Abstract
Finding relevant information from newly published scientific papers is becoming increasingly difficult due to the pace at which articles are published every year as well as the increasing amount of information per paper. Biocuration and model organism databases provide a map for researchers to navigate through the complex structure of the biomedical literature by distilling knowledge into curated and standardized information. In addition, scientific search engines such as PubMed and text-mining tools such as Textpresso allow researchers to easily search for specific biological aspects from newly published papers, facilitating knowledge transfer. However, digesting the information returned by these systems—often a large number of documents—still requires considerable effort. In this paper, we present Wormicloud, a new tool that summarizes scientific articles in a graphical way through word clouds. This tool is aimed at facilitating the discovery of new experimental results not yet curated by model organism databases and is designed for both researchers and biocurators. Wormicloud is customized for the Caenorhabditis elegans literature and provides several advantages over existing solutions, including being able to perform full-text searches through Textpresso, which provides more accurate results than other existing literature search engines. Wormicloud is integrated through direct links from gene interaction pages in WormBase. Additionally, it allows analysis on the gene sets obtained from literature searches with other WormBase tools such as SimpleMine and Gene Set Enrichment.
Database URL: https://wormicloud.textpressolab.com
Introduction
Given the overwhelming and constantly growing number of research papers published in biomedical research, finding relevant information from the scientific literature has become a challenging task. There are many strategies researchers have adopted over time in order to keep pace with recent scientific discoveries, such as subscribing to topic-based research blogs or setting up alerts on scientific search platforms such as PubMed MEDLINE or Europe PMC (Europe PubMed Central) (1). These platforms provide tools for querying relevant documents by free-text searches or controlled vocabulary to help researchers overcome the information overload by suggesting the most relevant list of articles. However, digesting information from the scientific articles returned by the queries may still be challenging, especially for large numbers of papers.
Biological data curation (biocuration) is aimed at extracting valuable knowledge from experimental results in the literature and making the extracted information readily available to researchers in an easy-to-interpret format (2). Curated data regarding specific model organisms are maintained by model organism databases such as WormBase, the main information resource for the nematode Caenorhabditis elegans (3), among others, and by the recently formed Alliance of Genome Resources (4). Although biocuration is vital for modern biological research, it is mostly a manual process performed by expert curators, and there is a considerable time lag between the publication of an article and the inclusion of curated data into model organism databases. Finding new scientific results in the literature in a timely fashion would be greatly beneficial for both bench scientists and biocurators.
One of the possible solutions to overcome the information overload is to automatically generate summaries of collections of scientific articles such that key aspects of new results can be easily digested without reading all the articles in the collection. Different summarization solutions have been proposed in the literature, including automated generation of text summaries using computational linguistic techniques (5) or graphical summary generation and visualization methods, such as graphical summaries based on word clouds.
Word clouds (also known as tag clouds or wordles) are visual representations of text data that depict the most important keywords in one or more textual documents such as cloud-shaped collections of words with different sizes, colors, orientations and fonts. The importance of each keyword is measured by its frequency of appearance in the source documents, and it is reflected in the different size of the words in the word cloud in order to make important keywords more visible than others.
Word clouds have been largely used on the web to summarize information and have become very popular with the advent of Web 2.0 and of social media such as Flickr (www.flickr.com) and Delicious (http://del.icio.us/, a social bookmarking service discontinued in 2019). In this context, they were mainly used to aid website navigation through visual representation of page content. However, overuse and the somewhat limited effectiveness of this specific application led to a gradual decline of their popularity. They have been recently rediscovered as powerful tools for data summarization and analysis, and their effectiveness has been formally analyzed by several research studies (6, 7).
A handful of tools that summarize research articles through word clouds have been proposed in the literature, even though, at the time of writing this paper, all of these were not accessible online or were not properly functioning. Perhaps the most promising of these tools in the context of biology is Genes2WordCloud (8). This tool generates word clouds from a list of genes or keywords by searching documents from different sources, including biological annotations from the Gene Ontology consortium (9, 10) and abstracts of scientific articles from PubMed (https://pubmed.ncbi.nlm.nih.gov/). The resulting word clouds summarize information related to the provided genes or keywords, highlighting prominent research topics in the related articles.
Another word cloud tool that was designed to summarize information in research articles from PubMed is LigerCat (11). This tool presents MeSH terms (the keywords in the controlled vocabulary used to index papers in PubMed) of articles retrieved through PubMed search API to build word clouds. Even though this technique proved to be effective in summarizing research aspects of a collection of articles, it excludes words not indexed as MeSH terms by PubMed and may therefore miss some important keywords not yet in the controlled vocabulary.
Kuo et al. (12) designed a tool that allows users to summarize the results of PubMed searches through word clouds based on words extracted from abstracts. This tool presents a simple interface but does not provide additional tools for data analysis such as word trends over time.
In this paper, we present Wormicloud (worm information cloud), a novel visual tool based on word clouds that summarizes knowledge about specific research topics from a large amount of textual documents and facilitates new discoveries from data not yet curated by model organism databases. Wormicloud uses keywords from abstracts and gene names mentioned in the full text of research articles to generate word clouds, thanks to the advanced search functionality provided by the Textpresso Central text-mining system (13). Textpresso allows fine-grained searches on keywords and categories specifically based for C. elegans and other model organisms (e.g. gene names and other biological entities extracted from articles). In particular, it provides searches on full text of articles, whereas PubMed searches exclusively on abstracts. This makes Wormicloud able to find more articles relevant to specific biological aspects than those that can be found via PubMed searches.
Wormicloud helps users dynamically refine their searches by adding keywords with a click on the displayed words to narrow down the original list of documents until the desired level of detail in the search is reached. Most importantly, Wormicloud does not depend on any manual curation. All the processes, from text mining to presentation of word clouds, are performed automatically, thereby ensuring that the data presented to the user is always up-to-date and complete. Wormicloud includes also a graphical word trends analysis tool that allows the user to trace the use of specific words in the obtained word clouds over time. Wormicloud is integrated into WormBase and can also be used in combination with other bioinformatics tools, such as SimpleMine (https://wormbase.org/tools/mine/simplemine.cgi) and Gene-set enrichment analysis tool (https://wormbase.org/tools/enrichment/tea/tea.cgi) from WormBase.
Methods
Wormicloud is structured into a backend and a frontend component, as depicted in Figure 1. The frontend allows users to perform keyword-based searches and displays the articles matching the search parameters in the form of word clouds. The interface also has an interactive reference list with the details about the articles used to build the word cloud, and a word trends analysis tool that displays the usage of specific words in the word cloud over time. More details on the frontend are provided in the Results section, where we describe the implementation of the UI and provide some use cases to show how it can be used for different research-oriented tasks.
Figure 1.
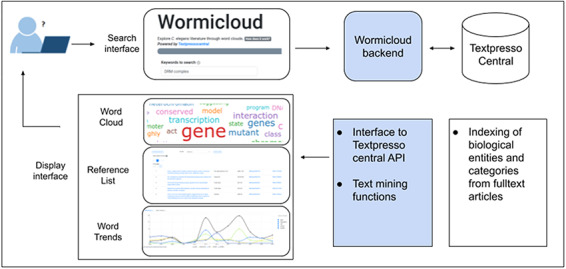
Wormicloud components and interactions with Textpresso Central through the Textpresso API.
In this section, we focus on the backend component, which searches for scientific articles by interfacing with the Textpresso Central Application Programmer Interface (API) and extracts lists of words from these articles with their respective frequency counters. The backend is geared towards C. elegans specific literature and nomenclature, but we plan to expand it to other organisms in the future, as explained in the Discussion section.
Textpresso (13) allows programmatic access to its functions through a public API (https://textpressoapi.readthedocs.io/en/latest/?badge=latest). This API allows keywords- and category-based full-text searches on scientific articles, including all C. elegans papers included in WormBase. The API returns abstracts and full text of the articles matching the search criteria, but it also provides the list of words in the text of each of these articles belonging to a specific category. Wormicloud backend uses the API to retrieve documents containing a list of user-provided keywords through full-text searches and combines the words in the abstracts of returned articles to build word clouds. Using abstracts only makes articles retrieval from Textpresso Central faster. In fact, even though searches are performed on the full text, the text of the matching articles is not retrieved automatically due to specific Textpresso internal optimization rules. Therefore, accessing the full text of a large set of articles would require too much time to provide an acceptable user experience. In addition, abstracts usually describe the core aspects of the research work, and the full text often includes related work and discussions on previous results that could add noise to the resulting word clouds. Wormicloud also uses the API to retrieve all genes, sequence names and protein names in the articles that match the search criteria, in order to build word clouds containing these entities, called ‘gene names only clouds’ in the user interface. Protein names are transformed to match their related gene names by converting them to lowercase. C. elegans gene names are standardized and defined by a specific nomenclature (https://wormbase.org/about/userguide/nomenclature). Textpresso identifies gene names in the full text of articles through regular expressions and matches approved gene names, sequence names and synonyms.
To extract words and counters from the list of abstracts obtained through the Textpresso API, the backend tokenizes the text in each of the abstracts—i.e. it breaks the text into individual linguistic units. We designed a custom tokenizer that considers particular biological entities as single words (e.g. C. elegans gene names, which often contain a dash, such as ‘daf-16’). Then, the resulting tokens are lemmatized in order to group together variations of the same base words. Lemmatization uses morphological analysis of words to identify their root, an approach that works well in the context of biology. Stemming is another possible technique that reduces different forms of a word to their common base form through a heuristic process that cuts off the ends of words. Note that stemming is used instead of lemmatization by all the other word cloud-based summarization tools in the literature. However, we decided to not apply stemming, as it turned out to flatten important differences between biological concepts (e.g. germ and germline). Note that lemmatization alone is not able to group all word variations, but it provides the best results for our use cases. A custom list of stopwords is also used to get rid of keywords considered noise in the C. elegans biological context. The lists of stopwords for the keyword-based clouds and gene only clouds are available in Supplemental Table S1.
The Textpresso API returns a list of articles sorted by a relevance score (hereinafter ‘Textpresso score’) that reflects how well the results match with the provided search parameters, limited to a maximum number that can be controlled through the Wormicloud user interface. This limit makes Textpresso Central searches faster and filters out less relevant articles. We decided to give the user the option to choose between 200, 400 and 1000 as the maximum result number. The default value for searches is set to 200, which is the fastest possible option (since Textpresso returns 200 maximum results per query). As supported by the statistical analysis below, this value is sufficient also for the accuracy of results returned by broad searches, even though the user can still manually set the maximum number of results through the interface to 400 or 1000 for more accurate results. The user can also decide whether to count words in abstracts by plain frequency or by frequency weighted by the Textpresso score received by each paper. In the latter case, words in papers with low Textpresso scores with respect to the search criteria count less than words in papers with higher scores.
Choosing the optimal parameters for Wormicloud searches
We performed a statistical analysis to choose the default maximum number of papers to be fetched from the Textpresso API looking for the best trade-off between search speed and accuracy of the results. We measured how increasing the maximum number of results impacts the retrieval time and accuracy of the resulting list of words and their respective frequencies used to build the word clouds. To do so, we performed searches through the Textpresso API for a broad biological term, which matches a large number of papers. We decided to use the term ‘meiosis’—which returned 2985 documents through a regular search on Textpresso Central (https://www.textpressocentral.org) at the time of writing—and we performed two separate analyses on these searches: (i) we measured the query time for searches with 200, 400 and 1000 maximum number of results; (ii) we calculated similarity measures between the lists of keywords obtained by the Wormicloud backend software processing pipeline and ranked by their frequencies (both plain frequency and weighted by the Textpresso score).
Query times
The Textpresso API has a caching mechanism that makes subsequent searches faster, so we measured the time required for the first query and we also measured the average time for subsequent queries. For non-cached queries, it took 16.18 s for the 200 results query, 68.82 s for 400 results and 123.47 s for the 1000 results one. With caching, the average query time, over 10 observations, was 13.33 s (±0.99 s) for 200 results maximum, 29.28 s (±1.73 s) for 400 and 75.86 s (±8.08 s) for 1000 results. These figures tell us that increasing the number of maximum results returned by Textpresso to more than 200 (the maximum number of results that are packed by Textpresso in a single query) can lead to long search times for the user and this significantly impacts user experience.
Similarity between lists of words obtained with different thresholds
We calculated indices to measure how similar the lists of words obtained by the searches with different maximum number of results are. To do so, we first considered the presence/absence of words across the three lists obtained for the search ‘meiosis’ with different thresholds and calculated the percentage of words in the list for 1000 maximum results which are also in the list with 400 and 200 maximum results. We calculated this percentage both for the full lists and by limiting the analysis to the first 100 words sorted by their counters, as the number of words included in the word clouds displayed in Wormicloud is limited to 100. For the latter analysis, we considered two cases, the first with counters obtained by plain word frequency in the returned documents, and the second with frequencies weighted by the Textpresso score assigned to each document. To further analyze the similarity between the lists of words, we calculated the correlation of the lists ranked by their counters, this time also taking into account variations in the position of each word across lists. Also in this case, we considered both plain frequencies and frequencies weighted by the Textpresso scores as counters. For this analysis, we calculated Kendall’s tau correlation coefficient, a standard coefficient for ranked lists.
Overlapping words between lists
Searches for ‘meiosis’ returned 3877 distinct words with the maximum number of results set to 200, 5960 with 400 and 11 085 with 1000 results. All of the words in the 200 and 400 lists are contained in the 1000 list. The 200 list has a 34.98% overlap with the 1000 list, whereas the 400 list has an overlap of 53.77% with the 1000 list. When taking only the first 100 entries per list into account, ranked by plain word counter, the overlap with the elements in the 1000 list is 74% for the 200 list and 83% for the 400 list. When ranked by counter weighted by Textpresso score, the overlap between the list with 1000 and 200 results is 74%, and the one between 1000 and 400 results is 84%. These results tell us that even though the full list of words with their counters obtained using 200 and 400 maximum results from the Textpresso API is significantly shorter than that obtained from 1000 results (especially the 200 results list), the first 100 words in the lists are quite similar and the resulting word clouds are comparable. Moreover, weighting the counters by the score returned by Textpresso for the relative papers does not significantly change the first 100 words in the lists. Figure 2 gives a visual representation of the word clouds obtained with the three thresholds and plain frequency counts. As can be noted from the figure, the main keywords related to ‘meiosis’ in C. elegans are all present in the three cases. This is another indication that limiting the number of results from Textpresso to 200 does not significantly alter the resulting word cloud. Nonetheless, users can manually select a higher number of results if they want to perform more accurate analyses, especially for word trends and to obtain a more accurate reference list.
Figure 2.
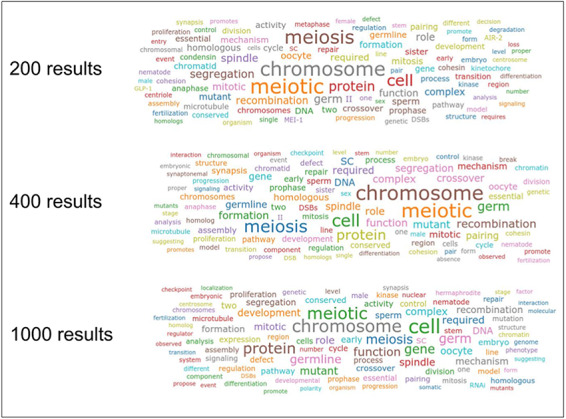
Word clouds for the keyword ‘meiosis’ obtained by combining 200, 400 and 1000 maximum results from the Textpresso API and with plain frequency word counts.
Correlation between ranked lists
For plain counters, the correlation between the list obtained with 1000 maximum results and the one obtained with 200 results is 0.61 (P < 0.01), and it is 0.72 (P < 0.01) for the lists obtained from 1000 and 200 maximum articles, respectively. The correlations do not change when considering the Textpresso score for ranking the words in the lists. When considering the first 100 words in the lists only, the correlation is 0.56 (P < 0.01) and 0.68 (P < 0.01) for the pairs of lists with 1000 and 200, and 1000 and 400 maximum results, respectively. Also, in this case, the correlations do not change significantly when the counters are weighted by Textpresso score. These correlations tell us that the lists of words returned using the different parameters for searches that match large numbers of papers are similar to each other not only in the specific words returned but also in their ranking within the lists. Nonetheless, in use cases where results from Textpresso have large differences in match score, advanced users (Textpresso users in particular) may still want to use counters weighted by Textpresso score. For this reason, we decided to leave the option of choosing which counters to use to the user.
Results
Wormicloud implementation
We developed Wormicloud as a web application available at https://wormicloud.textpressolab.com. The frontend component is written in JavaScript using the React framework, and the backend is a python program based on the Falcon framework (https://falcon.readthedocs.io/en/stable/). The Textpresso Central API, developed in a separate project, is written in C++ (https://textpressoapi.readthedocs.io/en/latest/?badge=latest).
Wormicloud is open source and available at https://github.com/WormBase/wormicloud.
Search interface
The Wormicloud frontend component (UI) allows users to insert a list of keywords and the desired word cloud format (all keywords from abstracts or gene names only from full text). Also, with the ‘Advanced options’ button, users can add additional selectors such as publication year range, author names, the maximum number of articles to be used to build the word clouds, and the method for counting word frequencies, a plain count or weighted by TextpressoCentral paper score. As depicted in Figure 3, keywords can be combined to search for documents containing at least one of them (‘OR’ option) or all of them (‘AND’ option). The ‘AND’ option generates word clouds containing only words that are present in all articles returned by searching each keyword separately instead of the union of all words contained in articles mentioning all the provided keywords. This makes the resulting word clouds more focused on aspects overlapping in all the returned articles. In addition, users can perform searches by author names only, without providing any specific keyword.
Figure 3.
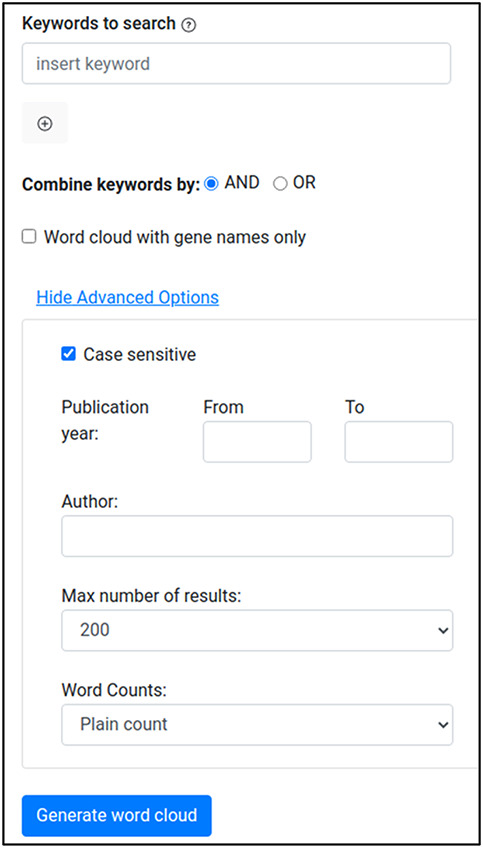
Wormicloud search interface with ‘advanced options’ menu expanded.
Wormicloud displays the results of searches with a combined view of word clouds (Figure 4), reference list (Figure 5) and word trends tool (Figure 6).
Figure 4.
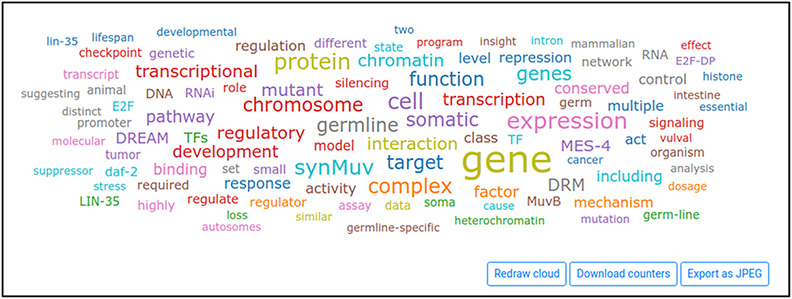
Word cloud displayed by Wormicloud for the query ‘DREAM complex’.
Figure 5.
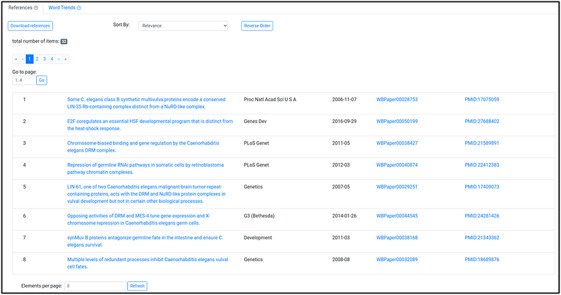
Reference list displaying the articles used to generate the word cloud in Figure 4.
Figure 6.
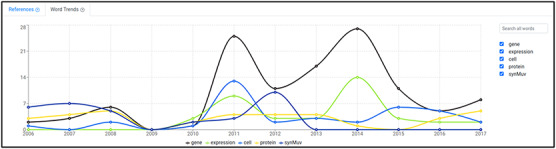
Word trends tool displaying the yearly usage of the first five words by number of mentions in the abstracts of the articles used to generate the word cloud in Figure 4. In this example, the query was ‘DREAM complex’.
Word cloud interface
Word clouds displayed by Wormicloud are based on a React word cloud package (https://www.npmjs.com/package/react-wordcloud). The package takes care of finding the best layout for words and displays them in different colors to maximize readability. The word clouds are interactive in that users can click on each word to add them back to the list of keywords in the search interface. In this way, users can refine their searches to find the most relevant research for their purposes. The word cloud component includes a set of buttons with different functions:
Redraw cloud button
Redraws the word cloud by re-applying the algorithm that positions the words and assigns colors.
Download counters
Downloads a csv file containing all the words obtained from the backend with their counters. Note that this file can contain more words than those displayed in the word cloud, which are limited to 100.
Export as JPEG
Downloads an image of the word cloud in jpeg format.
View on SimpleMine (for gene names word clouds only)
Opens SimpleMine search interface (https://wormbase.org/tools/mine/simplemine.cgi) with the field for the list of genes pre-filled with the gene names in the word cloud. SimpleMine is a WormBase tool for the retrieval of essential gene information.
View on Gene Set Enrichment tool (for gene names word clouds only)
Opens WormBase tissue enrichment analysis tool (https://wormbase.org/tools/enrichment/tea/tea.cgi) with the field for the list of genes pre-filled with the gene names in the word cloud.
Reference list interface
When a word cloud is displayed, users also see a list of the references used to build the word cloud at the bottom of the screen (Figure 5). This is an interactive component that allows users to sort the list of references by relevance (the score received by each article from Textpresso based on how well it matched the search), title, journal, date, and WormBase and PubMed IDs. The list is paginated for easier navigation. In addition, the user can download the list of references to file in csv format.
Word trends interface
The word trends interface (Figure 6) is accessible by clicking the ‘Word Trends’ tab that appears to the right of the ‘References’ tab when a word cloud is generated. This interface allows the user to visualize the number of mentions in abstracts per year of each of the words obtained from the backend and used to build the word cloud. These are displayed as an interactive graph. Note that the number of mentions included in the graph considers abstracts only, as for the word clouds. By default only the five words with the highest counters are displayed, but the user can add more words by searching them through the autocomplete text input on the right. The user can also remove words from the graph by unchecking the checkboxes near each word.
Finding new experimental results with Wormicloud
In the previous sections, we showed how Wormicloud generates a visual abstract starting from a combination of search keywords and helps users discover a common theme from the articles published in the research area related to each keyword. Here, we show some examples of how Wormicloud can be used to mine information on complex data types such as biological pathways or protein complexes from a simple gene pair. In most cases, protein–protein interaction data as a form of gene pair is not easy to understand, especially when the data are from high-throughput approaches such as mass-spectrometry and yeast two-hybrid screening. In fact, in this case, the importance of gene-gene interactions is usually not easy to assess if there is no further information on the context of the interactions. Using Wormicloud, if a user obtains a gene pair (lin-9 and lin-35) from interaction data, they can simply insert the gene names sequentially in the search interface and get a word cloud displaying the keywords ‘transcriptional’, ‘repression’, ‘DREAM’ and ‘complex’, which successfully capture the underlying biological relationship between the genes (Figure 7A) and are not easy to obtain with other methods. For the readers who are not familiar with these keywords, the DREAM complex comprises six additional proteins (LIN-37, LIN-52, LIN-53, LIN-54, DPL-1 and EFL-1) as well as LIN-9 and LIN-35. This protein complex functions as a transcriptional repressor complex that controls the expression of the key genes for cell cycle and development (14).
Figure 7.

Use cases of Wormicloud in mining complex data from literature and its analysis. (A) keyword cloud obtained by entering the keywords ‘lin-9’ and ‘lin-35’ in the Wormicloud search interface. Color highlighted entities show the biological function of lin-9 and lin-35. (B) Gene name cloud for ‘lin-9’ and ‘lin-35’ captures all the essential components in the DREAM complex, which are highlighted in color. (C) Gene ontology enrichment analysis of all genes obtained from the gene name word cloud in Figure 7B recapitulates the major information captured in Figure 7A. (Note that we have manually grayed out terms from Figure 7A and B to highlight the importance of some of the remaining terms in color and to improve readability, but since Wormicloud does not have a measure of ‘biological relevance’ of terms the results in the word clouds generated by the tool are all in color.)
By selecting the option to generate word clouds with only gene names, users can also get a whole set of gene names studied in the literature related to the queried keywords (Figure 7B). From this gene name word cloud, users can get essential information about what genes often studied with lin-9 and lin-35. The word cloud depicted in Figure 7B includes all the genes (lin-9, lin-35, lin-37, lin-52, lin-53, lin-54, dpl-1 and efl-1) that encode the protein components of ‘DRM (or DREAM) complex’ (14) as well as other involved genes. Therefore, gene name clouds successfully provide important information about the components of a protein complex or genes in a biological pathway, which are not easily obtained from biological databases.
Gene name clouds also provide further analysis options for the gene set through other bioinformatics tools in WormBase, namely Gene-set enrichment analysis and SimpleMine. Wormicloud sends the list of genes in gene name clouds to the gene-set enrichment analysis tool and redirects the user to the web pages of the tool which show three different enrichment analysis results for tissue, phenotype and gene ontology terms annotated in WormBase. This list of genes, combined with additional analyses provided by the WormBase Gene-set enrichment analysis tool, provide important clues to find what kind of biological processes or molecular functions are related to the queried keywords (Figure 7C). When comparing the results in Figure 7A and C, we found that the word cloud is well matched to the gene ontology enrichment result for the molecular functions which are based on the annotated data in WormBase. Users can also analyze the gene list by using a batch data-mining tool, SimpleMine. SimpleMine retrieves all essential bioinformatics data in at least 30 different topics from WormBase as shown in Supplemental Table S2. For the user convenience, we summarized all the procedures described above in the tutorial video (supplemental movie 1).
C. elegans is one of the most popular model systems for study of genes implicated in human diseases (15, 16). Therefore, it is useful to get a comprehensive list of genes used in the previous research articles related to a specific human disorder. By using the gene name cloud tool in Wormicloud and downloading the obtained list of keywords, users can easily obtain the list of all genes related to any disease names or disease phenotype terms of interest. For example, Alzheimer’s disease has been studied with 563 genes from the top 200 best matched research articles. In another example, human autism-related research articles have mentioned 1377 genes from the same number of best matched articles (Supplemental Table S3).
Discussion
Curation
Curation of certain data types such as lists of gene components of a protein complex or in biological pathways is not easy to keep up-to-date. Wormicloud can be a good alternative source of information for these data types, especially through gene name clouds. Wormicloud can also be used to get a summary of research articles from any type of keywords, including materials (e.g. drug and chemical names), phenotypes and allele names. In addition, understanding how information changes over time in a certain research area is not an easy task; Wormicloud can provide such longitudinal information with its word trends tool.
Current biological databases store a tremendous amount of information in diverse research areas. Therefore, finding the relevant information might be daunting for a naive user. In WormBase, automated gene descriptions make the key data easier to access by providing a detailed but easy to read summary of curated data for each gene (17). However, gene descriptions are related to single genes, and they do not help in the comparison of multiple genes to find any overlapping data. In this case, users still need to have advanced bioinformatics skills. Wormicloud can help solve this problem by generating a word cloud for multiple genes in WormBase.
Limitations and technical challenges
One of the main limitations of word clouds is that some displayed words may be closely related to each other. Grouping or clustering them can help improve the visualization and obtain more meaningful results. We plan to provide an option to group terms based on their distance in ontologies or other possible measures of distance as part of future improvements.
We designed Wormicloud to complement Textpresso Central with visual tools to explore the research literature and facilitate research, but Wormicloud has somewhat limited features compared to the main Textpresso search interface. In fact, Wormicloud cannot return more than 1000 results, and users should use Textpresso Central instead for large searches and for more advanced search options. Clearly, more results can provide a better chance of finding relevant papers. However, this goal can be achieved only when efficient filtering tools are available. Wormicloud is best suited for generating word clouds from up to 1000 papers at once, whereas the Textpresso search interface can return many more results, but it does not provide a graphical summary of the results.
Textpresso currently updates its corpus every month. Moreover, Textpresso includes only papers already present in the WormBase corpus. Therefore, some delay may occur between publication and inclusion in Textpresso and consequent availability in Wormicloud.
Wormicloud for other organisms
The flexible nature of Wormicloud is expected to make it easy to apply it to the literature of other organisms. In addition, Textpresso already covers several organisms through PubMed open access articles. Some components of the current Wormicloud implementation have been specifically designed for C. elegans. In particular, the Textpresso API used to return gene names works only with C. elegans genes, and the text-mining module in the Wormicloud backend component uses a list of stopwords specific to WormBase data. As part of our future work, we plan to extend Wormicloud to the literature of other organisms, starting from the Alliance of Genome Resources. To do this, we need to improve our text-mining module and expand searches on Textpresso.
Wormiclould is a useful tool for summarizing large and heterogeneous data from sources such as WormBase, and we think it would be applicable to a broad range of organisms and topics for which there are curated data. In particular, Wormicloud can be very useful to make a snapshot of the curated data from multiple gene pages of diverse model organisms such as those included in the Alliance of Genome Resources. This word cloud result can be interactive for filtering the original query and/or navigating the gene page related to a word or word group. From the single-cell level to the whole genomic level, integrating information from multiple sources has become vital for research. This more and more requires systematic approaches using comparative bioinformatics. Wormicloud, with its intuitive yet powerful interface, can be used to conveniently explore such comparative studies through word cloud images showing common topics among multiple genes.
Supplementary Material
Acknowledgements
We thank Ranjana Kishore, Daniela Raciti and Eduardo da Veiga Beltrame for their comments and suggestions on the manuscript.
Contributor Information
Valerio Arnaboldi, Division of Biology and Biological Engineering 156-29, California Institute of Technology, 1200 E California Blvd, Pasadena, CA 91125, USA.
Jaehyoung Cho, Division of Biology and Biological Engineering 156-29, California Institute of Technology, 1200 E California Blvd, Pasadena, CA 91125, USA.
Paul W Sternberg, Division of Biology and Biological Engineering 156-29, California Institute of Technology, 1200 E California Blvd, Pasadena, CA 91125, USA.
Supplementary data
Supplementary data are available at Database Online.
Funding
National Institutes of Health/National Human Genome Research Institute grants [U24HG002223 (WormBase) and U24HG010859 (Alliance Central)].
Conflict of interest.
The authors declare that they have no conflict of interest.
References
- 1. Landhuis E. (2016) Scientific literature: information overload. Nature, 535, 457–458. doi: 10.1038/nj7612-457a. [DOI] [PubMed] [Google Scholar]
- 2. International Society for Biocuration . (2018) Biocuration: distilling data into knowledge. PLoS Biol., 16, e2002846. doi: 10.1371/journal.pbio.2002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris T.W., Arnaboldi V., Cain S. et al. (2020) WormBase: a modern model organism information resource. Nucleic Acids Res., 48, D762–D767. doi: 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Alliance of Genome Resources Consortium . (2020) Alliance of Genome Resources Portal: unified model organism research platform. Nucleic Acids Res., 48, D650–D658. doi: 10.1093/nar/gkz813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Al Saied H., Dugué N. and Lamirel J. (2018) Automatic summarization of scientific publications using a feature selection approach. Int. J. Digit Libr., 19, 203–215. doi: 10.1007/s00799-017-0214-x. [DOI] [Google Scholar]
- 6. Felix C., Franconeri S. and Bertini E. (2018) Taking word clouds apart: an empirical investigation of the design space for keyword summaries. IEEE Trans. Vis. Comput. Graph., 24, 657–666. doi: 10.1109/TVCG.2017.2746018. [DOI] [PubMed] [Google Scholar]
- 7. Oesper L., Merico D., Isserlin R. et al. (2011) WordCloud: a Cytoscape plugin to create a visual semantic summary of networks. Source Code Biol. Med., 6, 7. doi: 10.1186/1751-0473-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baroukh C., Jenkins S.L., Dannenfelser R. et al. (2011) Genes2WordCloud: a quick way to identify biological themes from gene lists and free text. Source Code Biol. Med., 6, 15. doi: 10.1186/1751-0473-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashburner M., Ball C.A., Blake J.A. et al. (2000) Gene ontology: tool for the unification of biology. Nat. Genet., 25, 25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. The Gene Ontology Consortium . (2019) The Gene Ontology Resource: 20 years and still going strong. Nucleic Acids Res., 47, D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarkar I.N., Schenk R., Miller H. et al. (2009) LigerCat: using “MeSH Clouds” from journal, article, or gene citations to facilitate the identification of relevant biomedical literature. AMIA Annu. Symp. Proc., 2009: 563–567. [PMC free article] [PubMed] [Google Scholar]
- 12. Kuo B.Y.-L., Hentrich T., Good B.M. et al. (2007) Tag clouds for summarizing web search results. In: Proceedings of the 16th International Conference on World Wide Web (WWW ‘07). Association for Computing Machinery, New York, NY, USA. pp. 1203–1204. doi: 10.1145/1242572.1242766. [DOI] [Google Scholar]
- 13. Müller H.M., Van Auken K.M., Li Y. et al. (2018) Textpresso Central: a customizable platform for searching, text mining, viewing, and curating biomedical literature. BMC Bioinform., 19, 94. doi: 10.1186/s12859-018-2103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harrison M.M., Ceol C.J., Lu X. et al. (2006) Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc. Natl. Acad. Sci. USA, 103, 16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Apfeld J. and Alper S. (2018) What can we learn about human disease from the Nematode C. elegans? Methods Mol. Biol. (Clifton, N.J.), 1706, 53–75. doi: 10.1007/978-1-4939-7471-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Markaki M. and Tavernarakis N. (2020) Caenorhabditis elegans as a model system for human diseases. Curr. Opin. Biotechnol., 63, 118–125. doi: 10.1016/j.copbio.2019.12.011. [DOI] [PubMed] [Google Scholar]
- 17. Kishore R., Arnaboldi V., Van Slyke C.E. et al. (2020) Automated generation of gene summaries at the Alliance of Genome Resources. Database, 2020, baaa037. doi: 10.1093/database/baaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


