Dear Editor,
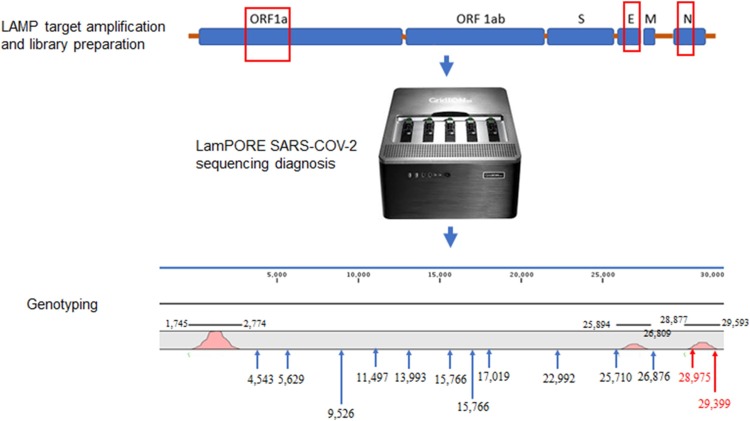
The direct diagnosis of COVID-19 is mainly based on the reverse-transcription PCR (RT-PCR) detection of the SARS-CoV-2 RNA [1,2]. Recently, the LamPORE process has been developed for detecting the viral genome using reverse-transcription loop-mediated amplification (RT-LAMP) and sequencing the RT-LAMP product, following the LamPORE protocol on an Oxford Nanopore GridION instrument (Oxford Nanopore, Oxford, UK) [3]. Here, 264 nasopharyngeal swabs routinely submitted to BGI’s Real-Time Fluorescent RT-PCR kit (BGI, Wuhan, China) (61 positive and 203 negative samples) at the IHU Méditerranée Infection, Marseille, France, were investigated in parallel using the LamPORE procedure. Briefly, the LamPORE procedure combines an isothermal amplification step for 30 min at 65 °C, performed in a 50-μL reaction containing 20 μL RNA previously extracted on a Kingfisher instrument, using the MagMax™Viral/Pathogen Kit (ThermoFisher, St. Austin, USA), 5 μL primer mix and 25 μL of LamPORE master mix, targeting three SARS-CoV-2 genes: ORF1a, the envelope (E) and nucleocapsid (N) genes in addition to the human actin mRNA as an internal control. In addition, 2 μL of each amplified sample was incorporated into the LamPORE library preparation, as previously described [3] and the library was sequenced on a GridION instrument for one hour. LamPORE assays incorporated water as negative control into the process, as provided for by the manufacturer. The results of the LamPORE analysis were interpreted as positive or negative and were automatically generated in a pdf file as follows: the test was considered positive when the sum of reads generated by the three targets ≥ 50, and negative when the number of reads < 20. A test was considered to be inconclusive when the number of reads was between 20 and 50. To eliminate any source of contamination, the positive and negative samples were separated by two empty columns on the plate within the library preparation. Using the strict interpretation algorithm proposed by the manufacturer, all 61 nasopharyngeal swabs which were routinely positive for SARS-CoV-2 were found to be positive by LamPORE (100 % sensitivity) but 23/203 of RT-PCR negative nasopharyngeal swabs were identified as being positive by LamPORE (88.7 % specificity). These 23 nasopharyngeal swabs exhibited RT-PCR Ct values > 40, reflecting a lack of SARS-CoV-2 viability [4,5]. Referring to the RT-PCR results, the LamPORE reaction was positive for the three targets in all the RT-PCR-positive samples, contrary to the 23 discordant swabs. Analyzing the 23 discordant results, we observed that for a result to be consistent with RT-PCR, all target genes should have at least one read mapped on them and the sum of the three targets should also read ≥50. Applying this new interpretation rule allowed us to achieve 100 % sensitivity and 100 % specificity, in the 61-sample collection we investigated. This new interpretation rule consisted in eliminating false positives by applying the “=IF(AND((ORFa1+N + E≥50);(ORFa1*N*E>0));"pos";"neg")” formula directly in the Excel file. In a second step, we used sequences to detect the SARS-CoV-2 strain Marseille genotype 4 also referred as 20AEU2, the most prevalent genotype in the Marseille area in the period under consideration. LamPORE theoretically generates substitutions G28,975 T and G29,399A in the nucleocapsid gene, among the 13 mutations specific for genotype 4 (20AEU2) (Fig. 1 ). Here, LamPORE detected two Marseille genotype 4, based on two sequences exhibiting only the G28,975 T mutation and one sequence exhibiting the mutation G29,399A.
Fig. 1.
LamPORE diagnosis strategy for SARS-COV-2 detection and genotyping. LAMP amplification and library preparation were followed by library sequencing on the LamPORE instrument. A genotyping step was added to specifically detect SARS-COV-2 genotype 4.
Although preliminary, the data here reported confirm that LamPORE is an appropriate method for the rapid direct detection of SARS-CoV-2 RNA in nasopharyngeal swabs, with a capacity of 480 tests per hour, depending on the adoption of the interpretation rule we report here. This method is also promising for one-shot genotyping of SARS-CoV-2 depending on further experimental improvements, and may offer a new, alternative way for detecting SARS-CoV-2 and conducting genomic surveillance.
Authors’ contribution
MM contributed to the experimental design, performance of the work, data analysis, interpretation and writing. AH collected samples, carried out data analysis and writing. BL organized the work. DJ conducted bioinformatic data analysis. BM contributed to the experimental design. PEF and DM contributed to critically reviewing the manuscript, interpreting the data, coordinating and directing the work. All authors declare that they have read and approved the manuscript.
Ethics
Only nasopharyngeal residual fluid left from standard-of-care clinical laboratory testing was used. All specimens had been referred to our laboratory for diagnostic purposes between 1 September and 1 November 2020. The study was approved by our Institute’s Ethics Committee under number 2020−016603.
Funding
Madjid Morsli is PhD student supported by the “Fondation Meditérranée Infection”. This work (lab material and reagents) was funded by the French Government under the Investissements d’Avenir (Investments in the Future) programme managed by the Agence Nationale de la Recherche (ANR,fr: National Agency for Research) [reference: Méditerranée Infection 10-IAHU-03]. This work was also supported by Région Le Sud (Provence Alpes-Côte d’Azur) and European funding [ERDF PA 0000320 PRIMMI].
Declaration of Competing Interest
Reagents for the LamPORE instrument were provided by Oxford Nanopore Technology, Oxford, UK. However, the supplier did not interfere in the experimental plan, data interpretation, manuscript preparation or submission.
Acknowledgements
This study was supported by Fondation Méditerranée Infection, IHU Méditerranée Infection, Marseille, France. MM receives a PhD grant from the Fondation Méditerranée Infection.
References
- 1.Chakraborty C., Sharma A.R., Sharma G., Bhattacharya M., Lee S.S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871. https://pubmed.ncbi.nlm.nih.gov/32329877/ Available from: [DOI] [PubMed] [Google Scholar]
- 2.Zheng Z., Yao Z., Wu K., Zheng J. The diagnosis of SARS-CoV2 pneumonia: a review of laboratory and radiological testing results. J. Med. Virol. 2020;92:2420–2428. doi: 10.1002/jmv.26081. https://pubmed.ncbi.nlm.nih.gov/32462770/ John Wiley and Sons Inc, Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James P., Stoddart D., Harrington E.D., Beaulaurier J., Ly L., Reid S.W., et al. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv. 2020 doi: 10.1101/2020.08.07.20161737. 08.07.20161737. Available from: [DOI] [Google Scholar]
- 4.La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P., Gautret P., Raoult D. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39(June 6):1059–1061. doi: 10.1007/s10096-020-03913-9. Epub 2020 Apr 27. PMID: 32342252; PMCID: PMC7185831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Hoang V.T., Colson P., Raoult D., La Scola B. Correlation between 3790 qPCR positives samples and positive cell cultures including 1941 SARS-CoV-2 isolates. Clin. Infect. Dis. 2020;(September) doi: 10.1093/cid/ciaa1491. Epub ahead of print. PMID: 32986798; PMCID: PMC7543373. [DOI] [Google Scholar]