Effects of gut microbiota on systemic immunity
Keywords: inflammatory disease, intestinal bacteria, metabolites, microbiome, systemic immune responses
Abstract
The mammalian intestine is colonized by trillions of microorganisms that have co-evolved with the host in a symbiotic relationship. Although the influence of the gut microbiota on intestinal physiology and immunity is well known, mounting evidence suggests a key role for intestinal symbionts in controlling immune cell responses and development outside the gut. Although the underlying mechanisms by which the gut symbionts influence systemic immune responses remain poorly understood, there is evidence for both direct and indirect effects. In addition, the gut microbiota can contribute to immune responses associated with diseases outside the intestine. Understanding the complex interactions between the gut microbiota and the host is thus of fundamental importance to understand both immunity and human health.
Introduction
Mammals are inhabited by a diverse community of symbionts composed of trillions of microorganisms that include viruses, bacteria, fungi and protozoa, known collectively as the microbiota. Microbes colonize mammalian hosts immediately after birth, resulting in a lifelong symbiotic relationship. Although these microorganisms are present at multiple sites including the skin, lung and gastrointestinal tract, the overwhelming majority of symbiotic bacteria reside in the distal intestine (1, 2). In healthy adults, the gut microbiota comprises four major phyla, namely Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria (3). The bacterial species belonging to these phyla vary along the intestinal tract, likely reflecting distinct microenvironments and nutrient availability in different parts of the intestine that favor the growth of particular bacterial taxa (3, 4). There is also significant heterogeneity in the composition of the microbiota among healthy individuals within and between populations in different geographical locations, which may contribute to the large variation in immune responses in healthy and diseased individuals (5, 6).
An important role for the gut microbiota in regulating host physiology and disease was postulated over one hundred years ago by the Russian zoologist Élie Metchnikoff (7). We know now that microbes that inhabit the gastrointestinal tract have a profound influence on host physiology including digestion and absorption of food, biosynthesis of micronutrients and protection against pathogen colonization (2, 8). In addition, symbionts can act locally to shape the composition and function of immune cells in intestinal tissues (1, 9). Mounting evidence indicates that intestinal bacteria can also act remotely to influence host immune responses important for host defense and disease pathogenesis. In this review, we discuss the direct and indirect mechanisms by which intestinal symbionts regulate immunity and immune-associated diseases at sites distal from the intestine, in particular, the liver, the lungs and the brain.
Gut microbiota-driven mechanisms in systemic immunity
The role of the gut microbiota in the development of both the intestinal and systemic immune system was first revealed in studies with germ-free animals that showed several immunological abnormalities not only in the intestinal mucosa, but also in lymphoid structures at systemic sites (10, 11). Germ-free mice also have defects in immunoglobulin production, expression of anti-microbial molecules, T-cell trafficking and pathogen clearance after systemic infection (12–15). Although the exact mechanism by which the gut microbiota contributes to immune responses at distant sites remains poorly understood, studies to date suggest, first, a direct mechanism via translocation of gut microbes, their components and/or their metabolites into the circulation and, second, an indirect mechanism in which stimulation of epithelial, stromal or immune cells within the gut results in downstream responses that are relayed systemically.
Direct microbial mechanisms
Translocation of bacteria and their components
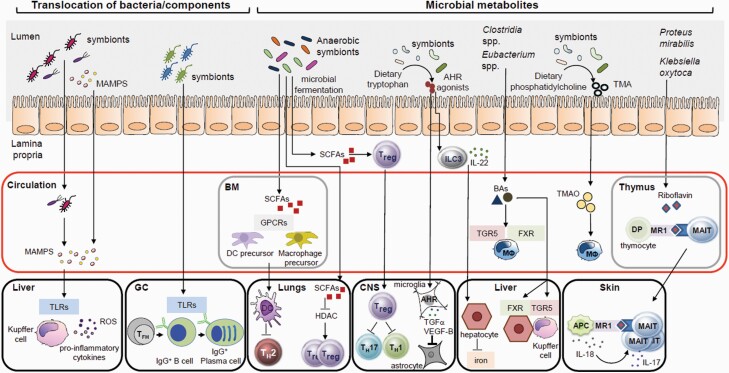
Despite residing within the intestinal lumen, gut symbionts and their components can translocate across the epithelium to regulate immune responses beyond the intestine (Fig. 1). In the colon, the thick mucus barrier consists of two clearly distinct layers which restrict the access of potentially harmful microorganisms to the mucosal surface (16, 17). The small intestine, however, lacks a well-demarcated mucus layer (18), which may explain the detection of small-intestinal microbes, such as Enterobacteriaceae and Lactobacillaceae members, in systemic tissues under homeostatic conditions, in both humans and animals (19, 20). The structurally conserved components of these microbes, referred as microbe-associated molecular patterns (MAMPs), such as lipopolysaccharide (LPS) and peptidoglycan (PGN), can also translocate across the intestinal barrier and stimulate pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain-like receptors (NLRs), to influence hematopoiesis and immune responses at distant tissues (21) (Fig. 2).
Fig. 1.
Potential direct microbial mechanisms in systemic immunity. The gut microbiota can contribute to immune responses at distant sites through direct mechanisms such as the translocation of gut microbes and/or their components or their metabolites to the blood circulation and systemic organs. BM, bone marrow; DP, double positive; MΦ, macrophage; ROS, reactive oxygen species; VEGF-B, vascular endothelial growth factor B.
Fig. 2.
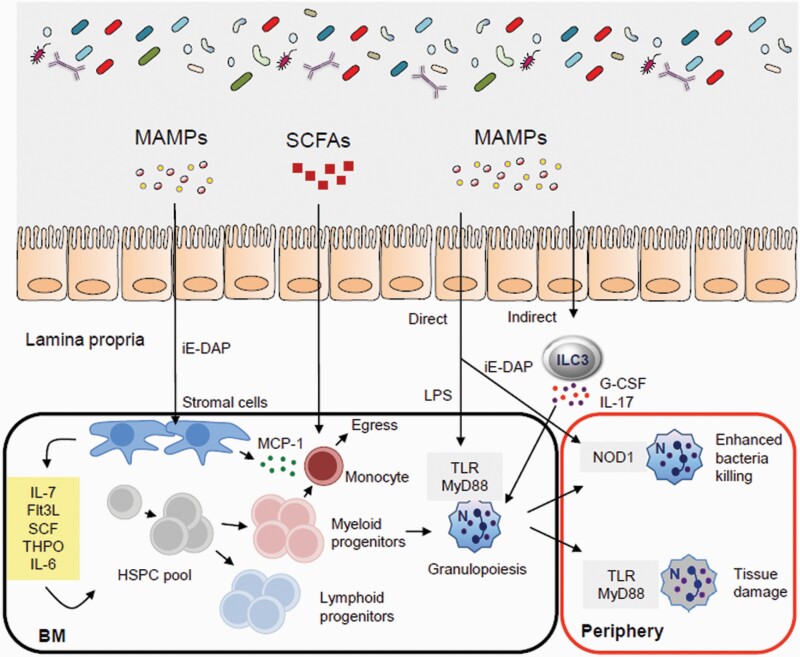
The gut microbiota regulates hematopoiesis and immunity. The microbiota controls bone marrow (BM) hematopoiesis through several processes. Flt3L, FMS-like tyrosine kinase 3 ligand; SCF, stem cell factor; THPO, thrombopoietin.
The bone marrow provides the primary niche that supports the function of hematopoietic stem and progenitor cells (HSPCs) as well as immune cells that mobilize and expand in response to infection (22, 23). Treatment of mice with broad-spectrum antibiotics led to partial depletion of the HSPC pool (24). Moreover, germ-free mice had diminished HSPC subsets in the bone marrow compared with specific pathogen-free (SPF) animals (25). HSPCs in germ-free mice can be restored by the systemic administration of γ-d-glutamyl-meso-diaminopimelic acid (iE-DAP), the bacterial dipeptide that stimulated nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and induced bone marrow stromal cells to secrete HSPC cell-supporting cytokines (Fig. 2) (25). In addition, myeloid cell development in the bone marrow is compromised in germ-free mice resulting in reduced numbers of monocytes, macrophages and neutrophils in the spleen and liver (26). Colonization of germ-free mice with the microbiota of SPF mice or oral feeding with MAMPs, but not short-chain fatty acids (SCFAs), can restore normal myelopoiesis (26).
In addition to the regulation of bone marrow myelopoiesis, systemic TLR ligands regulate monocyte egress from the bone marrow by inducing stroma expression of monocyte chemotactic protein-1 (MCP-1) (27). Furthermore, the gut microbiota sustains a population of splenic macrophages derived from myeloid progenitors of the embryonic yolk sac (26, 28). Circulating microbial components originating from the gut microbiota promote steady state granulopoiesis in the bone marrow through TLR signaling (Fig. 2) (29, 30). Likewise, neonatal colonization by maternal microbiota is required for normal frequencies of peripheral neutrophils and granulocyte/macrophage-restricted progenitors in the bone marrow (31). This regulation of neutrophil homeostasis by the microbiota in neonates was mediated through LPS-induced IL-17 production by intestinal group 3 innate lymphoid cells (ILC3s), which, in turn, enhanced levels of granulocyte colony-stimulating factor (G-CSF) for granulopoiesis (31). Consistent with these observations, LPS and CpG engagement with TLR4 and TLR9, respectively, efficiently induced the expression of IL-23 and IL-1β by intestinal C–X3–C chemokine receptor 1 (CX3CR1)+ mononuclear phagocytes required for ILC3 activity (32).
In addition, NOD1 stimulation by gut microbiota-derived PGN that entered the circulation enhanced the bacteria killing capacity of bone marrow-derived neutrophils to control sepsis from pneumococcal infection (33). The gut microbiota can also sustain a distinct subset of aged neutrophils characterized by enhanced pro-inflammatory activity (34). Neutrophil aging is, in part, driven by MAMPs and TLR–MyD88 (myeloid differentiation primary response 88) signaling pathways (Fig. 2). Depleting the microbiota reduced the frequency of circulating aged neutrophils and prevented inflammation-driven tissue damage during sickle cell disease and septic shock (34). Collectively, these data demonstrate that the complexity of an intact gut microbiota and innate sensing of systemic microbial signals are critical for neutrophil maintenance and function.
After translocation from the gut, bacterial symbionts and MAMPs can gain access to the liver through the portal vein (Fig. 1). Live, metabolically active bacteria can be detected in the peripheral tissues of healthy individuals, including mesenteric lymph nodes (MLNs), lung, ovary and breast (19, 35–37). Because MLNs and the liver act as a firewall to prevent systemic spread of a small subset of circulating microbes at the steady state (38, 39), the presence of live and metabolically active organisms in peripheral organs may be secondary to a disruption of the intestinal barrier. However, further work is required to address this issue.
Translocation of bacterial symbionts and their components, such as LPS, is enhanced in patients with chronic liver disorders, such as cirrhosis, alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD), that are associated with increased intestinal permeability (40, 41). LPS can be detected in the blood of both animals and patients with liver disease (42, 43). Furthermore, Tlr4–/– mice were protected from liver inflammation and hepatic lipid accumulation in a nutritional model of non-alcoholic steatohepatitis (NASH) (44, 45).
In the liver, MAMPs, such as LPS, appear to increase hepatic inflammation and disease progression via TLR stimulation in Kupffer cells (46–48) (Fig. 1). Stimulation of Kupffer cells via TLR4/TLR9 signaling can result in up-regulation of hepatic tumor necrosis factor α (TNFα) expression, which, in turn, promotes NASH progression in mice (49). Likewise, translocation of gut bacteria or MAMPs because of intestinal barrier disruption induced by chronic alcohol intake or other stimuli such as dietary factors has been linked to progression, in humans and in animals, of ALD and NAFLD, respectively (50, 51). Although the mechanism remains poorly understood, endotoxemia and subsequent TLR4-dependent Kupffer cell activation as well as activation of the NLRP3 inflammasome have been suggested to contribute to hepatic inflammation, steatosis and fibrosis (47, 52–54).
The link between the gut microbiota and liver disease, however, remains poorly understood. Bacterial symbionts appear to promote intestinal barrier dysfunction because treatment with antibiotics reduces intestinal permeability and subsequent liver damage, which was associated with enhanced expression of tight-junction proteins and attenuated hepatic stellate cell activation (55). Although this evidence suggests that impaired barrier function directly contributes to disease progression, liver injury can also lead to the loss of intestinal barrier integrity, even though the mechanism is not fully understood (56). Thus, further studies are needed to clarify the association between intestinal permeability and liver inflammation.
Microbial metabolites
Microbial metabolites generated in the gut can enter the circulation and affect host immune responses at distant sites (Fig. 1). Intestinal microbes produce a wide range of metabolites that can be broadly divided into three main groups: (i) metabolites produced by microbial fermentation/degradation of dietary components, (ii) host-derived metabolites that undergo microbial modification and (iii) de novo biosynthesis of microbial metabolites (57).
SCFAs, produced by microbial fermentation of plant-derived dietary polysaccharides, provide an energy source for intestinal epithelial cells, but also have immunomodulatory properties (9). The bulk of SCFAs produced in the gut are derived from anaerobic bacteria, such as members of Bacteroidaceae, Ruminococcaceae and Lachnospiraceae families (58). The most abundant gut SCFAs—propionate, butyrate and acetate—signal through multiple G protein-coupled receptors (GPCRs), including GPR43, GPR41 and GPR109A, that are expressed by both immune cells and epithelial cells (9). Whereas GPR43 recognizes all three SCFAs, GPR41 is activated by propionate and butyrate, and GPR109A only recognizes butyrate (59, 60). Both mucosal and peripheral inflammatory responses were dysregulated in germ-free and Gpr43–/– mice, suggesting that stimulation of GPR43 by SCFAs exerts crucial immunomodulatory properties under homeostatic conditions (61).
In animal studies, SCFAs regulate the expansion and suppressive function of colonic regulatory T (Treg) cells via GPR43 (62). These SCFA-mediated immune regulatory properties of Treg cells also extend to the central nervous system (CNS) (Fig. 1) (63). SCFAs can regulate mucosal immune responses at different barrier sites, including the intestine and the lungs, through stimulation of other GPCRs such as GPR109A and GPR41 (60, 64). SCFAs can also influence immune responses by inhibiting histone-deacetylases (HDACs) (65). In animal models and human cells in vitro, SCFA-mediated inhibition of HDACs can promote an anti-inflammatory phenotype in a variety of immune cells located in peripheral tissues (64–69). A role for SCFAs in promoting intestinal homeostasis through the regulation of the colonic FOXP3+ Treg cells is well established (62, 70–73), but SCFA-driven inhibition of HDACs also enhances the number and function of Treg cells in the lungs (74).
SCFAs can also influence B-cell responses in the intestine, MLNs and spleen (75). Although the mechanism by which SCFAs regulate B cells remains poorly understood, SCFAs can enhance B-cell metabolism, at least in part through the regulation of 5’ AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) (75).
Another important class of microbial metabolites derived from the metabolism of dietary substances are aryl hydrocarbon receptor (AHR)-activating tryptophan metabolites (Fig. 1). AHR is a ligand-inducible transcription factor that is expressed by several cell types, including immune cells and epithelial cells (76). AHR activation regulates postnatal development of intestinal lymphoid follicles and expansion of IL-22-producing retinoic acid receptor-related orphan receptor-γt (RORγt)+ ILC3s (77). Although IL-22 acts primarily in the intestine by promoting gut homeostasis and conferring resistance against enteric pathogens (77), IL-22 can also exert systemic effects in mice. For example, IL-22 can induce hepatocytes to produce anti-microbial molecules that protect against systemic bacterial infections (78). Thus, microbiota-derived tryptophan metabolites are crucial for the maintenance of the intestinal barrier and protection against mucosal and systemic infections.
Trimethylamine (TMA) is another diet-derived microbial metabolite. The gut microbiota metabolizes the dietary lipid, phosphatidylcholine, to TMA which is further metabolized by liver enzymes to generate trimethylamine N-oxide (TMAO) (Fig. 1) (79). This pathway is particularly relevant for the development of atherosclerosis, as diet supplementation with choline or TMAO promotes the formation of foamy macrophages and atherosclerotic plaques in atherosclerosis-prone Apoe–/– mice (80). These observations have significant clinical implications as administration of a small-molecule inhibitor of microbiota-derived TMA attenuated the formation of atherosclerosis in Apoe–/– mice (81).
Microbes can also modify host-derived metabolites that can influence systemic immune responses. For example, primary bile acids (BAs) are synthesized in the liver from cholesterol and further modified via conjugation to glycine or taurine (82). In response to food ingestion, conjugated primary BAs are released into the small intestine where they can undergo deconjugation by bile salt hydrolase (BSH)-expressing bacteria that include members of the genera Lactobacillus, Bifidobacterium, Clostridium and Bacteroides (83). The majority of BAs are absorbed in the ileum and transported into the liver via the enterohepatic circulation, whereas deconjugated BAs, which are not absorbed, reach the distal intestine where they undergo several modifications including 7α and/or β-dehydroxylation by a limited subset of bacterial species to generate secondary BAs (83, 84).
Although a main function of BAs is to promote the emulsification and absorption of dietary lipids (82), BAs can also regulate metabolic and immune responses in the intestine and at distant organs by stimulation of several host nuclear and GPCRs including farnesoid x receptor (FXR) and the Takeda G protein-coupled receptor 5 (TGR5) (Fig. 1) (85). FXR is expressed mostly in intestinal epithelial cells and hepatocytes and activated predominantly by primary BAs (86–88). In contrast, TGR5 is expressed by a variety of cells, including macrophages and Kupffer cells, and activated primarily by secondary BAs (89–92). Signaling via FXR and TGR5 can affect both immune processes and metabolic pathways through stimulation of epithelial cells, macrophages and Kupffer cells in animal models of liver disease and insulin resistance (93–98). Given the role of BAs in the regulation of metabolic and immune responses, microbiota-dependent BA signaling pathways represent a relevant area for improving health. However, the beneficial effects of targeting TGR5 and FXR activation require further investigation including large-scale human studies.
Microbes can also synthesize metabolites, such as riboflavin, a B group vitamin that is an essential component of cellular metabolism (Fig. 1) (99). The invariant T-cell antigen receptor of mucosal-associated invariant T (MAIT) cells recognizes microbial vitamin B2 metabolites such as the riboflavin precursor 5-A-RU bound to the antigen-presenting molecule MHC class I (MHCI)-related protein 1 (MR1) (100). Early in life, gut bacteria-derived riboflavin metabolites cross the mucosal barrier to reach the thymus where they stimulate thymocytes to drive the development of MAIT cells (101, 102). MAIT cells are innate-like lymphocytes highly enriched in the human and mouse skin where they respond rapidly to pathogens (101). Colonization of germ-free mice with specific intestinal bacterial species such as Proteus mirabilis and Klebsiella oxytoca that express genes that encode riboflavin-synthesizing enzymes can restore MAIT cell numbers in the skin (101). Although MAIT cells are involved in pathogen clearance, the contribution of microbiota-derived riboflavin metabolites in host defense responses remains to be clarified.
Indirect microbial mechanisms
The gut microbiota can act on intestinal cells, which, in turn, relay the microbial signals distally to affect peripheral organs via at least three mechanisms: (i) the gut microbiota can regulate the production of epithelial cell-derived, macrophage-derived or dendritic cell (DC)-derived factors that promote T-cell activation and polarization; (ii) intestinal microbes can regulate immune cell trafficking from the gut to distant tissues and (iii) enteric symbionts can promote antibody production systemically by modulating B-cell responses in lymphoid intestinal tissue.
T-cell polarization
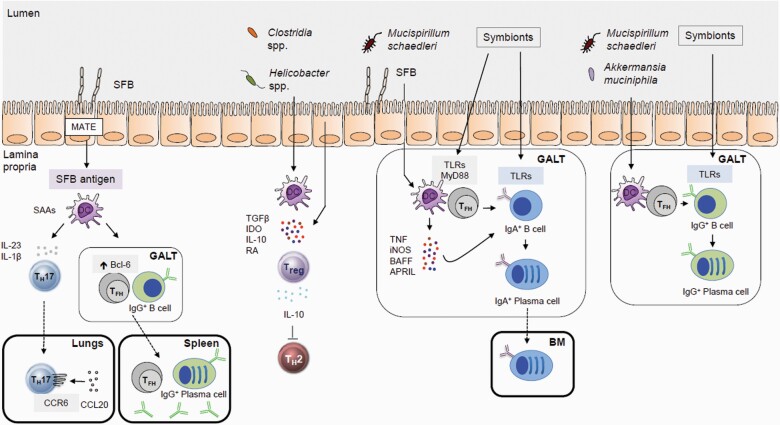
The best-characterized example of how gut symbionts can act on intestinal epithelial cells and antigen-presenting cells (APCs) to regulate T-cell activation and polarization is provided by segmented filamentous bacterium (SFB) (103, 104). Under homeostatic conditions, the intimate adhesion of SFB to the epithelia of the terminal ileum induces the production of molecules, such as serum amyloid A proteins (SAAs), and transfer of microbial proteins via endocytosis to adjacent intestinal epithelial cells (IECs), which primes local DCs and macrophages to promote T helper 17 (TH17) cell differentiation (105–109). Following SFB colonization, most TH17 cells in the lamina propria express T-cell antigen receptors specific for SFB antigens, which is mediated via MHCII-dependent presentation of SFB antigens through CD11c+ DCs.
Although microbiota-dependent TH17 cells act primarily in the gut, SFB-mediated induction of TH17 cells appears to protect the host against pulmonary infections (103, 110, 111). For example, mice harboring SFB+ complex microbiota are resistant to Staphylococcus aureus pneumonia compared with SFB– animals, which correlated with the presence of TH17-associated cytokines in the bronchoalveolar lavage fluid (110). Similarly, colonization with SFB led to the accumulation of TH17 cells in the lungs during fungal infections (111).
Although these findings implicate SFB in the susceptibility of C57BL/6 mice to lung infection, comparing mice from different vendors that may harbor some genetic variability represents a confounding factor in the interpretation of the results. Moreover, although it remains unclear whether gut-derived TH17 cells mediate the protection against bacterial and fungal lung infections in these models, intestinal C–C chemokine receptor 6 (CCR6)+ TH17 cells arising from SFB stimulation can be recruited to the lungs through robust local production of C–C chemokine ligand 20 (CCL20, which binds CCR6), where they promote lung pathology in an autoimmune arthritis model (112). Taken together, these findings suggest that the function of TH17 cells outside of the intestine is context-dependent and varies greatly according to their environment.
Select members of the gut microbiota play a critical role in the development of extrathymic FOXP3+ Treg (pTreg) cells as their T-cell antigen receptors recognize intestinal bacterial antigens including those expressed by Clostridium, Parabacteroides and Helicobacter species unlike Treg cells found in other tissues (113–115). In addition, colonization of Clostridium species in the intestinal mucus layer increased epithelial secretion of transforming growth factor β (TGFβ) and indolemine 2,3-dioxygenase (IDO), which promoted pTreg cell accumulation and restricted systemic IgE responses (72, 116). Furthermore, selective ablation of pTreg cells following deletion of the intronic Foxp3 enhancer conserved non-coding sequence 1 (CNS1) led to spontaneous type 2 immune-associated pathologies in the intestinal tract and the lungs (117). Although thymic Treg cells are sufficient for the control of systemic autoimmunity, peripherally educated Treg cells have some non-redundant immunosuppressive functions in mucosal barriers (117, 118).
FOXP3+ Treg cells are also maintained by retinoic acid (RA) synthesized by intestinal APCs through aldehyde dehydrogenase (ALDH) activity (119). Interestingly, perturbations of the vagal sensory afferents lead to reduced Treg cell numbers in the gut coinciding with attenuated ALDH expression by APCs (120). APCs can respond directly to neurotransmitters through muscarinic acetylcholine receptor engagement which promotes ALDH expression (120). The neural arc regulating colonic Treg cells begins in the liver where the hepatic vagal sensory afferents relay signals from the gut environment to the brainstem through the left nodose ganglion, and finally to the vagal efferents and enteric neurons that enhance APC ALDH activity (120). Importantly, the liver–brain–gut reflex arc that maintains intestinal Treg cells appears to involve microbial signals as susceptibility to colitis in hepatic-vagotomized mice is unaltered in antibiotic-treated mice and MyD88-deficient mice (120).
Immune cell trafficking
The gut microbiota can also affect systemic immune responses by regulating immune cell trafficking (Fig. 3). For example, a subset of inflammatory IL-25-induced or helminth-induced inflammatory ILC2s originating from the intestine can migrate to the lungs via sphingosine 1-phosphate-mediated chemotaxis (121). Inter-organ migration of ILC2s appears to be microbiota-dependent; during antibiotic treatment, the majority of lung ILC2s are maintained locally by self-renewal in response to Nippostrongylus brasiliens infection (122, 123). In the lung, ILC2s can contribute to anti-helminth defense and tissue repair (121).
Fig. 3.
Indirect effects of the gut microbiota on systemic immunity. The intestinal microbiota can modulate host systemic immunity through several indirect mechanisms via local stimulation of epithelial and immune cells which are then communicated to distal sites. GALT, gut-associated lymphoid tissue; MATE, microbial adhesion-triggered endocytosis.
Moreover, in a model of autoimmune arthritis, colonization of germ-free mice with TH17-inducing SFB resulted in trafficking of activated TH17 cells from the gut to the spleen, where they stimulate germinal center (GC) formation and production of auto-antibodies (124). However, the underlying mechanism by which a population of gut-imprinted TH17 cells is retained in the spleen and whether this occurs in conventionally housed animals remain unclear. SFB can also exacerbate systemic arthritis, in mice, by driving the differentiation and egress of Peyer’s patch T follicular helper (TFH) cells into systemic sites where they can elicit auto-antibody responses (125). In contrast to these observations, SFB-elicited homeostatic TH17 cells, unlike those induced by the gut pathogen Citrobacter rodentium, remained in a non-pathogenic state and did not participate in inflammatory responses (126).
Thus, the role of SFB in the induction of pathogenic TH17 cells and systemic immune responses remains controversial and not completely understood. SFB can also promote Peyer’s patch TFH cell differentiation by limiting the access of IL-2 to CD4+ T cells and enhancing the expression of B-cell lymphoma 6 (Bcl-6), a TFH cell master regulator, in DCs (125). In these studies, SFB induced the presence of Peyer’s patch-derived TFH cells in systemic lymphoid tissues (125), suggesting that Peyer’s patch TFH cells can relay microbial signals distally to regulate systemic immune responses.
Regulation of B-cell responses
The gut microbiota also regulates B-lineage maturation as germ-free mice display impaired lymphoid structures and reduced serum immunoglobulin concentrations (Fig. 3) (12). Intestinal symbionts stimulate intestinal epithelial cells and lamina propria DCs to secrete factors, such as TNFα, inducible nitric oxide synthase (iNOS), B-cell activating factor (BAFF) and a proliferation-inducing ligand (APRIL), that promote the induction of immunoglobulin A (IgA)+ B cells and plasma cell differentiation (127–130). Although the bulk of polymeric IgA is produced in gut-associated lymphoid tissue and transcytosed into the intestinal lumen, IgA+ cells instructed by gut-derived antigens have been detected at distal sites (131). In particular, several members of the Proteobacteria phylum can promote T-cell-dependent systemic IgA responses and induce IgA-secreting plasma cells in the bone marrow, which can confer protection against bacterial sepsis (131).
By modulating B-lineage maturation, the gut microbiota can also regulate microbe-reactive immunoglobulin repertoire diversification. Recombinase-activating gene (RAG)-expressing early B cells undergoing active V(D)J recombination can be tracked in the intestinal lamina propria during weaning, which coincided with the marked expansion of the gut microbial community (132). Colonization of germ-free animals with conventional microbiota during weaning resulted in enrichment of pro-B cells in the intestinal lamina propria and bone marrow (132). Notably, B-cell immunoglobulin diversification was elevated selectively in the gut lamina propria of conventionalized germ-free mice, but not at systemic sites, highlighting a critical period in which the gut microbiota shapes the local pre-immune B-cell repertoire (132). In addition, early-life exposure to symbionts also promotes the IgG repertoire diversity required for optimal systemic bacterial resistance (133). Thus, gut symbionts may benefit systemic host immunity by diversifying pre-immune B-cell repertoires with antibacterial reactivity.
Intestinal symbionts can elicit IgG antibodies that can be detected in the circulation. Whereas broadly reactive T-cell-independent commensal-specific IgG2b and IgG3 are generated following engagement of B cells via TLR signaling in mice (134), TFH cell-dependent IgG1 predominantly targets invasive mucus-dwelling bacteria (135). Induction of IgG against symbiotic bacteria is often triggered by members of the Enterobacteriaceae family that translocate to MLNs and spleen, under homeostatic conditions (20). These systemic IgG responses can be elicited against a highly conserved component of the outer membrane expressed by Gram-negative bacteria such as murein lipoprotein, which protects mice from pathogenic bacteremia (20). However, it remains unclear how symbiotic bacteria breach the epithelial barrier to induce protective IgG responses under homeostatic conditions.
Other effects on liver immunity
The liver is under constant exposure to gut microbial components and metabolites that traffic in the enterohepatic circulatory system (136). The liver contains immune cells, such as Kupffer cells, specializing in the detection and elimination of blood-borne bacteria and serves as a firewall to limit the systemic spread of intestinal symbionts (39, 136). Animal studies showed that gut microbial products, such as LPS, can gain access through the portal vein to the liver where they regulate the accumulation of Kupffer cells by promoting the expression of adhesion molecules in the sinusoidal endothelium (137). In addition, the bactericidal activity of murine Kupffer cells is regulated by the gut microbiota through commensal-derived D-lactate that reaches the liver via the portal vein (138). Colonization with D-lactate-producing symbionts or administration of purified D-lactate to germ-free mice can restore Kupffer cell-mediated pathogen clearance, thus preventing systemic bacteremia (138).
The gut microbiota also contributes to the regulation of invariant NKT (iNKT) cells, a population of ILCs that continuously patrol the sinusoids of the liver. iNKT cells recognize glycosphingolipid antigens, present in bacterial cell walls, which are presented by the MHCI-like molecule CD1d. In the absence of the intestinal microbiota, murine hepatic iNKT cells exhibit an immature phenotype and impaired activation, which is restored by colonization with bacteria expressing iNKT cell antigens (139).
In response to bacteria that have entered the portal circulation, Kupffer cells induce C–X–C chemokine receptor 3 (CXCR3)-dependent clustering of iNKT cells, and present antigens via CD1d, leading to iNKT cell activation and limiting bacterial systemic dissemination (140). The hepatic iNKT cell pool can be positively or negatively regulated by gut symbionts depending on the genetic background of the animals (141). However, the mechanism underlying these strain-dependent and microbiota-dependent differences in the regulation of hepatic iNKT cell numbers remains largely unknown. In contrast to mice, iNKT cells are found in small numbers in the healthy human liver (142), suggesting that these cells may be more important in animals than in humans.
The liver also contains a population of innate-like IL-17A-producing γδ T (γδT-17) cells, which are involved in pathogen clearance through the recruitment and activation of neutrophils. The presentation of gut microbiota-derived lipid antigens via hepatocyte expression of CD1d is required for γδT-17 cell homeostasis in the liver (143). The role of gut symbionts is further supported by the observation that germ-free mice or antibiotic-treated animals show reduced numbers of hepatic γδT-17 cells, and recolonization with a complex microbiota restored this cell population (143).
Although microbiota-elicited γδT-17 and iNKT cells promote protection against blood-borne infections, their aberrant activity can also fuel liver pathology. For instance, the microbiota can accelerate metabolic liver disease, such as NAFLD and NASH, via an increase in hepatic γδT-17 or iNKT cells in mice (143, 144). Several studies have reported aberrant changes in the composition of the gut microbiota in patients with ALD and NAFLD, the most common chronic liver disorders in Western countries (145, 146). However, the role of gut dysbiosis in ALD and NAFLD remains unclear because results are discordant and contradictory.
Other effects on lung immunity
Gut–lung cross-talk can be mediated directly and indirectly by intestinal symbionts (Figs 1 and 2). These effects can occur early in that maternal consumption of a high-fiber diet or SCFA supplementation during pregnancy can alter gene regulation in the fetal lung leading to enhanced immunosuppressive activity of Treg cells and protection against allergic asthma later in life (74). In addition, expansion of the gut microbiota during weaning is associated with a vigorous immune response that can limit susceptibility to allergic lung inflammation (147). Resistance to ‘pathological imprinting’ of the immune system in early life requires the induction of RORγt+ Treg cells by SCFAs and RA derived from the gut microbiota that is established during weaning (147).
In the absence of an intact intestinal microbiota or following antibiotic-mediated alterations in the microbiota during early life, mice are highly susceptible to type 2 immune pathologies characterized by increased eosinophil and CD4+ TH2 cell infiltration in the lungs and elevated IgE concentrations in the serum (148, 149). Increased systemic SCFAs can promote the generation of bone marrow DC precursors that subsequently traffic to the lungs and display enhanced phagocytic activity, but an attenuated capacity to induce allergic inflammation (64). The protective effects of propionate are mediated by GPR41, but not its related receptor GPR43 (64). However, Gpr43–/– mice exhibit more severe lung inflammation compared with wild-type littermates in an acute allergic airway inflammation model (61). The reason for these seemingly contradictory results is unclear, but one possibility is that these two SCFA receptors are expressed on distinct cell subsets, and thus, may differentially affect lung immune responses.
Antibiotic-induced intestinal dysbiosis can further promote allergic airway inflammation by shifting macrophage polarization in the lung toward the alternatively activated M2 phenotype (150). Moreover, the recognition of microbial sphingolipids early in life can further reduce susceptibility to type 2 immune diseases by suppressing C–X–C chemokine ligand 16 (CXCL16)-dependent accumulation of iNKT cells in the lung (141). However, these studies using antibiotic-treated mice did not rule out a role for the lung microbiota in the regulation of immune responses in the lung.
There is also evidence for a role of the gut microbiota in the regulation of protective immunity against lung infections. An intact gut microbiota is required for optimal anti-influenza CD4+ TH cell and CD8+ cytotoxic lymphocyte response in the lungs (151). Priming cells to express pro-IL-1β and pro-IL-18 by gut-derived bacterial components is a critical prerequisite for inflammasome-dependent cytokine release that promotes the migration of pulmonary DCs to the draining lymph nodes for T-cell activation (151). Commensal bacterial signals also contribute to anti-viral immunity in the lung by enhancing macrophage responsiveness to type I and type II interferons and by augmenting their capacity to restrict viral replication (152).
A high-fiber diet and SCFAs can further enhance the survival of influenza-infected animals by altering bone marrow hematopoiesis to promote the generation of Ly6C– monocytes with an attenuated capacity to secrete CXCL1 in the airways thus limiting neutrophil accumulation in the lungs (153). Meanwhile, SCFAs can boost anti-viral CD8+ T-cell effector responses by altering their metabolic responses (153). Notably, the metabolite desaminotyrosine (DAT), derived from the degradation of plant flavonoids by Clostridium orbiscindens, rescues mice from influenza-induced lethality due to antibiotic depletion of the microbiota (154). DAT can mediate host protection by augmenting type I interferon signaling and enhancing phagocytic activity in the lungs (154).
Although these studies utilize antibiotic manipulation of conventional microbiota or gnotobiotic animals to examine the effects of intestinal microbes on lung immunity, the use of feral or pet-store mice raised without barrier housing has provided a new strategy to examine the physiological impact of a ‘dirty’ gut microbiota (155). Free-living mice exhibit dramatic alterations in myeloid and lymphoid subsets, including a striking increase in differentiated effector memory CD8+ T cells in multiple tissue types including the lungs (155). Wild mouse microbiome-reconstituted (WildR) mice are more resistant to influenza infection, exhibiting reduced viral titers and attenuated immune-mediated lung pathology compared with conventional mice (156). The reported improvement in WildR mouse fitness was attributed to the abrogation of an excessive immune response during early stages of viral infection (156). However, the WildR symbionts or their respective products that confer protection from lung pathology and a survival advantage remain unclear.
Other effects on CNS immunity
Although the CNS has traditionally been considered an ‘immune-privileged organ’, there is bidirectional cross-talk between the gastrointestinal tract and the CNS system, referred to as the ‘microbiota–gut–brain axis’ (157, 158). The gut microbiome may impact brain health in numerous ways: (i) microbial components and metabolites can stimulate the CNS innate immune system; (ii) gut microbes can produce hormones and neurotransmitters that are transported in the blood to the brain; and (iii) circulating immune cells activated by the gut microbiota have the potential to traffic to the CNS, where they can produce cytokines and other inflammatory mediators (158).
CNS-resident cells are divided into two main groups: neuronal cells, and non-neuronal glial cells, a heterogeneous group of cells which is further classified into macroglia and microglial cells (157). Although virtually all CNS cell types can be influenced by microbial signals, we will only discuss here the role of the gut microbiota in the maturation and function of microglial cells and astrocytes, two major cell populations involved in brain immunity.
Microglial cells, the CNS-resident macrophages, originate from extra-embryonic erythromyeloid progenitors of the yolk sac (157). In contrast to other yolk sac-derived tissue macrophages that can continuously be replaced by bone marrow-derived short-lived macrophages, microglial cells are long-lived and self-renew postnatally throughout the whole life (157).
Under steady state conditions, the microbiota plays a central role in the development and maturation of the microglia (159, 160). In the absence of a complex microbiota, animals exhibit defects in microglial maturation, differentiation and function (159, 160). These defects can be explained by an arrest in their developmental maturation (159, 160). Similar to germ-free mice, antibiotic-treated SPF mice as well as mice colonized with a small number of symbionts, such as Bacteroides distasonis, Lactobacillus salivarius and Clostridium cluster XIV, exhibit microglial morphological abnormalities (159).
These findings suggest that signals derived from a complex microbiota are required for the homeostasis of microglia under steady state conditions. In addition, microglia in germ-free mice display limited responses towards viral and bacterial challenges, suggesting that gut microbiota can prime the brain to rapidly mount immune responses against infection. While multiple TLRs are not required for microglial maintenance under homeostatic conditions, administration of microbial SCFAs restored impaired microglial maturation and function in germ-free mice (159). However, the SCFA(s) that drive microglial maturation and function as well as the signaling pathway(s) involved remain unclear.
Besides SCFAs, microbiota-derived tryptophan ligands can also influence microglial activation and brain inflammation via AHR signaling (Fig. 1) (161). Microbial metabolites of tryptophan can also activate AHR signaling in astrocytes, the most abundant CNS glial cell type, which, in turn, led to suppression of brain inflammation in mice (162). Mechanistically, AHR signaling in astrocytes regulates transcriptional programs that controlled the recruitment of LY6C1hi inflammatory monocytes to the CNS and activation of microglia and monocytes (162).
Intestinal bacteria can also contribute to brain autoimmunity and inflammation in the experimental autoimmune encephalomyelitis (EAE) animal model of multiple sclerosis (MS) (163), a chronic degenerative disease caused by peripheral immune cell infiltration of the CNS and subsequent T-cell-dependent demyelination (164). Germ-free mice are protected from the onset of EAE because of a reduced capacity of DCs to induce both pathogenic TH1 and TH17 cell responses as well as decreased autoreactive B-cell responses (165, 166). Recolonization of germ-free mice with the conventional microbiota or SFB induced EAE in a TH17-dependent manner (165, 166). The use of germ-free mice, however, is a confounding factor in the interpretation of the results. Indeed, in mice harboring a diverse microbiota, that contains or lacks SFB, no differences in EAE susceptibility were observed despite the presence of intestinal TH17 cells (126). Recolonization of germ-free mice with two small-intestine-derived microbes, Allobaculum stercoricanis and Lactobacillus reuteri, which express peptides mimicking the myelin oligodendrocyte glycoprotein (MOG), enhances the responses of autoreactive MOG-specific TH17 cells and exacerbate EAE symptoms (167). However, further work is required to determine whether MOG-reactive T cells migrate from the small intestine to the CNS to exacerbate EAE.
Gut bacteria-induced TH17 cells have also been linked to the pathogenesis of neurodevelopmental disorders, such as autism spectrum disorder (ASD) (168, 169). Administration of polyinosinic-polycytidylic acid (poly I:C) to pregnant dams to mimic viral infection led to the development of ASD-like abnormalities in the offspring that are dependent on the presence of TH17-inducing intestinal bacteria in the mother (169).
In a model of amyotrophic lateral sclerosis (ALS), a neurodegenerative syndrome characterized by progressive loss of motor neurons in the brain and spinal cord leading to rapid paralysis, the presence of immunostimulatory microbes, such as Helicobacter species, was associated with inflammation and autoimmunity in mice deficient in the C9orf72 gene, the most common genetic variant of ALS (170, 171). Furthermore, microbial signals can regulate both microglial activation and the infiltration of myeloid cells in the CNS of C9orf72-deficient mice (171). In contrast, CNS disease was exacerbated in antibiotic-treated and germ-free ALS-prone Sod1 transgenic mice related to the reduced production of the microbial metabolite nicotinamide (172). Future studies are required to further elucidate the relative contribution of individual bacterial species to CNS disease and the relevance of these observations in humans.
Concluding remarks
It is becoming increasingly clear that symbionts that inhabit the gastrointestinal tract have a profound influence on immune cell responses in peripheral tissues beyond the intestine. How gut symbionts influence immune responses at distant sites remains partially understood, but there is evidence for both direct and indirect mechanisms. Expanding our understanding of the immunomodulatory capacity of individual bacterial species or minimal consortia will be fundamental to the rational design of microbiota-targeted therapies to prevent or treat systemic inflammatory disease and infection. The immunological impact of a particular microbe is dictated by its behavior in the context of a complex microbiota in addition to host genetics and disease state. Nonetheless, the precise therapeutic manipulation of the microbiota will need to account for the high inter-individual variability in the composition of the gut microbiota.
As microbial metabolites such as SCFAs or BAs have potent immune regulatory effects, interventions involving systemic supplementation of microbial metabolites or inhibition of their signaling pathways may be a more feasible alternative to stably altering microbiota composition or activity. Further understanding of the mechanisms by which microbial metabolites act to regulate host immunity will be critical for effective therapeutic intervention.
Lastly, as dietary regimens play an important role in shaping the composition and functionality of the intestinal symbiotic community, nutritional interventions provide an additional and accessible avenue for modulating host health outcomes. Although substantial challenges remain, strategies targeting the gut microbiota and their metabolites may hold tremendous promise for the treatment of systemic inflammatory disease.
Acknowledgement
The authors apologize to colleagues whose work was not cited or was cited through other review articles because of space limitations.
Funding
Work in the authors’ laboratories is supported by US National Institutes of Health grants (R01 DK121504 and R01 DK 095782 to G.N. and R01 DK 122812 to G.Y.C.). R.C. is supported by a Career Developments Award (545694) from the Crohn’s and Colitis Foundation and by the University of Michigan Center for Gastrointestinal Research (P30 DK034933). B.C.L. is supported by a Canadian Institutes of Health Research (CIHR) Fellowship.
Conflicts of interest statement: the authors declared no conflicts of interest.
References
- 1. Kamada, N., Seo, S. U., Chen, G. Y. and Núñez, G. 2013. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13:321. [DOI] [PubMed] [Google Scholar]
- 2. Hooper, L. V. and Macpherson, A. J. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159. [DOI] [PubMed] [Google Scholar]
- 3. Kamada, N., Chen, G. Y., Inohara, N. and Núñez, G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donaldson, G. P., Lee, S. M. and Mazmanian, S. K. 2016. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He, Y., Wu, W., Zheng, H. M.et al. . 2018. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 24:1532. [DOI] [PubMed] [Google Scholar]
- 6. Reitmeier, S., Kiessling, S., Clavel, T.et al. . 2020. Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28:258. [DOI] [PubMed] [Google Scholar]
- 7. Underhill, D. M., Gordon, S., Imhof, B. A., Nunez, G. and Bousso, P. 2016. Elie Metchnikoff (1845–1916): celebrating 100 years of cellular immunology and beyond. Nat. Rev. Immunol. 16:651. [DOI] [PubMed] [Google Scholar]
- 8. Pickard, J. M., Zeng, M. Y., Caruso, R. and Núñez, G. 2017. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belkaid, Y. and Harrison, O. J. 2017. Homeostatic immunity and the microbiota. Immunity 46:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Macpherson, A. J. and Harris, N. L. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4:478. [DOI] [PubMed] [Google Scholar]
- 11. Bauer, H., Horowitz, R. E., Levenson, S. M. and Popper, H. 1963. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am. J. Pathol. 42:471. [PMC free article] [PubMed] [Google Scholar]
- 12. Round, J. L. and Mazmanian, S. K. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benveniste, J., Lespinats, G., Adam, C. and Salomon, J. C. 1971. Immunoglobulins in intact, immunized, and contaminated axenic mice: study of serum IgA. J. Immunol. 107:1647. [PubMed] [Google Scholar]
- 14. Moreau, M. C., Ducluzeau, R., Guy-Grand, D. and Muller, M. C. 1978. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 21:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Inagaki, H., Suzuki, T., Nomoto, K. and Yoshikai, Y. 1996. Increased susceptibility to primary infection with Listeria monocytogenes in germfree mice may be due to lack of accumulation of L-selectin+ CD44+ T cells in sites of inflammation. Infect. Immun. 64:3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johansson, M. E., Phillipson, M., Petersson, J., Velcich, A., Holm, L. and Hansson, G. C. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105:15064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johansson, M. E., Larsson, J. M. and Hansson, G. C. 2011. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl Acad. Sci. USA 108(Suppl. 1):4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Atuma, C., Strugala, V., Allen, A. and Holm, L. 2001. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G922. [DOI] [PubMed] [Google Scholar]
- 19. Sedman, P. C., Macfie, J., Sagar, P.et al. . 1994. The prevalence of gut translocation in humans. Gastroenterology 107:643. [DOI] [PubMed] [Google Scholar]
- 20. Zeng, M. Y., Cisalpino, D., Varadarajan, S.et al. . 2016. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ignacio, A., Morales, C. I., Câmara, N. O. and Almeida, R. R. 2016. Innate sensing of the gut microbiota: modulation of inflammatory and autoimmune diseases. Front. Immunol. 7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burberry, A., Zeng, M. Y., Ding, L.et al. . 2014. Infection mobilizes hematopoietic stem cells through cooperative NOD-like receptor and Toll-like receptor signaling. Cell Host Microbe 15:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takizawa, H., Fritsch, K., Kovtonyuk, L. V.et al. . 2017. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 21:225. [DOI] [PubMed] [Google Scholar]
- 24. Josefsdottir, K. S., Baldridge, M. T., Kadmon, C. S. and King, K. Y. 2017. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood 129:729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iwamura, C., Bouladoux, N., Belkaid, Y., Sher, A. and Jankovic, D. 2017. Sensing of the microbiota by NOD1 in mesenchymal stromal cells regulates murine hematopoiesis. Blood 129:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khosravi, A., Yáñez, A., Price, J. G.et al. . 2014. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shi, C., Jia, T., Mendez-Ferrer, S.et al. . 2011. Bone marrow mesenchymal stem and progenitor cells induce monocyte emigration in response to circulating toll-like receptor ligands. Immunity 34:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schulz, C., Gomez Perdiguero, E., Chorro, L.et al. . 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336:86. [DOI] [PubMed] [Google Scholar]
- 29. Bugl, S., Wirths, S., Radsak, M. P.et al. . 2013. Steady-state neutrophil homeostasis is dependent on TLR4/TRIF signaling. Blood 121:723. [DOI] [PubMed] [Google Scholar]
- 30. Balmer, M. L., Schürch, C. M., Saito, Y.et al. . 2014. Microbiota-derived compounds drive steady-state granulopoiesis via MyD88/TICAM signaling. J. Immunol. 193:5273. [DOI] [PubMed] [Google Scholar]
- 31. Deshmukh, H. S., Liu, Y., Menkiti, O. R.et al. . 2014. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 20:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Longman, R. S., Diehl, G. E., Victorio, D. A.et al. . 2014. CX(3)CR1(+) mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 211:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clarke, T. B., Davis, K. M., Lysenko, E. S., Zhou, A. Y., Yu, Y. and Weiser, J. N. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang, D., Chen, G., Manwani, D.et al. . 2015. Neutrophil ageing is regulated by the microbiome. Nature 525:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urbaniak, C., Cummins, J., Brackstone, M.et al. . 2014. Microbiota of human breast tissue. Appl. Environ. Microbiol. 80:3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dickson, R. P. and Huffnagle, G. B. 2015. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 11:e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nejman, D., Livyatan, I., Fuks, G.et al. . 2020. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Macpherson, A. J. and Smith, K. 2006. Mesenteric lymph nodes at the center of immune anatomy. J. Exp. Med. 203:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balmer, M. L., Slack, E., de Gottardi, A.et al. . 2014. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci. Transl. Med. 6:237ra66. [DOI] [PubMed] [Google Scholar]
- 40. Tiniakos, D. G., Vos, M. B. and Brunt, E. M. 2010. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu. Rev. Pathol. 5:145. [DOI] [PubMed] [Google Scholar]
- 41. Miele, L., Marrone, G., Lauritano, C.et al. . 2013. Gut-liver axis and microbiota in NAFLD: insight pathophysiology for novel therapeutic target. Curr. Pharm. Des. 19:5314. [PubMed] [Google Scholar]
- 42. Carpino, G., Del Ben, M., Pastori, D.et al. . 2020. Increased liver localization of lipopolysaccharides in human and experimental NAFLD. Hepatology 72:470. [DOI] [PubMed] [Google Scholar]
- 43. Sharifnia, T., Antoun, J., Verriere, T. G.et al. . 2015. Hepatic TLR4 signaling in obese NAFLD. Am. J. Physiol. Gastrointest. Liver Physiol. 309:G270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rivera, C. A., Adegboyega, P., van Rooijen, N., Tagalicud, A., Allman, M. and Wallace, M. 2007. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J. Hepatol. 47:571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fei, N., Bruneau, A., Zhang, X.et al. . 2020. Endotoxin producers overgrowing in human gut microbiota as the causative agents for nonalcoholic fatty liver disease. mBio 11:e03263-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye, D., Li, F. Y., Lam, K. S.et al. . 2012. Toll-like receptor-4 mediates obesity-induced non-alcoholic steatohepatitis through activation of X-box binding protein-1 in mice. Gut 61:1058. [DOI] [PubMed] [Google Scholar]
- 47. Miura, K., Yang, L., van Rooijen, N., Brenner, D. A., Ohnishi, H. and Seki, E. 2013. Toll-like receptor 2 and palmitic acid cooperatively contribute to the development of nonalcoholic steatohepatitis through inflammasome activation in mice. Hepatology 57:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miura, K., Kodama, Y., Inokuchi, S.et al. . 2010. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology 139:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henao-Mejia, J., Elinav, E., Jin, C.et al. . 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Llopis, M., Cassard, A. M., Wrzosek, L.et al. . 2016. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 65:830. [DOI] [PubMed] [Google Scholar]
- 51. Chu, H., Duan, Y., Yang, L. and Schnabl, B. 2019. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut 68:359. [DOI] [PubMed] [Google Scholar]
- 52. Elamin, E. E., Masclee, A. A., Dekker, J. and Jonkers, D. M. 2013. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr. Rev. 71:483. [DOI] [PubMed] [Google Scholar]
- 53. Uesugi, T., Froh, M., Arteel, G. E., Bradford, B. U. and Thurman, R. G. 2001. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology 34:101. [DOI] [PubMed] [Google Scholar]
- 54. Wree, A., McGeough, M. D., Peña, C. A.et al. . 2014. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J. Mol. Med. (Berl.) 92:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Douhara, A., Moriya, K., Yoshiji, H.et al. . 2015. Reduction of endotoxin attenuates liver fibrosis through suppression of hepatic stellate cell activation and remission of intestinal permeability in a rat non-alcoholic steatohepatitis model. Mol. Med. Rep. 11:1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luther, J., Garber, J. J., Khalili, H.et al. . 2015. Hepatic injury in nonalcoholic steatohepatitis contributes to altered intestinal permeability. Cell. Mol. Gastroenterol. Hepatol. 1:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sharon, G., Garg, N., Debelius, J., Knight, R., Dorrestein, P. C. and Mazmanian, S. K. 2014. Specialized metabolites from the microbiome in health and disease. Cell Metab. 20:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Koh, A., De Vadder, F., Kovatcheva-Datchary, P. and Bäckhed, F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332. [DOI] [PubMed] [Google Scholar]
- 59. Blad, C. C., Tang, C. and Offermanns, S. 2012. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat. Rev. Drug Discov. 11:603. [DOI] [PubMed] [Google Scholar]
- 60. Singh, N., Gurav, A., Sivaprakasam, S.et al. . 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maslowski, K. M., Vieira, A. T., Ng, A.et al. . 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith, P. M., Howitt, M. R., Panikov, N.et al. . 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haghikia, A., Jörg, S., Duscha, A.et al. . 2015. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43:817. [DOI] [PubMed] [Google Scholar]
- 64. Trompette, A., Gollwitzer, E. S., Yadava, K.et al. . 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20:159. [DOI] [PubMed] [Google Scholar]
- 65. Chang, P. V., Hao, L., Offermanns, S. and Medzhitov, R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl Acad. Sci. USA 111:2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Usami, M., Kishimoto, K., Ohata, A.et al. . 2008. Butyrate and trichostatin A attenuate nuclear factor kappaB activation and tumor necrosis factor alpha secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr. Res. 28:321. [DOI] [PubMed] [Google Scholar]
- 67. Vinolo, M. A., Rodrigues, H. G., Hatanaka, E., Sato, F. T., Sampaio, S. C. and Curi, R. 2011. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 22:849. [DOI] [PubMed] [Google Scholar]
- 68. Kendrick, S. F., O’Boyle, G., Mann, J.et al. . 2010. Acetate, the key modulator of inflammatory responses in acute alcoholic hepatitis. Hepatology 51:1988. [DOI] [PubMed] [Google Scholar]
- 69. Singh, N., Thangaraju, M., Prasad, P. D.et al. . 2010. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 285:27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Furusawa, Y., Obata, Y., Fukuda, S.et al. . 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446. [DOI] [PubMed] [Google Scholar]
- 71. Arpaia, N., Campbell, C., Fan, X.et al. . 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Atarashi, K., Tanoue, T., Oshima, K.et al. . 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232. [DOI] [PubMed] [Google Scholar]
- 73. Tao, R., de Zoeten, E. F., Ozkaynak, E.et al. . 2007. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13:1299. [DOI] [PubMed] [Google Scholar]
- 74. Thorburn, A. N., McKenzie, C. I., Shen, S.et al. . 2015. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6:7320. [DOI] [PubMed] [Google Scholar]
- 75. Kim, M., Qie, Y., Park, J. and Kim, C. H. 2016. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gutiérrez-Vázquez, C. and Quintana, F. J. 2018. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 48:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kiss, E. A., Vonarbourg, C., Kopfmann, S.et al. . 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334:1561. [DOI] [PubMed] [Google Scholar]
- 78. Sakamoto, K., Kim, Y. G., Hara, H.et al. . 2017. IL-22 controls iron-dependent nutritional immunity against systemic bacterial infections. Sci. Immunol. 2:eaai8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. al-Waiz, M., Mikov, M., Mitchell, S. C. and Smith, R. L. 1992. The exogenous origin of trimethylamine in the mouse. Metabolism 41:135. [DOI] [PubMed] [Google Scholar]
- 80. Wang, Z., Klipfell, E., Bennett, B. J.et al. . 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang, Z., Roberts, A. B., Buffa, J. A.et al. . 2015. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163:1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ridlon, J. M., Harris, S. C., Bhowmik, S., Kang, D. J. and Hylemon, P. B. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wahlström, A., Sayin, S. I., Marschall, H. U. and Bäckhed, F. 2016. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24:41. [DOI] [PubMed] [Google Scholar]
- 84. Ridlon, J. M., Kang, D. J. and Hylemon, P. B. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47:241. [DOI] [PubMed] [Google Scholar]
- 85. Liu, H. X., Keane, R., Sheng, L. and Wan, Y. J. 2015. Implications of microbiota and bile acid in liver injury and regeneration. J. Hepatol. 63:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Makishima, M., Okamoto, A. Y., Repa, J. J.et al. . 1999. Identification of a nuclear receptor for bile acids. Science 284:1362. [DOI] [PubMed] [Google Scholar]
- 87. Wang, H., Chen, J., Hollister, K., Sowers, L. C. and Forman, B. M. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3:543. [DOI] [PubMed] [Google Scholar]
- 88. Ryan, K. K., Tremaroli, V., Clemmensen, C.et al. . 2014. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 509:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kawamata, Y., Fujii, R., Hosoya, M.et al. . 2003. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278:9435. [DOI] [PubMed] [Google Scholar]
- 90. Katsuma, S., Hirasawa, A. and Tsujimoto, G. 2005. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem. Biophys. Res. Commun. 329:386. [DOI] [PubMed] [Google Scholar]
- 91. Klindt, C., Reich, M., Hellwig, B.et al. . 2019. The G protein-coupled bile acid receptor TGR5 (Gpbar1) modulates endothelin-1 signaling in liver. Cells 8:1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lou, G., Ma, X., Fu, X.et al. . 2014. GPBAR1/TGR5 mediates bile acid-induced cytokine expression in murine Kupffer cells. PLoS ONE 9:e93567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pols, T. W., Nomura, M., Harach, T.et al. . 2011. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 14:747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Keitel, V., Donner, M., Winandy, S., Kubitz, R. and Häussinger, D. 2008. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 372:78. [DOI] [PubMed] [Google Scholar]
- 95. Ali, A. H., Carey, E. J. and Lindor, K. D. 2015. Recent advances in the development of farnesoid X receptor agonists. Ann. Transl. Med. 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ferrell, J. M., Pathak, P., Boehme, S., Gilliland, T. and Chiang, J. Y. L. 2019. Deficiency of both farnesoid X receptor and Takeda G protein-coupled receptor 5 exacerbated liver fibrosis in mice. Hepatology 70:955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lee, C. G., Kim, Y. W., Kim, E. H.et al. . 2012. Farnesoid X receptor protects hepatocytes from injury by repressing miR-199a-3p, which increases levels of LKB1. Gastroenterology 142:1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fiorucci, S., Biagioli, M., Zampella, A. and Distrutti, E. 2018. Bile acids activated receptors regulate innate immunity. Front. Immunol. 9:1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Thakur, K., Tomar, S. K. and De, S. 2016. Lactic acid bacteria as a cell factory for riboflavin production. Microb. Biotechnol. 9:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kjer-Nielsen, L., Patel, O., Corbett, A. J.et al. . 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491:717. [DOI] [PubMed] [Google Scholar]
- 101. Constantinides, M. G., Link, V. M., Tamoutounour, S.et al. . 2019. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366:eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Legoux, F., Bellet, D., Daviaud, C.et al. . 2019. Microbial metabolites control the thymic development of mucosal-associated invariant T cells. Science 366:494. [DOI] [PubMed] [Google Scholar]
- 103. Ivanov, I. I., Atarashi, K., Manel, N.et al. . 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gaboriau-Routhiau, V., Rakotobe, S., Lécuyer, E.et al. . 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677. [DOI] [PubMed] [Google Scholar]
- 105. Atarashi, K., Tanoue, T., Ando, M.et al. . 2015. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sano, T., Huang, W., Hall, J. A.et al. . 2016. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 164:324. [DOI] [PubMed] [Google Scholar]
- 107. Yang, Y., Torchinsky, M. B., Gobert, M.et al. . 2014. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Goto, Y., Panea, C., Nakato, G.et al. . 2014. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ladinsky, M. S., Araujo, L. P., Zhang, X.et al. . 2019. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363:eaat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gauguet, S., D’Ortona, S., Ahnger-Pier, K.et al. . 2015. Intestinal microbiota of mice influences resistance to Staphylococcus aureus pneumonia. Infect. Immun. 83:4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. McAleer, J. P., Nguyen, N. L., Chen, K.et al. . 2016. Pulmonary Th17 antifungal immunity is regulated by the gut microbiome. J. Immunol. 197:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bradley, C. P., Teng, F., Felix, K. M.et al. . 2017. Segmented filamentous bacteria provoke lung autoimmunity by inducing gut-lung axis Th17 cells expressing dual TCRs. Cell Host Microbe 22:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lathrop, S. K., Bloom, S. M., Rao, S. M.et al. . 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Xu, M., Pokrovskii, M., Ding, Y.et al. . 2018. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554:373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chai, J. N., Peng, Y., Rengarajan, S.et al. . 2017. Helicobacter species are potent drivers of colonic T cell responses in homeostasis and inflammation. Sci. Immunol. 2:eaal5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Atarashi, K., Tanoue, T., Shima, T.et al. . 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Josefowicz, S. Z., Niec, R. E., Kim, H. Y.et al. . 2012. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ohnmacht, C., Park, J. H., Cording, S.et al. . 2015. Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349:989. [DOI] [PubMed] [Google Scholar]
- 119. Tanoue, T., Atarashi, K. and Honda, K. 2016. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 16:295. [DOI] [PubMed] [Google Scholar]
- 120. Teratani, T., Mikami, Y., Nakamoto, N.et al. . 2020. The liver-brain-gut neural arc maintains the Treg cell niche in the gut. Nature 585:591. [DOI] [PubMed] [Google Scholar]
- 121. Huang, Y., Mao, K., Chen, X.et al. . 2018. S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gasteiger, G., Fan, X., Dikiy, S., Lee, S. Y. and Rudensky, A. Y. 2015. Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mjösberg, J. and Rao, A. 2018. Lung inflammation originating in the gut. Science 359:36. [DOI] [PubMed] [Google Scholar]
- 124. Wu, H. J., Ivanov, I. I., Darce, J.et al. . 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Teng, F., Klinger, C. N., Felix, K. M.et al. . 2016. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of peyer’s patch T follicular helper cells. Immunity 44:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Omenetti, S., Bussi, C., Metidji, A.et al. . 2019. The intestine harbors functionally distinct homeostatic tissue-resident and inflammatory Th17 cells. Immunity 51:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tezuka, H., Abe, Y., Iwata, M.et al. . 2007. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature 448:929. [DOI] [PubMed] [Google Scholar]
- 128. Tezuka, H., Abe, Y., Asano, J.et al. . 2011. Prominent role for plasmacytoid dendritic cells in mucosal T cell-independent IgA induction. Immunity 34:247. [DOI] [PubMed] [Google Scholar]
- 129. Mora, J. R., Iwata, M., Eksteen, B.et al. . 2006. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science 314:1157. [DOI] [PubMed] [Google Scholar]
- 130. Uematsu, S., Fujimoto, K., Jang, M. H.et al. . 2008. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 9:769. [DOI] [PubMed] [Google Scholar]
- 131. Wilmore, J. R., Gaudette, B. T., Gomez Atria, D.et al. . 2018. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe 23:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Wesemann, D. R., Portuguese, A. J., Meyers, R. M.et al. . 2013. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature 501:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chen, Y., Chaudhary, N., Yang, N.et al. . 2018. Microbial symbionts regulate the primary Ig repertoire. J. Exp. Med. 215:1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Koch, M. A., Reiner, G. L., Lugo, K. A.et al. . 2016. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Ansaldo, E., Slayden, L. C., Ching, K. L.et al. . 2019. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kubes, P. and Jenne, C. 2018. Immune responses in the liver. Annu. Rev. Immunol. 36:247. [DOI] [PubMed] [Google Scholar]
- 137. Corbitt, N., Kimura, S., Isse, K.et al. . 2013. Gut bacteria drive Kupffer cell expansion via MAMP-mediated ICAM-1 induction on sinusoidal endothelium and influence preservation-reperfusion injury after orthotopic liver transplantation. Am. J. Pathol. 182:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. McDonald, B., Zucoloto, A. Z., Yu, I. L.et al. . 2020. Programing of an intravascular immune firewall by the gut microbiota protects against pathogen dissemination during infection. Cell Host Microbe 28:660. [DOI] [PubMed] [Google Scholar]
- 139. Wingender, G., Stepniak, D., Krebs, P.et al. . 2012. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology 143:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lee, W. Y., Moriarty, T. J., Wong, C. H.et al. . 2010. An intravascular immune response to Borrelia burgdorferi involves Kupffer cells and iNKT cells. Nat. Immunol. 11:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Olszak, T., An, D., Zeissig, S.et al. . 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Kenna, T., Golden-Mason, L., Porcelli, S. A.et al. . 2003. NKT cells from normal and tumor-bearing human livers are phenotypically and functionally distinct from murine NKT cells. J. Immunol. 171:1775. [DOI] [PubMed] [Google Scholar]
- 143. Li, F., Hao, X., Chen, Y.et al. . 2017. The microbiota maintain homeostasis of liver-resident gammadeltaT-17 cells in a lipid antigen/CD1d-dependent manner. Nat. Commun. 7:13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Macpherson, A. J., Heikenwalder, M. and Ganal-Vonarburg, S. C. 2016. The liver at the nexus of host-microbial interactions. Cell Host Microbe. 20:561. [DOI] [PubMed] [Google Scholar]
- 145. Loomba, R. and Sanyal, A. J. 2013. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 10:686. [DOI] [PubMed] [Google Scholar]
- 146. Rehm, J., Samokhvalov, A. V. and Shield, K. D. 2013. Global burden of alcoholic liver diseases. J. Hepatol. 59:160. [DOI] [PubMed] [Google Scholar]
- 147. Al Nabhani, Z., Dulauroy, S., Marques, R.et al. . 2019. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50:1276. [DOI] [PubMed] [Google Scholar]
- 148. Herbst, T., Sichelstiel, A., Schär, C.et al. . 2011. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 184:198. [DOI] [PubMed] [Google Scholar]
- 149. Russell, S. L., Gold, M. J., Hartmann, M.et al. . 2012. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kim, Y. G., Udayanga, K. G., Totsuka, N., Weinberg, J. B., Núñez, G. and Shibuya, A. 2014. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE2. Cell Host Microbe 15:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ichinohe, T., Pang, I. K., Kumamoto, Y.et al. . 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl Acad. Sci. USA 108:5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Abt, M. C., Osborne, L. C., Monticelli, L. A.et al. . 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Trompette, A., Gollwitzer, E. S., Pattaroni, C.et al. . 2018. Dietary fiber confers protection against flu by shaping Ly6c- patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity 48:992. [DOI] [PubMed] [Google Scholar]
- 154. Steed, A. L., Christophi, G. P., Kaiko, G. E.et al. . 2017. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Beura, L. K., Hamilton, S. E., Bi, K.et al. . 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Rosshart, S. P., Vassallo, B. G., Angeletti, D.et al. . 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171:1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Erny, D. and Prinz, M. 2020. How microbiota shape microglial phenotypes and epigenetics. Glia 68:1655. [DOI] [PubMed] [Google Scholar]
- 158. Cryan, J. F., O’Riordan, K. J., Cowan, C. S. M.et al. . 2019. The microbiota-gut-brain axis. Physiol. Rev. 99:1877. [DOI] [PubMed] [Google Scholar]
- 159. Erny, D., Hrabě de Angelis, A. L., Jaitin, D.et al. . 2015. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Matcovitch-Natan, O., Winter, D. R., Giladi, A.et al. . 2016. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353:aad8670. [DOI] [PubMed] [Google Scholar]
- 161. Rothhammer, V., Borucki, D. M., Tjon, E. C.et al. . 2018. Microglial control of astrocytes in response to microbial metabolites. Nature 557:724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Rothhammer, V., Mascanfroni, I. D., Bunse, L.et al. . 2016. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Constantinescu, C. S., Farooqi, N., O’Brien, K. and Gran, B. 2011. Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 164:1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Dendrou, C. A., Fugger, L. and Friese, M. A. 2015. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 15:545. [DOI] [PubMed] [Google Scholar]
- 165. Lee, Y. K., Menezes, J. S., Umesaki, Y. and Mazmanian, S. K. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 108(Suppl. 1):4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Berer, K., Mues, M., Koutrolos, M.et al. . 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479:538. [DOI] [PubMed] [Google Scholar]
- 167. Miyauchi, E., Kim, S. W., Suda, W.et al. . 2020. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585:102. [DOI] [PubMed] [Google Scholar]
- 168. Lai, M. C., Lombardo, M. V. and Baron-Cohen, S. 2014. Autism. Lancet 383:896. [DOI] [PubMed] [Google Scholar]
- 169. Kim, S., Kim, H., Yim, Y. S.et al. . 2017. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. van Es, M. A., Hardiman, O., Chio, A.et al. . 2017. Amyotrophic lateral sclerosis. Lancet 390:2084. [DOI] [PubMed] [Google Scholar]
- 171. Burberry, A., Wells, M. F., Limone, F.et al. . 2020. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature 582:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Blacher, E., Bashiardes, S., Shapiro, H.et al. . 2019. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572:474. [DOI] [PubMed] [Google Scholar]