Abstract
A recent outbreak of coronavirus disease 2019 (COVID-19) caused by the single-stranded enveloped RNA virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has developed into a global pandemic, after it was first reported in Wuhan in December 2019. SARS-CoV-2 is an emerging virus, and little is known about the basic characteristics of this pathogen, the underlying mechanism of infection, and the potential treatments. The immune system has been known to be actively involved in viral infections. To facilitate the development of COVID-19 treatments, the understanding of immune regulation by this viral infection is urgently needed. This review describes the mechanisms of immune system involvement in viral infections and provides an overview of the dysregulation of immune responses in COVID-19 patients in recent studies. Furthermore, we emphasize the role of gut microbiota in regulating immunity and summarized the impact of SARS-CoV-2 infection on the composition of the microbiome. Overall, this review provides insights for understanding and developing preventive and therapeutic strategies by regulating the immune system and microbiota.
Keywords: COVID-19, SARS-CoV-2, immune system, microbiome
Introduction
Coronaviruses are RNA viruses that cause respiratory and intestinal infections in animals and humans. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a new group of animal RNA viruses, is the pathogen responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic. On December 31, 2019, Chinese local hospitals and the Center for Disease Control and Prevention in China reported several cases of pneumonia with unknown etiology, and later found a seafood market in China's Wuhan, Hubei Province as the primary location of the infection.1,2 The infecting pathogen was named 2019-novel coronavirus, which was later changed into SARS-CoV-2, and cases have been reported in 47 countries (Figure 1). On January 30, 2020, the World Health Organization declared the novel coronavirus outbreak as an “Emergency of International Concern” and announced it as pandemic on March 11, 2020.3 Up to the end of July 15, 2020, SARS-CoV-2 caused over 15 million confirmed cases and 0.6 million confirmed deaths over the 216 countries,4 which led to huge public health concerns as well as economic burdens worldwide.
Figure 1.

A timeline of COVID-19 development in 2020. Based on information provided by the American Journal of Managed Care (AJMC) website105 and the World Health Organization (WHO).106
It has been suggested that immune responses may contribute to disease severity and progression. However, the mechanism of SARS-CoV-2 virus infection and the process of immune responses against SARS-CoV-2 are still unclear. Other factors that may interfere with immune responses, such as the microbiome, also require more attention. This article aimed to provide a review of current knowledge regarding SARS-CoV-2 on the immune system and how dysregulation of the immune response may contribute to the pathogenesis of COVID-19. Furthermore, we discussed the dysbiosis caused by the virus-related immune system disruption and provide insights into the therapeutic potentials of regulating the immune system and microbiota on the prevention, care management, and treatment of COVID-19.
SARS-CoV-2
According to the phylogenetic classification,5,6 SARS-CoV-2 belongs to the betacoronaviruses out of the four genera of coronaviruses, which includes the alphacoronaviruses, betacoronaviruses, gammacoronaviruses, and deltacoronaviruses.7 Coronaviruses are common pathogens of humans and animals. The word “corona” was named by a similar shape of the outside of the virus as the solar corona of the sun.8 Coronavirus is about 120 nanometers in diameter and is highly diverse. Common cold coronaviruses can cause upper respiratory tract infections in adults as well as diarrhea in infants and children.9
The known hosts for the former two genera are mammals whereas for the latter two are mostly avian.10 The genome structure of the coronavirus has been shown by the Zhang Lab.11 The genome of the coronavirus consists of (1) structural proteins, including envelope glycoproteins spike (S), envelope (E), membrane (M) and nucleocapsid (N), and (2) non-structure proteins (nsps), which are responsible for viral replication and host invasion.12
SARS-CoV-2 shares 88% identity with two bat-derived SARS-like coronaviruses, and about 76% identity with SARS-CoV,10 which was responsible for the outbreak of atypical pneumonia in 2003.13 Homology modeling demonstrated that SARS-CoV-2 and SARS-CoV share a similar receptor-binding domain, indicating that they may share the same receptor.14 Indeed, several groups have identified that the SARS-CoV receptor angiotensin-converting enzyme 2 (ACE2) serves as the receptor for SARS-CoV-2.15 ACE2 is a transmembrane protein and shows high expression on various human tissues, including the small intestine, testis, heart, and lung.16 Blocking the virus entry is one of the common strategies for developing therapeutics against viral infections. According to this strategy, angiotensin-converting enzyme inhibitors/angiotensin receptor blockers are considered as potential therapeutics to prevent infection, accompanied with concerns raised due to the possibility to attenuate the tissue protective function of ACE2.17,18
Apart from the receptor, the serine protease TMPRSS2 has been shown to play an essential role in the S protein cleavage, which primes the S protein and facilitates the entry of SARS-CoV-2 into host cells.19,20 Additionally, another protease, furin, has also been reported to play a role in cleaving between the S1/S2 subunits and priming the S protein.21
Like most of the coronaviruses, SARS-CoV-2 infection causes inflammation within the respiratory track as well as the intestines in humans.7 Common clinical features of SARS-CoV-2 include fever, cough, myalgia, and/or fatigue.22 Lymphopenia is also commonly observed in COVID-19 patients.22 Among severe cases, a high level of proinflammatory cytokines can be found in patient serum,23,24 indicating dysregulated immune responses in severe COVID-19 patients. Up to now, there is no specific therapeutics against SARS-CoV-2, and vaccines are not commercially available yet. Considering the imbalanced immune responses in patients and the essential role of the immune system in fighting against viral infections, it is reasonable to believe that regulating the immune system can potentially contribute to the control and elimination of the viral infection. Therefore, understanding the innate and adaptive immune response in COVID-19 will be important in patient-care management and treatment.
The immune response against viral infections
An effective immune response can be defined as a biological reaction of the host's body to the invasion of foreign substances (eg, viral antigens). The immune responses include innate immune responses and adaptive immune responses. The response time, target antigens, and the mechanism of viral antigen recognition vary between innate and adaptive immune response, and provide a dynamic and broad protection of the host (Figure 2).
Figure 2.
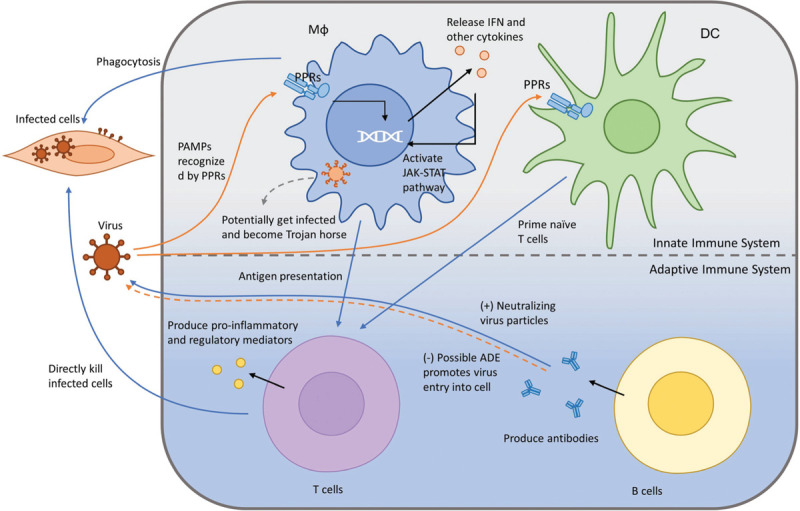
The immune response against viral infections. This figure shows how the immune system response to viral infections. The PRRs from Mϕ recognize PAMPs from the virus and release IFN and other cytokines. Mϕ can potentially get infected and become Trojan horses. PPRs from Mϕ and DC present viral antigens to T cells and B cells. Activated T cells produces pro-inflammatory and regulatory mediators, which kill infected cells. Activated B cells produce neutralizing virus particles to prevent the virus from cellular entry. DC: dendritic cell; IFN: interferon; Mϕ: macrophage; PAMP: pathogen-associated molecular patterns; PPRs: pattern-recognition receptors.
The activation of innate immune responses involves the pattern-recognition receptors (PRRs), including toll-like receptors (TLRs), and nucleotide-binding receptors.25,26 PRRs can recognize the pathogen-associated molecular patterns (PAMPs) such as viral RNA and oxidized phospholipids.27 Once PPRs detect PAMPSs, the intracellular signaling cascades will be activated, which leads to the secretion of proinflammatory effector molecules, including type I interferon (IFN), that play a crucial role in orchestrating antiviral infection immune responses and elimination of the virus.28 Type I IFN then activates the Janus kinase-signal transducer and activator of transcription pathway and promotes the transcription of interferon-stimulated genes, which are involved in host defense, inflammation, and immune modulation.28
Apart from the production of proinflammatory molecules and IFN signaling, the cellular compartments of the innate immune system, including dendritic cells (DCs) and macrophages (Mϕ), also play a vital important role in combating the virus. As the most potent professional antigen presenting cells, DCs can present processed antigen to and prime naive T cells, which are crucial for the initiation of the adaptive immune system.29 Mϕ can also present viral antigens to T cells, beside its function in phagocytosis, releasing cytokines to modulate immune responses as well as tissue repairing.30 However, the functions of Mϕ could be utilized for viral replication. By being engulfed into Mϕ, infected Mϕ becomes a reservoir and help viruses disseminate throughout the body during cellular circulation.29 In addition, chronic and systemic inflammation induced by Mϕ is likely to contribute to fatality as well.29
The adaptive immune system kicks in after the activation of the innate immune system. One of the major players, T cells, is activated after encountering antigen presenting cells bearing virus antigens, and naive and memory T cells undergo a series of events including activation, proliferation, and differentiation. They eventually become effector T cells, which mediate the cellular immune response against viruses.31 The antiviral effects of T cells are mainly in two forms: (1) direct killing of the virus-infected cells and (2) release of regulatory and proinflammatory mediators. The humoral immune response mediated by B cells makes up the other part of the adaptive immune responses. The neutralizing antibodies (NAbs) produced by B cells play a crucial role in restricting the virus life cycle. NAbs can function by binding to the virus particles and preventing its entry into the target cells or binding to the viral glycoprotein on the host cells to inhibit virus budding.32 On the other hand, the virus-specific antibody may facilitate the virus entering the host cells expressing Fc or complementary receptors by interacting with them and getting internalized, which is referred to as antibody-dependent enhancement (ADE). In short, because of the specificity and the immunological memory provided by the adaptive immune system, it is of vital importance that we understand the role it plays in SARS-CoV-2 infection in order to develop vaccines as well as treatments.
The impact of SARS-CoV-2 on the innate immune system
It is common to observe a dysregulation of the innate immune responses in COVID-19 patients. Upregulated IL-6, potentially produced by Mϕ or monocytes, has been reported in COVID-19 patients in multiple studies.22 IL-6 is known to recruit immune mediators and can drive the cytokine release syndromes (CRS), which can cause local tissue damage and systemic non-protective inflammation.33 As CRS has been detected in SARS-CoV-2-infected patients, and it has been previously known to contribute to the morbidity in patients with SARS-CoV or MERS-CoV, clinical trials have been proposed and conducted on the blockade of IL-6 or its upstream activators, such as the blockade of GM-CSF, which activates myeloid cells and promotes the production as IL-6.33 Additionally, inflammatory cytokines and chemokines including IFNγ, MCP1, IP-10, TNF-α, and IL-10 are reported to be evaluated in COVID-19 patients.22 The pyroptosis marker lactate dehydrogenase is also reported to be evaluated in COVID-19 patients, and is thought to be an indicator for disease severity and mortality.34
Although an activated innate immune response was observed in SARS-CoV-2-infected patients, their immune system failed to launch robust type I and type III IFN responses. The lack of IFN responses could potentially lead to deficiency in restricting the establishment of viral infection at early stages33 and contribute to the pathogenicity of COVID-19. An in vitro study indeed revealed that insufficient IFN responses result in higher virus loads in host cells and vice versa supplying exogenous type I IFN results in decreased virus titers in Vero cells infected with SARS-CoV-2.35,36 Interestingly, the virus receptor ACE2 has been reported as an interferon-stimulated gene that upregulates upon IFN I and II stimulation in the human airway epithelial cells,37 which adds complexity to the host-virus interaction. There are limited reports regarding the mechanisms underlying the inhibition of IFN responses by SARS-CoV-2. A few studies suggested that ORF6, ORF8, and nucleocapsid proteins potentially suppress the IFN production.38 Considering that SARS-CoV, which shares 76% similarity with SARS-CoV-239 encodes various IFN antagonists, including the structural proteins membrane and nucleocapsid, nonstructural proteins, Papain-Like Proteases, and accessory proteins,40 the potential antagonism of SARS-CoV-2 needs to be further investigated.
Despite its role in anti-viral immune responses, Mϕ has been reported as susceptible to SARS-CoV-2 infection. The expression of the SARS-CoV-2 receptor ACE2 on the surface of Mϕ may contribute to the pathogenicity of COVID-19.41,42 Indeed, SARS-CoV-2 structural proteins are detectable in macrophages isolated from the spleen, lymph nodes, and lung tissue samples of COVID-19 patients43 and SARS-CoV-2 spike proteins can interact with macrophages.44 The susceptibility of Mϕ to SARS-CoV-2 infection indicates that Mϕ may serve as a potential viral reservoir and CD68+CD169+ Mϕ have been detected to contain SARS-CoV-2 antigens during SARS-CoV-2 infection.45
The impact of SARS-CoV-2 on the adaptive immune system
Humoral immunity plays a vital role in the host immune response against viruses. Particularly, the NAbs can potentially block the interaction between the SARS-CoV-2 spike protein and ACE2 on the cell membrane and thus prevent the entry of the virus. Antibody responses can be detected as early as the first-week post symptom onset, and most patients show antibody responses within 2 weeks after symptom onset.46 Reports showed that immunoglobulin G (IgG) and immunoglobulin M (IgM) against SARS-CoV-2 nucleocapsid (NP) protein and membrane protein (M) have been detected in patients.47 Seroconversion for IgG and IgM are observed simultaneously or sequentially within 20 days post symptom onset, 6 days after which both IgG and IgM titers reach their plateau.46 IgG is found to persist longer compared to IgM and it is also widely observed in discharged patients.47 NAbs have been detected in patients within their third week after disease onset,48,49 and most of the discharged patients show persistent NAb titers.47 Of note, the titer of NAbs is found to be positively correlated with disease severity,50 and the correlation between NAb levels and the number of viral-specific T cell counts is significant.47 However, there are various levels of antibody responses between different patients, indicating the individual difference in immune responses to viral infection. Considering the similarity of SARS-CoV to SARS-CoV-2, cross-reactive NAbs have been reported by several groups.14 This is highly instructive for vaccine development.
While NAbs provide antiviral immunity, non-NAbs could enhance SARS-CoV-2 infection through ADE, which can induce sustained inflammation, lymphopenia, and CRS.51 Research on SARS-CoV showed that ADE contributes to the severity of the disease.51 Although ADE has not been well-studied in COVID-19 patients, there is a possibility that ADE could contribute to the disease progression.
Another aim of adaptive immunity is cellular immunity against viral infections, with CD4+ T cells releasing cytokines, which help B cells and cytotoxic T cells. After being activated, the CD8+ T cells eliminate the infected cells. T cell reactivity against SARS-CoV-2 can be detected about 1-week post symptom onset.21 Specific T cell reactions were detected against the S, M, NP as well as non-structural proteins.47,52 Similar to the antibody response, specific T cell reactivity against SARS-CoV-2 was detected in some unexposed healthy donors, indicating a potential cross-reactivity in some individuals.52
Peripheral CD4+ and CD8+ T cell depletion has been observed in COVID-19 patients and especially in severe cases. For instance, IFNγ produced by CD4+ T cells is decreased in severe patients.53,54 The decrease of peripheral CD4+ and CD8+ T cells has also been reported in SARS-CoV infections and was related to the onset of disease.55 However, whether T cell depletion correlates with severity of COVID-19 needs to be further addressed. In addition, functional exhaustion of T cells has been reported in COVID-19 patients, indicated by the upregulation of exhaustion markers, including PD-1, CTLA-4, TIGIT, and TIM-3 and downregulation of IFN-γ, TNF-α, granzyme B, and IL-2.56 Of note, PD-1 is upregulated in both CD4+ and CD8+ T cell subsets in severe patients.56 A potential cause of the T cell exhaustion could be the inflammatory cytokines, such as IL-6, which is known to induce T cell exhaustion and has been shown to be elevated in COVID-19 patients.56 Taking together, targeting T cell depletion and exhaustion may help to provide appropriate immunity and bring therapeutic benefits in terms of fighting SARS-CoV-2 infection.
Role of microbiota in innate and adaptive immunity and SARS-CoV-2 infection
Microbiota consists of various microorganisms, including bacteria, viruses, fungi, and protozoa.57–59 They have been found in high density in the intestine and closely regulate homeostasis of the innate and adaptive immune system.60 Gut microbiota provides signals to adjust the immune cells for inducing or repressing an immune response.60 Such signals are essential for functional activation of the immune system under certain circumstances. For example, germ-free mice showed impaired development of lymphocytes61 and failed to sustain production of proinflammatory cytokines after repeated TLR stimulation.62 Therefore, attention has been drawn on how the microbiome is interacting with the immune system, and how we can modulate such interactions.
A functional antiviral immune response requires the participation of microbiota. Antibiotics-treated mice are unable to generate robust antibody response against influenza infection.63,64 Also, the absence of TLR5 recognizing flagellin from bacteria leads to reduced antibody production post viral infection.65 Such findings imply that, for B cells to produce healthy antibodies against viral infections, the microbiome is required. The underlining mechanism of such relationship might be explained by a recent study that showed that selection and diversity of the B cell repertoire is regulated by microbiota,66,67 and this regulation might be species-specific, as indicated by another multi-omics study.68 On the other hand, an important cause of COVID-19 related deaths is secondary respiratory bacterial infections,22 and there are evidences that microbiome immune interactions are regulating such secondary infections.69,70 Therefore, we believe that the microbiota is an essential component that not only regulates the antiviral immune response, but also closely affects the disease outcome.
Role of microbiota in functions of innate immunity
Gut commensal microorganisms are normally colonizing the mucous layer of the small and large intestines, which are formatted by specialized epithelial cells and create a physical barrier for the microbiota.71 A change of the components or injuries of the layer would let the microbiota enter the intestinal lamina propria through an opening of the barrier or by uptake by Mϕ, DCs, or M cells. DCs or Mϕ patrolling epithelia of barrier organs would recognize the microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs) from bacteria by their surface PRRs and then the phagocytosis process begins.26
There are different MAMPs or PAMPs from bacteria that have been identified affecting the differentiation and function of lymphoid lineage innate cells in various mechanisms.62 One type of MAMPs is from the microbial components, which act as ligands that can initiate PRR signaling. For example, bacterial flagellin can induce nucleotide-binding receptor signaling through recognition of TLR-5 signaling.72,73 Another type of MAMPs are the microbiota-derived products/metabolites, such as short chain fatty acids (SCFAs),74 and interestingly certain bacteria appear to be major producers of SCFAs. For example, Faecalibacterium prausnitzii, is responsible for the majority of butyrate production.75 These SCFAs affect the activation of innate cells and signaling by inhibiting histone deacetylation.76
Despite the important role of microbiota in regulating innate immunity, several studies have provided evidences that the microbiota tunes the functions of the adaptive immune response. One of the underlying mechanism is through activated Mϕ and DCs carrying engulfed bacteria or epithelium-adhering bacteria to draining lymph nodes, where they initiate the differentiation of naive CD4+ T cells into specialized T cells such as IL-17+CD4+ (Th17) cells.77 Th17 cells are dominated in the lamina propria of the small intestine (30%–40% of differentiated memory CD4+ T cells78), but were absent in germ-free mice or mice treated with antibiotics. Certain species of bacteria were reported to induce differentiation to Th17 cells, including segmented filamentous bacteria (SFB)79 and Bifidobacterium adolescentis.80 Regulatory T cells are mucosal related T cells in the intestine and are known to maintain commensal tolerance.81,82 The development and IL-10 secretion of regulatory T cells is influenced by commensal bacteria Bacteroides fragilis, via microbial product polysaccharide A.83 Therefore, the composition of the intestinal microbiota regulates the balance between Th17 and Treg cells, and influences mucosal immunity and the susceptibility to mucosal related diseases such as bowel diseases.79
Microbiota also talks to the host through unconventional T cells, invariant natural killer T cells, and mucosal-associated invariant T cells. They are less common compared to conventional T cells, but highly abundant in the gut mucosa. These unconventional T cells encode different T cell receptors and recognize antigens that may not be recognized by conventional T cells. Invariant natural killer T cells respond to glycolipid antigen and phospholipids,84 whereas mucosal-associated invariant T cells recognize riboflavin (vitamin B2) metabolic derivatives.85 These unconventional T cells add another layer of complexity to the relationship between microbiota and host immunity.
Humoral mucosal immunity is also affected by microbiota as germ-free mice showed a reduced IgA response, which could be restored rapidly by microbial colonization.78 Gut microbiota are required for a functional B cells in marginal zone86 and can directly regulate B cell activation and differentiation.87 In addition, the process of class switch recombination can also be affected by microbiota.88
Changes in the microbiota during SARS-CoV-2 infection
There are evidences suggesting that the gut microbiota affects pulmonary health through gut-lung cross talk.89 Such crosstalk is bidirectional, which means microbial products/metabolites can influence the homeostasis of the lung, and the pathology of the lung would also affect the gut microbiota.90 Several studies identified the presence of lung microbiota and revealed a distinct microbiome that was relevant to the chronic progression of lung diseases.91 Understanding such crosstalk will provide important knowledge on the role of microbiota regarding viral infection and host defenses.
Although it is widely known that viral infection could affect the gut microbiome,92 the number of current studies on determination of the changes in composition of microbiota in COVID-19 patients is limited. COVID-19 infection is associated with acute respiratory distress syndrome,93 for which microbiota is surely involved in disease progression.94 Also, it has been confirmed that SARS-CoV-2 could infect gut tissue,95 and viral RNA was detected in COVID-19 patient fecal samples.96 More direct measurements of gut microbiota composition showed that gut microbial communities were significantly changed in COVID-19 patients compared to healthy controls.97 One study pointed out that the abundance of the Ruminococcaceae family and several genera from the Lachnospiraceae family were dramatically reduced in COVID-19 patients. Also, compared to H1N1 patients, COVID-19 patients had a unique gut microbiota signature, suggesting potential biomarkers for COVID-19 infection and potential targets for future therapy. Another study linked the anti-inflammatory bacteria F. prausnitzii to be negatively correlated with disease severity. Several studies pointed out the increase of opportunistic pathogens in gut microbiota post SARS-CoV-2 infections,98 which are proposed to be the cause or complications for gut related disorders, such as inflammatory bowel disease.99 Several insights already pointed out the potential mechanisms underlying this gut-lung axis signal cross-talking in COVID-19 patients,100 including the modulation of the immune system through SCFA,101 or mitochondrial oxidative stress related dysbiosis.102 Of note, one review pointed out the age-related dysbiosis might worsen the disease outcome of COVID-19 patients.103 In summary, understanding the role of microbiota will not only help understanding the SARS-CoV-2 infection process, but also provide guidance on disease treatment, such as the use of probiotics.104
Conclusions
SARS-CoV-2 has become a global threat to health and economics. The immune response in SARS-CoV-2 infection plays an important role and significant dysregulation of innate and adaptive immune responses may contribute to disease severity and progression. However, the mechanism of interaction between virus infection and immune response regulation and the relationship of immune response to other medical complications (eg, health and lung failure) are still unclear. In addition, other factors that may regulate immune response and disease onset such as microbiome need further investigation. In this review, we focus on the current knowledge regarding how the immune system reacts after SARS-CoV-2 infection, and emphasize the importance of understanding the role of microbiota in this infection process. We hope this review will provide information and guidance for future research related to the understanding and treatment of COVID-19 patients.
References
- [1].Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis 2020;94:44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet 2020;395(10223):470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. Published July 20, 2020. Available from: https://covid19.who.int/. Accessed November 4, 2020. [Google Scholar]
- [5].Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall 2017;1(1):33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].GISAID. Genomic epidemiology of hCoV-19. Published 2020. Available from: https://www.gisaid.org/epiflu-applications/phylodynamics/. Updated November 11, 2020. Accessed November 4, 2020. [Google Scholar]
- [7].Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].The Centre for Evidence-Based Medicine Develops padbefh. Coronaviruses – A General Introduction. Published 2020. Available from: https://www.cebm.net/covid-19/coronaviruses-a-general-introduction/. Updated March 25, 2020. Accessed November 4, 2020. [Google Scholar]
- [9].Date Ut. Coronaviruses. Published 2020. Available from: https://www.uptodate.com/contents/coronaviruses. Updated June 11, 2020. Accessed November 4, 2020. [Google Scholar]
- [10].Tang Q, Song Y, Shi M, et al. Inferring the hosts of coronavirus using dual statistical models based on nucleotide composition. Sci Rep 2015;5:17155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Lab. Genome-wide Structure and Function Modeling of SARS-COV-2. Available from: https://zhanglab.ccmb.med.umich.edu/COVID-19/. Updated October 29 2020. Accessed November 4, 2020. [Google Scholar]
- [12].Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect 2020;31. S1684-1182 (20)30082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zheng BJ, Wong KH, Zhou J, et al. SARS-related virus predating SARS outbreak, Hong Kong. Emerg Infect Dis 2004;10(2):176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tai W, He L, Zhang X, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020;17(6):613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020;581(7807):221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li MY, Li L, Zhang Y, et al. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 2020;9(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kuba K, Imai Y, Rao S, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 2005;11(8):875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Guo J, Huang Z, Lin L, et al. Coronavirus disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. J Am Heart Assoc 2020;9(7):e016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zang R, Gomez Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 2020;5(47):eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].di Mauro G, Cristina S, Concetta R, Francesco R, Annalisa C. SARS-CoV-2 infection: response of human immune system and possible implications for the rapid test and treatment. Int Immunopharmacol 2020;84:106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Girija ASS, Shankar EM, Larsson M. Could SARS-CoV-2-induced hyperinflammation magnify the severity of coronavirus disease (COVID-19) leading to acute respiratory distress syndrome? Front Immunol 2020;11:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012;12(4):496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bingula R, Filaire M, Radosevic-Robin N, et al. Desired turbulence? Gut-lung axis, immunity, and lung cancer. J Oncol 2017;2017:5035371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol 2014;14(5):315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol 2014;14(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rossi M, Young JW. Human dendritic cells: potent antigen-presenting cells at the crossroads of innate and adaptive immunity. J Immunol 2005;175(3):1373–1381. [DOI] [PubMed] [Google Scholar]
- [30].Nikitina E, Larionova I, Choinzonov E, et al. Monocytes and macrophages as viral targets and reservoirs. Int J Mol Sci 2018;19(9):2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 2012;12(4):295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Murin CD, Wilson IA, Ward AB. Antibody responses to viral infections: a structural perspective across three different enveloped viruses. Nat Microbiol 2019;4(5):734–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang C, Wu Z, Li JW, et al. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents 2020;55(5):105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shi J, Li Y, Zhou X, et al. Lactate dehydrogenase and susceptibility to deterioration of mild COVID-19 patients: a multicenter nested case-control study. BMC Med 2020;18(1):168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181(5):1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mantlo E, Bukreyeva N, Maruyama J, et al. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antiviral Res 2020;179:104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181(5):1016–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li JY, Liao CH, Wang Q, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 2020;286:198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jaimes JA, Andre NM, Chappie JS, et al. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 2020;432(10):3309–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2012;2(3):264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Keidar S, Strizevsky A, Raz A, et al. ACE2 activity is increased in monocyte-derived macrophages from prehypertensive subjects. Nephrol Dial Transplant 2007;22(2):597–601. [DOI] [PubMed] [Google Scholar]
- [42].Zhang C, Zhao YX, Zhang YH, et al. Angiotensin-converting enzyme 2 attenuates atherosclerotic lesions by targeting vascular cells. Proc Natl Acad Sci U S A 2010;107(36):15886–15891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bostanciklioglu M. SARS-CoV2 entry and spread in the lymphatic drainage system of the brain. Brain Behav Immun 2020;87:122–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang C, Xie J, Zhao L, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine 2020;57:102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Park MD. Macrophages: a Trojan horse in COVID-19? Nat Rev Immunol 2020;20(6):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 2020;71(16):2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ni L, Ye F, Cheng ML, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 2020;52(6):971–977.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26(6):845–848. [DOI] [PubMed] [Google Scholar]
- [49].Sun B, Feng Y, Mo X, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect 2020;9(1):940–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Choe PG, Kang CK, Suh HJ, et al. Antibody responses to SARS-CoV-2 at 8 weeks postinfection in asymptomatic patients. Emerg Infect Dis 2020;26(10):2484–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect 2020;22(2):72–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181(7):1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;130(5):2620–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Diao B, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19). Front Immunol 2020;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].He Z, Zhao C, Dong Q, et al. Effects of severe acute respiratory syndrome (SARS) coronavirus infection on peripheral blood lymphocytes and their subsets. Int J Infect Dis 2005;9(6):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zheng HY, Zhang M, Yang CX, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol 2020;17(5):541–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009;136(1):65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature 2016;535(7610):56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lazar V, Ditu LM, Pircalabioru GG, et al. Aspects of gut microbiota and immune system interactions in infectious diseases, immunopathology, and cancer. Front Immunol 2018;9:1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Negi S, Pahari S, Bashir H, et al. Gut microbiota regulates mincle mediated activation of lung dendritic cells to protect against Mycobacterium tuberculosis. Front Immunol 2019;10:1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bauer H, Horowitz RE, Levenson SM, et al. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol 1963;42(4):471–483. [PMC free article] [PubMed] [Google Scholar]
- [62].Weaver LK, Minichino D, Biswas C, et al. Microbiota-dependent signals are required to sustain TLR-mediated immune responses. JCI Insight 2019;4(1):e124370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 2011;108(13):5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bradley KC, Finsterbusch K, Schnepf D, et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep 2019;28(1):245–256. [DOI] [PubMed] [Google Scholar]
- [65].Oh JZ, Ravindran R, Chassaing B, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014;41(3):478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chen H, Zhang Y, Ye AY, et al. BCR selection and affinity maturation in Peyer's patch germinal centres. Nature 2020;582(7812):421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li H, Limenitakis JP, Greiff V, et al. Mucosal or systemic microbiota exposures shape the B cell repertoire. Nature 2020;584(7820):274–278. [DOI] [PubMed] [Google Scholar]
- [68].Hagan T, Cortese M, Rouphael N, et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 2019;178(6):1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Deriu E, Boxx GM, He X, et al. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog 2016;12(5):e1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Lee KH, Gordon A, Shedden K, et al. The respiratory microbiome and susceptibility to influenza virus infection. PLoS One 2019;14(1):e0207898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Brown EM, Kenny DJ, Xavier RJ. Gut microbiota regulation of T cells during inflammation and autoimmunity. Annu Rev Immunol 2019;37:599–624. [DOI] [PubMed] [Google Scholar]
- [72].Hajam IA, Dar PA, Shahnawaz I, et al. Bacterial flagellin-a potent immunomodulatory agent. Exp Mol Med 2017;49(9):e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Kinnebrew MA, Buffie CG, Diehl GE, et al. Intestinal CD103+ CD11b+ lamina propria dendritic cells instruct intestinal epithelial cells to express antimicrobial proteins in response to Toll-like receptor 5 activation. Immunity 2012;36(2):276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Singh N, Thangaraju M, Prasad PD, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem 2010;285(36):27601–27608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhou L, Zhang M, Wang Y, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis 2018;24(9):1926–1940. [DOI] [PubMed] [Google Scholar]
- [76].Luhrs H, Gerke T, Muller JG, et al. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol 2002;37(4):458–466. [DOI] [PubMed] [Google Scholar]
- [77].Ivanov II, McKenzie BS, Zhou L, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006;126(6):1121–1133. [DOI] [PubMed] [Google Scholar]
- [78].Hapfelmeier S, Lawson MA, Slack E, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 2010;328(5986):1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008;4(4):337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tan TG, Sefik E, Geva-Zatorsky N, et al. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A 2016;113(50):E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cebula A, Seweryn M, Rempala GA, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013;497(7448):258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Kamada N, Nunez G. Role of the gut microbiota in the development and function of lymphoid cells. J Immunol 2013;190(4):1389–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kayama H, Takeda K. Polysaccharide A of Bacteroides fragilis: actions on dendritic cells and T cells. Mol Cell 2014;54(2):206–207. [DOI] [PubMed] [Google Scholar]
- [84].Burdin N, Brossay L, Koezuka Y, et al. Selective ability of mouse CD1 to present glycolipids: alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. J Immunol 1998;161(7):3271–3281. [PubMed] [Google Scholar]
- [85].Gold MC, Cerri S, Smyk-Pearson S, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol 2010;8(6):e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Wei B, Su TT, Dalwadi H, et al. Resident enteric microbiota and CD8+ T cells shape the abundance of marginal zone B cells. Eur J Immunol 2008;38(12):3411–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kim M, Kim CH. Regulation of humoral immunity by gut microbial products. Gut Microbes 2017;8(4):392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Choi JH, Wang KW, Zhang D, et al. IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc Natl Acad Sci U S A 2017;114(7):E1196–E1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol 2012;5(1):7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dumas A, Bernard L, Poquet Y, et al. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol 2018;20(12):e12966. [DOI] [PubMed] [Google Scholar]
- [91].Cabrera-Rubio R, Garcia-Nunez M, Seto L, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol 2012;50(11):3562–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bartley JM, Zhou X, Kuchel GA, Weinstock GM, Haynes L. Impact of age, caloric restriction, and influenza infection on mouse gut microbiome: an exploratory study of the role of age-related microbiome changes on influenza responses. Front Immunol 2017;8:1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Fan E, Beitler JR, Brochard L, et al. COVID-19-associated acute respiratory distress syndrome: is a different approach to management warranted? Lancet Respir Med 2020;8(8):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Dickson RP. The lung microbiome and ARDS. It is time to broaden the model. Am J Respir Crit Care Med 2018;197(5):549–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020;69(6):1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wong MC, Huang J, Lai C, et al. Detection of SARS-CoV-2 RNA in fecal specimens of patients with confirmed COVID-19: a meta-analysis. J Infect 2020;81(2):e31–e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020;159(3):944–955.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zuo T, Liu Q, Zhang F, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2020;70(2):276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Trottein F, Sokol H. Potential causes and consequences of gastrointestinal disorders during a SARS-CoV-2 infection. Cell Rep 2020;32(3):107915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dhar D, Mohanty A. Gut microbiota and COVID-19 - possible link and implications. Virus Res 2020;285:198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Onishi JC, Haggblom MM, Shapses SA. Can dietary fatty acids affect the COVID-19 infection outcome in vulnerable populations? mBio 2020;11(4):e01723–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Saleh J, Peyssonnaux C, Singh KK, Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion 2020;54:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Viana SD, Nunes S, Reis F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities - role of gut microbiota dysbiosis. Ageing Res Rev 2020;62:101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Giannoni E, Baud D, Agri VD, Gibson GR, Reid G. Probiotics and COVID-19. Lancet Gastroenterol Hepatol 2020;5(8):720–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].American Journal of Managed Care. A Timeline of COVID-19 Developments in 2020. Published 2020. Available from: https://www.ajmc.com/view/a-timeline-of-covid19-developments-in-2020. Updated July 3, 2020. Accessed November 4, 2020. [Google Scholar]
- [106].World Health Organization. Timeline of WHO's Response to COVID-19. Published 2020. Available from: https://www.who.int/news/item/29-06-2020-covidtimeline. Updated September 9, 2020. Accessed November 06, 2020. [Google Scholar]


