Abstract
Background
Strategies for monitoring the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are crucial for combating the pandemic. Detection and mutation surveillance of SARS-CoV-2 and other respiratory viruses require separate and complex workflows that rely on highly specialized facilities, personnel, and reagents. To date, no method can rapidly diagnose multiple viral infections and determine variants in a high-throughput manner.
Methods
We describe a method for multiplex isothermal amplification-based sequencing and real-time analysis of multiple viral genomes, termed nanopore sequencing of isothermal rapid viral amplification for near real-time analysis (NIRVANA). It can simultaneously detect SARS-CoV-2, influenza A, human adenovirus, and human coronavirus and monitor mutations for up to 96 samples in real time.
Findings
NIRVANA showed high sensitivity and specificity for SARS-CoV-2 in 70 clinical samples with a detection limit of 20 viral RNA copies per μL of extracted nucleic acid. It also detected the influenza A co-infection in two samples. The variant analysis results of SARS-CoV-2-positive samples mirror the epidemiology of coronavirus disease 2019 (COVID-19). Additionally, NIRVANA could simultaneously detect SARS-CoV-2 and pepper mild mottle virus (PMMoV) (an omnipresent virus and water-quality indicator) in municipal wastewater samples.
Conclusions
NIRVANA provides high-confidence detection of both SARS-CoV-2 and other respiratory viruses and mutation surveillance of SARS-CoV-2 on the fly. We expect it to offer a promising solution for rapid field-deployable detection and mutational surveillance of pandemic viruses.
Funding
M.L. is supported by KAUST Office of Sponsored Research (BAS/1/1080-01). This work is supported by KAUST Competitive Research Grant (URF/1/3412-01-01; M.L. and J.C.I.B.) and Universidad Catolica San Antonio de Murcia (J.C.I.B.). A.M.H. is supported by Saudi Ministry of Education (project 436).
Keywords: SARS-CoV-2, nanopore sequencing, influenza, wastewater, isothermal amplification, adenovirus, mutation surveillance, variant virus sequencing, real-time pathogen detection, co-infection
Graphical abstract
Context and significance
It is challenging to simultaneously identify severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and detect viral mutations. Bi et al. develop a convenient sequencing-based method that can simultaneously detect SARS-CoV-2, influenza A (FluA), human adenovirus, and human coronavirus and monitor viral mutations for up to 96 samples in real time. The method not only showed high sensitivity and specificity for SARS-CoV-2 in 70 clinical samples but also detected cases of FluA co-infection. Additionally, it could simultaneously detect SARS-CoV-2 and other viruses in municipal wastewater samples. This rapid and field-deployable method promises to help contain the spread of coronavirus disease 2019 (COVID-19) and ensure the effectiveness of current vaccines.
Bi et al. develop a convenient sequencing-based method that can simultaneously detect SARS-CoV-2, influenza A, human adenovirus, and human coronavirus and monitor viral mutations for up to 96 samples in real time. This rapid and field-deployable method promises to help contain the spread of COVID-19 and ensure the effectiveness of vaccines.
Introduction
The novel coronavirus disease (COVID-19) pandemic is one of the most serious challenges to public health and the global economy in modern history. SARS-CoV-2 is a positive-sense RNA betacoronavirus that causes COVID-19.1 It was identified as the pathogenic cause of an outbreak of viral pneumonia of unknown etiology in Wuhan, China by the Chinese Center for Disease Control and Prevention (CCDC) on January 7, 2020.2 To date, real-time reverse transcription-polymerase chain reaction (rRT-PCR) assays of various designs, including one approved by the US Centers for Disease Control and Prevention (US CDC) under emergency use authorization (EUA), have remained the predominant diagnostic method for SARS-CoV-2. Although proven sensitive and specific for providing a positive or negative answer, rRT-PCR provides little information on the genomic sequence of the virus, knowledge of which is crucial for monitoring how SARS-CoV-2 is evolving and spreading and for ensuring successful development of new diagnostic tests and vaccines. To this end, samples need to go through a separate workflow—typically Illumina shotgun metagenomics or targeted next-generation sequencing (NGS).3 Because NGS requires complicated molecular biology procedures and high-value instruments in centralized laboratories, it is performed in <1% as many cases as rRT-PCR, as evidenced by the number of genomes in the GISAID (Global Initiative on Sharing Avian Influenza Data) database (39,954) and confirmed global cases tallied by the Wold Health Organization (WHO) (6,535,354) as of June 5, 2020.
SARS-CoV-2 infections often cause symptoms similar to other respiratory viruses, thus making it challenging to distinguish co-infections, especially in the flu season. Several studies have reported co-infection of SARS-CoV-2 and other respiratory viruses—respiratory syncytial virus (RSV) and influenza being the most common viral pathogens identified,4 , 5 and influenza was particularly high in deceased patients.6 Even though the overall co-infection rate may be low, and non-COVID-19 respiratory infections have experienced a sharp decline,7 presumably due to public health interventions, the detection of co-infection is potentially useful for monitoring the pandemic and may benefit how patients are treated. To date, no method can rapidly diagnose multiple viral infections in a high-throughput manner. Ideally, such methods should be field deployable to allow timely assessment of outbreaks anywhere anytime.
Both rRT-PCR and NGS are sophisticated techniques whose implementation is contingent on the availability of highly specialized facilities, personnel, and reagents. These limitations could translate into long turnaround times and inadequate access to tests, even in developed countries. To address these issues, several PCR-free nucleic acid detection assays have been proposed as point-of-care replacements of rRT-PCR. Chief among them is RT coupled loop-mediated isothermal amplification (RT-LAMP),8 which has been used for rapid detection of SARS-CoV-2 RNA.9, 10, 11, 12 On the sequencing front, the pocket-sized Oxford Nanopore MinION sequencer has been used for rapid pathogen identification in the field.13 , 14 Because MinION offers base calling on the fly, it is an attractive platform for consolidating viral nucleic acid detection by PCR-free rapid isothermal amplification and viral mutation monitoring by sequencing. However, there are several challenges for an integrated point-of-care solution based on RT-LAMP and Nanopore sequencing. RT-LAMP requires a complex mixture of primers that increases the chance of non-specific amplification and makes it difficult to multiplex. Additionally, LAMP amplicons used for SARS-CoV-2 detection are short.10 , 11 Sequencing singleplex short amplicons not only fails to take advantage of longreads and sequencing throughput (∼10 Gb) of the MinION flow cell, it is also prone to false negative reporting due to amplification failure. Nanopore sequencing has its own caveats too. Due to its relatively high base-calling error, new bioinformatics tools based on dedicated algorithms15 , 16 are needed to accurately identify the presence of viral sequences (substituting for rRT-PCR) and analyze virus mutations (substituting for NGS).
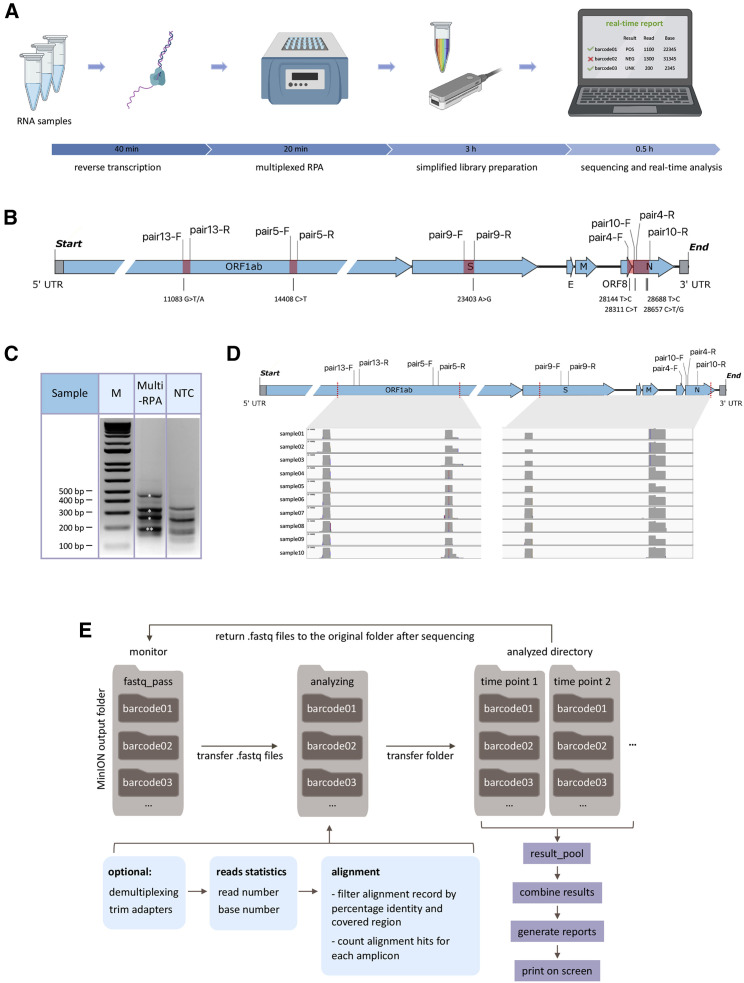
Here, we developed isothermal recombinase polymerase amplification (RPA) assays to simultaneously amplify multiple regions (up to 2,184 bp) of the SARS-CoV-2 genome and genomic sequences of three common respiratory viruses. This forms the basis for an integrated workflow to detect the presence of viral sequences and monitor mutations in multiple regions of the SARS-CoV-2 genome in up to 96 patients at a time (Figure 1 A). We developed a bioinformatics pipeline for on-the-fly analysis to reduce the time to diagnosis and sequencing cost by stopping the sequencing run when data are sufficient to provide confident answers.
Figure 1.
Multiplex RPA workflow for SARS-CoV-2 detection and Nanopore sequencing
(A) Schematic representation of NIRVANA. RNA samples were subjected to reverse transcription, followed by multiplex RPA to amplify multiple regions of the SARS-CoV-2 genome. The amplicons were purified and prepared to the Nanopore library using an optimized barcoding library preparation protocol. In the end, the sequencing was performed in the pocket-sized Nanopore MinION sequencer and sequencing results were analyzed by our algorithm termed RTNano on the fly. Created with BioRender.com.
(B) The RPA primers used in this study were plotted in the SARS-CoV-2 genome. The RPA amplicons are highlighted in red. The corresponding prevalent variants were labeled under the genome.
(C) Agarose gel electrophoresis results of multiplex RPA. All of the five amplicons were shown in the gel with correct size (asterisks, note that pairs 5 and 13 have similar sizes). The no template control (NTC) showed a different pattern of non-specific amplicons. M, molecular size marker.
(D) IGV plots showing Nanopore sequencing read coverage of the SARS-CoV-2 genome. All samples showed reads covering all of the targeted regions.
(E) Pipeline of RTNano real-time analysis. RTNano monitors the Nanopore MinION sequencing output folder. Once newly generated fastq files are detected, it moves the files to the analyzing folder and makes a new folder for each sample. If the Nanopore demultiplexing tool guppy is provided, RTNano will do additional demultiplexing to make sure reads are correctly classified. The analysis will align reads to the SARS-CoV-2 reference genome, filter, and count alignment records and assign result mark (POS, NEG, or UNK) for each sample. As sequencing proceeds, RTNano will merge the newly analyzed results with existing ones to update the current sequencing statistics.
Results
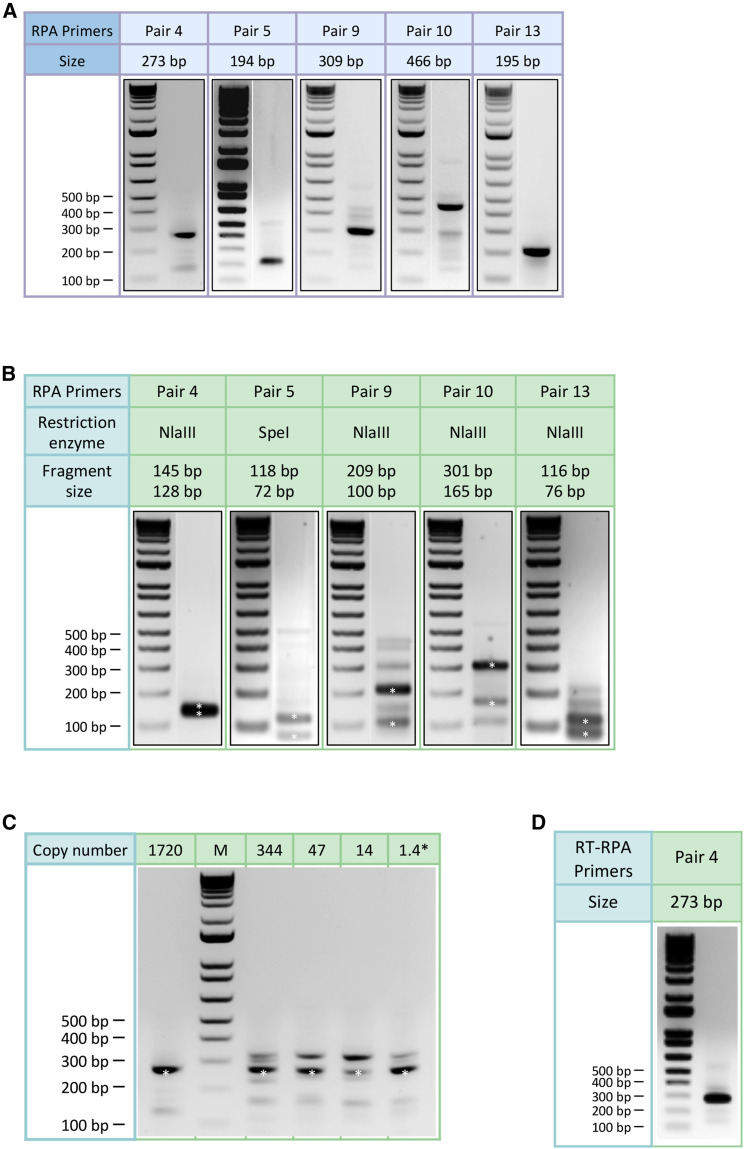
Singleplex RPA was first performed to test its ability to amplify the SARS-CoV-2 genome from a nasopharyngeal swab sample tested positive for the virus using the US CDC assays (cycle threshold or CT value = 21).17 Sixteen primers were tested in 12 combinations to amplify 5 regions harboring either reported signature mutations18 , 19 useful for strain classification or mutation hotspots (GISAID, as of March 15, 2020). Five pairs of primers showed robust amplification of DNA of predicted size (range: 194–466 bp) in a 20-min isothermal reaction at 39°C (Figures 1B and 2A). The specificity of all 5 RPA products was verified by restriction enzyme digestion (Figure 2B). The limit of detection of RPA reaches below 10 copies per reaction (Figure 2C). Furthermore, we showed that these RPA reactions could be multiplexed to amplify the five regions of the SARS-CoV-2 genome in a single reaction (Figure 1C), thus significantly simplifying the workflow.
Figure 2.
Agarose gel electrophoresis results of singleplex RPA
(A) Agarose gel electrophoresis results of singleplex RPA with selected primers shown next a molecular size marker. The amplicons range from 194 bp to 466 bp.
(B) Agarose gel electrophoresis results of restriction enzyme digestion. The amplicon of pair 5 was digested by SpeI although the others were digested by NlaIII. The digested DNA bands (asterisks) were of expected sizes.
(C) Agarose gel electrophoresis results showing the sensitivity of RPA in amplifying the SARS-CoV-2 genome. Primer pair 4 was used in the experiment. Reliable amplification can be achieved with 1.4 copies (calculated from dilution) of the SARS-CoV-2 genome.
(D) Agarose gel electrophoresis result of one-pot reverse transcription and RPA reaction using primer pair 4.
We next performed multiplex RPA using ten SARS-CoV-2-positive samples (SARS-CoV-2+; determined by US CDC assays;17 CT value range: 15–27.9). Multiplex RPA products of the 10 samples were individually barcoded, pooled, and prepared into a Nanopore sequencing library using an optimized protocol to save time (see STAR Methods). The whole workflow from RNA to sequencing took approximately 4 h (Figure 1A). The barcoded library was sequenced on a Nanopore MinION using a R9.4.1 flow cell. After 12-h sequencing, we acquired a total of 1.7 million reads from this barcoded library (Figure S1A). The demultiplexed reads were distributed relatively evenly among the barcodes (samples). We further aligned the reads to the SARS-CoV-2 reference genome and found that all RPA amplicons were covered by thousands of reads in all samples, suggesting that barcoded multiplexed RPA sequencing worked effectively (Figures 1D and S1B).
The identification of a SARS-CoV-2+ sample can be achieved by surveying the existence of targeted amplicons via sequencing. However, the determination of negative samples needs to rule out potential sample collection failure, thus requiring a sample quality validation control. We used the existence of transcripts of the human housekeeping gene ACTB as a quality check of sample collection. The sequencing results showed that the ACTB gene could be effectively amplified without significantly affecting the amplification of SARS-CoV-2 (Figure S1C).
Co-infection with other respiratory pathogens in COVID-19 patients has been evaluated, though current detection of co-infections requires additional rRT-PCR assays6 , 20 , 21 or NGS.5 Such ad hoc tests are not amenable to large-scale screening of co-infections. On the other hand, multiplexed RPA of different viruses could be a promising solution to detect co-infections in a timely and efficient manner.
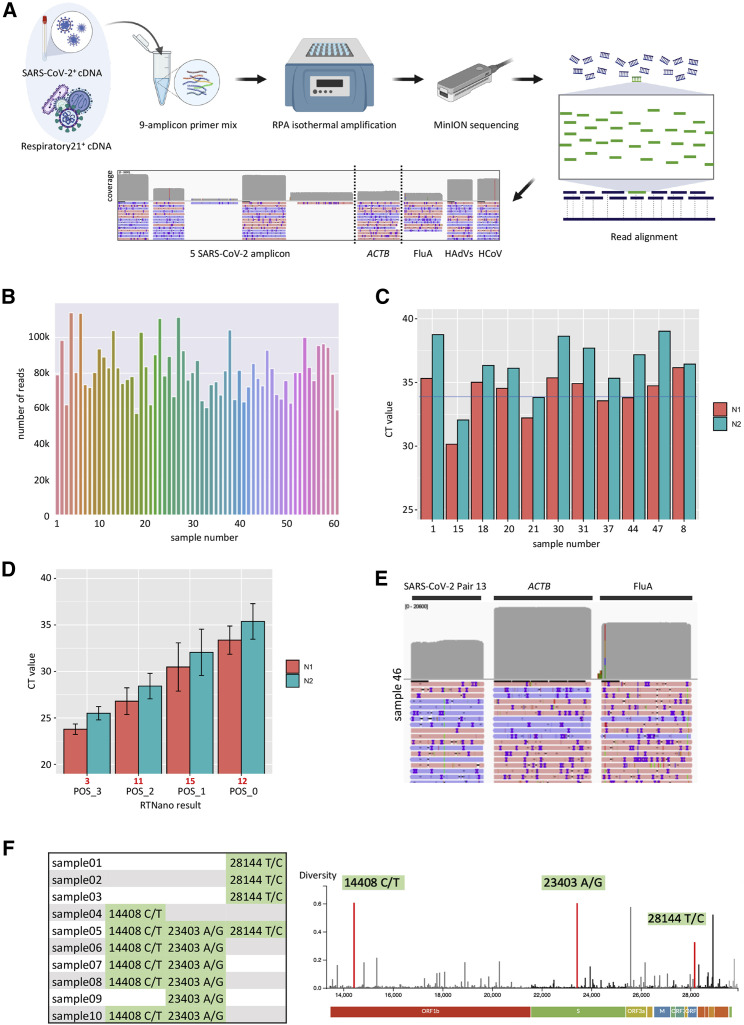
As a proof of principle of multiplex RPA of multiple human viral pathogens, we screened and validated RPA primers that robustly amplify FluA, HAdV, and non-SARS-CoV-2 human coronavirus (HCoV), respectively (data not shown). We added the three pairs of respiratory virus primers to the multiplex SARS-CoV-2 RPA panel to achieve simultaneous isothermal amplification of four viral pathogens (Table S1). To ensure effective amplification of all targets, we adjusted the concentration of each primer pair based on the read depth of the cognate genomic region when sequencing a contrived co-infection sample made from a SARS-CoV-2+ sample spiked with a control panel of 21 respiratory viruses, including FluA, HAdV, and HCoV (Respiratory21+). The final primer mix achieved amplification of all targeted amplicons (Figure 3 A).
Figure 3.
Real-time detection of multiple viral pathogens and mutational analysis of SARS-CoV-2
(A) Experimental design of multiple virus detection by one-pot NIRVANA. A mixture of SARS-CoV-2+ and Respiratory21+ samples was used as positive control to adjust the primer concentration. The final primer mix could amplify all targeted viral regions. Created with BioRender.com.
(B) The sequencing throughput of 60 clinical samples (1–60) and NTC (61). A total of 6.3 million reads were acquired in a 24-h sequencing run.
(C) CT values of potentially false-negative samples by RTNano analysis. The average CT value of the N1 assay was indicated by the blue line.
(D) The average rRT-PCR CT values of SARS-CoV-2 RTNano+ samples (PCR+ of both N1 and N2 assays) of different confidence level using 9-amplicon NIRVANA. The sample number is shown in red under the graph. The error bars represent the standard deviation of CT values. RTNano confidence level inversely correlates with CT value.
(E) IGV plots showing the read alignment to the SARS-CoV-2, ACTB, and FluA amplicon in sample 46 using 9-amplicon NIRVANA.
(F) The SNVs detected in multiplex RPA sequencing and their position as shown in the Nextstrain data portal (https://nextstrain.org). A total of 16 SNVs were detected from 10 SARS-CoV-2-positive samples.
The presence of SARS-CoV-2 has been reported in municipal sewage, of which the viral load was found to be correlated to the reported COVID-19 prevalence.22 This suggests that wastewater surveillance could be a sensitive indicator of the total COVID-19 caseload (including asymptomatic cases) in the population. To evaluate the possibility of using multiplex RPA to detect SARS-CoV-2 in wastewater, we determined the primer mix to simultaneously amplify five regions of SARS-CoV-2 and one region of pepper mild mottle virus (PMMoV) (an omnipresent indicator of water quality).23 We spiked the wastewater concentrate (see STAR Methods) with RNA of SARS-CoV-2+ samples and used this as a positive control to test the multiplex RPA. Two primer mixes with different concentration of PMMoV primers were tested. Both primer mixtures could detect the SARS-CoV-2 and PMMoV within the positive control (Figure S1D). These results suggested that multiplex RPA could be used to monitor the presence of SARS-CoV-2 and other viruses in municipal wastewater. In addition, due to the simultaneous acquisition of viral sequences, this method could also survey the strains circulating in the population (see below).
We next performed multi-virus multiplex RPA assay followed by Nanopore sequencing in 60 clinical samples suspected of SARS-CoV-2 infection. Following RT and RPA (Figure 1A), amplicons of each sample were barcoded and sequenced in one Nanopore MinION flow cell. To take advantage of the unique feature of real-time base calling offered by Nanopore sequencing, we developed a bioinformatics algorithm termed Real-Time Nanopore sequencing monitor (RTNano) (Figure 1E). RTNano continuously monitors the output folder of base-called data during the sequencing run and generates analysis reports in a matter of seconds. After detecting new fastq files (base-called sequence output format of Nanopore sequencing), RTNano quickly aligns the reads to the targeted amplicons of the viruses. To confidently determine the existence of a virus, RTNano filters the alignment records by percentage identity and amplicon coverage and provides the number of positive records for each targeted viral amplicon, which is then used to evaluate the infection status of the sample.
When demultiplexing a large number of samples, barcode misclassification could potentially happen due to base-calling errors in the barcode. RTNano included an additional round of demultiplexing using stringent parameters to reduce the chance of barcode misclassification (see STAR Methods). Furthermore, a no template control (NTC) was included to monitor barcode misclassification. The reads of targeted amplicons assigned to the NTC barcode likely represented background errors in demultiplexing. The positive record number of the NTC was subtracted from the individual sample analysis to further reduce false positive identification. After the real-time analysis, RTNano generated a report for each of the 60 samples, including current read number, base number, and details of alignment-positive records (Figure 1E). To simplify the interpretation of the results, RTNano provided a summary score of SARS-CoV-2 based on a set of predefined rules (Table S2). SARS-CoV-2+ samples (POS) were assigned a confidence level ranging from 0 to 3 (lowest to highest) based on the number of covered amplicons and corresponding positive records. When there is no record of SARS-CoV-2, the sample could be categorized as either negative (NEG), if there were enough records (>1,000) of ACTB, or unknown (UKN), if there were insufficient records of ACTB. The introduction of the confidence level could improve the accuracy of diagnosis, which is in principle similar to the CT value of rRT-PCR but based on quantitative information of multiple amplicons. Because base calling and demultiplexing may lead to a brief delay, we refer to the workflow as nanopore sequencing of isothermal rapid viral amplification for near real-time analysis (NIRVANA) (Figure 1A).
RTNano identified 35 SARS-CoV-2+ cases in the 60 samples after 2 h of Nanopore sequencing. Ten more positives were identified by RTNano (RTNano+) as the data output increased in the next 22 h of sequencing (Figure 3B; Data S1). An identical aliquot of each sample was analyzed in parallel using the US CDC rRT-PCR SARS-CoV-2 assays. Three samples were inconclusive after two rounds of rRT-PCR tests (Data S2) and were excluded in the downstream analysis. Among the 45 RTNano+ samples, 43 had rRT-PCR confirmed status, in which 41 (95.35%) were positive by the rRT-PCR assay (PCR+). The two remaining weakly RTNano+ sample (POS_0) had high CT values of RNase P and failed in PCR amplification of N1 and N2. Three of the RTNano− samples were also PCR− due to no amplification of N1 and/or N2, although the rest of the 11 RTNano− were PCR+. These 11 false-negative samples had high CT values (N1: 34.16 ± 0.36; N2: 36.27 ± 0.47; Figure 3C). The incorrect identification of these samples could be due to amplification failure of multiplex RPA or insufficient sequencing throughput. In addition, high CT values in SARS-CoV-2 rRT-PCR tests have a higher chance of being false positive.24 , 25
We calculated the average CT value of RTNano+ samples with different confidence levels. RTNano+ samples with a high confidence level correlated with lower CT values, which proved the reliability of RTNano results (Figure 3D). Based on the N1 standard curve, we estimated the limit of detection (LoD) of NIRVANA was ∼29 viral RNA copies/μL of extracted nucleic acid (86 copies per reaction, based on average N1 CT value of 33.37 of POS_0; see STAR Methods), which was comparable to current rRT-PCR assays.26
We next surveyed co-infection of three common respiratory viruses in the 60 samples. FluA co-infection was determined by two independent commercial rRT-PCR assays (Resp’Easy in vitro diagnostic kit [CE-IVD] and IDT influenza A,B/RSV identification kit). Four samples were identified as FluA+ in both assays (CT value range: 33.97 to 39.33 by Resp’Easy; Figure S1E). RTNano detected the FluA+ sample with the lowest CT value (sample 46; CT 33.97) with high confidence (Figure 3E). HAdVs and HCoV co-infection was examined using the Resp’Easy kit, and only one HCoV+ sample was identified (Resp’Easy CT = 35.18). RTNano reported no HAdVs co-infection and failed to detect the HCoV+ sample.
A high level of multiplexing reactions could negatively affect RPA efficiency of each amplicon. To test whether a less-complex primer combination could improve the performance of NIRVANA, we removed the less-robust SARS-CoV-2 primers (pairs 9 and 10) and performed a new NIRVANA sequencing. Twenty-nine of the 60 samples were resequenced, including 11 RTNano−/PCR+, 4 FluA+, and 1 HCoV+. Nine of the previous 11 RTNano−/PCR+ samples were identified as SARS-CoV-2+, suggesting an improved sensitivity (Data S3). To test whether the remaining two RTNano−/PCR+ samples were true positives, we performed Nanopore sequencing of the PCR products of the two samples. The results showed that both samples contained high-confidence reads mapped to the expected amplicon sequences (data not shown). Thus, we concluded that these two samples were true positives. The average N1 CT value of POS_1 was improved from 30.48 to 32.06 (Figure S1F). Using the new data, we calculated the LoD of SARS-CoV-2 of 7-amplicon NIRVANA to be ∼20 viral RNA copies/μL of extracted nucleic acid (61 copies per reaction, based on average N1 CT value of 33.37 of POS_0). It is worth noting that the sensitivity of SARS-CoV-2 detection of multi-virus multiplex NIRVANA is lower than that of singleplex rRT-PCR assays achieved in our hands.27 Further optimization of the RPA primer combination may improve the sensitivity, and the ability of NIRVANA to identify multiple viruses and to detect variants could partially offset the reduced sensitivity.
The sensitivity of FluA detection was also improved as two FluA+ samples (Resp’Easy CT value of 33.97 and 36.03) were correctly identified (samples 46 and 58 in Figures S1E and S1G). The HCoV+ sample was still not identified. Because NIRVANA showed a robust ability in detecting HCoV in the positive control sample (Figure 3A), we suspected that the HCoV in the clinical sample may belong to a different strain that could not be amplified by our HCoV primers. Taken together, these results showed that NIRVANA provided highly confident SARS-CoV-2 detection for virus loads above 20 copies/μL of extracted nucleic acid and reliable detection of potential co-infections.
RTNano has an integrated function to quickly analyze variants in each sample during sequencing. We used it to analyze sequence variants in ten SARS-CoV-2+ samples (Figure 1D). It detected 16 single-nucleotide variants (SNVs) in the ten samples, and all of them had been reported in GISAID (Figure 3F; as of June 7, 2020). The reported SNVs suggested that the strains in samples 01–03 are close to clade 19B (nt28144 T/C), first identified in Wuhan, China, although the strains in samples 04–10 are close to clade 20 (nt14408 C/T; 23403 A/G), first becoming endemic in Europe. Given the fact that samples 01–03 were collected early in the pandemic, although the others were later (around May), the SNV signature of the strains revealed by NIRVANA is consistent with the pattern of COVID-19 case importation, which initially came from China and shifted to Europe after a ban of flights from China. Prospectively, collecting such data regularly could guide public health policy making to better control the pandemic.
To validate the SNVs and compare NIRVANA with conventional RT-PCR amplicon sequencing,28 we chose three samples (01–03) to perform multiplex RT-PCR amplicon sequencing using the MinION. Variant calling was done by RTNano using the same parameters, and the results showed that RT-PCR sequencing confirmed all 3 SNVs detected by NIRVANA (Figure S2A). We further compared the SNVs of samples 01–03 with their corresponding assembled genome from Illumina sequencing published in GISAID (GISAID: EPI_ISL_437459 for sample01, GISAID: EPI_ISL_437460 for sample02, and GISAID: EPI_ISL_437461 for sample03). All of the three SNVs existed in the assembled genome.
Discussion
Taken together, NIRVANA provides high-confidence detection of both SARS-CoV-2 and other respiratory viruses and mutation surveillance of SARS-CoV-2 on the fly. Compared to Oxford Nanopore’s official method termed LAMPore,29 which is based on RT-LAMP, NIRVANA offers several advantages, including longer amplicons and a higher level of multiplexing. LAMPore generated short amplicons (∼80 bp), of which half was composed of primer sequences. Thus, the read alignment may be more prone to amplification artifacts that lead to false-positive results. In NIRVANA, significantly longer amplicons (up to 466 bp in this study) combined with an alignment record filter can improve the accuracy of positive results by minimizing the influence of amplification artifacts. In addition, LAMP-based LAMPore requires multiple primers (at least 4) to amplify one amplicon, which is a challenge to multiplexing. In contrast, we showed that RPA-based NIRVANA is able to amplify and detect nine amplicons in one pot (Figure 3A).
The recent worldwide spread of the SARS-CoV-2 B.1.1.7 variant with an increased infectivity reveals an urgent need for rapid and field-deployable methods to monitor mutations in hopes of containing the spread of COVID-19 and ensuring the effectiveness of current vaccines. The 9-amplicon NIRVANA covers three B.1.1.7-specific mutations and is potentially usable for the identification of the variant. The framework of NIRVANA has a built-in flexibility in choosing what viral sequences to target and the level of multiplexing. The screening of new RPA primers can be performed at a low cost and with little effort. The whole workflow of NIRVANA can be further shortened by performing one-pot RT-RPA (Figure 2D) so that the time from RNA to result can be as short as 3.5 h. All molecular biology reactions in the workflow can be done in a simple heating block, and all necessary supplies fit into a briefcase (Figure S2B). We expect it to provide a promising solution for rapid field-deployable detection and mutational surveillance of pandemic viruses.
Limitations of study
Although our method offers unique advantages, some limitations should be noted. First, NIRVANA relies on efficient primers to amplify the targeted viral sequences. The clinical samples used in the development of NIRVANA are all from Saudi Arabia and collected before June 2020. It is possible that the primers presented in this study may need to be re-validated or re-designed because of the evolution of SARS-CoV-2 variants. The same also applies to the design of primers targeting the seasonal influenza viruses. Second, the LoD of the 7-amplicon NIRVANA for SARS-CoV-2 is still lower than that of rRT-PCR in this study. Further improvements in sensitivity may be required for clinical application. Third, NIRVANA entails multiplex Nanopore sequencing. Although several optimizations have been done to reduce sample crosstalk, the frequency of residual sample misclassification error requires further characterization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Respiratory (21 targets) control panel | Microbiologics | Cat. No 8217 |
| Biological samples | ||
| RNA samples | Ministry of Health (MOH) hospitals in the western region in Saudi Arabia | N/A |
| Wastewater samples | The wastewater equalization tank in KAUST, Thuwal, Saudi Arabia | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol | Invitrogen | Cat. No 15596018 |
| Critical commercial assays | ||
| Direct-Zol RNA Miniprep kit | Zymo Research | Cat. No R2070 |
| NEB ProtoScript II reverse transcriptase | New England Biolabs | Cat. No M0368 |
| Invitrogen SuperScript IV reverse transcriptase | Thermo Fisher Scientific | Cat. No 18090010 |
| RNase H | New England Biolabs | Cat. No M0523S |
| Respiratory Virus PCR Panel kit | Diagenode diagnostics | DDGR-90-L048 |
| AMPure XP beads | Beckman Coulter | Cat. No A63882 |
| TwistAmp® Basic kit | TwistDx | Cat. No MSPPTABAS03KIT |
| QIAquick PCR purification kit | QIAGEN | Cat. No 28106 |
| Ligation sequencing kit | Oxford Nanopore Technologies | Cat. No SQK-LSK109 |
| Native Barcoding Expansion 96 | Oxford Nanopore Technologies | Cat. No EXP-NBD196 |
| Flow cell (R9.4.1) | Oxford Nanopore Technologies | Cat. No FLO-MIN106D |
| Deposited data | ||
| Raw and analyzed data | This paper | SRA database (accession ID PRJNA638039) |
| Additional supplemental files | This paper | Mendeley Data (https://dx.doi.org/10.17632/35g7t55h4c.1) |
| Oligonucleotides | ||
| rRT-PCR assay for SARS-CoV-2 | Integrated DNA Technologies | Cat. No 10006770 |
| rRT-PCR assay for FluA | Integrated DNA Technologies | Cat. No 1079729 |
| PMMoV-F: TCAAATGAGAGTGGTTTGACC TTAACGTTTGA |
Integrated DNA Technologies | N/A |
| PMMoV-R: AACTCATCGGACACTGTGTTGCC TGTTAGAC |
Integrated DNA Technologies | N/A |
| pair4-F: GCTGGTTCTAAATCACCCATTCAGT | Integrated DNA Technologies | N/A |
| pair4-R: TCTGGTTACTGCCAGTTGAATCTG | Integrated DNA Technologies | N/A |
| pair5-F: TTGGGATCAGACATACCACCCA | Integrated DNA Technologies | N/A |
| pair5-R: CAACACCTAGCTCTCTGAAGTGG | Integrated DNA Technologies | N/A |
| pair9-F: CCAGCAACTGTTTGTGGACCT | Integrated DNA Technologies | N/A |
| pair9-R: AGCAACAGGGACTTCTGTGC | Integrated DNA Technologies | N/A |
| pair10-F: GACCCCAAAATCAGCGAAAT | Integrated DNA Technologies | N/A |
| pair10-R: TGTAGCACGATTGCAGCATTG | Integrated DNA Technologies | N/A |
| pair13-F: CCAGAGTACTCAATGGTCTTTGTTC | Integrated DNA Technologies | N/A |
| pair13-R: ACCCAACTAGCAGGCATATAGAC | Integrated DNA Technologies | N/A |
| ACTB-F: CCCAGCCATGTACGTTGCTATCCAGGC | Integrated DNA Technologies | N/A |
| ACTB-R: ACAGCTTCTCCTTAATGTCACGCACGAT | Integrated DNA Technologies | N/A |
| influA-F: ATGAGYCTTYTAACCGAGGTCGAAACG | Integrated DNA Technologies | N/A |
| influA-R: TGGACAAANCGTCTACGCTGCAG | Integrated DNA Technologies | N/A |
| HAdVs-F: GCCGAGAAGGGCGTGCGCAGGTA | Integrated DNA Technologies | N/A |
| HAdVs-R: TACGCCAACTCCGCC CACGCGCT |
Integrated DNA Technologies | N/A |
| HCoV-F: ATGGTCAAGGAGTTCCCAT TGCTTTCGGAGTA |
Integrated DNA Technologies | N/A |
| HCoV-R: GGGCCGGTACCGAGATAGT AGAAATACCATCTCG |
Integrated DNA Technologies | N/A |
| Software and algorithms | ||
| RTNano | This paper | https://github.com/milesjor/RTNano |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Mo Li (mo.li@kaust.edu.sa).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RTNano and sample data in this study are accessible at GitHub (https://github.com/milesjor/RTNano). Raw sequencing data are available in the SRA database (SRA: PRJNA638039), which are accessible with the link (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA638039/).
Experimental model and subject details
RNA samples and primers
Anonymized RNA samples were obtained from Ministry of Health (MOH) hospitals in the western region in Saudi Arabia. The use of clinical samples in this study is approved by the institutional review board (IRB# H-02-K-076-0320-279) of MOH and KAUST Institutional Biosafety and Bioethics Committee (IBEC). Oropharyngeal and nasopharyngeal swabs were carried out by physicians and samples were steeped in 1 mL of TRIzol (Invitrogen Cat. No 15596018) to inactivate virus during transportation. Total RNA extraction of the samples was performed following instructions as described in the CDC EUA-approved protocol using the Direct-Zol RNA Miniprep kit (Zymo Research Cat. No R2070) or TRIzol reagent (Invitrogen Cat. No 15596026) following the manufacturers’ instructions. The Respiratory (21 targets) control panel (Microbiologics Cat. No 8217) was used as positive control in the amplification of FluA, HAdVs and HCoV. A list of primers used in this study can be found in Table S1. The PMMoV primers used in wastewater samples were PMMoV-F: TCAAATGAGAGTGGTTTGACCTTAACGTTTGA and PMMoV-R: AACTCATCGGACACTGTGTTGCCTGTTAGAC.
Wastewater samples
Raw sewage was collected at 9 AM and 4 PM on 7 June 2020 from the wastewater equalization tank in KAUST, and then mixed together to constitute a composite sample. A 300-500 mL of sewage mixture was concentrated by electronegative membrane in the present of cation as previously described.30 The eluate of viruses was recovered in a tube with 50 μL of 100 mM H2SO4 (pH 1.0) and 100 μL of 100 × Tris–EDTA buffer (pH 8.0) for neutralization. Centripep YM-50 (Merck Millipore) was used to further concentrate the samples to a volume of 600-700 μl.
Method details
Reverse transcription
Reverse transcription of RNA samples was done using either NEB ProtoScript II reverse transcriptase (NEB Cat. No M0368) or Invitrogen SuperScript IV reverse transcriptase (Thermo Fisher Scientific Cat. No 18090010), following protocols provided by the manufacturers. After reverse transcription, 5 units of thermostable RNase H (New England Biolabs Cat. No M0523S) was added to the reaction, which was incubated at 37°C for 20 min to remove RNA. The final reaction was diluted to be used as templates in RPA. All of the web-lab experiments in this study were conducted in a horizontal flow clean bench to prevent contaminations. The bench was decontaminated with 70% ethanol, DNAZap (Invitrogen, Cat no. AM9890) and RNase AWAY (Invitrogen, Cat no. 10328011) before and after use. The filtered pipette tips (Eppendorf epT.I.P.S.® LoRetention series) and centrifuge tubes (Eppendorf DNA LoBind Tubes, Cat. No 0030108051) used in this study were PCR-clean grade. All of the operations were performed carefully following standard laboratory operating procedures.
rRT-PCR
The rRT-PCR assay of SARS-CoV-2 was purchased from IDT (Cat. No 10006770). The rRT-PCR assay for FluA was purchase from IDT (Cat. No 1079729). Respiratory Virus PCR Panel kit (Diagenode diagnostics, DDGR-90-L048) was used for the determination of FluA, HAdVs and HCoV. All reactions were run on a CFX384 Touch Real-Time PCR Detection System (Bio-rad) following the instruction of manufacturers. The copy number of SARS-CoV-2 is determined by the IDT SARS-CoV-2 rRT-PCR assay based on the standard curve of CT values of known copies of synthetic SARS-CoV-2 N gene RNA (nt28,287-29,230 in NC_045512.2, DNA template purchased from IDT). A serial dilution of the synthetic RNA is performed to obtain final concentrations of 10, 102, 103, 104, 105, 106, 107 copies/μl. The reverse transcription is done using 1 μl of diluted RNA, followed by rRT-PCR using 2.5 μl of 5-fold diluted cDNA.
Singleplex RPA
Singleplex RPA was performed using TwistAmp® Basic kit following the standard protocol. Thirteen pairs of SARS-CoV-2 primers covering N gene, S gene, ORF1ab and ORF8 were tested and the corresponding amplicons were purified by 0.8X Beckman Coulter AMPure XP beads (Cat. No A63882) and eluted in 40 μl H2O. The purified amplicons were first analyzed by running DNA agarose gel to check the specificity and efficiency. The most robust five pairs of primers with correct size were further analyzed by NlaIII (NEB Cat. No R0125L) and SpeI (NEB Cat. No R0133L) digestion following standard protocols. For one-pot RT-RPA, 10 U of AMV Reverse Transcriptase (NEB Cat no. M0277S) and 20 U of SUPERase⋅In RNase Inhibitor (Invitrogen Cat no. AM2694) were added to a regular RPA reaction mix. The RT-RPA was carried out per the manufacturer protocol. For the RPA of respiratory viruses, reverse-transcribed cDNA from the Respiratory (21 targets) control panel (Microbiologics Cat. No 8217) was used as the template.
Multiplex RPA
Multiplex RPA was done by add all of the primers in the same reaction. The total final primer concentration is set to 2 μM. To achieve an even and robust amplification, we empirically determined the final concentration of the five-amplicon primers (for the initial test of 10 SARS-CoV-2 positive samples) as follows: 0.166 μM for each pair-4 primer, 0.166 μM for each pair-5 primer, 0.242 μM for each pair-9 primer, 0.26 μM for each pair-10 primer, 0.166 μM for each pair-13 primer, 29.5 μl of primer free rehydration buffer, 1 μl of 10-fold diluted cDNA, 7μl H2O. In the multiplex RPA of 9-amplicon and 7-amplicon (remove pairs 9 & 10) primers, the primer mixtures were obtained by combining different amount of 10 μM primers according to the ratios in Table S1, and 2.5 μl of 5-fold diluted cDNA were used as template. The FluA and HAdV primers target regions that are expected to be shared by all subtypes of the corresponding viruses, while the HCoV primers are specific for the HKU1 subtype. The reaction was incubated at 39°C for 4 min, then vortexed and spin down briefly, followed by a 16-min incubation at 39°C.
Library preparation and sequencing
The RPA library preparation was done using Native barcoding expansion kit (Oxford Nanopore Technologies EXP-NBD114 and EXP-NBD196) following Nanopore PCR tiling of COVID-19 Virus protocol (Ver: PTC_9096_v109_revE_06Feb2020) with a few changes to save time. The RPA reaction was purified using QIAGEN QIAquick PCR purification kit (QIAGEN Cat No. 28106) and elute in 30 μl H2O. The end-prep reaction was done separately in 15 μl volume using 5 μl of each multiplex RPA samples. After that, we followed the same procedures as described in the official protocol. The RT-PCR library preparation was done using Native barcoding expansion kit (Oxford Nanopore Technologies EXP-NBD104) according to the standard native barcoding amplicons protocol. The sequencing runs were performed on an Oxford Nanopore MinION sequencer using R9.4.1 flow cells.
Quantification and statistical analysis
RTNano scanned the sequencing folder repeatedly based on user defined interval time. Once newly generated fastq files were detected, it moved the files to the analysis folder and made a new folder for each sample. If the Nanopore demultiplexing tool guppy is provided, RTNano will do additional demultiplexing to make sure reads are correctly classified. The analysis part utilized minimap231 to quickly align the reads to the SARS-CoV-2 reference genome (GenBank: NC_045512). After alignment, RTNano will filter the alignment records based on defined thresholds of parentage identity and amplicon coverage, followed by counting the alignment records of each amplicon. A read with > = 89% alignment identity and > = 96% amplicon coverage will be counted as one positive record. If an NTC barcode number was provided, RTNano will subtract this number in individual sample analysis to further ensure confident demultiplexing. In the end, RTNano will assign samples with different result marks (POS, NEG and UNK) based on the number of alignment records of each amplicon. With the sequencing continuing, RTNano will merge the newly analyzed result with completed ones to update the current sequencing statistics. RTNano is ultra-fast, a typical analysis with additional guppy demultiplexing of 5 fastq files (containing 4000 reads each and sequenced with 12-barcode kit) will take ∼10 s using one thread in a MacBook Pro 2016 15-inch laptop. Variant calling was performed using samtools (v1.9) and bcftools (v1.9).32 The detected variants were filtered by position (within the targeted regions) and compared with the data in Nextstrain.org as of Jun 2, 2020.
Acknowledgments
We thank members of the Li laboratory, Baolei Yuan, Xuan Zhou, Yingzi Zhang, and Samhan Alsolami for helpful discussions and Marie Krenz Y. Sicat for administrative support. We thank members of the Pain lab, Rahul Salunke, and Amit Subudhi for technical assistance. We thank members of the Izpisua Belmonte lab, Yanjiao Shao, Yasuo Ouchi, and Pradeep Reddy for generously sharing ideas of simultaneous pathogen detection. We thank Professor Peiying Hong and her lab members, Dr. Andri Rachmadi, and Dr. David Mantilla-Calderon for kindly sharing wastewater samples and technical assistance. We thank KAUST Rapid Research Response Team (R3T) for supporting our research during the COVID-19 crisis. We thank members of the KAUST R3T for generously sharing materials and advices. M.L. is supported by KAUST Office of Sponsored Research (OSR) under award number BAS/1/1080-01. The work is supported by a KAUST Competitive Research Grant (award number URF/1/3412-01-01) given to M.L. and J.C.I.B. This work is supported by Universidad Catolica San Antonio de Murcia (J.C.I.B.). This work is partially supported by Fundación Séneca, Agencia de Ciencia y Tecnología de la Región de Murcia under the project 00009/COVI/20. A.M.H. is supported by funding from the deputyship for Research and Innovation, Ministry of Education in Saudi Arabia (project number 436).
Author contributions
M.L. and C.B. performed the majority of the experiments related to Nanopore sequencing. G.R.-M. and C.B. performed diagnosis assays for various viral pathogens. C.B. wrote the code and performed the bioinformatics analysis. M.L., C.B., J.X., and C.R.E. designed and performed molecular biology experiments. F.S.A., A.K., A.M.H., and N.A.M.A. collected clinical samples. S.M. extracted RNA and performed molecular assays. S.H. and A.P. coordinated the clinical samples and molecular testing. C.B., G.R.-M., Y.T., E.N.D., J.C.I.B., and M.L. analyzed the data and wrote the manuscript. M.L. and C.B. conceived the study. J.C.I.B. and M.L. supervised the study.
Declaration of interests
A patent application based on methods described in this paper has been filed by King Abdullah University of Science and Technology, in which C.B. and M.L. are listed as inventors. The authors declare no other competing interests.
Published: March 31, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2021.03.015.
Supplemental information
References
- 1.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(2020). Li, Q.; The nCo-V Outbreak Joint Field Epidemiology Investigation Team (2020). An outbreak of NCIP (2019-nCoV) infection in China — Wuhan, Hubei Province, 2019−2020. China CDC Weekly 2, 79–80. [PMC free article] [PubMed]
- 3.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81:266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babiker A., Bradley H.L., Stittleburg V.D., Ingersoll J.M., Key A., Kraft C.S., Waggoner J.J., Piantadosi A. Metagenomic sequencing to detect respiratory viruses in persons under investigation for COVID-19. J. Clin. Microbiol. 2020;59:e02142-20. doi: 10.1128/JCM.02142-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashemi S.A., Safamanesh S., Ghasemzadeh-Moghaddam H., Ghafouri M., Azimian A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J. Med. Virol. 2021;93:1008–1012. doi: 10.1002/jmv.26364. [DOI] [PubMed] [Google Scholar]
- 7.Parry M.F., Shah A.K., Sestovic M., Salter S. Precipitous fall in common respiratory viral infections during COVID-19. Open Forum Infect. Dis. 2020;7:ofaa511. doi: 10.1093/ofid/ofaa511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang, Y., Odiwuor, N., Xiong, J., Sun, L., Nyaruaba, R.O., Wei, H., and Tanner, N.A. (2020). Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv, 10.1101/2020.02.26.20028373 (2020).
- 10.Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joung J., Ladha A., Saito M., Segel M., Bruneau R., Huang M.W., Kim N.-G., Yu X., Li J., Walker B.D., et al. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 13.Gardy J.L., Loman N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greninger A.L., Naccache S.N., Federman S., Yu G., Mbala P., Bres V., Stryke D., Bouquet J., Somasekar S., Linnen J.M., et al. Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med. 2015;7:99. doi: 10.1186/s13073-015-0220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi C., Wang L., Yuan B., Zhou X., Li Y., Wang S., Pang Y., Gao X., Huang Y., Li M. Long-read individual-molecule sequencing reveals CRISPR-induced genetic heterogeneity in human ESCs. Genome Biol. 2020;21:213. doi: 10.1186/s13059-020-02143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Wang S., Bi C., Qiu Z., Li M., Gao X. DeepSimulator1.5: a more powerful, quicker and lighter simulator for Nanopore sequencing. Bioinformatics. 2020;36:2578–2580. doi: 10.1093/bioinformatics/btz963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention, Respiratory Viruses Branch, Division of Viral Diseases . 2020. Real-time RT-PCR panel for detection 2019-novel coronavirus.https://www.who.int/docs/default-source/coronaviruse/uscdcrt-pcr-panel-for-detection-instructions.pdf?sfvrsn=3aa07934_2 [Google Scholar]
- 18.Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112:3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R.C., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323:2085–2086. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L., Wang W., Le Grange J.M., Wang X., Du S., Li C., Wei J., Zhang J.N. Coinfection of SARS-CoV-2 and other respiratory pathogens. Infect. Drug Resist. 2020;13:3045–3053. doi: 10.2147/IDR.S267238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 23.Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. npj Clean Water. 2018;1:19. [Google Scholar]
- 24.Surkova E., Nikolayevskyy V., Drobniewski F. False-positive COVID-19 results: hidden problems and costs. Lancet Respir. Med. 2020;8:1167–1168. doi: 10.1016/S2213-2600(20)30453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katz A.P., Civantos F.J., Sargi Z., Leibowitz J.M., Nicolli E.A., Weed D., Moskovitz A.E., Civantos A.M., Andrews D.M., Martinez O., Thomas G.R. False-positive reverse transcriptase polymerase chain reaction screening for SARS-CoV-2 in the setting of urgent head and neck surgery and otolaryngologic emergencies during the pandemic: clinical implications. Head Neck. 2020;42:1621–1628. doi: 10.1002/hed.26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Muenker M.C., Moore A.J., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos-Mandujano G., Salunke R., Mfarraj S., Rachmadi A.T., Hala S., Xu J., Alofi F.S., Khogeer A., Hashem A.M., Almontashiri N.A.M., et al. A robust, safe, and scalable magnetic nanoparticle workflow for RNA extraction of pathogens from clinical and wastewater samples. Global Challenges. 2021 doi: 10.1002/gch2.202000068. Published online February 22, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.2020). COVID-19 Investigation Team (2020). Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat. Med. 26, 861–868. [DOI] [PubMed]
- 29.James P., Stoddart D., Harrington E.D., Beaulaurier J., Ly L., Reid S.W., Turner D.J., Juul S. LamPORE: rapid, accurate and highly scalable molecular screening for SARS-CoV-2 infection, based on nanopore sequencing. medRxiv. 2020 doi: 10.1101/2020.08.07.20161737. [DOI] [Google Scholar]
- 30.Haramoto E., Katayama H., Ohgaki S. Detection of noroviruses in tap water in Japan by means of a new method for concentrating enteric viruses in large volumes of freshwater. Appl. Environ. Microbiol. 2004;70:2154–2160. doi: 10.1128/AEM.70.4.2154-2160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RTNano and sample data in this study are accessible at GitHub (https://github.com/milesjor/RTNano). Raw sequencing data are available in the SRA database (SRA: PRJNA638039), which are accessible with the link (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA638039/).