29. Storm A. J., Chen J. H., Zandbergen H. W., Dekker C., Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2005, 71, 051903. [DOI] [PubMed] [Google Scholar]
- 30. Cabodi M., Turner S. W. P., Craighead H. G., Anal. Chem. 2002, 74, 5169. [DOI] [PubMed] [Google Scholar]
- 31. Bell N. A. W., Muthukumar M., Keyser U. F., Phys. Rev. E 2016, 93, 022401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bonthuis D. J., Meyer C., Stein D., Dekker C., Phys. Rev. Lett. 2008, 101, 108303. [DOI] [PubMed] [Google Scholar]
- 33. Maffeo C., Aksimentiev A., Nucleic Acids Res. 2020, 48, 5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilson J., Sarthak K., Si W., Gao L., Aksimentiev A., ACS Sens. 2019, 4, 634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zwanzig R., Proc. Natl. Acad. Sci. USA 1988, 85, 2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plesa C., Verschueren D., Pud S., van der Torre J., Ruitenberg J. W., Witteveen M. J., Jonsson M. P., Grosberg A. Y., Rabin Y., Dekker C., Nat. Nanotechnol. 2016, 11, 1093. [DOI] [PubMed] [Google Scholar]
- 37. Sharma R. K., Agrawal I., Dai L., Doyle P. S., Garaj S., Nat. Commun. 2019, 10, 4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Esfandiar A., Radha B., Wang F. C., Yang Q., Hu S., Garaj S., Nair R. R., Geim A. K., Gopinadhan K., Science 2017, 358, 511. [DOI] [PubMed] [Google Scholar]
- 39. Boukhvalov D. W., Katsnelson M. I., Nano Lett. 2008, 8, 4373. [DOI] [PubMed] [Google Scholar]
- 40. Garaj S., Hubbard W., Reina A., Kong J., Branton D., Golovchenko J. A., Nature 2010, 467, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Banhart F., Kotakoski J., Krasheninnikov A. V., ACS Nano 2011, 5, 26. [DOI] [PubMed] [Google Scholar]
- 42. Schneider G. F., Xu Q., Hage S., Luik S., Spoor J. N. H., Malladi S., Zandbergen H., Dekker C., Nat. Commun. 2013, 4, 2619. [DOI] [PubMed] [Google Scholar]
- 43. Plesa C., Dekker C., Nanotechnology 2015, 26, 084003. [DOI] [PubMed] [Google Scholar]
- 44. Comer J., Aksimentiev A., J. Phys. Chem. C 2012, 116, 3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supplemental Video 1
Supplemental Video 2
Supplemental Video 3
Supplemental Video 4
Supplemental Video 5
Supplemental Video 6


