Abstract
Background
COVID‐19 has caused an unprecedented global health emergency. The strains of such a pandemic can overwhelm hospital capacity. Efficient clinical decision‐making is crucial for proper healthcare resource utilization in this crisis. Using observational study data, we set out to create a predictive model that could anticipate which COVID‐19 patients would likely be admitted and developed a scoring tool that could be used in the clinical setting and for population risk stratification.
Methods
We retrospectively evaluated data from COVID‐19 patients across a network of 6 hospitals in northeastern Pennsylvania. Analysis was limited to age, gender, and historical variables. After creating a variable importance plot, we chose a selection of the best predictors to train a logistic regression model. Variable selection was done using a lasso regularization technique. Using the coefficients in our logistic regression model, we then created a scoring tool and validated the score on a test set data.
Results
A total of 6485 COVID‐19 patients were included in our analysis, of which 707 were hospitalized. The biggest predictors of patient hospitalization included age, a history of hypertension, diabetes, chronic heart disease, gender, tobacco use, and chronic kidney disease. The logistic regression model demonstrated an AUC of 0.81. The coefficients for our logistic regression model were used to develop a scoring tool. Low‐, intermediate‐, and high‐risk patients were deemed to have a 3.5%, 26%, and 38% chance of hospitalization, respectively. The best predictors of hospitalization included age (odds ratio [OR] = 1.03, confidence interval [CI] = 1.02–1.03), diabetes (OR = 2.08, CI = 1.69–2.57), hypertension (OR = 2.36, CI = 1.90–2.94), chronic heart disease (OR = 1.53, CI = 1.22–1.91), and male gender (OR = 1.32, CI = 1.11–1.58).
Conclusions
Using retrospective observational data from a 6‐hospital network, we determined risk factors for admission and developed a predictive model and scoring tool for use in the clinical and population setting that could anticipate admission for COVID‐19 patients.
Keywords: COVID, machine learning, predictive model, risk of admission
1. INTRODUCTION
1.1. Background
COVID‐19, declared a global pandemic by the World Health Organization (WHO) in March 2020, 1 has caused an unprecedented global health emergency with more than 23.7 million confirmed cases globally as of August 25, 2020. 2 In the United States alone, there have been 5.7 million confirmed cases, with over 177,000 deaths—numbers that increase by the day. 2 The virus displays person‐to‐person transmission, 3 , 4 and to cope with the tremendous burden this places on the healthcare system, governments worldwide have instituted quarantine measures to slow the spread. Although the typical intensive care unit (ICU) occupancy is 60%–80%, 5 the strains of a pandemic, such as that of COVID‐19, can overwhelm hospital capacity. 6 Furthermore, COVID‐19 demonstrates widespread symptomology with varying degrees of illness and inconsistent radiologic findings. Although the majority of patients present with fever, cough, shortness of breath, and respiratory distress, 7 , 8 gastrointestinal symptoms, such as nausea and vomiting, have also been reported, 9 as well as the asymptomatic patient. 10 Although COVID‐19 has a diverse presentation, clinical variables, such as age, race, or comorbidities can help inform disease course. Data show higher prevalence of COVID‐19 in patients with features of the metabolic syndrome, hypertension, cardiovascular disease, and positive smoking history, 11 , 12 , 13 and these factors have also been correlated with a greater rate of ICU admission 14 , 15 and mortality. 16 , 17
1.2. Importance
Predictive analytics that use algorithms to identify patterns in large amounts of data 18 have been gaining popularity in healthcare research to provide researchers with the ability to harness data to inform clinical decision making. 19 COVID‐19 has proven no exception, as the global pandemic has spurred the research community to recommend using predictive modelling techniques with the data gathered to expedite clinical decision making. 20 , 21 , 22 This call has been answered, as clinicians are using emerging patterns in disease presentation and prognosis in combination with predictive modelling to improve hospital and patient management. Modelling tools have been developed that predict ICU admission, 23 detect COVID‐19 patients using routine blood tests, 24 combine clinical history and computed tomograpy (CT) images for accurate diagnosis, 25 and act as a COVID‐19 mortality predictor using clinical features. 26 Although data have been used to inform decision making and provide a prediction of prognosis and hospitalization, 27 , 28 these models rely on clinical data that have to be obtained on a hospital visit (ie, lab values, vital signs, and chest X‐ray findings). An accurate model has not yet been introduced that predicts whether a patient who tested positive for COVID‐19 will be admitted to the hospital using only age and historical variables. Such a model can be used both in the clinical and population level.
1.3. Goals of investigation
We created a logistic regression model using retrospective observational study data to try to predict which patients will likely be admitted to the hospital that test positive for COVID‐19. We also understand that, on a practical level, incorporating a predictive model into an electronic health record for decision support can be challenging, and, thus, we also used variables from our logistic regression model to develop a practical scoring tool.
2. METHODS
2.1. Study design/setting
This study is an observational retrospective study that includes description and analysis of COVID‐19 patients across our 6‐hospital network in northeastern Pennsylvania who had data collected in a COVID‐19 registry database. The registry was developed to be used as an analytical tool as a part of the organization's quality improvement COVID initiatives. It was built in the Epic electronic health record system (Epic Systems, Verona, WI) after being developed and maintained by the network's enterprise analytics team in the information support (IS) department. The data were extracted from the database with de‐identified patient data, and variables within the database were used in our analysis.
2.2. Selection of participants
A COVID‐19 patient was defined as a patient who had a positive SARS‐COV2 PCR test. Patients younger than 18 years old and older than 90 years old were excluded from the analysis based on our network institutional review board requirements to maintain a quality improvement initiative and preserve patient confidentiality. We limited our analysis to age, gender, and historical variables because our goal was to develop an ambulatory predictive tool to predict which patients likely will be hospitalized.
2.3. Measurements
The demographic and historical variables were defined using Epic electronic health record groupers that generally are used in our electronic health record to capture patient clinical history. The initial independent variables that were extracted include age, hypertension, diabetes, chronic heart disease, gender, smoking history, chronic kidney disease, whether the patient was taking an ACE inhibitor, a history of cancer, chronic obstructive pulmonary disease, asthma, chronic liver disease, chronic renal failure, corticosteroid use, whether the patient was taking an immunosuppressive, and history of chronic bronchitis and HIV status. The groupers aggregate together ICD‐10 codes on problem lists that fall into a particular category.
2.4. Analysis
We first split the data into a training set that consisted of 80% of the data. Exploratory data analyses and model developments were conducted on the training set, and validation was done on a test set (20% of the data) not seen by the model. A separate 20% test set to analyze model performance is standard practice for model development to detect overfitting. We created a variable importance plot to examine which variables were most associated with admission. We also used a least absolute shrinkage and selection operator (lasso) regularization technique to select the variables to include in our final logistic regression model. Lasso performs variable selection to enhance the prediction accuracy and interpretability of the statistical model by penalizing model complexity such as the number of variables used.
2.5. Outcomes
The definition of hospitalization for COVID‐19 was a positive test, defined as a positive SAR‐COV2 PCR test, and hospitalization, defined as time of admission order placement, within 7 days of each other. We used the best predictors to train a logistic regression model to predict which patients will likely be hospitalized. We reported odds ratios (ORs) and confidence intervals (CIs) for those variables in our model. The performance of the model was validated on the 20% test set data that the model did not see as mentioned above. We used the coefficient values from our logistic regression model with binned age to develop a manual scoring tool that allows a manual calculation of the risk of hospitalization. We then validated the score on the test set data. The R statistical software was used to conduct all statistical analysis, and the stats and randomForest package were used for model development. This study proposal was reviewed by our institutional review board and deemed to be non‐human research.
The Bottom Line
This retrospective study used data from 6,485 patients with COVID‐19 at 6 hospitals in Pennsylvania to develop a logistic regression model and scoring tool to predict hospitalization using easily‐obtained input variables. This model has potential to support resource planning for patients with COVID‐19.
3. RESULTS
A total of 6485 patients were included in our analysis. Of these, 707 patients were defined as being hospitalized for COVID‐19. There was a clear difference in age between those who were hospitalized compared to those who were not with a mean of 64 and 48 years of age, respectively. Table 1 shows the variable differences between hospitalized versus non‐hospitalized patients. The best predictors of hospitalization were age, a history of hypertension, diabetes, chronic heart disease, gender, tobacco use, and chronic kidney disease (see Figure 1, variable importance plot). The receiver operating characteristic (ROC) curve for the model tested on the 20% validation data is presented in Figure 2. The area under the curve (AUC) for the logistic regression model was 0.81. The AUC is a measure of how much better the model performs, compared with a random guess. A perfect model would give an AUC of 1, and an uninformed model would give an AUC of 0.5. This indicated that this model performed overall well in predicting which patients will need hospitalization for COVID‐19. The model had a sensitivity of 80%, a specificity of 71%, a positive predictive value of 25%, and a negative predictive value of 97%. The OR for our logistic regression model is shown in Table 2.
TABLE 1.
Historical variable differences between hospitalized versus non‐hospitalized patients for COVID‐19 (%)
| Comparison of history for non‐hospitalized versus hospitalized patients (%) | ||
|---|---|---|
| No | Yes | |
| Male | 44 | 50 |
| Has HIV | 0.49 | 0.75 |
| Tobacco smoker | 22 | 37 |
| On ACE inhibitor | 8.7 | 16 |
| Inhaled steroids use | 1.9 | 3.6 |
| Diabetes mellitus | 11 | 39 |
| Hypertension | 22 | 64 |
| Chronic kidney disease | 4.8 | 17 |
| Has cancer | 3.4 | 8.9 |
| Asthma | 6.4 | 8 |
| Chronic bronchitis | 0.49 | 1.5 |
| Chronic heart disease | 11 | 41 |
| Chronic lung disease | 6.6 | 18 |
| Chronic liver disease | 3 | 6.6 |
| On immunosuppression | 0.72 | 2.4 |
| Chronic obstructive pulmonary disease | 2.4 | 11 |
FIGURE 1.
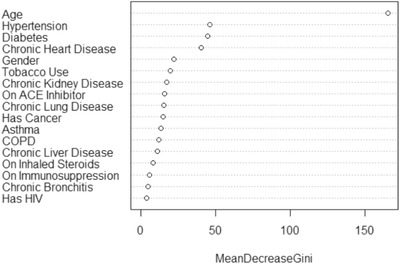
Variable importance plot for predicting hospitalization for COVID‐19
FIGURE 2.
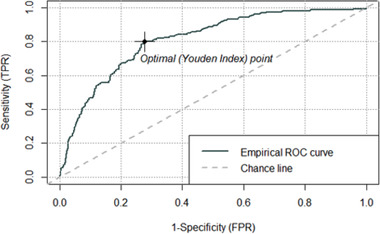
ROC curve for predicting hospitalization for COVID‐19. ROC, receiver operating characteristic
TABLE 2.
OR of logistic regression model coefficients
| OR | CI | |
|---|---|---|
| Age | 1.026 | 1.021–1.032 |
| Diabetes mellitus | 2.083 | 1.685–2.572 |
| Hypertension | 2.357 | 1.892–2.937 |
| Chronic heart disease | 1.530 | 1.222–1.912 |
| Male gender | 1.324 | 1.107–1.583 |
| Tobacco use | 1.060 | 0.871–1.288 |
| Chronic kidney disease | 0.875 | 0.664–1.148 |
CI, confidence interval; OR, odds ratio.
The distribution of those admitted versus not admitted were evaluated and ranged from a score ≤3 (37 hospitalized vs 1019 not hospitalized) to a score ≥8 (78 hospitalized vs 123 not hospitalized). Based on the way the model weighted each variable, we developed the score shown in Table 3. Patients were considered intermediate risk if they had a score between 4–7 and a 26% chance of hospitalization. Patients with scores ≥8 had a 38% chance of hospitalization.
TABLE 3.
COVID‐19 risk of hospitalization score
| Points | |
|---|---|
| Diabetes mellitus | +2 |
| Hypertension | +3 |
| Male | +1 |
| Has chronic heart disease | +2 |
| Age 55–65 y | +2 |
| Age 66–75 y | +3 |
| Age >75 y | +4 |
| Total score | |
| ≥8 | High risk, 38% chance of hospitalization |
| 4–7 | Intermediate risk, 26% chance of hospitalization |
| ≤3 | Low risk, 3.5% chance of hospitalization |
4. LIMITATIONS
This study was performed at a single hospital network with a convenience sample limited by retrospective data. We did not study those under 18 or over 90 years old, and we did not study the severity of illness at admission, either of which may be an area of interest for future research. Our calculations were based on a 7‐day interval between when the test is positive and admission date; other institutions might prefer 10 or 14 days—for this and other reasons, our scoring needs validation at other sites to be generalizable. Additionally, this scoring predictor has not been compared to hospitalizations in patients without COVID‐19. Next steps should involve validating our model in other institutions across a range of settings and gathering information on hospitalization rates. Furthermore, this study is unique in that it synthesizes these data in the COVID‐19 population to provide a predictor for hospitalization using age and clinical history only.
5. DISCUSSION
In this study, we used observational data to develop a predictive model to calculate the probability of hospital admission of a COVID‐19 patient. Predictive model algorithms were applied to 6485 patients with a 10.9% hospital admission rate, which is comparable to other published data. 27 We then used differences in historical variable data to inform our variable importance plot and used the top features to train a logistic regression model. Our model was internally validated on a test set of data. We then created an easy‐to‐use risk stratification score based on our statistical analysis. There are other models that predict the risk of hospitalization and prognosis. 28 , 29 However, one of the more recently published relies heavily on data that have to be collected on a hospital or office visit including vital signs, lab data, and clinical imaging. 29 Our model relies only on easy to collect clinical history and age. This indicates that this model can be used at both the clinical and population level. Incorporating a predictive model into the electronic health record for decision support is challenging and not immediately feasible. Therefore, we also developed a manual scoring tool using coefficients from our logistic regression model. Our study demonstrated that the greatest predictors of hospitalization included age, hypertension, chronic heart disease, diabetes, gender, tobacco use, and chronic kidney disease. This is consistent with other studies demonstrating these variables have high impact on ICU transfer, poor prognosis, and mortality. 14 , 15 , 16 , 17 Given the heterogeneity in disease presentation, having a model that uses historical clinical characteristics, such as these, allows for use in a variety of clinical settings. Some examples include selecting individuals to undergo more aggressive preventative measures and even vaccination prioritization.
Unsurprisingly, the model found age as the highest predictor, which is consistent with previous data. 30 , 31 , 32 , 33 In addition to age, the other variables included in the model are consistent with the literature. Several studies published have found patients with chronic heart disease, hypertension, and diabetes have worse prognosis and higher mortality 34 , 35 , 36 and thus have a higher likelihood of hospitalization. Given the high prevalence of these chronic diseases 37 and potential role they play in disease progression, it is important that our model and future models incorporate these diagnoses.
We are currently using this model to inform referral for our network's remote home monitoring program to allow early remote intervention. Our model can be calculated without an office visit needed to collect clinical visit data, which other models would require. Therefore, referrals can be made once patients’ SARS‐COV2 test results are back. This could allow us to potentially reduce ER visits and hospitalizations, and we hope to publish on the results of our program.
In summary, we have described the predictors of hospital admission from our observational data and created a tool that predicts hospitalization rates in COVID‐19 patients using common clinical variables and comorbidities without collecting vital signs or laboratory values. This information can be used to guide clinical decision making and increase efficiency and prioritization of patient care in an era where hospital resources are being pushed to their limits.
DECLARATIONS OF INTEREST
None.
AUTHORS CONTRIBUTION
ZC, MM, RM developed the study concept and design and participated in acquisition of the data. ZC performed the analysis and all authors (ZC, NR, MM, RM, DB) participated in the described interpretation of the data; NR and ZC drafted their portion of the manuscript, and all participated in the critical revision of the manuscript for important intellectual content. All authors take final responsibility for the manuscript as a whole.
ACKNOWLEDGMENTS
Authors would like to acknowledge Erin Shigo, BA and Marna Greenberg for their scholarly work in formatting and editing.
Biography
Zhe Chen, MD, is Associate Director of Medical Informatics at Lehigh Valley Health Network, Allentown, PA.

Chen Z, Russo NW, Miller MM, Murphy RX, Burmeister DB. An observational study to develop a scoring system and model to detect risk of hospital admission due to COVID‐19. JACEP Open. 2021;2:e12406. 10.1002/emp2.12406
Funding and support: By JACEP Open policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). The authors have stated that no such relationships exist.
Supervising Editor: Nathan R. Hoot, MD, PhD.
REFERENCES
- 1. World Health Organization . WHO Director‐General's Opening Remarks at the Media Briefing on COVID‐19 ‐ 20 March 2020. Geneva: World Health Organization; 2020. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---20-march-2020. Accessed March 17, 2021. [Google Scholar]
- 2. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20(5):533‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phan LT, Nguyen TV, Luong QC, et al. Importation and human‐to‐human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382:1199‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wunsch H, Wagner J, Herlim M, Chong DH, Kramer AA, Halpern SD. ICU occupancy and mechanical ventilator use in the United States. Crit Care Med. 2013;41(12):2712‐2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weissman GE, Crane‐Droesch A, Chivers C. Locally informed simulation to predict hospital capacity needs during the COVID‐19 pandemic. Ann Intern Med. 2020;173(8):680–681. 10.7326/M20-1260. Epub ahead of print 7 April 2020. [DOI] [PubMed] [Google Scholar]
- 7. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of Coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hui DS, IA E, Madani TA, et al. The continuing 2019‐nCoV epidemic threat of novel coronaviruses to global health ‐ the latest 2019 novel coronavirus outbreak inWuhan, China. Int J Infect Dis. 2020;91:264‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929‐936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Day M. Covid‐19: identifying and isolating asymptomatic people helped eliminate virus in italian village. BMJ. 2020;368:m1165. [DOI] [PubMed] [Google Scholar]
- 11. Costa FF, Rosário WR, Ribeiro Farias AC, de Souza RG, Duarte Gondim RS, Barroso WA. Metabolic syndrome and COVID‐19: an update on the associated comorbidities and proposed therapies. Diabetes Metab Syndr. 2020;14(5):809‐814.[published online ahead of print, 2020 Jun 11]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Emami A, Javanmardi F, Pirbonyeh N, Akbari A. Prevalence of underlying diseases in hospitalized patients with COVID‐19: a systematic review and meta‐analysis. Arch Acad Emerg Med. 2020;8(1):e35. [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson S, Hirsch JS, Narasimhan M, et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID‐19 in the New York City area. JAMA. 2020;323(20):2052‐2059. [published online ahead of print, 2020 Apr 22] [published correction appears in. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. CDC COVID‐19 Response Team . Severe outcomes among patients with Coronavirus disease 2019 (COVID‐19)—United States, February 12–March 16, 2020. MMWRMorb Mortal Wkly Rep. 2020;69:343‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model‐based analysis. Lancet. 2020;20:669‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Espinosa OA, Zanetti ADS, Antunes EF, Longhi FG, Matos TA, Battaglini PF. Prevalence of comorbidities in patients and mortality cases affected by SARS‐CoV2: a systematic review and meta‐analysis. Rev Inst Med Trop Sao Paulo. 2020;62:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanyaolu A, Okorie C, Marinkovic A, et al. Comorbidity and its impact on patients with COVID‐19. SN Compr Clin Med. 2020:1‐8. [published online ahead of print, 2020 Jun 25]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rowe M. An introduction to machine learning for clinicians. Acad Med. 2019;94(10):1433‐1436. [DOI] [PubMed] [Google Scholar]
- 19. Kolachalama VB, Garg PS. Machine learning and medical education. NPJ Digit Med. 2018;1:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Santosh KC. AI‐Driven tools for coronavirus outbreak: need of active learning and cross‐population train/test models on multitudinal/multimodal Data. J Med Syst. 2020;44(5):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mei X, Lee H, Diao K, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID‐19. Nat Med. 2020;26:1224‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Debnath S, Barnaby DP, Coppa K, et al. Machine learning to assist clinical decision‐making during the COVID‐19 pandemic. Bioelectron Med. 2020;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng FY, Joshi H, Tandon P, et al. Using machine learning to predict ICU transfer in hospitalized COVID‐19 patients. J Clin Med. 2020;9(6):1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brinati D, Campagner A, Ferrari D, Locatelli M, Banfi G, Cabitza F. Detection of COVID‐19 infection from routine blood exams with machine learning: a feasibility study. J Med Syst. 2020;44(8):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mei X, Lee HC, Diao K, et al. Artificial intelligence‐enabled rapid diagnosis of COVID‐19 patients. Preprint. medRxiv. 2020. 10.1101/2020.04.12.20062661. Published April 17, 2020. Accessed March 17, 2021. [DOI] [Google Scholar]
- 26. Yadaw AS, Li YC, Bose S, Iyengar R, Bunyavanich S, Pandey G. Clinical predictors of COVID‐19 mortality. The Lancet Digital Health. 2020;2(10):E16–E525. https://www.thelancet.com/journals/landig/article/PIIS2589-7500(20)30217-X/fulltext. Published October 1, 2020. Accessed March 17, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory‐confirmed Coronavirus disease 2019 ‐ COVID‐NET, 14 States, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(15):458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jehi L, Ji X, Milinovich A, et al. Development and validation of a model for individualized prediction of hospitalization risk in 4,536 patients with COVID‐19. PLoS One. 2020;15(8):e0237419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun H, Jain A, Leone MJ, et al. CoVA: an acuity score for outpatient screening that predicts Coronavirus disease 2019 prognosis. J Infect Dis. 2020;223(1):38‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giammaria D, Pajewski A. Can early treatment of patients with risk factors contribute to managing the COVID‐19 pandemic. J Glob Health. 2020;10(1):010377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid‐19 in critically ill patients in the Seattle Region ‐ Case Series. N Engl J Med [Internet]. 2020;382(21):2012‐2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS‐CoV‐2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323(16):1574‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng Z, Peng F, Xu B, et al. Risk factors of critical & mortal COVID‐19 cases: a systematic literature review and meta‐analysis. J Infect. 2020;81(2):e16‐e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kumar A, Arora A, Sharma P, et al. Clinical features of COVID‐19 and factors associated with severe clinical course: a systematic review and meta‐analysis. SSRN. 2020;3566166. 10.2139/ssrn.3566166. Preprint. Published April 21, 2020 . [DOI] [Google Scholar]
- 36. Tian W, Jiang W, Yao J, et al. Predictors of mortality in hospitalized COVID‐19 patients: a systematic review and meta‐analysis. J Med Virol. 2020;92(10):1875‐1883.[published online ahead of print, 2020 May 22]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raghupathi W, Raghupathi V. An empirical study of chronic diseases in the United States: a visual analytics approach. Int J Environ Res Public Health. 2018;15(3):431. [DOI] [PMC free article] [PubMed] [Google Scholar]


