Abstract
Introduction
The SARS-CoV-2 pandemic has been associated with the occurrence since summer 2020 of several viral variants that overlapped or succeeded each other in time. Those of current concern harbor mutations within the spike receptor binding domain (RBD) that may be associated with viral escape to immune responses. In our geographical area a viral variant we named Marseille-4 harbors a S477 N substitution in this RBD.
Materials and methods
We aimed to implement an in-house one-step real-time reverse transcription-PCR (qPCR) assay with a hydrolysis probe that specifically detects the SARS-CoV-2 Marseille-4 variant.
Results
All 6 cDNA samples from Marseille-4 variant strains identified in our institute by genome next-generation sequencing (NGS) tested positive using our Marseille-4 specific qPCR, whereas all 32 cDNA samples from other variants tested negative. In addition, 39/42 (93 %) respiratory samples identified by NGS as containing a Marseille-4 variant strain and 0/26 samples identified as containing non-Marseille-4 variant strains were positive. Finally, 2018/3960 (51%) patients SARS-CoV-2-diagnosed in our institute, 10/277 (3.6 %) respiratory samples collected in Algeria, and none of 207 respiratory samples collected in Senegal, Morocco, or Lebanon tested positive using our Marseille-4 specific qPCR.
Discussion
Our in-house qPCR system was found reliable to detect specifically the Marseille-4 variant and allowed estimating it is involved in about half of our SARS-CoV-2 diagnoses since December 2020. Such approach allows the real-time surveillance of SARS-CoV-2 variants, which is warranted to monitor and assess their epidemiological and clinical characterics based on comprehensive sets of data.
Keywords: SARS-CoV-2, Covid-19, Variant, Marseille-4, qPCR, Diagnosis, Molecular epidemiology
1. Introduction
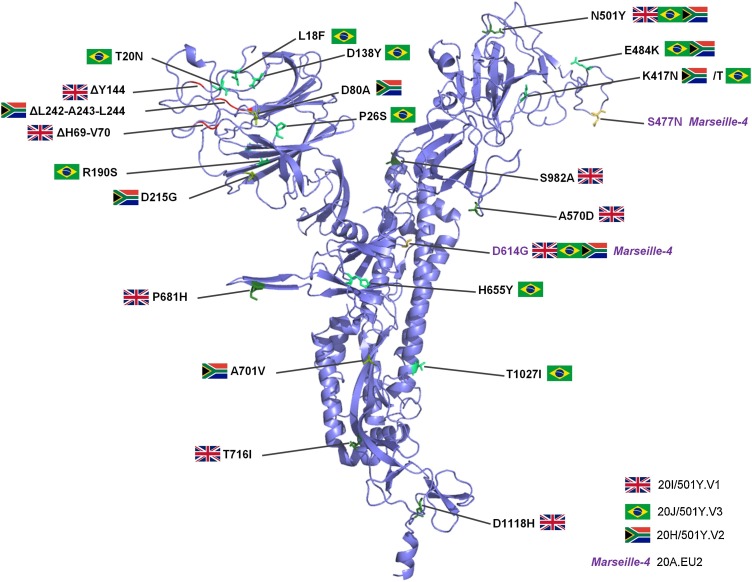
Since the onset of the SARS-CoV-2 pandemic in December 2019 in China, almost 100 million cases have been reported worldwide as on January 28th, 2021 (https://www.ecdc.europa.eu/en/covid-19-pandemic). This has been associated with the occurrence since summer 2020 of several viral variants that overlapped or succeeded each other in time [[1], [2], [3]]. Those of current concern harbor mutations within the spike glycoprotein, particularly within the spike receptor binding domain (RBD) that leads to viral entry into host cells by binding to the ACE2 receptor (Fig. 1 ) [4]. Such SARS-CoV-2 variants include the 20I/501Y.V1 [3], 20 H/501Y.V2 [5], and 20 J/501Y.V3 [6] strains that harbor a N501Y substitution in the spike RBD and were reported in the UK and in South Africa, as highly transmissible, and in Brazil, respectively. In our geographical area we detected 10 viral variants since June 2020 [1]. One of them, we named Marseille-4, harbors a S477 N substitution in the spike RBD that has been associated with an improved binding affinity to ACE2 [6] and a broad resistance to monoclonal neutralizing antibodies [7]. It predominates in Marseille since August 2020, has been reported to spread in Europe since early summer and was classified as the Nextstrain 20A.EU2 lineage [1,2]. The continuous emergence of new SARS-CoV-2 variants, including some of substantial concern regarding their transmissibility and their possible ability to evade immune responses [[8], [9], [10]], warrants to set up strategies for their detection and surveillance. SARS-CoV-2 incidence is currently substantial in several countries including France, and in our institute we for instance diagnose >100 new cases daily. Therefore, alternative strategies to sequencing are useful for variant screening. We aimed to implement an in-house one-step real-time reverse transcription-PCR (qPCR) assay that specifically detects the SARS-CoV-2 Marseille-4 variant.
Fig. 1.
Three-dimensional structure of the SARS-CoV-2 spike protein showing amino acid substitutions and deletions for major SARS-CoV-2 variants including the Marseille-4 variant.
Structure prediction was performed using the Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index) and visualized using the Pymol tool v.1.8 (https://pymol.org/2/). Amino acid substitutions and deletions are showed with a country flag for the variants first detected in UK (20I/501Y.V1), South Africa (20 H/501Y.V2) and Brazil (20 J/501Y.V3), or with the label “Marseille-4.
2. Material and methods
SARS-CoV-2 genomes from our institute sequence database and from the GISAID database (https://www.gisaid.org/ [11]) were used to design a primer pair and a hydrolysis probe. These sequences target a fragment of the nsp4 gene that contains nucleotide position 9,526 of the viral genome [in reference to genome GenBank Accession no. NC_045512.2 (Wuhan-Hu-1 isolate)] where is located a hallmark mutation G > U of the SARS-CoV-2 Marseille-4 variant. The sequences of the qPCR primers and probe are shown in Table 1 . The qPCR was performed by adding 5 μL of extracted viral RNA to 15 μL of reaction mixture containing 5 μL of 4X TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher Scientific, Grand Island, NY, USA), 0.5 μL of forward primer (10 pmol/μL), 0.5 μL of reverse primer (10 pmol/μL), 0.4 μL of probe (10 pmol/μL), and 8.6 μL of water. PCR conditions were as follows: reverse transcription at 50 °C for 10 min, then a hold at 95 °C for 20 s followed by 40 cycles comprising a denaturation step at 95 °C for 15 s and a hybridization-elongation step at 60 °C for 60 s. This qPCR was run on a LC480 thermocycler (Roche Diagnostics, Mannheim, Germany).
Table 1.
Primers and probe of the Marseille-4 variant-specific qPCR.
| Name | Sequence (5′-3′) | Positions* |
|---|---|---|
| Primers: | ||
| Pri_IHU_ C4_5_MBF | GAGGTTTAGAAGAGCTTTTGGTGA | 9,460−9,483 |
| Pri_IHU_ C4_5_MBR | CCAGGTAAGAATGAGTAAACTGGTG | 9,549−9,573 |
| Probe (6FAM-labelled): | ||
| Pro_IHU_ C4_5_MBP | CCTTATTTCATTCACTGTACTCTG | 9,520−9,543 |
In reference to SARS-CoV-2 genome GenBank Accession no. NC_045512.2 (Wuhan-Hu-1 isolate). The nucleotide carrying the mutation specific to the Marseille-4 variant is covered by the probe and underlined.
3. Results
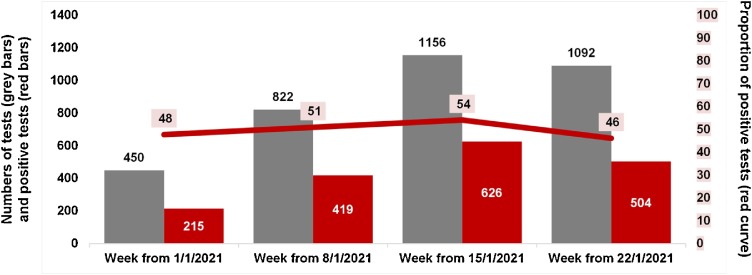
Firstly, we tested a panel of 38 cDNA samples from each of the 10 variants named Marseille-1 to Marseille-10 that we identified by genome next-generation sequencing (NGS) and that circulated since summer 2020 in our geographical area (6 from Marseille-4 strains, 5 from Marseille-5 strains, 5 from Marseille-3 strains, 4 from Marseille-1 strains, and 3 from strains classified in each of the variants Marseille-2, Marseille-6, Marseille-7, Marseille-8, Marseille-9, and Marseille-10) [1]. All 6 Marseille-4 samples tested positive whereas all 32 samples from other variants tested negative. Secondly, we tested 42 respiratory samples identified in our institute by genome NGS [1] as being from patients infected with a SARS-CoV-2 Marseille-4 variant: 39 of them (93 %) were positive using our Marseille-4 specific qPCR. Thirdly, we tested 26 samples identified by next-generation genome sequencing as containing SARS-CoV-2 strains that were not Marseille-4 variants (including 17 N501Y variants, 5 Marseille-2 variants, 3 clade 20A strains and 1 clade 20C strain): none of them were positive using our Marseille-4 specific qPCR. Positive and negative predictive values of Marseille-4 detection by our qPCR were 100 % and 90 %, respectively. Finally, we tested with our Marseille-4 specific qPCR the respiratory samples from 4,339 patients SARS-CoV-2-diagnosed in our institute. None of 22 patients’ samples collected in June, 20 (5.6 %) of 357 patients’ samples collected in July, and 2,018 (51 %) of 3,960 patients’ samples collected in December 2020 and January 2021 tested positive (Fig. 2 ). These results are congruent with those obtained based on genome NGS that showed that the Marseille-4 variant emerged in our geographical area in July and has been predominant since August 2020 [1]. In addition, we found that 10 (3.6 %) of 277 respiratory samples collected in Algeria in September-October tested positive using our Marseille-4 specific qPCR, while none of 94 respiratory samples collected in Senegal in September-October, of 94 samples collected in Morocco in November 2020, and of 19 samples collected in Lebanon in October 2020, tested positive.
Fig. 2.
Number and proportion of Marseille-4 variants detected by qPCR in respiratory samples from patients diagnosed with SARS-CoV-2 in our institute.
The graph shows the weekly numbers of patients tested by our Marseille-4 specific qPCR assay (grey bars) and the weekly numbers (red bars) and proportions (red curve) of positive tests in January 2021. Systematic testing of RNA extracts obtained from nasopharyngeal samples of patients newly-diagnosed with SARS-CoV-2 was implemented in our institute from January 1 st, 2021 using our Marseille-4 specific qPCR assay (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
4. Discussion
Our in-house qPCR system was found reliable to detect specifically the Marseille-4 variant and allowed estimating it is involved in about half of our SARS-CoV-2 diagnoses since December 2020. This assay is currently routinely used in our clinical microbiology and virology laboratory to screen systematically all samples found SARS-CoV-2-positive using the first-line qPCR diagnosis assay, which allows the real-time classification of viral strains in about half of the diagnoses (Fig. 2). In case of negativity of this Marseille-4 specific qPCR, samples are tested using alternative qPCR assays that are specific to other variants that circulate at a lower incidence level than the Marseille-4 variant, or they are submitted to next-generation sequencing in case of cycle threshold value (Ct) <18 with the SARS-CoV-2 qPCR diagnosis test [1,12]. Such approach based on qPCR assays targeting specifically SARS-CoV-2 variants allows their real-time surveillance, which is warranted to monitor and assess their epidemiological and clinical characterics based on comprehensive sets of data. In addition, in-house qPCR assays can be implemented rapidly, easily and at low cost on various open qPCR microplate platforms, which may allow adapting continuously the diagnosis strategies to the emergence and dynamics of SARS-CoV-2 variants.
Credit authorship contribution statement
Conceived and designed the experiments: DR, PC. Contributed materials/analysis tools: MB, PEF, LH, AL, JD, LP, AP, AC, HTD, PB, CS, EA, RS, SM, IB, PC. Analyzed the data: MB, PC, DR. Wrote the paper: PC, MB, DR.
Funding
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU-03 and was also supported by Région Provence Alpes Côte d’Azur and European funding, FEDER PRIMMI (Fonds Européen de Développement Régional-Plateformes de Recherche et d'Innovation Mutualisées Méditerranée Infection), FEDER PA 0000320 PRIMMI.
Funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Ethics
This study has been approved by the ethics committee of our institution (N°2020−029).
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
This manuscript text has been edited by a native English speaker.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jcv.2021.104814.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Fournier P.E., Colson P., Levasseur A., et al. IHU Pre-prints; 2021. Emergence and Outcome of the SARS-CoV-2 “Marseille-4” Variant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodcroft E.B., Zuber M., Nadeau S., et al. 2020. Emergence and Spread of a SARS-CoV-2 Variant Through Europe in the Summer of 2020. medRxiv. [DOI] [PubMed] [Google Scholar]
- 3.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro. Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegally H., Wilkinson E., Giovanetti M., et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;9(March) doi: 10.1038/s41586-021-03402-9. Epub ahead of print. [DOI] [Google Scholar]
- 6.Mejdani M., Haddadi K., Pham C., Mahadevan R. 2021. SARS-CoV-2 Receptor Binding Mutations and Antibody-Mediated Immunity. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., VanBlargan L.A., Bloyet L.M., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell. Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington D., Kele B., Pereira S., et al. Confirmed reinfection with SARS-CoV-2 variant VOC-202012/01. Clin. Infect. Dis. 2021:ciab014. doi: 10.1093/cid/ciab014. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colson P., Finaud M., Levy N., Lagier J.C., Raoult D. Evidence of SARS-CoV-2 re-infection with a different genotype. J. Infect. 2020 doi: 10.1016/j.jinf.2020.11.011. S0163-4453(20)30706-4. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J.I., Burbelo P.D. Reinfection with SARS-CoV-2: implications for vaccines. Clin. Infect. Dis. 2020:ciaa1866. doi: 10.1093/cid/ciaa1866. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haddad G., Bellali S., Fontanini A., et al. Rapid scanning electron microscopy detection and sequencing of severe acute respiratory syndrome coronavirus 2 and other respiratory viruses. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.596180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.