Abstract
Ameloblastin is the second most abundant enamel matrix protein, and is thought to be essential for ameloblast cell polarization, cell adhesion, and enamel mineralization. However, studies of ameloblastin’s function and its molecular mechanism have been limited due to difficulty in obtaining recombinant ameloblastin in vitro. Here, we present a protocol for successful ameloblastin expression and purification in E. coli.
Keywords: Ameloblastin, Enamel, Matrix protein, Expression, purification
1. Introduction
Enamel extracellular matrix is composed of proteins, including amelogenin, ameloblastin, enamelin, and amelotin, and proteinases matrix metalloproteinase-20 (MMP-20 or enamelysin) and kallikrein-4 (KLK-4) [1]. Ameloblastin (AMBN), which is encoded by a secretory calcium-binding phosphoprotein (SCPP) gene located on chromosome 4q21 [2], is the second most abundant enamel matrix protein, accounting for roughly 5% of the matrix [3]. It is a two-domain, intrinsically disordered protein with one specific and several non-specific calcium-binding regions [4].
Ameloblastin is crucial for enamel mineralization. Ameloblastin-mutant mice develop severe enamel hypoplasia [5], and mice with overexpression of ameloblastin exhibit imperfections in their enamel that are evident on the nanoscale [6]. It has also been reported recently that deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta [7]. However, the exact function of ameloblastin and its molecular mechanism is still unclear. It has been suggested that ameloblastin is a cell adhesion molecule which adheres ameloblasts to the enamel extracellular matrix [5], that it may interact with calcium ions [4, 8], that it could act as a signal molecule [9], that serine phosphorylation of ameloblastin is important for enamel formation [10], and that the self-assembly of ameloblastin is crucial for the organization of enamel extracellular matrix and formation of properly structured enamel [11].
Ameloblastin is highly expressed by ameloblasts during the secretory stage of amelogenesis [8]. It has several identified or putative phosphorylation, O-glycosylation, and hydroxylation sites [4, 12]. Soon after its secretion, ameloblastin is cleaved by matrix metalloproteinase 20 [13]. The N-terminal cleavage products of ameloblastin are stable and accumulate in the enamel prism sheaths, while the C-terminal cleavage products are successively cleaved into smaller peptides and ultimately lost [8,14]. Full-length ameloblastin is only found adjacent to the secretory face of Tomes’ process. Thus, intact ameloblastin is a trace component of developing enamel and has never been isolated in vivo. Mouse ameloblastin has been successfully expressed in Drosophila melanogaster expression system using Schneider 2 cells in DES system [15]. However, this is a slow and low-yielding process. For these reasons, we developed a technique to express mouse ameloblastin (GenBank No. AAB93765.1) with cleavable Thioredoxin, Histidine Trx-, His-, and S-tags in E. coli. We describe here how this can be achieved, after which the protein can be purified using nickel affinity chromatography, and the tags can be cleaved by enterokinase (NEB). As shown in Fig. 1, the final product of this protocol was of sufficient quality for biochemical and biophysical experiments, and mass spectra confirmed that this product was ameloblastin. The protein expressed in E. coli lacks the posttransitional modifications of glycosylation and phosphorylation. The advantages of the E. coli expression system are the relative high yield and ease of expression and purification. This recombinant ameloblastin is suitable for secondary and tertiary structural studies.
Fig. 1.
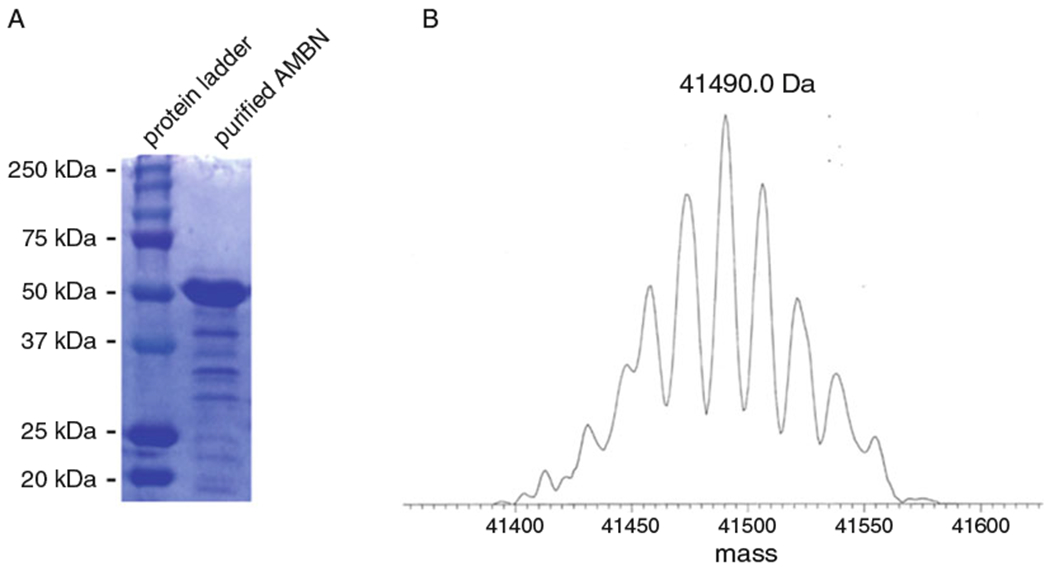
SDS-PAGE and mass spectra of purified mouse AMBN. (a) SDS-PAGE showed that the purity of the AMBN obtained by this technique was of sufficient quality for biochemical and biophysical experiments and the apparent molecular weight was not higher than the theoretical value. (b) Mass spectra of the band around 50 kDa in SDS-PAGE showed that the exact molecular weight of the purified protein was close to the theoretical value (41459.8 Da), suggesting the purified protein was AMBN
2. Materials
E. coli strain BL21(DE3) pLysS (Stratagene).
500 mL ×4 and 50 mL LB broth (1.25 g LB broth powder in 50 mL deionized water).
LB/NZCYM agar plates with ampicillin.
100 mg/mL ampicillin 1 g sodium salt of ampicillin in 10 mL dI water.
1 M IPTG (2.383 g of IPTG in 10 mL dI water Sigma-Aldrich).
0.5 M EDTA in water, pH 8.5.
0.1 M benzamidine HCl (156.62 mg benzamidine HCl in 10 mL deionized water).
1 M PMSF in DMSO (1.74 g PMSF in 10 mL DMSO Sigma-Aldrich).
Ni-NTA Agarose (Qiagen).
Imidazole (Sigma-Aldrich).
Phenomenex C4 High Performance Liquid Chromatography (HPLC) column (10 × 250 mm, 5 μm).
16,000 units/mL enterokinase (New England Biolabs).
8 M urea (4.8 g urea in 10 mL dI water).
- Buffers for affinity chromatography:
- Lysis buffer (binding buffer): pH 8.0, 50 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole.
- Washing buffer: pH 7.2, 50 mM NaH2PO4, 500 mM NaCl, 50 mM imidazole.
- Elution buffer: pH 7.2, 50 mM NaH2PO4, 500 mM NaCl, 500 mM Imidazole.
- Buffers for HPLC:
- Buffer A: 0.1% TFA in water, filtered with 0.45 um filter.
- Buffer B: 0.1% TFA, 80% acetonitrile in water, filtered with 0.45 um filter.
3. Methods
3.1. Day 1 Make Bacterial Culture Plates
Make LB agar plates with 1:1000 dilution of 100 mg/mL ampicillin.
Prepare recombinant BL21 E coli. with pET-32a (Novagen) plasmid inserted with mouse ameloblastin gene (GenBank No. AAB93765.1) having thioredoxin, histidine, and S-tags using standard methods of bacterial cloning.
Plate the recombinant E coli on ampicillin agar plates and culture overnight at 37 °C.
3.2. Day 2 Make Starter Culture
Prepare 2 L LB media in 4 L flasks, 500 mL in each flask.
Prepare 50 mL LB media in a separate 250 mL flask.
Autoclave all media at 121 °C and allow it to cool. Store these flasks at 4 °C until used.
Add 50 μL of 100 mg/mL ampicillin to the 50 mL media.
Inoculate the 50 mL LB media supplemented with ampicillin with a single colony from the BL21 agar plate. Seal and save the plate for later use at 4 °C.
Incubate 50 mL culture overnight in a shaking incubator at 37 °C. This is the starter culture.
3.3. Day 3 Protein Expression
Remove the starter culture from the shaker-incubator and keep at 4 °C until used.
Add 500 μL of 100 mg/mL ampicillin to each flask of 500 mL LB media.
Measure optical density (OD) of the culture using a UV-Vis spectrophotometer at 595 nm. This will serve as the baseline or “blank” measurement. Keep this for later reading.
Inoculate each 500 mL LB broth supplemented with ampicillin with one fourth of the starter culture (day 2 step 6).
Take OD reading at 595 nm immediately after inoculation. Further readings should be taken periodically to keep track of the growth. Read the blank each time you measure the OD.
Induce each flask with 500 μL 1 MIPTG mentioned in materials, no need to say again when the bacterial growth reaches ~0.75–0.8 OD at 595 nm.
Continue to take OD readings periodically. Bacterial growth is usually slow right after induction, so it is best to check OD 2 h after induction. Harvest the bacteria after 4 h.
Pour the contents of the flask into centrifuge bottles. Do not overfill the bottles. Balance the weight and centrifuge for 6 min at 8000 rpm (9700 × g) at 4 °C.
Discard the supernatant and keep the bacterial pellets at −20 °C overnight.
3.4. Day 4 Protein Purification
All steps from this point forward should be done on ice or in a cold room (see Note 1).
Resuspend the bacterial pellets in lysis buffer (20 mL lysis buffer/500 mL culture pellet).
Add 1 mM EDTA (400 μL of 0.5 M EDTA solution for 80 mL lysis buffer), 1 mM benzamidine, and 1 mM PMSF (200 μL each) (see Note 2).
Sonicate the bacteria for 30 min at amplitude 20%, 1 s on 1 s off, with a precooled sonicator tip using an ultrasonicator like Branson Sonifier (Branson Ultrasonics, US).
Centrifuge twice at 12,000 rpm (20,000 × g) for 15 min at 4 °C.
Prepare Ni-NTA columns by washing with 15 mL elution buffer, followed by 20 mL lysis buffer.
Decant supernatant in clean Ni-NTA agarose gel tubes, and place on a rocker for the proteins to bind for 1 h at 4 °C (see Note 3).
Let the supernatant pass through the columns. Add 50 mL binding buffer (25 mL twice) and let it drip through the Ni-NTA tube.
Add 50 mL washing buffer and let it pass through the Ni-NTA tubes.
Elute proteins using 5 mL elution buffer and prepare for dialysis as described below.
3.5. Days 4 and 5 Dialysis
Make 3 L dialysis buffer by diluting 30 mL pH 7.4 1 M NaH2PO4 in 2.970 L water, and cool it at 4 °C.
Use 1 L dialysis buffer and add 1 mL 1 M PMSF (final concentration 0.1 mM), 1 mL 0.1 M benzamidine (final concentration 0.01 mM), and 1 mL 0.5 M EDTA (final concentration 0.5 mM) and stir.
Take the eluted protein solution, and carefully place it in a 10,000 kDa dialysis membrane clamped at one end. Clamp the other end and suspend in the dialysis buffer stirring overnight. Change the buffer twice: once in the morning and then again 4 h later. Collect the dialyzed protein 4 h after the second change.
Determine the concentration of protein after dialysis using a Pierce BCA or other appropriate method kit.
3.6. Day 6 Cleave the Trx-, His-, and S-Tags
At this point the protein is in pH 7.4 10 mM NaH2PO4 buffer containing EDTA, PMSF, and benzamidine.
Add 8 M urea to the protein solution such that the final concentration of urea is 1 M.
Enterokinase is used to cleave the tags to obtain Ameloblastin in its native state. For 1 mg protein, 0.8 μL enterokinase (12.8 units) is added. Calculate the amount of enterokinase needed based on the concentration of protein after dialysis and incubate the protein solution with enterokinase and urea at 37 °C for 6 h with gentle mixing. Stop the reaction by adding 10% of the total volume of acetic acid, and store at −20 °C overnight.
3.7. Day 7 Remove the Cleaved Tags Using HPLC
To separate the fragments of cleaved tags from full-length ameloblastin, HPLC (Varian) system is used. The system is prepared by removing bubbles following the standard protocol.
Clean the C4 column Phenomenex (10 × 250 mm, 5 μm) following the standard column cleaning protocol.
Centrifuge the cleaved ameloblastin and keep the supernatant to remove any solid particles. The amount of ameloblastin injected in the HPLC column depends upon to the maximum sample loop volume.
Elute with a gradient increasing from 40 to 90% buffer B for 80 min, at a flow rate of 1.5 mL/min.
Collect all the peaks. The first peak appears around 17 min. There will typically be a four-peak pattern in which the second peak is ameloblastin at around 30 min (see Note 4).
Lyophilize the collected peaks and dissolve them in distilled water to run an SDS-PAGE.
Final protein should appear at around 50 kDa in 12% SDS-PAGE gel.
Abbreviations
- AMBN
Ameloblastin
- DMSO
Dimethyl sulfoxide
- IPTG
Isopropyl β-d-1-thiogalactopyranoside
- LB broth
Luria-Bertani broth
- PMSF
Phenylmethylsulfonyl fluoride
- His-tag
An amino acid motif in proteins that consists of at least six histidine (His) residues,6 aa
- S-tag
An oligopeptide derived from pancreatic ribonuclease A, 15 aa
- Trx-tag
Thioredoxin protein, 105 aa
Footnotes
Ameloblastin is not stable at room temperature, particularly when the purity is low. For this reason, do not keep the protein or protein mixture at room temperature.
Use freshly prepared benzamidine and PMSF stock can be stored for several weeks at −20 °C.
Ni-NTA can be reused up to 5 times. Follow manufacturer’s recommendations for cleaning and regenerating the Ni-NTA resin.
Ameloblastin is intrinsically disordered, and its aggregation behavior is sensitive to the buffer conditions. The position of peaks for ameloblastin in high-performance liquid chromatography may therefore change slightly. It is advisable to run SDS-PAGE to confirm the final product each time.
References
- 1.Moradian-Oldak J (2012) Protein-mediated enamel mineralization. Front Biosci 17:1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacDougall M, DuPont BR, Simmons D, Reus B, Krebsbach P, Karrman C, Holmgren G, Leach RJ, Forsman K (1997) Ameloblastin gene (AMBN) maps within the critical region for autosomal dominant amelogenesis imperfecta at chromosome 4q21. Genomics 41(1):115–118. 10.1006/geno.1997.4643 [DOI] [PubMed] [Google Scholar]
- 3.Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y (1996) Full-length sequence, localization, and chromosomal mapping of ameloblastin a novel tooth-specific gene. J Biol Chem 271(8):4431–4435 [DOI] [PubMed] [Google Scholar]
- 4.Vymětal J, Slabý I, Spahr A, Vondrášek J, Lyngstadaas SP (2008) Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur J Oral Sci 116(2):124–134 [DOI] [PubMed] [Google Scholar]
- 5.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y (2004) Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167(5):973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML (2003) A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem 278 (21):19447–19452. 10.1074/jbc.M300445200 [DOI] [PubMed] [Google Scholar]
- 7.Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell Aj (2014) Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet 23(20):5317–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murakami C, Dohi N, Fukae M, Tanabe T, Yamakoshi Y, Wakida K, Satoda T, Takahashi O, Shimizu M, Ryu O (1997) Immunochemical and immunohistochemical study of the 27-and 29-kDa calcium-binding proteins and related proteins in the porcine tooth germ. Histochem Cell Biol 107(6):485–494 [DOI] [PubMed] [Google Scholar]
- 9.Zeichner-David M, Chen LS, Hsu Z, Reyna J, Caton J, Bringas P (2006) Amelogenin and ameloblastin show growth-factor like activity in periodontal ligament cells. Eur J Oral Sci 114(Suppl 1):244–253.; discussion 254–246, 381–242. 10.1111/j.1600-0722.2006.00322.x [DOI] [PubMed] [Google Scholar]
- 10.Ma P, Yan W, Tian Y, He J, Brookes SJ, Wang X (2016) The importance of serine phosphorylation of ameloblastin on enamel formation. J Dent Res 95(12):1408–1414. 10.1177/0022034516661513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wald T, Spoutil F, Osickova A, Prochazkova M, Benada O, Kasparek P, Bumba L, Klein OD, Sedlacek R, Sebo P, Prochazka J, Osicka R (2017) Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif. Proc Natl Acad Sci U S A 114(9): E1641–E1650. 10.1073/pnas.1615334114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delsuc F, Gasse B, Sire JY (2015) Evolutionary analysis of selective constraints identifies ameloblastin (AMBN) as a potential candidate for amelogenesis imperfecta. BMC Evol Biol 15:148. 10.1186/s12862-015-0431-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun YH, Yamakoshi Y, Yamakoshi F, Fukae M, Hu JC, Bartlett JD, Simmer JP (2010) Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res 89(8):785–790. 10.1177/0022034510366903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uchida T, Murakami C, Wakida K, Satoda T, Dohi N, Takahashi O (1997) Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem 45(10):1329–1340 [DOI] [PubMed] [Google Scholar]
- 15.Ravindranath HH, Chen LS, Zeichner-David- M, Ishima R, Ravindranath RM (2004) Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem Biophys Res Commun 323(3):1075–1083. 10.1016/j.bbrc.2004.08.207 [DOI] [PubMed] [Google Scholar]


