Abstract
Coronavirus (CoV) nucleocapsid (N) protein is a highly phosphorylated protein required for viral replication, but whether its phosphorylation and the related kinases are involved in the viral life cycle is unknown. We found the severe acute respiratory syndrome CoV N protein to be an appropriate system to address this issue. Using high resolution PAGE analysis, this protein could be separated into phosphorylated and unphosphorylated isoforms. Mass spectrometric analysis and deletion mapping showed that the major phosphorylation sites were located at the central serine-arginine (SR)-rich motif that contains several glycogen synthase kinase (GSK)-3 substrate consensus sequences. GSK-3-specific inhibitor treatment dephosphorylated the N protein, and this could be recovered by the constitutively active GSK-3 kinase. Immunoprecipitation brought down both N and GSK-3 proteins in the same complex, and the N protein could be phosphorylated directly at its SR-rich motif by GSK-3 using an in vitro kinase assay. Mutation of the two priming sites critical for GSK-3 phosphorylation in the SR-rich motif abolished N protein phosphorylation. Finally, GSK-3 inhibitor was found to reduce N phosphorylation in the severe acute respiratory syndrome CoV-infected VeroE6 cells and decrease the viral titer and cytopathic effects. The effect of GSK-3 inhibitor was reproduced in another coronavirus, the neurotropic JHM strain of mouse hepatitis virus. Our results indicate that GSK-3 is critical for CoV N protein phosphorylation and suggest that it plays a role in regulating the viral life cycle. This study, thus, provides new avenues to further investigate the specific role of N protein phosphorylation in CoV replication.
The causative pathogen for the epidemic severe acute respiratory syndrome (SARS)2 was identified as the SARS coronavirus (SCoV) in 2003 (1, 2). Its genome consists of a ∼30-kilobase positive-sense single-stranded RNA which encodes a 3′ co-terminal set of nine subgenomic mRNAs with a common leader sequence at their 5′ ends (3, 4). These subgenomic RNAs encode various structural and nonstructural proteins required to produce progeny virions, including the viral nucleocapsid (N) protein.
The SCoV N protein is the most abundant viral structural protein. During the viral life cycle multiple copies of the N protein interact with the viral genome to form the ribonucleoprotein complex, which is subsequently packaged by a lipid envelope during viral budding, possibly through its interaction with the viral structure membrane (M) protein (5). In addition to its structural role, the N protein is also implicated in regulating the synthesis of viral RNA and protein (4, 6, 7). Using reverse genetics, the critical role of N protein in the replication of coronaviruses has been identified in HCoV-229E, TGEV (transmissible gastroenteritis coronavirus), and IBV (infectious bronchitis virus) (8, 9, 10). However, the molecular mechanisms in N protein participation in viral replication and the cellular gene(s) involved in regulating the process remain unknown.
The typical CoV N protein (∼400 amino acids, ∼50 kDa) is highly basic and is composed of three distinct domains. The N-terminal domain (∼130 residues) folds similarly to the U1A RNA-binding protein and is suggested to bind RNA (11, 12). The C-terminal domain contributes to its di- or multimerization assembly (13, 14), and the central region contains a serine/arginine (SR)-rich motif with unknown function but which is possibly also involved in the regulation of its multimerization (15). Notably, N proteins are highly phosphorylated in infected cells (16, 17). Surjit et al. (18) provided evidence that SCoV N protein undergoes phosphorylation in cells, mainly at the serine residues. The phosphorylation residues of the TGEV, IBV, MHV A59, and SCoV N proteins have recently been identified as Ser and Thr (16, 19, 20, 21). However, the putative kinase(s) responsible for N protein phosphorylation and the effects of phosphorylation on viral life cycle have not yet been conclusively elucidated.
The biological effects of N protein phosphorylation could influence its RNA binding activity and its subcellular localization (17, 18, 19, 22). For IBV, Chen et al. (19) demonstrated that phosphorylated N protein was bound to viral RNA with a higher affinity than nonviral RNA. In JHMV the nonphosphorylated N protein was found exclusively in the cytosol, whereas the phosphorylated N protein was mainly associated with the cellular membrane fraction (17). Mohandas and Dales (23) demonstrated that dephosphorylation of JHMV N protein by an endosomal-associated cellular protein phosphatase might facilitate viral infections. The phosphorylated SCoV N was recently shown to translocate from the nucleus to the cytoplasm by binding with the 14-3-3 protein, as a mechanism for phosphorylation-dependent nucleocytoplasmic shuttling (18). Recently, Peng et al. (24) reported that phosphorylation of SCoV N at the SR-rich motif could modulate its translation inhibitory activity and also its multimerization activity. Based on these observations, the phosphorylation of N protein has long been proposed to participate in regulating viral replication, but currently there is a lack of conclusive evidence supporting its critical involvement.
We aimed to explore this issue by identification of the cellular kinase(s) for phosphorylation of SCoV N protein. We applied mass spectrometric analysis and deletion mapping to localize the putative phosphorylation sites of SCoV N to the central SR-rich motif, which contains several consensus substrate sequences for the GSK-3 kinase. The authentic role of GSK-3 in N protein phosphorylation was confirmed by evidence from both in vitro and in vivo experiments. The involvement of GSK-3 in N phosphorylation has also been shown in another coronavirus, JHMV. Finally, we found that inhibition of GSK-3 could suppress the replication of both coronaviruses. The results not only indicate that GSK-3 is critical for N phosphorylation but also suggest its involvement in regulating viral replication.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—The full-length SCoV N and JHMV N were constructed by PCR amplification of cDNA template reverse-transcribed from the virus RNA. The template virus used for SCoV is strain TW1 (GenBank™ accession no. AY291451) (25) and the neurotropic JHM strain of mouse hepatitis virus, JHMV, was kindly provided by Prof. Michael M. C. Lai (National Cheng Kung University) (26). The detailed procedure for virus preparation, RNA extraction, and reverse transcription was described previously (25). The amplified cDNA was cloned into the pcDNA3.1, pCMV-Tag2B (with FLAG tag), and pGEX-4T vectors for transfection and GST fusion protein purification experiments. Introduction of specific mutations into the SCoV N plasmids was conducted by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The SCoVΔSR-N-FLAG construct, containing SCoV N with deletion of SR-rich motif (amino acids 178∼213), was constructed in the vector of pcDNA3.1 and kindly provided by Dr. Woan-Yuh Tarn (Institute of Biomedical Sciences of Taiwan Academia Sinica, Taipei, Taiwan). The plasmid constructs for the constitutive active form of pHA-GSK-3β and pHA-MEK were kindly provided by Prof. Junichi Sadoshima (Department of Molecular Cellular Physiology, Pennsylvania State University College of Medicine) and Dr. Ruey-Hwa Chen (Institute of Biological Chemistry of Taiwan Academia Sinica, Taipei, Taiwan).
Cell Culture and Transfection Experiment—The VeroE6 and 293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and 1% glutamine and 1% penicillin/streptomycin. The DBT mouse astrocytoma cell line were cultured in minimum Eagle's complete medium (Invitrogen) supplemented with 7% heat-inactivated fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin and 10% tryptose phosphate broth solution. All of these cells were incubated in 37 °C incubator with 5% CO2. When cells were grown to 80-90% confluence, the transfection experiments were conducted by using Lipofectamine 2000 (Invitrogen) according to the detailed procedures described as previously (27).
Cell Lysate Preparation, High Resolution NuPAGE, and Western Blot Analysis—Cells were lysed with immunoprecipitation buffer (0.25% Triton X-100, 0.025 m Tris pH7.4, 0.15 m NaCl) containing protease inhibitor (Complete Protease Inhibitor Mixture Tablets, Roche Diagnostics) and phosphatase inhibitor (Phosphatase Inhibitor Mixture Set II, Calbiochem). The lysates were separated by 10% NuPAGE Bis-Tris 1.0-mm gel (Invitrogen) with constant voltage of 70 volts until the 37-kDa protein marker moved to the bottom of gel (at 4 °C). The gel was then electrotransferred onto the nitrocellulose membrane and blocked at 5% skimmed milk in TBST buffer (10 mm Tris/pH 7.5, 150 mm NaCl, 0.25% Tween 20) at room temperature for 1 h, probed with first antibodies and horseradish peroxidase-conjugated secondary antibody, and then the signal was detected by the ECL assay system (Pierce). The Abs used in the current study include rabbit anti-SCoV N (generated in our laboratory (28)), rabbit anti-GSK-3α (Cell Signaling, Danvers, MA), rabbit anti-GSK-3β (Cell Signaling), rabbit anti-JHMV N (kindly provided by Prof. Eric J. Snijder, Leiden University), and horseradish peroxidase-linked donkey anti-rabbit IgG (Amersham Biosciences). The Abs specific against the phosphorylated N proteins of SCoV (Ser-177) and JHMV (Ser-197) was raised by injection of chemically synthesized phosphopeptides into mice (LTK Laboratories, Taipei, Taiwan). The amino acid sequence for the two peptides is FYAEGSRGGSQ (for SCoV N-Ser-177) and EGSGRSAPASR (for JHMV N-Ser-197).
CIP Treatment of N Protein—The cells were washed twice with phosphate-buffered saline and lysed by 1000 μl of immunoprecipitation buffer. After centrifugation at 13,000 rpm for 10 min at 4 °C, the supernatant was incubated with 15 μl of anti-FLAG M2 beads (Sigma-Aldrich) at 4 °C for 2 h. The beads were washed three times with 1000 μl of immunoprecipitation buffer without phosphatase inhibitors and then processed for the CIP reaction with 1 unit of calf alkaline phosphatase (New England Biolabs, Beverly, MA) in 20 μl of 1× reaction buffer (100 mm NaCl, 50 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol, pH 7.9) at 37 °C for 1 h. The beads were then washed 3 times with 1000 μl of immunoprecipitation buffer and then eluted by FLAG peptide (Sigma-Aldrich) for subsequent analysis.
Mass Spectrometric Analysis of SCoV N Protein—The cell lysate prepared from 293T cells transfected with pCMV-FLAGSCoVN construct (in pCMV-Tag2B vector) was used for purification of the FLAG-tagged N protein of SCoV by immunoprecipitation with M2 beads (Sigma-Aldrich). The purified proteins were eluted with FLAG peptide and were separated by 10% SDS-PAGE. After staining with Coomassie Blue, the protein band corresponding to FLAG-N was harvested for in-gel tryptic digestion. The gel slice was lyophilized and incubated in 10 μl of 10 mg/ml trypsin solution at 37 °C for 8 h and then analyzed by a liquid chromatography-MS/MS system consisting of Agilent 1200 nanoflow high performance liquid chromatography and LTQ-Orbitrap hybrid tandem mass spectrometer. TurboSequest and several in-house programs were used to interpret the liquid chromatography-MS/MS data.
Treatment of Cells with Inhibitors against Specific Kinases—To evaluate the effect of specific kinases on N phosphorylation, the cells transfected with N expression constructs or infected with coronaviruses were treated with inhibitors against specific kinases, including LiCl and kenpaullone for GSK-3, wortmannin for phosphatidylinositol 3-kinase; 5,6-Dichlorobenzimidazole riboside for casein kinase 2, olomoucine for cyclin-dependent kinase, H89 for protein kinase A (all from Sigma-Aldrich), and U0126 for MEK (Calbiochem). The inhibitors were added into the culture medium with proper/effective concentrations 1 h before transfection (or virus infection) until the cytopathic effect (CPE) was recorded or the supernatant or lysates were harvested for subsequent analysis.
In Vitro Kinase Assay—The substrates used for the in vitro GSK-3 kinase assay are either the GST fusion proteins of wild type SCoV N and ΔSR-N or the FLAG-tagged N proteins purified from the 293T cells. The GST fusion proteins were purified from Escherichia coli following the procedures as described previously (28). The reactions were performed in a 20-μl reaction mixture containing 1× kinase reaction buffer (50 mm HEPES, pH 7.4, 0.5 mm dithiothreitol, 5% glycerol, and 800 mm MgCl2), 1 μl of purified recombinant active human GSK-3α or GSK-3β (Upstate Biotechnology, Charlottesville, VA), 10 μCi of [γ-32P]ATP, 10 μm ATP, and 5 μl of purified substrates. The mixtures were incubated at 30 °C for 30 min, and stopped by the addition of 5× SDS sample buffer, and separated by 10% SDS-PAGE. The gels were dried by vacuum dryer (SGD5040 Slab Gel Dryer, ThermoSavant, Holbrok, NY) and then processed to autoradiography.
Two-dimensional Gel Electrophoresis (SDS-PAGE)—We have also tried to separate the phosphorylated versus unphosphorylated N protein by the two-dimensional SDS-PAGE. The protein samples were prepared in sample buffer (8 m urea, 2 m dithiothreitol, 0.0025% Triton X-100, 2% immobilized pH gradient (IPG) buffer pH 7-11, 0.05% bromphenol blue) and separated by the isoelectric focusing method using immobilized linear gradient pH 7-11 7-cm polyacrylamide strips (Amersham Biosciences) which were first rehydrated overnight with the rehydration buffer (8 m urea, 40 mm dithiothreitol, 0.0025% Triton X-100, 2% IPG buffer, 0.05% bromphenol blue). The isoelectric focusing reaction was performed at 20 °C with conditions of 500 V for 30 min, 1500 V for 30 min, and 3000 V for 17 h. After isoelectric focusing the strips were equilibrated with the equilibration buffer (50 mm Tris, pH 8.8, 6 m urea, 30% glycerol, 2% SDS, 1% dithiothreitol), separated by 4-12% polyacrylamide gels (Invitrogen), and processed for the subsequent immunoblot analysis.
Determination of Viral Titer by Plaque Assay, TCID50, and Real Time Quantitative PCR—The TW1 strain of SCoV was propagated in Vero E6 cells in Biosafety level 3 laboratory (25), and the JHMV strain of mouse hepatitis virus was propagated with the murine DBT astrocytoma cells as previously described (26). The viral titer of SCoV was determined as the unit of 50% tissue culture-infective dose (TCID50)/ml, recorded as log10 TCID50 units with detailed procedures as described previously (29) and also by the quantitative PCR. The viral titer of JHMV was determined by plaque assay in DBT cells as previously described (26) and also by quantitative PCR.
For quantitative PCR, the viral RNA was first reverse-transcribed into cDNA by the SuperScript cDNA synthesis system (Invitrogen) (25). Quantitative PCR was done with the LightCycler FastStart DNA SYBR Green kit (Roche Diagnostics). The primer set designed for JHMV quantification contains JHMV-N-F2 (5′-ACACAACCGACGTTCC-3′) and JHMV-N-R2 (5′-GCAATACCGTACCGGG-3′), and the primer set designed for SCoV quantification contains SARS-N-F (5′-GTATTCAAGGCTCCCTCAGTG-3′) and SARS-N-R (5′-TGGCTACTACCGAAGAGCTACC-3′). The PCR reaction was performed in a total volume of 20 μl containing 2 μl of viral cDNA template, 0.5 μm forward and reverse primers, 3 mm MgCl2, and 2 μl of 10× FastStart SYBR Master Mix. The PCR reaction was performed with LightCycler (Roche Diagnostics) as an initial hot start denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 3 s, annealing at 56 °C for 5 s, and extension at 72 °C for 15 s, and the fluorescence was measured at the end of the annealing phase for each cycle. To verify the specificity of the amplification, a melting curve analysis was done at the end of amplification by holding the reaction at 95 °C for 60 s and then lowering the temperature to 65 °C with the transition rate 0.1 °C/s and holding for 120 s followed by heating slowly at transition rate 0.1 °C/s to 95 °C with continuous collection of fluorescence. To quantify the viral load, we used the plasmids pCMV-Tag2B-SCoVN and pCMMV-Tag2B-JHMVN to generate the standard curves (in copy number). The plasmid DNA was purified and processed for the subsequent generation of the standard curve for quantification as described previously (30).
Northern Blot Analysis—Total cellular RNA was extracted using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The RNA (1 μg/lane) was denatured and fractionated by electrophoresis (70 V, 6 h) with formaldehyde, 0.8% agarose gels in 1× MOPS buffer (20 mm MOPS, pH 7.0, 5 mm sodium acetate, 1 mm EDTA), capillary-transferred to nylon membranes (Hybond-N+; Amersham Biosciences), and cross-linked by UV cross-linker (Stratagene). The membrane was processed for the subsequent hybridization using the DIG Northern Starter kit (Roche Diagnostics) by following the manufacturer's instruction. The probe used for hybridization was labeled with DIG-dUTP during PCR amplification (PCR DIG probe synthesis kit, Roche Diagnostics). The primer sets used for amplification of probes are listed as followed. SARS-N-F (5′-GTATTCAAGGCTCCCTCAGTTG-3′) and SARS-N-R (5′-TGGCTACTACCGAAGAGCTACC-3′) were used for amplification of SCoV N probe. JHMV-N-F2 (5′-ACACAACCGACGTTCC-3′) and JHMV-N-R2 (5′-GCAATACCGTACCGGG-3′) were used for amplification of JHMV N probe, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward (5′-GAAGGTGAAGGTCGGAGTC-3′) and GAPDH reverse (5′-GAAGATGGTGATGGGATTTC-3′) were used for amplification of the GAPDH probe. The hybridization signals were visualized with chemiluminescence which is recorded on x-ray films.
RESULTS
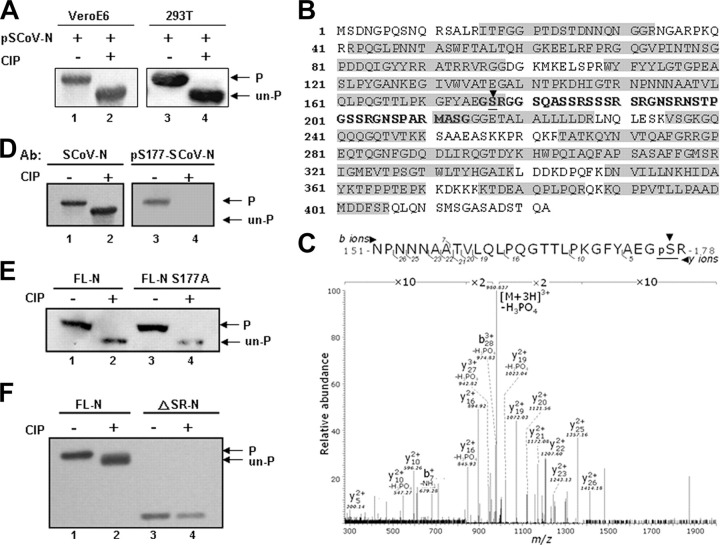
The N Protein of SCoV Is Phosphorylated in VeroE6 and 293T Cells—To analyze the phosphorylation status of the SCoV N protein, we first used high resolution NuPAGE gels to differentiate the phosphorylated from unphosphorylated forms. Lysates extracted from VeroE6 or 293T cells were treated or not treated with CIP protein phosphatase and then run in high resolution gels. Western blotting identified a faster migrating N protein from the CIP-treated cell lysate (Fig. 1A , lanes 2 and 4) compared with that from the untreated cell lysate (Fig. 1A, lanes 1 and 3). Thus, most of the N protein in both cell lines was phosphorylated, and this phosphorylation could retard the mobility of this protein in NuPAGE analysis.
FIGURE 1.
The N protein of SCoV is hyperphosphorylated over the central SR-rich motif.A, the N expression plasmid pSCoV-N was transfected into VeroE6 (lanes 1 and 2) and 293T (lanes 3 and 4) cells. The cell lysates, either untreated (lanes 1 and 3) or treated with CIP enzyme (lanes 2 and 4), were assayed by Western blot analysis (probed with an Ab against N protein). The phosphorylated and unphosphorylated N proteins are indicated. B, the purified FLAG-tagged N protein of SCoV from 293T cells was subjected to mass spectrometric analysis. The sequences recovered by the analysis are marked within gray boxes. The serine residue (Ser-177) identified to be phosphorylated is underlined and identified with an arrowhead. C, the results of mass spectrometric analysis, showing the peptide containing Ser-177 to be ∼100% phosphorylated, marked by underlining and an arrowhead. The lysates from 293T cells, transfected with wild type N protein (D), wild type N and mutant S177A N protein (E), or with wild type N and ΔSR-N protein expression constructs (F) were treated with CIP and processed for Western blot analysis. Except for the right panel of D (probed with an Ab recognizing the phosphorylated Ser-177 N protein), the blots of D-E were probed with an Ab against N protein.
The Ser-177 Residue Appears to Be Fully Phosphorylated in Vivo—To map the phosphorylated residues we transfected the FLAG-tagged N (FLAG-N) construct into 293T cells and purified the N proteins by immunoprecipitation with anti-FLAG antibody-coated beads. Mass spectrometric analysis showed that >72% of the protein sequence could be recovered (Fig. 1B). We have detected phosphorylation in one peptide, 151NPNNNAATVLQLPQGTTLPKGFYAEGSR178.
Tandem MS analysis of this peptide revealed that there was one prominent 980.837 m/z peak, which corresponds to the precursor ion with a neutral loss of phosphoric acid. This feature is an MS signature of many peptides containing a phosphorylated Ser or Thr residue. Based on the observed masses of fragment ions y5 and y10, we concluded that the phosphate group should be positioned over the C-terminal five residues. Because Ser-177 is the only residue that can be phosphorylated within this stretch, we inferred that this must be the one (Fig. 1C). It is intriguing that we did not detect any unmodified counterpart for this phosphopeptide, suggesting that it is fully phosphorylated in vivo. The results from Western blot analysis probed with Ab specifically raised for the phosphorylated form of N-Ser-177 evidently supported the phosphorylation of this residue of N protein in 293T cells (Fig. 1D). Because some other minor phosphorylation sites have much lower modification percentages (typically <1%), including Thr-92, Thr-363, and Thr-367, we first studied whether Ser-177 phosphorylation was likely to be that associated with the observed retardation of N protein in gel mobility.
The Phosphorylation Sites of N Protein Cluster at the Central SR-rich Region—To test the contribution of Ser-177 in the retardation of N protein in a gel mobility assay, one mutant N construct with Ser-177 substituted by Ala was used for CIP treatment and the subsequent NuPAGE analysis. The mobility of this mutant N did not differ from that of wild type N protein (Fig. 1E, lanes 2 and 4), suggesting that in addition to Ser-177, there are some other major phosphorylation residues that remained unidentified.
One major region remains unrecovered and unanalyzed by our current MS analysis and is located at the central SR-rich motif (amino acids 179-210, Fig. 1B, black bold characters), which are also likely candidate residues for phosphorylation. Recently, aided by the [32P]orthophosphate labeling, Peng et al. (24) demonstrated this region as the major phosphorylation region for SCoV N in HEK293 cells. To test this, we constructed a SR region-deleted N construct (ΔSR-N, deletion of amino acids 178-213), expressed the mutant protein, and immunoprecipitated it from 293T cells for analysis. Interestingly, although the mobility of wild-type N protein shifted after CIP treatment (Fig. 1F, lane 2), that of ΔSR-N did not change by CIP treatment (Fig. 1F, lane 4). Apparently, the SR-rich motif is the major phosphorylation region of N proteins in both 293T and VeroE6 cells.
GSK-3 Is the Putative Kinase Contributing to N Phosphorylation at the SR-rich Region—The NetPhos software predicted several putative kinases recognizing this motif (Table 1 ), supporting the idea that the SR-rich motif (amino acids 177-213) is the likely substrate site for cellular kinase(s). To investigate which kinase might contribute to N protein phosphorylation, we treated cells expressing N with inhibitors specific against each of the candidate kinases, including protein kinase A, GSK-3, MEK, cyclin-dependent kinase, and phosphatidylinositol 3-kinase. We anticipated that any inhibitor for the putative kinase involved in N phosphorylation would suppress N phosphorylation and lead to a faster migrating band on NuPAGE gels.
TABLE 1.
The phosphorylation sites (Ser residues) and the corresponding kinases (predicted by the NetPhos program) for the SR-rich motif in the SCoV N protein
Characteristics are underlined in bold. PKA, protein kinase A; PKB, protein kinase B; Cdk, cyclin-dependent kinase; DNAPK, DNA-dependent serine/threonine protein kinase; ATM, ataxia telangiectasia mutated; RSK, ribosomal s6 kinase; PKC, protein kinase C; CK, casein kinase.
| Position | Amino acid sequence | Potential kinase(s) |
|---|---|---|
| Ser-177a,b | YAEGSRGGS | GSK-3 |
| Ser-181a | SRGGSQASS | DNAPK, ATM, PKA, GSK-3 |
| Ser-184 | GSQASSRSS | PKC, Cdc2, GSK-3, CK1 |
| Ser-185a | SQASSRSSS | Cdc2, GSK-3 |
| Ser-187c | ASSRSSSRS | Cdc2, GSK-3, CK1 |
| Ser-188 | SSRSSSRSR | PKC, PKA, CK1, Cdc2 |
| Ser-189a | SRSSSRSRG | GSK-3 |
| Ser-191c | SSSRSRGNS | CK1, GSK-3 |
| Ser-195c | SRGNSRNST | RSK, PKB, PKA |
| Ser-198 | NSRNSTPGS | Cdc2, CK1 |
| Ser-202 | STPGSSRGN | PKC |
| Ser-203c | TPGSSRGNS | GSK-3 |
| Ser-207c | SRGNSPARM | GSK-3, Cdk5 |
| Ser-213 | ARMASGGGE | PKA |
The Ser residues for the series 1 consensus sequences of GSK-3 in the N protein.
Ser-177 was not predicted by the NetPhos program, but it matches the GSK-3 consensus sequence.
The Ser residues for the series 2 consensus sequences of GSK-3 in the N protein.
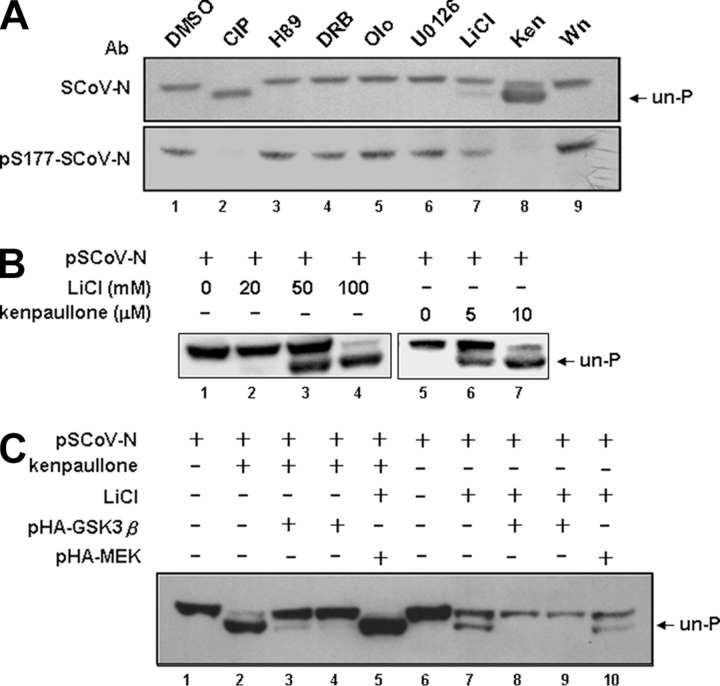
Only LiCl and kenpaullone-specific inhibitors of GSK-3 kinase resulted in the appearance of faster migrating bands (Fig. 2A , lanes 7 and 8) in a dose-dependent manners (Fig. 2B). Other kinase inhibitors did not inhibit N protein phosphorylation (Fig. 2A). Therefore, GSK-3 appeared to be the putative kinase for N protein phosphorylation. Supportively, the hybridization signal for LiCl and kenpaullone-treated N protein by probing with Ab against phosphorylated N-Ser-177 was significantly decreased (Fig. 2A, lower panel), suggesting that phosphorylation of this residue is also regulated by GSK-3. Indeed, Ser-177 also matched the consensus phosphorylation site of GSK-3.
FIGURE 2.
GSK-3 kinase is involved in the phosphorylation of SCoV N protein.A, by treatment with specific inhibitors against kinases predicted by the NetPhos software (for SR-rich motifs), N protein-expressing 293T cell lysates were processed for Western blot analysis (probed with an Ab against N protein and an Ab against phosphorylated Ser-177 N protein). The unphosphorylated form is indicated. The lysate subjected to CIP treatment was used as the control showing the position of unphosphorylated N protein. Wn, wortmannin; Ken, kenpaullone; olo, olomoucine; DRB, 5,6-dichlorobenzimidazole riboside. B, the dose-dependent effects of two specific inhibitors for GSK-3, LiCl and kenpaullone, on the phosphorylation of N were evaluated, with the position of the unphosphorylated N indicated. C, dephosphorylated N resulting from treatment with LiCl and kenpaullone could be recovered by the constitutively active form of GSK-3β (pHA-GSK-3β, an S9A mutant). The control used pHA-MEK, the constitutively active form of MEK, showing the specificity of recovery. The position of the unphosphorylated N protein is indicated.
To further examine the involvement of GSK-3 in regulating N protein phosphorylation, we used a “gain of function” approach. One constitutively active GSK-3β construct, S9A, was transfected into 293T cells treated with kenpaullone or LiCl to study whether it could restore the phosphorylation of N protein. As a control we included a constitutively active MEK. In Fig. 2C, the dephosphorylated N treated by both inhibitors was restored by GSK-3β (lanes 3, 4, 8, and 9) but not by MEK (lanes 5 and 10). These results, thus, provided further evidence that GSK-3 is the cellular kinase for N protein phosphorylation.
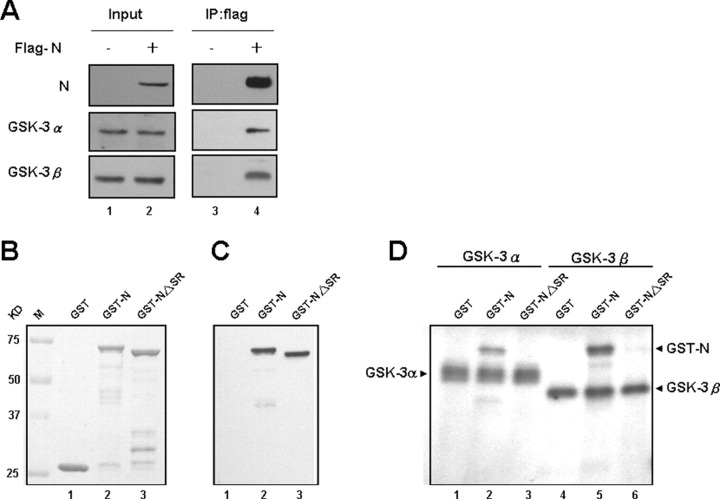
SCoV N Can Interact with GSK-3α and GSK-3β in Vivo and Can Be Phosphorylated by GSK-3α and GSK-3β in Vitro—We conducted coimmunoprecipitation analysis to test whether GSK-3 interacts with SCoV N protein in vivo. By transfecting FLAG-N into 293T cells, the lysates were subjected to immunoprecipitation with anti-FLAG beads and then processed for Western blot analysis using antibodies against either GSK-3α or GSK-3β. Both coprecipitated in the same protein complex as the FLAG-N (Fig. 3A , lane 4).
FIGURE 3.
Characterization of the interaction between SCoV N and GSK-3 α and β forms and a direct phosphorylation effect of GSK-3 kinases on N by in vitro kinase assays.A, cell lysates from 293T cells transfected with FLAG-N protein construct were processed for coimmunoprecipitation analysis. Immunoprecipitates (IP) were analyzed by Western blot analysis and probed with Abs for N and GSK-3 (α and β) as indicated. B, the input control of GST-fusion proteins used for the in vitro kinase assay was shown by Coomassie Blue staining. C, the control for GST fusion proteins was done by immunoblotting with an Ab against SCoV N protein. D, the in vitro GSK-3α and GSK-3β kinase assays using the GST fusion proteins as substrates. The positions of autophosphorylated GSK-3 kinases are marked, which can serve as the positive controls for the reaction.
In addition, we conducted an in vitro kinase assay using recombinant GSK-3 kinase to phosphorylate the GST-N protein. The GST-NΔSR protein (deletion of residues 178-213) was used as a negative control. Both GSK-3α and GSK-3β could phosphorylate the full-length N protein (Fig. 3D, lanes 2 and 5) but not truncated NΔSR in vitro (Fig. 3D, lanes 3 and 6). Because NΔSR could not be phosphorylated by either kinase, the results reinforce our contention that the SR-rich motif of N protein is the major phosphorylation site for GSK-3.
Identification of the Critical Serine Residues Phosphorylated by GSK-3 on SCoV N Protein—It is known that GSK-3 phosphorylates serine or threonine residues separated by three residues (i.e. (S/T)XXX(S/T)) and always requires the priming phosphorylation of a serine residue located four amino acids C-terminal to the main phosphorylation site (n + 4) (31). Within the SR-rich motif, 9 of 14 Ser residues conform to the consensus GSK-3 substrate phosphorylation site (Table 1; characteristics are underlined in bold), which can be divided into two series based on their continuous (S/T)XXX(S/T) sequences (Fig. 4A and Table 1). The first series contains the sequence Ser-177—Ser-181—Ser-185—Ser-189, with the Ser-189 position as the likely candidate priming phosphorylation site. The second series contains Ser-187-Ser-191-Ser-195—Thr-199-Ser-203-Ser-207, with Ser-207 as the likely candidate priming phosphorylation site. Notably, Lin et al. (21) recently reported that both Ser-189 and Ser-207 of SCoV-N are phosphorylated in eukaryotic cells, which was revealed by their refining liquid chromatography-MS/MS analysis. Phosphorylation of the priming sites presumably can initiate a subsequent phosphorylation cascade at the N-terminal S/T residues by GSK-3. Therefore, blocking of the initial priming phosphorylation would prohibit subsequent phosphorylation events by GSK-3. If GSK-3 is indeed the kinase responsible for phosphorylation of the SR-rich motif, we predicted that mutation of either Ser-189 and/or Ser-207 (to alanine) would dramatically decrease phosphorylation of the N protein.
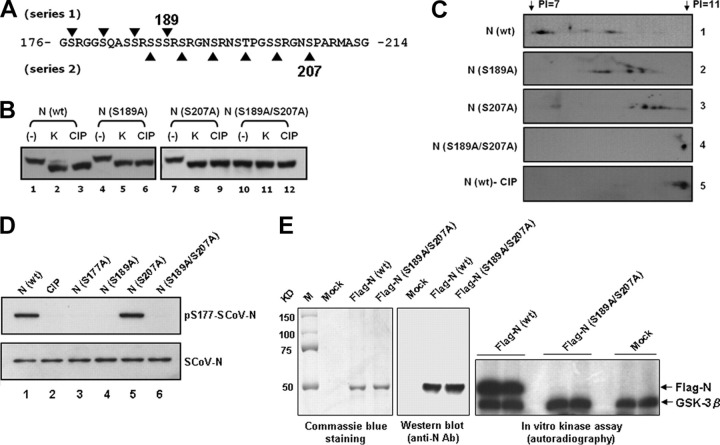
FIGURE 4.
The amino acid residues Ser-189 and Ser-207 of the SR-rich motif are critical for GSK-3 mediated phosphorylation of the SCoV N protein.A, two series of GSK-3 consensus phosphorylation sites were identified within the SR-rich motif (residues 176-214). B, lysates from 293T cells transfected with wild type or mutant N protein expression constructs (mutations at residues 189 and/or 207) were pretreated with kenpaullone (10 μm) or with DMSO (control) and subjected to high resolution NuPAGE gels and Western blot analysis (probed with an Ab against N protein). Lysates from control cells were treated with CIP as a control to mark the position of the dephosphorylated N protein. C, the five protein lysates from panel B as indicated were processed for two-dimensional gel electrophoresis and Western blot analysis (pI range 7-11; probed with an Ab against SCoV N protein). D, wild type (wt) N (with or without CIP treatment) and mutant N proteins containing S177A, S189A, and/or S207A mutations were subjected to Western blotting analysis probed with Ser(P)-177-N and SCoV-N Abs as indicated. E, the wild type FLAG-N and FLAG-S189A/S207A N proteins were used as substrates for GSK-3β in vitro kinase assay. The expression of protein substrates was shown by Coomassie Blue staining (left panel) and Western blotting analysis (middle panel). The positions of phosphorylated FLAG-N and autophosphorylated GSK-3β are marked (right panel, duplicate experiments for each substrate).
Four His-tagged N protein constructs were used to test this hypothesis, including wild type, S189A, S207A, and S189A/S207A N proteins. They were transfected into 293T cells treated either with DMSO (controls) or with kenpaullone. The S189A/S207A N protein showed similar mobility to the CIP-treated wild type (Fig. 4B, lane 10), suggesting that it was hypophosphorylated. Additionally, treatment with CIP or kenpaullone did not further change the mobility of this double mutant N protein (Fig. 4B, lanes 11 and 12). However, no significant mobility change was identified in either S189A or S207A N proteins (as compared with the wild type N).
To evaluate the effect of the two series of GSK-3 consensus sites on the resulting N phosphorylation by a more sensitive method, S189A, S207A, and S189A/S207A N proteins from 293T cells were subjected to two-dimensional gel analysis. As predicted, the wild type N was highly phosphorylated and resulted in multiple isoforms (the same molecular weight but with different pI values; Fig. 4C, lane 1). They were indeed phosphorylated proteins, as the lower-pI N protein spots were reduced after CIP treatment (Fig. 4C, lane 5). Intriguingly, N proteins with mutations either at S189A or at S207A shifted to higher pI values than did the wild type, suggesting some decrease in phosphorylation (Fig. 4C, lanes 2 and 3). For the N proteins with mutations at S189A or S207A, the pI was similar to that of the wild type treated with CIP (Fig. 4C, lane 4). The results further supported our contention that both series of GSK-3 consensus sequences contribute to N protein phosphorylation and that mutations of Ser-189 and Ser-207 within the SR-rich motif can effectively block the phosphorylation of N protein by GSK-3. The presence of two series of GSK-3 substrate phosphorylation sites has been further supported by the observation that only S207A, but not S189A, mutant N could be recognized by SCoV-N-Ser(P)-177 Ab (Fig. 4D, lane 5 versus l and 4).
Finally, the even more direct evidence comes from our in vitro GSK-3β kinase assay using the S189A/S207A N protein as the substrate. The phosphorylation signal of N was fully abolished in the double mutant N compared with that of wild type N protein (Fig. 4E, right panel). The results fully supported that these two residues are critical for GSK-3 to phosphorylate the SCoV-N protein.
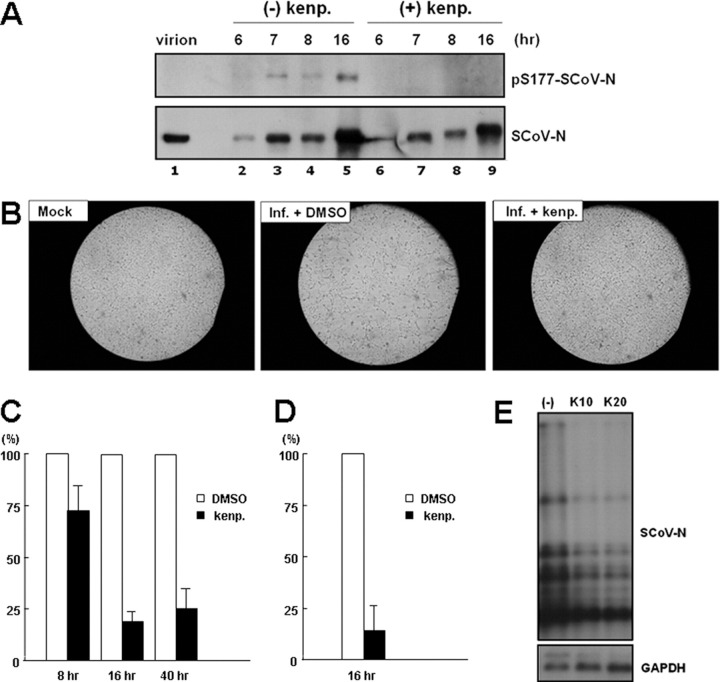
GSK-3 Inhibitor Suppressed the Replication of SCoV—To further investigate whether GSK-3 could affect N phosphorylation in the infectious system and regulate the viral life cycle, we studied the effect of kenpaullone on SCoV replicating in VeroE6 cells. We studied its effects on N phosphorylation, virus-induced CPE, viral titers, and viral RNA synthesis. We first noted that most N protein in the virion was hypophosphorylated, as it could not be recognized by SCoV-N-Ser(P)-177 Ab (Fig. 5A , lane 1) and also showed mobility similar to that of the CIP-treated N protein in VeroE6 cells (data not shown). However, in the virus-infected cells, certain N protein underwent phosphorylation (Fig. 5A, lanes 2-5). After pretreatment with kenpaullone (1 h before infection), the level of phosphorylated N protein in the infected cells had diminished (Fig. 5A, lanes 6-9), suggesting that this GSK-3 inhibitor could also reduce N phosphorylation in the infectious system. No toxicity effect on cell viability was noted for the kenpaullone treatment (after 24 h of treatment).
FIGURE 5.
The effects of the GSK-3 inhibitor, kenpaullone, on N protein phosphorylation and the virology of SCoV in the VeroE6 infectious system.A, the phosphorylation status of N proteins in SCoV virions and in virus-infected (Inf.) cells (treated or untreated with kenpaullone (kenp.)) was evaluated by Western blot analysis (probed with Abs against Ser(P)-177-SCoV-N and SCoV-N Abs as indicated). After pretreatment with 10 μm kenpaullone or with DMSO (control), the cells were infected with virus (multiplicity of infection 1), and cell lysates were harvested at different time points for Western blot analysis. B, the effect of kenpaullone pretreatment on virus-induced CPE was evaluated at 16 h after SCoV infection (multiplicity of infection 1) by microscopy. Moreover, the effect of kenpaullone pretreatment on viral titer was evaluated by quantitative PCR (C) and by TCID50 assay (D), with virus harvested from the medium at time as indicated after virus infection. The viral titers from medium treated with DMSO control was set to a value of 1 and the virus titers from kenpaullone pretreatment medium were normalized to this value. E, the effect of kenpaullone pretreatment (10 and 20 μm) on the synthesis of virus RNA was evaluated by Northern blot analysis (probed with DIG-labeled N or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes) with cellular RNA harvested 16 h after infection.
We further found that kenpaullone treatment could reduce virus-induced CPE (Fig. 5B, right panel, compared with middle panel, 16 h after infection). To evaluate the effect of GSK-3 on virus production, the supernatant from infected cells treated with DMSO (control) and kenpaullone was harvested 16 h after infection for quantitative PCR and for the estimation of TCID50. Quantitative PCR showed that viral titer was reduced to around 20% with kenpaullone treatment compared with that from DMSO controls as 100% (Fig. 5C). Moreover, the TCID50 assay further showed an ∼85% decrease for the infectious viral titer after kenpaullone treatment compared with that of the DMSO control (Fig. 5D). Northern blot analysis (Fig. 5E) revealed that the viral RNA level was reduced to be around 50% by kenpaullone treatment.
GSK-3 Is Also Involved in Regulating the Phosphorylation of JHMV N Protein—It was noted that at least one stretch of continuous consensus sequence for GSK-3, SXXXS, was also present at the central SR-rich region of MHV, BCV, OC43, TGEV, 229E, and IBV (22), suggesting the possible involvement of GSK-3 in regulating the phosphorylation of N in other coronaviruses. To test this possibility, we evaluated the effect of GSK-3 on the phosphorylation of N from another coronavirus, JHMV (a strain of MHV).
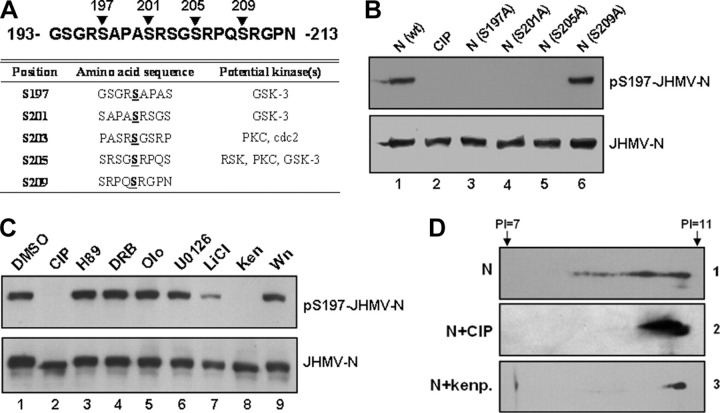
The putative GSK-3 substrate phosphorylation sites within SR-rich region of JHMV contain Ser-197-Ser-201-Ser-205-Ser-209, with Ser-209 as the candidate priming phosphorylation site (Fig. 6A ). To test if GSK-3 is also involved in regulating the phosphorylation of these putative sites in a similar way with that of SCoV-N, we raised the Ab against the phosphorylated Ser-197 of JHMV N for the following studies. Phosphorylation of this residue of JHMV-N was first demonstrated by Western blot analysis (Fig. 6B, lane 1 versus lane 2). Next, to investigate if Ser-209 functions as the priming site for this series of Ser phosphorylation, the S209A mutant N was constructed, but it still can be recognized by N-Ser(P)-197 Ab (Fig. 6B, lane 6). The S205A and S201A mutant N was, thus, constructed to help identify the true priming site. Mutant N cannot be recognized by N-Ser(P)-197 Ab (Fig. 6B, lanes 4 and 5), suggesting that the initial priming site locates at residue Ser-205. The GSK-3 substrate phosphorylation sites in JHMV were, thus, consisted of Ser-197—Ser-201—Ser-205.
FIGURE 6.
GSK-3 kinase is also involved in the phosphorylation of JHMV N protein.A, only one series of GSK-3 consensus phosphorylation site was identified in SR-rich motif of JHMV N protein (residues 193-213). The potential kinase(s) of each serine residue within this region predicted by NetPhos program was summarized. PKC, protein kinase C. B, wild type (wt) N (with or without CIP treatment) and mutant N proteins containing S197A, S201A, S205A, and S209A mutations isolated from 293T cells were subjected to Western blotting analysis probed with Ser(P)-197-JHMV-N and JHMV-N Abs as indicated. C, by treatment with specific inhibitors against various kinases, N protein-expressing 293T cell lysates were processed for Western blot analysis (probed with Abs against Ser(P)-197-JHMV-N and JHMV-N proteins as indicated). The lysate subjected to CIP treatment was used as the control showing the specificity of Ser(P)-197-JHMV-N Ab. Wn, wortmannin; Ken, kenpaullone; olo, olomoucine; DRB, 5,6-Dichlorobenzimidazole riboside. D, three protein lysates from panel C were processed for two-dimensional gel electrophoresis (pI range 7-11) and for subsequent immunoblotting analysis (probed with Ab against JHMV-N protein).
The phosphorylation of this series of JHMV-N was further shown to be specifically down-regulated by GSK-3 inhibitors, kenpaullone and LiCl (Fig. 6C, lanes 7 and 8), but not affected by several other kinase inhibitors (Fig. 6C, lanes 3-6 and 9). Only three Ser residues contained in this series of phosphorylation sites makes the phosphorylated N not show gel mobility shift compared with the dephosphorylated N even in our high resolution gel electrophoresis. The more sensitive two-dimensional gel electrophoresis was, thus, used for validation of the effect of kenpaullone on N phosphorylation. A series of spot signals of N at pI < 11 were detected in the lysate from 293T cells transfected with a JHMV-N construct (Fig. 6D). The signals with lower pI values were significantly diminished either by CIP or by kenpaullone treatment (Fig. 6D), suggesting that kenpaullone could induce dephosphorylation of JHMV-N to a level similar to that produced by CIP. Therefore, GSK-3 is also the major kinase involved in phosphorylation of JHMV-N as well as that of SCoV-N.
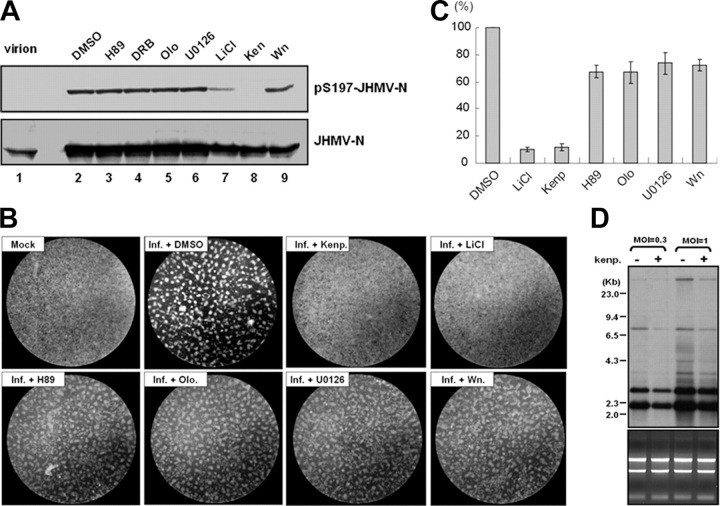
The effect of GSK-3 inhibitors on JHMV-N phosphorylation at Ser-197 residue has been further demonstrated in the virus infectious system. Both kenpaullone and LiCl treatment can decrease the phosphorylation of N at Ser-197 (Fig. 7A , lanes 7 and 8). We again noted that the N in virion cannot be recognized by N-Ser(P)-197 Ab (Fig. 7A, lane 1). The specificity of GSK-3 inhibitors on phosphorylation of N at Ser-197 has been well demonstrated in the infectious system by including other kinase inhibitors for our analysis (Fig. 7A, lanes 3-6, and lane 9).
FIGURE 7.
Effects of the GSK-3 inhibitors kenpaullone and LiCl on N protein phosphorylation and on the virology of JHMV in the infectious system.A, the phosphorylation status of N proteins in JHMV virions and in virus-infected cells (treated with DMSO control or various kinase inhibitors as indicated) was evaluated by Western blot analysis (probed with Abs against Ser(P)-197-JHMV-N and JHMV-N Abs). The DBT cells were pretreated with the inhibitors for 1 h before JHMV infection (multiplicity of infection 1), and then the cell lysates were harvested at 16 h post-infection. Wn, wortmannin; Ken, kenpaullone; olo, olomoucine; DRB, 5,6-Dichlorobenzimidazole riboside. The effects of specific kinase inhibitors on virus-induced CPE (by microscopy observation) (B) and viral titer (by quantitative PCR assay) (C) were also evaluated at 16 h post-infection. The viral titers from medium treated with DMSO control was set to a value of 1, and the virus titers from inhibitors pretreatment medium were normalized to this value. D, the effect of kenpaullone pretreatment on viral RNA synthesis was evaluated by Northern blot analysis (probed with DIG-labeled JHMV-N protein probes) with cellular RNA extracted at 12 h after infection (multiplicity of infection 1 and 0.3).
Finally, we also studied the effect of GSK-3 inhibitors on the replication of JHMV. Both JHMV-induced CPE (Fig. 7B) and the viral titer could be inhibited by kenpaullone and LiCl treatment (Fig. 7C), similar to that shown for SCoV. Moreover, the specificity of the GSK-3 inhibitors on the viral-induced CPE and the viral titer has been well demonstrated by including the other kinase inhibitors in the experiments (Fig. 7, B and C). And the Northern blot analysis also confirmed that kenpaullone inhibited viral RNA synthesis in JHMV (Fig. 7D).
DISCUSSION
Our studies clearly demonstrated GSK-3 kinase is involved in the phosphorylation of the N protein of coronaviruses, at least for SCoV and JHMV. Based on the presence of similar continuous consensus GSK-3 sequences at the SR-rich region of N protein in most coronaviruses, the involvement of GSK-3 for regulating N protein phosphorylation in other coronaviruses is highly likely. Consistent with this idea, we noted that the phosphorylation site determined by MS analysis in TGEV (at Ser-152) was also a consensus sequence for GSK-3 (with a position equivalent to Ser-177 in the SCoV N protein).
Despite these persuasive data, our studies have two limitations. First, although the central SR regions are likely the phosphorylation sites, we have not yet identified the exact phosphorylated residues within the region except for Ser-177. A more comprehensive MS analysis by partially digestion of N protein with trypsin (or other proteases) is under way, which was successfully applied to detect the phosphorylation at Ser-189 and Ser-207 SCoV-N previously (21). Second, although the mutations of the priming serine residues can significantly block N phosphorylation, the kinase(s) responsible for phosphorylating the priming residues is still unknown. Notably, some other kinases have been reported to be associated with N phosphorylation using in vitro phosphorylation assay, including cyclin-dependent kinase, mitogen-activated protein kinase (18), and SR protein kinase 1 (24). However, when we tested if these putative kinases are responsible for the phosphorylation of the priming residues by treating the cells with specific inhibitors for these kinases, none of them caused a mobility change in the N protein, which thus excluded such a possibility. Therefore, we need to search for other cellular kinases responsible for phosphorylating the priming residues.
The effect of GSK-3 on the phosphorylation of N protein was confirmed both in SCoV-infected VeroE6 cells and in JHMV-infected DBT cells in which kenpaullone and LiCl treatment led to a decrease in phosphorylation. Kenpaullone and LiCl treatment could also suppress cytopathy and viral titers. The results, although suggesting that the inhibition of GSK-3-mediated N phosphorylation by kenpaullone and LiCl might be critical for coronavirus replication, cannot exclude the possibility that GSK-3-mediated phosphorylation of a host protein(s) or a viral protein(s) other than N contributes. One more conclusive approach for evaluating the effect of N phosphorylation on viral life cycle would be to use reverse genetics by specifically mutating the phosphorylation priming sites, Ser-189 and Ser-207 for SCoV and Ser-205 for JHMV, in the viral genome, which would make the N protein to be produced mainly as the unphosphorylated form. The effects on viral life cycle could then be evaluated, including RNA transcription and replication levels, viral titers, and cytopathic effects. Indeed, Spencer et al. (10) used a reverse genetics analysis and showed that the wild-type N protein had a higher activity to recover the production of IBV virus than the hypophosphorylated form. These results reinforce the idea that N protein phosphorylation plays a significant role in viral replication.
Although we expect that such a reverse genetics approach can provide more evidence supporting the functional role of N phosphorylation in the viral life cycle, this will raise another argument that the resulting effect is not directly caused by the dephosphorylation of N but, rather, by changes in either amino acids or the underlying RNA. Therefore, some more extensive studies are required to assess the functional relevance of GSK-3-mediated N phosphorylation for the viral life cycle. An alternative approach to this may come either from dissecting the effect of GSK-3 inhibitors or from comparing the different effects of wild type and S189A/S207A SCoV-N or S205A JHMV-N proteins on individual events regulating viral life cycle. Previous studies have already pointed out the possible involvement of N phosphorylation in regulating its RNA binding affinity, subcellular localization patterns, or its multimerization ability (17, 18, 19, 22, 24). These are assumed critical for controlling various processes of the virus life cycle, including uncoating, assembly, and RNA replication/transcription. However, there is still no conclusive evidence on this, mainly because of the lack of knowledge about how to block the phosphorylation of N protein. To this point our current results provide valuable information that using either the GSK-3 inhibitors or mutating the priming sites of GSK-3 can block the phosphorylation of N protein significantly, and therefore, the possible effect of N phosphorylation on these events can be investigated conclusively. Besides, several recent publications documented that the N protein can also modulate several cellular biological processes, including the activation of cellular signaling pathways (6, 32), regulate the cell cycle progression (33), and function as a type I interferon antagonist (34, 35). These might also participate in regulating viral replication and, thus, are additional candidate processes for investigating the functional effect of N phosphorylation in the viral life cycle.
The great specificity of the Abs we raised for recognition of phosphorylated coronaviral N proteins presumably can help study the kinetics of N phosphorylation in the viral life cycle. Indeed, we found that the N proteins both in the SCoV and JHMV virion exist mainly as the hypophosphorylated form. However, some N proteins are only phosphorylated inside infected cells (Figs. 5A and 7A). This observation raises two directions for the future study of the role of N protein phosphorylation in the viral life cycle. First, it might affect certain events of viral entry or nucleocapsid assembly. In the case of Rubella virus, Law et al. (36, 37) reported that phosphorylation of capsid protein can help the disassembly of nucleocapsids during early infection, which facilitates the release of viral genomes for subsequent viral replication. However, at the late infection stage and before virus assembly the capsid proteins need to be dephosphorylated, which can greatly increase the binding affinity of capsid with the viral genomic RNA and help for effective mature virus assembly (36, 37). Therefore, the effect of coronaviral N phosphorylation on binding affinity with viral RNA and regulating the nucleocapsid assembly is worthy of investigation. Second, as our Northern blot analysis showed that kenpaullone treatment reduced the synthesis of viral RNA, phosphorylated N might, thus, participate in viral RNA synthesis. In JHMV, the phosphorylated N protein is mainly associated with the intracellular membrane fraction (17), the position where viral RNA replicates actively (38). This, thus, raises the possibility that the phosphorylated form of coronaviral N protein might target to the viral replication complex in virus-induced double membrane vesicles and thereby participate in viral RNA synthesis. In support of this, N protein has been shown to colocalize with coronavirus replication complexes in the cytoplasm during viral infection (39, 40). The association of phosphorylated N with the double membrane and with the replication complex can be investigated by using the Abs recognizing the phosphorylated N proteins.
In conclusion, we have shown that a cellular kinase, GSK-3, is able to phosphorylate both SCoV and JHMV N proteins. This suggests a possible role for the GSK-3 kinase in regulating the viral life cycle. Our results, thus, identify a novel potential antiviral target for the treatment of coronavirus infections and provide new avenues to investigate the specific role of N protein phosphorylation in coronavirus life cycles.
Acknowledgments
We greatly appreciate the gift of rabbit anti-MHV N antibody from Prof. Eric J. Snijder (Molecular Virology Laboratory, Dept. of Medical Microbiology, Leiden University Medical Center, Leiden, The Netherlands).
Footnotes
The abbreviations used are: SARS, severe acute respiratory syndrome; SCoV, severe acute respiratory syndrome coronavirus; N protein, nucleocapsid protein; CPE, cytopathic effect; JHMV, JHM strain of mouse hepatitis virus; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; pI, isoelectric point; SR, serine-arginine; GSK, glycogen synthase kinase; GST, glutathione S-transferase; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; Ab, antibody; MS, mass spectrometry; MOPS, 4-morpholinepropanesulfonic acid; DIG, digoxigenin; CIP, calf intestinal alkaline phosphatase; TGEV, transmissible gastroenteritis coronavirus; IBV, infectious bronchitis virus.
References
- 1.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai M.M., Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang X., Ye L., Timani K.A., Li S., Zen Y., Zhao M., Zheng H., Wu Z. J. Biochem. Mol. Biol. 2005;38:381–385. doi: 10.5483/bmbrep.2005.38.4.381. [DOI] [PubMed] [Google Scholar]
- 6.He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahara S.M., Dietlin T.A., Nelson G.W., Stohlman S.A., Manno D.J. Adv. Exp. Med. Biol. 1998;440:313–318. doi: 10.1007/978-1-4615-5331-1_41. [DOI] [PubMed] [Google Scholar]
- 8.Almazan F., Galan C., Enjuanes L. J. Virol. 2004;78:12683–12688. doi: 10.1128/JVI.78.22.12683-12688.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schelle B., Karl N., Ludewig B., Siddell S.G., Thiel V. J. Virol. 2005;79:6620–6630. doi: 10.1128/JVI.79.11.6620-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer K.A., Dee M., Britton P., Hiscox J.A. Virology. 2008;370:373–381. doi: 10.1016/j.virol.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X., Lescar J. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., Hajduk P., Mack J., Fesik S.W., Olejniczak E.T. Biochemistry. 2004;43:6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 13.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W., Lescar J., Prasad B.V. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo H., Chen J., Chen K., Shen X., Jiang H. Biochemistry. 2006;45:11827–11835. doi: 10.1021/bi0609319. [DOI] [PubMed] [Google Scholar]
- 15.Luo H., Ye F., Chen K., Shen X., Jiang H. Biochemistry. 2005;44:15351–15358. doi: 10.1021/bi051122c. [DOI] [PubMed] [Google Scholar]
- 16.Calvo E., Escors D., Lopez J.A., Gonzalez J.M., Alvarez A., Arza E., Enjuanes L. J. Gen. Virol. 2005;86:2255–2267. doi: 10.1099/vir.0.80975-0. [DOI] [PubMed] [Google Scholar]
- 17.Stohlman S.A., Fleming J.O., Patton C.D., Lai M.M. Virology. 1983;130:527–532. doi: 10.1016/0042-6822(83)90106-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surjit M., Kumar R., Mishra R.N., Reddy M.K., Chow V.T., Lal S.K. J. Virol. 2005;79:11476–11486. doi: 10.1128/JVI.79.17.11476-11486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H., Gill A., Dove B.K., Emmett S.R., Kemp C.F., Ritchie M.A., Dee M., Hiscox J.A. J. Virol. 2005;79:1164–1179. doi: 10.1128/JVI.79.2.1164-1179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White T.C., Yi Z., Hogue B.G. Virus Res. 2007;126:139–148. doi: 10.1016/j.virusres.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin L., Shao J., Sun M., Liu J., Xu G., Zhang X., Xu N., Wang R., Liu S. Int. J. Mass Spectrom. 2007;268:296–303. doi: 10.1016/j.ijms.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson G.W., Stohlman S.A., Tahara S.M. J. Gen. Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 23.Mohandas D.V., Dales S. FEBS Lett. 1991;282:419–424. doi: 10.1016/0014-5793(91)80528-B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng T.Y., Lee K.R., Tarn W.Y. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeh S.H., Wang H.Y., Tsai C.Y., Kao C.L., Yang J.Y., Liu H.W., Su I.J., Tsai S.F., Chen D.S., Chen P.J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stohlman S.A., Brayton P.R., Fleming J.O., Weiner L.P., Lai M.M. J. Gen. Virol. 1982;63:265–275. doi: 10.1099/0022-1317-63-2-265. [DOI] [PubMed] [Google Scholar]
- 27.Chiu C.M., Yeh S.H., Chen P.J., Kuo T.J., Chang C.J., Chen P.J., Yang W.J., Chen D.S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2571–2578. doi: 10.1073/pnas.0609498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang L.R., Chiu C.M., Yeh S.H., Huang W.H., Hsueh P.R., Yang W.Z., Yang J.Y., Su I.J., Chang S.C., Chen P.J. J. Med. Virol. 2004;73:338–346. doi: 10.1002/jmv.20096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darnell M.E., Taylor D.R. Transfusion. 2006;46:1770–1777. doi: 10.1111/j.1537-2995.2006.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh S.H., Tsai C.Y., Kao J.H., Liu C.J., Kuo T.J., Lin M.W., Huang W.L., Lu S.F., Jih J., Chen D.S., Chen P.J. J. Hepatol. 2004;41:659–666. doi: 10.1016/j.jhep.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 31.Fiol C.J., Mahrenholz A.M., Wang Y., Roeske R.W., Roach P.J. J. Biol. Chem. 1987;262:14042–14048. [PubMed] [Google Scholar]
- 32.Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Surjit M., Liu B., Chow V.T., Lal S.K. J. Biol. Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Y., Hauns K., Langland J.O., Jacobs B.L., Hogue B.G. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Law L.J., Ilkow C.S., Tzeng W.P., Rawluk M., Stuart D.T., Frey T.K., Hobman T.C. J. Virol. 2006;80:6917–6925. doi: 10.1128/JVI.01152-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law L.M., Everitt J.C., Beatch M.D., Holmes C.F., Hobman T.C. J. Virol. 2003;77:1764–1771. doi: 10.1128/JVI.77.3.1764-1771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denison M.R., Spaan W.J., van der Meer Y., Gibson C.A., Sims A.C., Prentice E., Lu X.T. J. Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meer Y., Snijder E.J., Dobbe J.C., Schleich S., Denison M.R., Spaan W.J., Locker J.K. J. Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]