Abstract
Cytokines play pleiotropic, antagonistic, and collaborative in viral disease. The high morbidity and mortality of coronavirus disease 2019 (COVID-19) make it a significant threat to global public health. Elucidating its pathogenesis is essential to finding effective therapy. A retrospective study was conducted on 71 patients hospitalized with COVID-19. Data on cytokines, T lymphocytes, and other clinical and laboratory characteristics were collected from patients with variable disease severity. The effects of cytokines on the overall survival (OS) and event-free survival (EFS) of patients were analyzed. The critically severe and severe patients had higher infection indexes and significant multiple organ function abnormalities than the mild patients (P < 0.05). IL-6 and IL-10 were significantly higher in the critically severe patients than in the severe and mild patients (P < 0.05). IL-6 and IL-10 were closely associated with white blood cells, neutrophils, T lymphocyte subsets, D-D dimer, blood urea nitrogen, complement C1q, procalcitonin C-reactive protein. Moreover, the IL-6 and IL-10 levels were closely correlated to dyspnea and dizziness (P < 0.05). The patients with higher IL-10 levels had shorter OS than the group with lower levels (P < 0.05). The older patients with higher levels of single IL-6 or IL-10 tended to have shorter EFS (P < 0.05), while the patients who had more elevated IL-6 and IL-10 had shorter OS (P < 0.05). The Cox proportional hazard model revealed that IL-6 was the independent factor affecting EFS. IL-6 and IL-10 play crucial roles in COVID-19 prognosis.
Keywords: COVID-19, Cytokines, T lymphocyte, Disease severity, Survival
1. Introduction
The high morbidity and mortality of COVID-19 make it a significant threat to global public health. The latest data reported more than one hundred million laboratory-confirmed cases with over two million deaths worldwide. Internationally, cases have been reported in over 200 countries and five continents [1]. The SARS-CoV-2 predominantly infects the lower respiratory tract and intestinal tract, presenting fever, non-productive cough, dyspnea, myalgia, fatigue, normal or decreased lymphocyte counts, and radiographic evidence pneumonia at diagnosis. Organ dysfunction (for example, shock, acute respiratory distress syndrome [ARDS], acute cardiac injury, and acute kidney injury), and death frequently occur in severe cases [2].
The patients were divided into four types according to disease severity: no symptoms, mild/moderate, severe, and critically severe. The disease severity was influenced by the initial viral titers, age, and comorbid conditions of the infected individual. Older age is generally considered an essential element in disease severity. In Severe Acute Respiratory Syndrome (SARS) or Middle East Respiratory Syndrome (MERS), younger individuals experience mild to moderate clinical illness, while elderly individuals exhibit worse outcomes after infection [3], [4], [5]. In COVID-19, similar median patient age was reported in different clinical cohorts [2], [6]; however, severe or critically severe COVID-19 was also observed in young people, and some young individuals experienced dramatic disease deterioration, even in cases that had previously demonstrated few comorbidities. Though the factors causing COVID-19’s unusually high morbidity and mortality remain incompletely understood, mechanisms like thromboembolic risk have been learned over the past several months. Clotting factors like factor V [7], [8], thrombin-antithrombin complex (TAT), α2-plasmin inhibitor-plasmin Complex (PIC), thrombomodulin (TM), t-PA/PAI-1 Complex (t-PAIC) and other coagulation parameters [9], links to venous thromboembolism and mortality risk, and nominate a candidate biomarker to investigate for guiding anticoagulation therapy COVID-19. More importantly, a few recent studies revealed that cytokine storm (inflammatory cytokines, such as TNF-α, IL-1β, VEGF and IL-6) could increase microvascular permeability and induce intravascular coagulation with embolization of different organs in COVID-19 [10], [11], [12].
The complex interplay between the host antiviral immune responses and the viral ability to modify these responses are critical for disease severity. Previous studies elucidated a pivotal role for virus-associated immunopathological events in causing fatal pneumonia in MERS and SARS patients that are often associated with massive inflammatory cell infiltration and elevated proinflammatory cytokine responses called hypercytokinemia in patients [13]. Cytokine storms are an important underlying cause of mortality in patients infected with severe emerging viruses, including Ebola, Marburg, MERS-CoV, pandemic influenza, and others [14]. SARS patients with worse outcomes had high levels of proinflammatory cytokines [15], [16], indicating that immune response-associated lung immunopathology might determine the outcome of infected patients. The effects of cytokine storms coupled with acute viral infection in the host can be complicated, overwhelming, and potentially fatal. This raised a question about the role of cytokine storms in the pathogenesis of COVID-19. In this study, we demonstrate the change in cytokines and T lymphocytes together with the clinical symptoms and laboratory parameters of COVID-19 patients with different disease status. Our results may explain the pathogenesis of COVID-19.
2. Materials and methods
2.1. Patients
The Ethics committee of Zhongnan Hospital of Wuhan University approved the study (No. 2020051 K). Written consent was obtained from all enrolled patients. From January 20 to February 27, 2020, patients suspected of having COVID-19 were enrolled at Zhongnan Hospital. Among 71 confirmed cases, there were 26 critically severe patients, 15 severe patients, and 30 mild/moderate patients. The classification criteria were according to the New Coronavirus Pneumonia Prevention and Control Program (7th edition) published by the National Health Commission of China. The mild type included patients with fever, respiratory symptoms, and CT scans showing pneumonia. The severe type (meeting any of the following criteria) included respiratory distress, RR more than 30 times/min, finger oxygen saturation less than 93% at rest, and partial arterial oxygen pressure (PaO2)/oxygen absorption (FiO2) concentration less than or equal to 300 mmHg. The critically severe type included respiratory failure requiring mechanical ventilation, shock, and a combination of other organ failures requiring treatment in the intensive care unit. The patients' medical records, including clinical data, laboratory results, radiological characteristics, treatments, and outcome data, were obtained with data collection forms from the electronic medical records at Zhongnan Hospital [6].
2.2. Cytokine measurement
The serum cytokines, including IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, and IFN-γ, were measured using BD FACSCalibur flow cytometry. The protocol was followed according to the cytometric bead array (CBA) reagent manufacturer's instructions (Node Company, Jiangxi, China).
2.3. T lymphocyte determination via flow cytometry
A total of 2 ml of blood was collected and anticoagulated using heparin sodium or EDTA-k2. Human peripheral blood lymphocyte differentiation antigens (CD) CD3, CD4, CD8, CD19, CD16, and CD56 were detected using flow cytometry according to the manufacturer's instructions (Becton, Dickinson and Company, BD Biosciences, San Jose, CA, USA).
2.4. Real-time reverse transcription-polymerase chain reaction (RT-PCR) assay for SARS-CoV-2
Throat swab samples were collected to extract SARS-CoV-2 RNA. from the suspected patients. The real-time RT-PCR assay was conducted using a SARS-CoV-2 nucleic acid detection kit according to the manufacturer's protocol (Shanghai Biogerm Medical Technology, Co. Ltd.). A cycle threshold value (Ct value) less than 37 was defined as a positive test result, and a Ct value of 40 or more was identified as a negative test. (http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html).
2.5. Statistical analyses
Data are presented as mean ± standard deviation (SEM). Three-group comparisons were conducted using one-way ANOVA, Kruskal-Wallis H, Mann-Whitney U, and chi-squared tests. All of the biomarkers were selected from the initial measurement when the patients were admitted to the hospital. Associations among the variables were assessed by Partial Correlations, with control variables for age, sex, and chronic history. The overall survival (OS) was defined as the time from onset to the date of death, and event-free survival (EFS) was defined as the time from start to the first event (respiratory distress) or until the last follow-up. The Kaplan-Meier estimation was used to plot the survival curves, and the log-rank test was used to test the differences among groups. Multivariate regression analysis using the Cox proportional hazard model was applied to determine independent factors affecting survival. SPSS 20.0 was used for statistical analyses (SPSS Incorporated, Chicago, IL, USA). P < 0.05 was statistically significant. All of the statistical tests were two-sided.
3. Results
3.1. General patient characteristics
All of the enrolled patients met the COVID-19 diagnostic criteria. Among them, 70 patients demonstrated bilateral involvement in their chest CT scans, while 63 were positive for SARS-CoV-2 RNA ( Table 1 ). The median ages in the three groups were 65 (55–74), 56 (48–65), and 56 (42–67), respectively. The ratio of females to males was 1.63:1. Of the 71 patients, the most prevalent comorbidities were hypertension (23 [32.4%]), cardiovascular disease (15 [21.1%]), diabetes (10 [14.1%]), digestive diseases (9 [12.7%]), chronic lung disease (7 [9.9%]), and malignant disease (4 [5.6%]). The most common symptoms at disease onset were fever (65 [91.5%]), cough (47 [66.2%]), expectoration (10 [14.1%]), dyspnea (37 [52.1%]), fatigue (31 [43.7%]), myalgia (18 [25.4%]), and anorexia (11 [15.5%]). Less common symptoms (fewer than 10 cases) included diarrhea (7 [9.9%]), syncope (4 [5.6%]), dizziness (4 [5.6%]), pharyngalgia (4 [5.6%]), headache (2 [2.8%%]), hematuria (2 [2.8%%]), coryza (2 [2.8%]), vomiting (2 [2.8%]), and oliguria (1 [1.4%]). Overall, 36 cases had ARDS (50.7%), 37 cases had abnormal liver function (52.1%), 28 cases had abnormal renal function (39.4%), 22 cases had acute cardiac injury (31.0%), and 6 cases had shock (8.5%) ( Table 1 ).
Table 1.
Baseline characteristics of patients infected by SARS-CoV-2 with various severity.
| NO.(%) | Group 1 | Group 2 | Group 3 | P Value | |||
|---|---|---|---|---|---|---|---|
| Total N = 71 |
Critically severe N = 26 |
Severe N = 15 |
Mild N = 30 |
1 vs 2 | 1 vs 3 | 2 vs 3 | |
| Characteristics | |||||||
| Age, median (IQR), years | 60(47–68) | 65(55–74) | 56(48–65) | 56(42–67) | 0.28 | 0.05 | 1.00 |
| Sex | |||||||
| Female | 44(62.0) | 18(69.2) | 13(86.7) | 13(43.3) | 0.03 | 0.05 | 0.01 |
| Male | 27(38.0) | 8(30.8) | 2(13.3) | 17(56.7) | |||
| Confirmed patient exposure | 11(15.5) | 6(12.1) | 2(13.3) | 3(10.0) | 0.22 | 0.05 | 1.00 |
| CT positive | 70(98.6) | 26(100.0) | 15(100.0) | 29(96.7) | – | 1.00 | 1.00 |
| SARS-CoV-2 RNA positive | 63(88.7) | 21(80.8) | 13(86.7) | 29(96.7) | 1.00 | 0.09 | 0.25 |
| Signs and symptoms | |||||||
| T, median (IQR), ℃ | 38.5(38.0–39.0) | 38.9(38.3–39.4) | 38.6(37.5–39.0) | 38.0(38.0–38.5) | 0.25 | 0.01 | 1.00 |
| HR, median (IQR) | 82.0(78.0–90.0) | 84.5(78.0–99.2) | 80.0(76.0–88.0) | 82.0(77.7–92.2) | 0.06 | 0.30 | 0.54 |
| RR, median (IQR) | 20.0(18.0–22.0) | 22.5(20.0–26.0) | 20.0(18.0–20.0) | 20.0(18.0–20.0) | 0.01 | <0.01 | 0.67 |
| BP, median (IQR), mmHg | 130.0 (115.0–142.0) |
127.0 (110.2–138.2) |
131.0 (115.0–139.0) |
130.0 (120.0–145.0) |
1.00 | 0.80 | 1.00 |
| Fever | 65(91.5) | 25(96.2) | 13(86.7) | 27(90.0) | 0.54 | 0.62 | 1.00 |
| Cough | 47(66.2) | 18(69.2) | 11(73.3) | 18(60.0) | 1.00 | 0.47 | 0.38 |
| Dyspnea | 37(52.1) | 19(73.1) | 11(73.9) | 7(23.3) | 1.00 | <0.01 | <0.01 |
| Fatigue | 31(43.7) | 13(50.0) | 9(60.0) | 9(30.0) | 0.54 | 0.13 | 0.05 |
| Myalgia | 18(25.4) | 7(26.9) | 4(26.7) | 7(23.3) | 1.00 | 0.76 | 1.00 |
| Anorexia | 11(15.5) | 6(23.1) | 3(20.0) | 2(6.7) | 1.00 | 0.13 | 0.32 |
| Expectoration | 10(14.1) | 3(11.5) | 2(13.3) | 5(16.7) | 1.00 | 0.71 | 1.00 |
| Diarrhea | 7(9.9) | 6(23.1) | 1(6.7) | 0 | 0.23 | 0.01 | 0.33 |
| Pharyngalgia | 4(5.6) | 1(3.8) | 0 | 3(10.0) | 1.00 | 0.62 | 0.54 |
| Dizziness | 4(5.6) | 2(7.7) | 1(6.7) | 1(3.3) | 1.00 | 0.59 | 1.00 |
| Syncope | 4(5.6) | 4(3.8) | 0 | 0 | 0.28 | 0.04 | – |
| Headache | 2(2.8) | 1(3.8) | 1(6.7) | 0 | 1.00 | 0.46 | 0.33 |
| Coryza | 2(2.8) | 1(3.8) | 1(6.7) | 0 | 1.00 | 0.46 | 0.33 |
| Vomiting | 2(2.8) | 1(3.8) | 1(6.7) | 0 | 1.00 | 0.46 | 0.33 |
| Hematuria | 2(2.8) | 2(7.7) | 0 | 0 | 0.52 | 0.21 | – |
| Oliguria | 1(1.4) | 1(3.8) | 0 | 0 | 1.00 | 0.46 | – |
| Comorbidities | |||||||
| Hypertension | 23(32.4) | 13(50.0) | 3(20.0) | 7(23.3) | 0.06 | 0.04 | 1.00 |
| Diabetes | 10(14.1) | 5(19.2) | 3(20.0) | 2(6.7) | 1.00 | 0.23 | 0.32 |
| Cardiovascular disease | 15(21.1) | 11(42.3) | 1(6.7) | 3(10.0) | 0.03 | 0.01 | 1.00 |
| Chronic lung disease | 7(9.9) | 2(7.7) | 2(13.3) | 3(10.0) | 0.62 | 1.00 | 1.00 |
| Malignancy | 4(5.6) | 1(3.8) | 1(6.7) | 2(6.7) | 1.00 | 1.00 | 1.00 |
| Digestive diseases | 9(12.7) | 3(11.5) | 3(20.0) | 3(10.0) | 0.65 | 1.00 | 0.38 |
| Treatment | |||||||
| Antiviral therapy | 69(97.2) | 25(96.2) | 15(100.0) | 29(96.7) | 1.00 | 1.00 | 1.00 |
| Antibiotic treatment | 64(90.1) | 26(100.0) | 15(100.0) | 23(76.7) | – | 0.01 | 0.08 |
| Glucocorticoid therapy | 50(70.4) | 24(92.3) | 13(86.7) | 13(43.3) | 0.62 | <0.01 | 0.01 |
| Immunoglobulin therapy | 19(26.8) | 9(34.6) | 5(33.3) | 5(16.7) | 0.93 | 0.12 | 0.26 |
| Oxygen inhalation | 47(66.2) | 19(73.1) | 15(100.0) | 13(43.3) | 0.04 | 0.03 | <0.01 |
| NIV | 28(39.4) | 22(84.6) | 6(40.0) | 0 | 0.01 | <0.01 | <0.01 |
| IMV | 22(31.0) | 22(84.6) | 0 | 0 | <0.01 | <0.01 | – |
| ECMO | 8(11.3) | 8(30.8) | 0 | 0 | 0.02 | <0.01 | – |
| CRRT | 10(14.1) | 10(38.5) | 0 | 0 | 0.01 | <0.01 | – |
| CPR | 12(16.9) | 12(46.2) | 0 | 0 | <0.01 | <0.01 | – |
| Blood transfusion | 13(18.3) | 13(50.0) | 0 | 0 | <0.01 | <0.01 | – |
| Bronchial lavage | 5(7.0) | 5(19.2) | 0 | 0 | 0.14 | 0.02 | – |
| Clinical trial | 8(11.3) | 3(11.5) | 5(33.3) | 0 | 0.12 | 0.09 | <0.01 |
| Complications | |||||||
| ARDS | 36(50.7) | 26(100.0) | 10(66.7) | 0 | <0.01 | <0.01 | <0.01 |
| Abnormal liver function | 37(52.1) | 17(65.4) | 13(86.7) | 7(23.3) | 0.17 | <0.01 | <0.01 |
| Acute cardiac injury | 22(31.0) | 20(76.9) | 1(6.7) | 1(3.3) | <0.01 | <0.01 | 1.00 |
| Acute kidney injury | 28(39.4) | 21(80.8) | 5(33.3) | 2(6.7) | <0.01 | <0.01 | 0.03 |
| Shock | 6(8.5) | 6(23.1) | 0 | 0 | 0.07 | 0.01 | – |
| Clinical outcome | |||||||
| Died | 10(14.1) | 10(38.5) | 0 | 0 | 0.01 | <0.01 | – |
| Remained in ICU | 8(11.3) | 8(30.8) | 0 | 0 | 0.02 | <0.01 | – |
| Condition improve | 31(43.7) | 5(19.2) | 8(53.3) | 18(60.0) | 0.27 | <0.01 | 0.21 |
| Hospital discharge | 22(31.0) | 3(11.5) | 7(46.7) | 12(40.0) | 0.02 | 0.02 | 0.67 |
Data are n (%), n/N (%), mean (SEM), and median (1QR). P values comparing the critically severe, severe, and mild patients are from one-way analysis of variance, Pearson’s chi-squared test, or Fisher's exact test. A P value < 0.05 was considered statistically significant. Data were available for 71 patients. IQR: interquartile range; T: temperature; HR: heart rate; RR: respiratory rate; BP: blood pressure; NIV: non-invasive ventilation; IMV: intermittent mandatory ventilation; ECMO: extracorporeal membrane oxygenation; CRRT: continuous renal replacement therapy; CPR: cardiopulmonary resuscitation; ARDS: Acute Respiratory Distress Syndrome.
3.2. Patient treatment and outcomes
Among the therapeutic strategies given to the patients, antiviral therapy (arbidol, oseltamivir, remdesivir, or traditional Chinese medicine used) was administered to 69 (97.2%), antibiotic treatment to 64 (90.1%), glucocorticoid therapy to 50 (70.4%), and immunoglobulin therapy to 19 (26.8%). Oxygen inhalation was needed in 47 (66.2%) patients, non-invasive ventilation (NIV) in 28 (39.4%), invasive mechanical ventilation (IMV) in 22 (31.0%), ECMO in 8 (11.3%), and CRRT in 10 (14.1%). A total of 12 (16.9%) patients underwent cardiopulmonary resuscitation (CPR), and 13 (18.3%) received blood transfusions. By the end of February 27, 2020, a total of 22 (31.0%) patients had been discharged, 31 (43.7%) improved, 8 (11.2%) remained in the ICU, and 10 (14.1%) died ( Table 1 ).
3.3. Laboratory parameters
Compared to the mild patients, the critically severe patients had significantly higher levels of white blood cells (WBCs), neutrophils (NEUT), procalcitonin (PCT), C-reactive protein (CRP), prothrombin time (PT), D-D dimer (D-D), alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), creatinine, creatine kinase-MB (CK-MB), lactate dehydrogenase (LDH), HSTNI, and glucose (P < 0.05), while significantly lower levels of lymphocytes, platelet (PLT), total protein (TP), albumin (ALB), CO2, K+, Na+, Ca2+, and C1q (P < 0.05) ( Table 2 ). Our data showed that the critically severe patients had liver/renal function abnormalities, abnormal myocardial zymograms, and high infection indexes (P < 0.05). Comparing the critically severe and severe patients, WBC, NEUT, PT, HSTNI, and PCT was significantly higher, while CO2 was markedly lower in the critically severe group than in the severe group (P < 0.05). There were significantly higher levels of GLU, ALT, CK-MB, and CRP and lower levels of lymphocytes, Ca2+, ALB, and TP in the severe group than in the mild group (P < 0.05) ( Table 2 ).
Table 2.
Laboratory characteristics of the COVID-19 patients with various disease severity.
| Group 1 | Group 2 | Group 3 | P Value | ||||
|---|---|---|---|---|---|---|---|
| Clinical Indicator | Normal Range | Critically severe N = 26 |
Severe N = 14 |
Mild N = 31 |
1 VS 2 | 1 VS 3 | 2 VS 3 |
| WBC, ×109/L | 3.5–9.5 | 12.40 ± 1.50 | 6.81 ± 1.00 | 6.20 ± 0.51 | 0.01 | <0.01 | 0.85 |
| NEUT, ×109/L | 1.8–6.3 | 11.05 ± 1.44 | 5.76 ± 1.04 | 4.56 ± 0.49 | 0.01 | <0.01 | 0.56 |
| NEUT,% | 40–75 | 88.50 ± 1.47 | 79.78 ± 4.03 | 70.21 ± 2.19 | 0.17 | <0.01 | 0.05 |
| LY, ×109/L | 1.1–3.2 | 0.62 ± 0.07 | 0.57 ± 0.10 | 1.11 ± 0.08 | 1.00 | <0.01 | <0.01 |
| LY, % | 20–50 | 6.76 ± 1.07 | 11.36 ± 2.82 | 20.91 ± 1.87 | 0.31 | <0.01 | 0.02 |
| HGB, g/L | 115–150 | 125.23 ± 4.19 | 135.20 ± 3.41 | 136.04 ± 2.41 | 0.23 | 0.05 | 1.00 |
| RBC, ×1012/L | 3.8–5.1 | 3.98 ± 0.13 | 4.24 ± 0.12 | 4.26 ± 0.10 | 0.62 | 0.27 | 1.00 |
| HCT,% | 35–45 | 36.35 ± 1.22 | 39.16 ± 1.11 | 38.81 ± 0.76 | 0.29 | 0.21 | 1.00 |
| PLT, ×109/L | 125–350 | 174.19 ± 9.60 | 195.93 ± 23.47 | 242.03 ± 18.19 | 0.67 | 0.01 | 0.28 |
| PT, s | 9.4–12.5 | 14.43 ± 0.48 | 12.92 ± 0.34 | 12.54 ± 0.34 | 0.04 | 0.01 | 0.71 |
| APTT, s | 25.1–36.5 | 40.80 ± 7.41 | 28.78 ± 0.98 | 30.78 ± 0.76 | 0.26 | 0.38 | 0.26 |
| TT-E, s | 10.3–16.6 | 32.55 ± 9.69 | 15.89 ± 0.41 | 15.76 ± 0.63 | 0.22 | 0.21 | 0.98 |
| FIB, mg/dL | 238–498 | 445.38 ± 26.14 | 479.14 ± 28.89 | 424.58 ± 22.66 | 1.00 | 1.00 | 0.58 |
| DD, ng/mL | 0–500 | 5431.92 ± 1989.89 | 1771.07 ± 1042.10 | 2759.05 ± 2250.63 | 0.12 | <0.01 | 0.97 |
| ALT, U/L | 7–45 | 50.88 ± 7.44 | 62.50 ± 13.53 | 32.65 ± 5.46 | 1.00 | 0.01 | 0.03 |
| AST, U/L | 13–35 | 72.46 ± 12.63 | 44.29 ± 7.39 | 30.26 ± 3.64 | 0.15 | 0.01 | 0.23 |
| TP, g/L | 65–85 | 60.59 ± 0.93 | 62.74 ± 1.89 | 69.94 ± 1.22 | 0.89 | <0.01 | <0.01 |
| ALB, g/L | 40–55 | 30.74 ± 0.79 | 31.20 ± 1.34 | 39.40 ± 0.81 | 1.00 | <0.01 | <0.01 |
| GLB, g/L | 20–30 | 29.85 ± 0.90 | 31.54 ± 1.82 | 30.54 ± 0.84 | 0.69 | 0.84 | 0.87 |
| GGT, U/L | 8–57 | 61.62 ± 13.35 | 87.00 ± 26.75 | 40.58 ± 6.37 | 0.68 | 0.34 | 0.24 |
| ALP, U/L | 30–120 | 73.65 ± 4.58 | 80.50 ± 8.94 | 84.13 ± 4.97 | 1.00 | 0.47 | 1.00 |
| TBA, umol/L | 0–15 | 4.32 ± 0.48 | 4.50 ± 0.94 | 3.21 ± 0.38 | 0.98 | 0.18 | 0.43 |
| BUN, mmol/L | 2.8–7.6 | 7.31 ± 0.56 | 6.33 ± 0.68 | 4.62 ± 0.31 | 0.48 | <0.01 | 0.07 |
| CREA, umol/L | 49–90 | 81.87 ± 6.13 | 70.84 ± 4.30 | 65.07 ± 2.73 | 0.28 | 0.04 | 0.51 |
| UA, umol/L | 155–357 | 298.52 ± 32.32 | 242.60 ± 25.96 | 304.56 ± 17.51 | 0.30 | 0.99 | 0.09 |
| CO2, mmol/L | 21–29 | 20.62 ± 0.69 | 24.09 ± 0.83 | 25.00 ± 0.52 | <0.01 | <0.01 | 1.00 |
| CYSC, mg/L | 0–1.2 | 1.20 ± 0.08 | 1.17 ± 0.11 | 1.00 ± 0.06 | 1.00 | 0.14 | 0.51 |
| K+, mmol/L | 3.5–5.3 | 3.88 ± 0.10 | 4.21 ± 0.13 | 4.39 ± 0.08 | 0.25 | <0.01 | 0.46 |
| Na+, mmol/L | 137–147 | 136.23 ± 1.28 | 138.34 ± 0.90 | 138.98 ± 0.50 | 0.36 | 0.02 | 1.00 |
| Cl−, mmol/L | 99–110 | 102.23 ± 1.14 | 103.42 ± 1.03 | 102.30 ± 0.82 | 0.83 | 1.00 | 0.81 |
| Ca2+, mmol/L | 2.11–2.52 | 1.96 ± 0.03 | 2.03 ± 0.03 | 2.24 ± 0.03 | 0.65 | <0.01 | <0.01 |
| CK, U/L | <145 | 426.96 ± 184.91 | 159.96 ± 51.66 | 120.88 ± 27.35 | 0.36 | 0.25 | 0.78 |
| CKMB, U/L | 0–25 | 53.79 ± 17.84 | 49.69 ± 24.74 | 12.50 ± 0.63 | 1.00 | <0.01 | 0.08 |
| LDH, U/L | 125–243 | 527.13 ± 52.61 | 354.38 ± 73.33 | 222.63 ± 17.55 | 0.33 | <0.01 | 0.22 |
| MYO, ng/mL | <140.1 | 262.18 ± 72.13 | 135.11 ± 33.80 | 117.73 ± 34.58 | 0.26 | 0.19 | 0.93 |
| HSTNI, pg/mL | 0–26.2 | 966.33 ± 793.33 | 35.42 ± 30.30 | 8.98 ± 5.96 | 0.03 | 0.01 | 1.00 |
| GLU, mmol/L | 3.9–6.1 | 10.26 ± 1.10 | 8.57 ± 0.82 | 6.67 ± 0.51 | 1.00 | <0.01 | 0.04 |
| C1q, mg/L | 159–233 | 175.50 ± 8.76 | 185.96 ± 10.00 | 210.90 ± 7.74 | 1.00 | 0.01 | 0.22 |
| PCT, ng/ml | <0.05 | 1.60 ± 0.67 | 0.12 ± 0.03 | 0.06 ± 0.01 | 0.01 | <0.01 | 0.41 |
| CRP, mg/L | 0.0–3.0 | 105.99 ± 15.33 | 77.31 ± 30.04 | 25.21 ± 5.88 | 0.44 | <0.01 | 0.03 |
P value < 0.05 was considered statistically significant.
Blood RT: WBC: white blood cell; NEUT: neutrophils; LY: lymphocytes; HGB: hemoglobin; RBC: red blood cell; HCT: hematocrit; PLT: platelet.
Coagulogram: PT: prothrombin time; APTT: activated partial thromboplastin time; TT-E: thrombin time (extended); FIB: fibrinogen content; DD: D-dimer.
Liver function: ALT: alanine aminotransferase; AST: aspartate aminotransferase; TP: total protein; ALB: albumin; GLB: globin; GGT: alpha-glutamyl transpeptidase; ALP: alkaline phosphatase; TBA: total bile acid.
Renal function: BUN: blood urea nitrogen; CREA: creatinine; UA: uric acid; CO2: carbon dioxide; CYSC: cystatin C.
Cardiac function: CK: creatine kinase; LDH: lactate dehydrogenase; CKMB: creatine kinase-MB; MYO: myoglobin; HSTNI: hypersensitive troponin I.
Inflammatory immune index: C1q: complement C1q; PCT: procalcitonin; CRP: C-reactive protein; GLU: glucose.
3.4. Cytokine levels in mild, severe, and critically severe patients
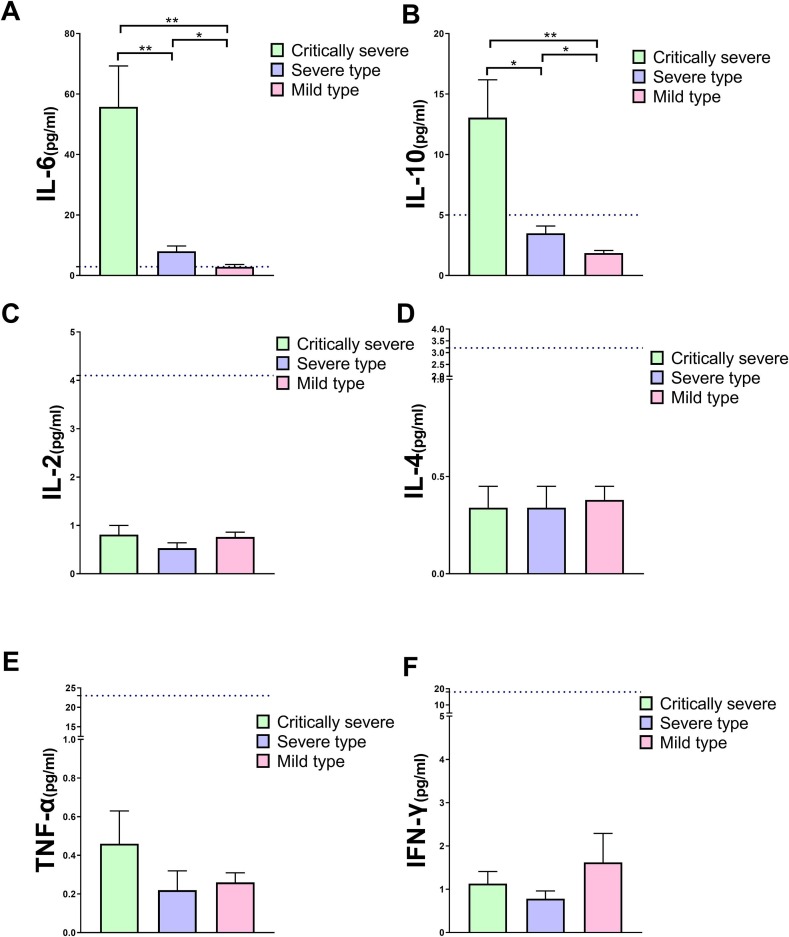
The IL-6 concentration in the critically severe group was 19-fold higher than in the mild group and 7-fold higher than in the severe group at disease onset. IL-6 in the severe type was 2.8-fold higher than in the mild group (P < 0.05) ( Fig. 1 a). The IL-10 level in the critically severe group was 7-fold higher than in the mild group and 3.7-fold higher than in the severe group. IL-10 in the severe group was 1.9-fold higher than in the mild group (P < 0.05) ( Fig. 1 b). There was no significant difference in IL-2, IL-4, TNF-α, and IFN-γ among the three groups (P > 0.05) ( Fig. 1 ).
Fig. 1.
Difference in the serum cytokine (IL-6, IL-10, IL-2, IL-4, TNF-α, and IFN-γ) levels among the critically severe, severe, and mild groups. (a) IL-6 in the critically severe group was significantly higher than in the severe (P < 0.01) or mild groups (P < 0.01), and higher levels of IL-10 were also found in the severe group than in the mild group (P < 0.05). (b) IL-10 showed the same trend as IL-6 among the three groups (P < 0.05). (c), (d), (e), and (f) represent IL-2, IL-4, TNF-α, and IFN-γ, respectively, all of which showed no significant differences among the three groups. The blue dashed line represents the upper limit of the reference range.
3.5. Dynamic changes in cytokines during the disease course
A longitudinal analysis of the progressive changes in IL-6 was conducted in the critically severe patients. The fluctuating changes chart typically showed one single peak of IL-6 in 8 patients (Fig. 2 a). Specific treatments also influenced the IL-6 level. For example, patient No. 9 received plasma (from convalescent COVID-19 patients) infusion 25 days after disease onset, and their IL-6 level decreased significantly to the normal range over the following two days. Patient No. 12 demonstrated a slight decrease in IL-6 in the up-slope phase from days 9 to 24 following remdesivir treatment (Fig. 2 a). No particular trend was found in IL-10 (Fig. 2 b).
Fig. 2.
IL-6 and IL-10 levels changed as the disease progressed. (a) A longitudinal analysis of the dynamic changes in IL-6 was conducted in 10 patients from whom three or more samples were collected. The linear chart typically showed one single peak in 8 patients. (b) No significant regularity was found in IL-10. The blue dashed line represents the upper limit of the reference range. The arrows indicate when the patient received clinical trial treatment.
3.6. T lymphocytes in the mild, severe, and critically severe patients
There was no significant difference in the percentage of T lymphocyte (CD3+%), helper/inducible T lymphocyte (CD3 + CD4+%), and NK. cell (CD16 + CD56+%) among the three groups (P > 0.05), but the percentage of inhibitory/cytotoxic T lymphocyte (CD3 + CD8+%) in the critically severe group was significantly lower than in the mild group (P < 0.05) ( Fig. 3 a). Patients in the critically severe group also showed a marked decrease in CD3 + lymphocyte (Lym) and inhibitory/cytotoxic T lymphocyte (CD3 + CD8 + Lym) absolute count (Abs) in comparison with the mild group (P < 0.05). Conversely, the percentage of B lymphocyte (CD19+%) in the critically severe group was significantly higher than in the severe and mild groups (P < 0.05). NK cell absolute count (CD16 + CD56 + Lym Abs) in the mild group was considerably higher than in the critically severe group (P < 0.05) (Fig. 3 b).
Fig. 3.
T lymphocyte (Lym) and subset (CD3+%Lym, CD3 + CD4+%Lym, CD3 + CD8+%Lym, CD4/CD8 ratio, CD19+%Lym, and CD16 + 56+%Lym) among the critically severe, severe, and mild groups (absolute counting [Abs] and percentage [%]). (a) CD3 + 8+%Lym was substantially lower in the critically severe group than in the severe and mild groups (P < 0.05), but the CD4/CD8 ratio and CD19+%Lym were the opposite. (b) The Abs of CD3+, CD3 + 8+, and CD16 + 56 + were significantly lower in the critically severe group than in the mild group (P < 0.05). CD3+: T lymphocyte absolute count; CD3 + 4+: helper/inducible T lymphocyte absolute count; CD3 + 8+: inhibitory/cytotoxic T lymphocyte absolute count; 4/8 ratio: CD4/CD8; CD19+: B lymphocyte absolute count; CD16 + 56+: NK cell absolute count. *P < 0.05 was considered statistically significant.
3.7. Correlations among all of the parameters
IL-6 levels were closely associated with IL-10 (r = 0.56, P < 0.01), CD16 + CD56 + Abs (r = 0.35, P = 0.03), NEUT (r = 0.43, P = 0.01), PT (r = 0.40, P = 0.01), BUN (r = 0.47, P = 0.01), PCT (r = 0.63, P < 0.01), and CRP (r = 0.48, P < 0.01). IL-10 was closely associated with IL-6, PT (r = 0.34, P < 0.01) and PCT (r = 0.42, P < 0.01) (Table 3 ). IFN-γ levels were closely associated with TNF-α (r = 0.46, P < 0.01), IL-4 (r = 0.29, P = 0.01), IL-2 (r = 0.5, P < 0.01), CD3 + CD4 + Abs (r = 0.30, P = 0.04), NEUT (r = 0.31, P = 0.05) and D-D (r = 0.29, P = 0.02). IL-4 levels were closely associated with IFN-γ, TNF-α, and IL-2 (r = 0.42, P < 0.01). IL-6 were closely related to dyspnea (r = 0.323, P < 0.01), fatigue (r = 0.326, P < 0.01), Anorexia (r = 0.266, P = 0.025) and Diarrhea (r = 0.267, P = 0.024), and. IL-10 were closely related to dyspnea (r = 0.242, P = 0.042), dizziness (r = 0.255, P = 0.032), and Diarrhea (r = 0.345, P < 0.01). An interrelationship was also demonstrated among T and B lymphocytes and T subsets. CD16 + CD56 + Abs correlated with IL-6, IL-2 (r = −0.34, P = 0.03), NEUT (r = 0.31, P = 0.05), BUN (r = −0.33, P = 0.04), CD3+ (r = 0.40, P = 0.01), CD3 + CD4+ (r = 0.31, P = 0.05), CD3 + CD8+ (r = 0.58, P < 0.01), 4/8ratio (r = −0.32, P = 0.04) (Table 3).
Table 3.
Correlation between cytokines and other lab parameters in COVID-9 patients.
| IFN-γ | IL-10 | TNF-α | IL-4 | IL-6 | IL-2 | CD3 | CD3 + 4+ | CD3 + 8+ | 4/8 Ratio | CD19 | CD16 + 56+ | NEUT | PT | DD | ALT | BUN | C1q | PCT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IFN-γ | |||||||||||||||||||
| IL-10 | r = −0.01 | ||||||||||||||||||
| P = 0.91 | |||||||||||||||||||
| TNF-α |
r = 0.46 ** |
r = −0.16 | |||||||||||||||||
| P < 0.01 | P = 0.18 | ||||||||||||||||||
| IL-4 | r = 0.29* | r = −0.18 |
r = 0.32 ** |
||||||||||||||||
| P = 0.01 | P = 0.12 | P < 0.01 | |||||||||||||||||
| IL-6 | r = 0.09 |
r = 0.56 ** |
r = 0.21 | r = −0.14 | |||||||||||||||
| P = 0.36 | P < 0.01 | P = 0.19 | P = 0.39 | ||||||||||||||||
| IL-2 |
r = 0.50 ** |
r = −0.08 |
r = 0.39 ** |
r = 0.42 ** |
r = −0.06 | ||||||||||||||
| P < 0.01 | P = 0.52 | P < 0.01 | P < 0.01 | P = 0.71 | |||||||||||||||
| CD3 | r = 0.03 | r = −0.14 | r = 0.02 | r = 0.18 | r = −0.19 | r = 0.00 | |||||||||||||
| P = 0.88 | P = 0.39 | P = 0.93 | P = 0.26 | P = 0.26 | P = 0.99 | ||||||||||||||
| CD3 + 4+ | r = 0.30* | r = −0.06 | r = 0.09 | r = 0.13 | r = −0.06 | r = 0.22 |
r = 0.94 ** |
||||||||||||
| P = 0.04 | P = 0.68 | P = 0.54 | P = 0.38 | P = 0.73 | P = 0.14 | P < 0.01 | |||||||||||||
| CD3 + 8+ | r = −0.06 | r = −0.18 | r = −0.05 | r = 0.25 | r = −0.30 | r = −0.08 |
r = 0.91 ** |
r = 0.73 ** |
|||||||||||
| P = 0.71 | P = 0.27 | P = 0.75 | P = 0.12 | P = 0.07 | P = 0.64 | P < 0.01 | P < 0.01 | ||||||||||||
| 4/8 Ratio | r = 0.17 | r = 0.19 | r = −0.07 | r = 0.12 | r = 0.22 | r = 0.17 | r = 0.01 | r = 0.20 | r = −0.28 | ||||||||||
| P = 0.29 | P = 0.25 | P = 0.69 | P = 0.48 | P = 0.17 | P = 0.31 | P = 0.97 | P = 0.22 | P = 0.09 | |||||||||||
| CD19 | r = 0.06 | r = −0.02 | r = 0.16 | r = 0.09 | r = 0.19 | r = 0.27 | r = 0.23 | r = 0.34* | r = 0.08 | r = 0.33* | |||||||||
| P = 0.70 | P = 0.90 | P = 0.32 | P = 0.57 | P = 0.25 | P = 0.09 | P = 0.16 | P = 0.03 | P = 0.61 | P = 0.04 | ||||||||||
| CD16 + 56+ | r = −0.20 | r = −0.24 | r = 0.10 | r = 0.07 | r = −0.35* | r = −0.34* | r = 0.40* | r = 0.31* |
r = 0.58 ** |
r = −0.32* | r = 0.02 | ||||||||
| P = 0.21 | P = 0.14 | P = 0.52 | P = 0.66 | P = 0.03 | P = 0.03 | P = 0.01 | P = 0.05 | P < 0.01 | P = 0.04 | P = 0.93 | |||||||||
| NEUT | r = 0.31* | r = 0.24 | r = 0.29 | r = −0.01 |
r = 0.43 ** |
r = 0.18 | r = −0.03 | r = 0.19 | r = −0.18 | r = 0.36* | r = 0.33* | r = −0.31* | |||||||
| P = 0.05 | P = 0.13 | P = 0.07 | P = 0.96 | P = 0.01 | P = 0.27 | P = 0.86 | P = 0.23 | P = 0.26 | P = 0.02 | P = 0.03 | P = 0.05 | ||||||||
| PT | r = 0.11 |
r = 0.34 ** |
r = 0.12 | r = 0.03 |
r = 0.40 ** |
r = 0.19 | r = −0.03 | r = 0.06 | r = −0.11 | r = 0.15 | r = 0.40* | r = −0.12 |
r = 0.53 ** |
||||||
| P = 0.41 | P < 0.01 | P = 0.35 | P = 0.84 | P = 0.01 | P = 0.15 | P = 0.84 | P = 0.72 | P = 0.48 | P = 0.34 | P = 0.01 | P = 0.47 | P < 0.01 | |||||||
| DD | r = 0.29* | r = 0.25 | r = 0.17 | r = −0.03 | r = 0.11 | r = 0.31* | r = 0.05 | r = −0.04 | r = −0.01 | r = 0.03 | r = 0.07 | r = −0.22 | r = 0.19 |
r = 0.42 ** |
|||||
| P = 0.02 | P = 0.06 | P = 0.19 | P = 0.84 | P = 0.52 | P = 0.01 | P = 0.76 | P = 0.77 | P = 0.97 | P = 0.86 | P = 0.67 | P = 0.17 | P = 0.22 | P < 0.01 | ||||||
| ALT | r = 0.06 | r = −0.01 | r = 0.07 | r = 0.12 | r = 0.22 | r = 0.14 | r = −0.30 | r = −0.11 | r = −0.28 | r = −0.05 | r = 0.07 | r = 0.13 | r = 0.08 | r = 0.08 | r = −0.02 | ||||
| P = 0.73 | P = 0.95 | P = 0.66 | P = 0.47 | P = 0.18 | P = 0.39 | P = 0.06 | P = 0.48 | P = 0.08 | P = 0.78 | P = 0.66 | P = 0.42 | P = 0.63 | P = 0.64 | P = 0.91 | |||||
| BUN | r = 0.00 | r = 0.02 | r = 0.08 | r = −0.18 |
r = 0.47 ** |
r = 0.07 | r = −0.10 | r = 0.01 | r = −0.20 | r = 0.16 | r = 0.17 | r = −0.33* | r = 0.30 | r = −0.02 | r = 0.37* | r = 0.23 | |||
| P = 0.99 | P = 0.91 | P = 0.64 | P = 0.28 | P < 0.01 | P = 0.69 | P = 0.55 | P = 0.97 | P = 0.22 | P = 0.32 | P = 0.30 | P = 0.04 | P = 0.06 | P = 0.92 | P = 0.02 | P = 0.15 | ||||
| C1q | r = −0.28 | r = −0.30 | r = −0.08 | r = 0.19 | r = −0.30 | r = −0.10 | r = −0.22 | r = −0.30 | r = −0.04 | r = −0.10 | r = 0.01 | r = 0.28 | r = −0.33* | r = −0.28 | r = −0.15 | r = 0.13 | r = −0.13 | ||
| P = 0.08 | P = 0.06 | P = 0.61 | P = 0.23 | P = 0.06 | P = 0.52 | P = 0.17 | P = 0.06 | P = 0.82 | P = 0.53 | P = 0.94 | P = 0.08 | P = 0.04 | P = 0.07 | P = 0.34 | P = 0.44 | P = 0.44 | |||
| PCT | r = 0.16 |
r = 0.42 ** |
r = −0.01 | r = −0.05 |
r = 0.63 ** |
r = 0.23 | r = −0.03 | r = 0.02 | r = −0.13 | r = 0.22 | r = 0.42* | r = −0.21 |
r = 0.74 ** |
r = 0.53 ** |
r = 0.53 ** |
r = 0.27 | r = 0.35* | r = −0.31* | |
| P = 0.23 | P < 0.01 | P = 0.93 | P = 0.72 | P < 0.01 | P = 0.09 | P = 0.87 | P = 0.88 | P = 0.41 | P = 0.18 | P = 0.01 | P = 0.19 | P < 0.01 | P < 0.01 | P < 0.01 | P = 0.09 | P = 0.03 | P = 0.02 | ||
| CRP | r = 0.10 | r = 0.20 | r = 0.27 | r = −0.18 |
r = 0.48 ** |
r = −0.09 | r = −0.05 | r = −0.04 | r = −0.21 | r = 0.15 | r = −0.20 | r = −0.23 | r = 0.42* |
r = 0.52 ** |
r = 0.16 | r = 0.03 | r = 0.01 | r = −0.15 |
r = 0.57 ** |
| P = 0.56 | P = 0.22 | P = 0.09 | P = 0.26 | P < 0.01 | P = 0.58 | P = 0.76 | P = 0.80 | P = 0.20 | P = 0.35 | P = 0.21 | P = 0.16 | P = 0.01 | P < 0.01 | P = 0.33 | P = 0.85 | P = 0.97 | P = 0.36 | P < 0.01 | |
*P < 0.05 and **P < 0.01 were considered statistically significant. IFN-γ: interferon-γ; IL-10: interleukin-10; TNF-α: tumor necrosis factor-α; CD3: absolute T lymphocyte count; CD3 + 4+: helper/inducible T lymphocyte absolute count; CD3 + 8+: inhibitory/cytotoxic T lymphocyte absolute count; 4/8 ratio: CD4/CD8; CD19: B lymphocyte count; CD16 + 56+: absolute NK cell count; NEUT: neutrophils; PT: prothrombin time; DD: D-dimer; ALT: alanine aminotransferase; BUN: blood urea nitrogen; C1q: complement C1q; PCT: procalcitonin; CRP: C-reactive protein.
3.8. Survival analyses
In the critically severe group, ten patients died, eight remained in the ICU, and five improved; three were discharged (out of the total of all critically severe patients and not include in the improved group). In the severe group, eight patients improved, and seven were discharged. In the mild group, 18 improved, and 12 were discharged (Table 1). The parameters' mean values were used as cutoff points to divide the patients into high- and low-risk groups. The patients with low levels of IL-10 had longer OS (high vs. low: 40.84 ± 4.17 vs. 42.75 ± 1.21, P = 0.02) (Fig. 4 b), while those who had doubly higher IL-6 and IL-10 had shorter OS (high vs. low: 39.14 ± 4.31 vs. 49.31 ± 1.63, P = 0.01) (Fig. 4 c). The other parameters played no significant role in the patients' OS (Table 4 ). Regarding EFS, the elderly (high vs. low: 20.12 ± 2.60 vs. 29.11 ± 2.63, P = 0.02) and patients with comorbidities (with vs. without 18.91 ± 2.23 vs. 29.38 ± 2.70, P = 0.02) tended to have shorter EFS (Table 4). The patients with higher IL-6 levels had shorter EFS (high vs. low: 14.54 ± 1.94 vs. 34.69 ± 2.13, P < 0.001) (Fig. 4 d), and the patients with higher IL-10 levels had shorter EFS (high vs. low: 15.41 ± 2.22 vs. 33.14 ± 2.25, P < 0.001) (Fig. 4 e). The patients who had doubly high IL-6 and IL-10 had shorter EFS than the lower group (high vs. low: 8.10 ± 0.77 vs. 31.11 ± 2.07, P < 0.001) (Fig. 4 f). The other parameters played no significant role in the patients' EFS (Table 4). Using the parameters (sex, age, comorbidities, index of organ function, inflammatory factors, and cytokines) as covariables, the Cox proportional hazard model with forwarding stepwise selection revealed that only doubly high IL-6 and IL-10 were independent factors affecting OS (hazard ratio: 9.009, 95% CI: 1.139–71.229, P = 0.037) and EFS ((hazard ratio:5.746, 95% CI: 2.538–13.009, P < 0.001). If IL-6 + IL-10 was removed from the covariables, IL-6 was an independent factor affecting EFS (hazard ratio: 1.012, 95% CI: 1.005–1.018, P = 0.001).
Fig. 4.
Overall survival (OS) and event-free survival (EFS) in patients with different levels of IL-6 and IL-10. (a) No significant difference existed in IL-6 (P > 0.05). (b) The OS was shorter in patients with high IL-10 (P = 0.020). (c) Both IL-6 and IL-10 above the normal range had shorter OS than the other conditions (P = 0.01). (d), (e), and (f) IL-6 and IL-10 were elevated individually or both abnormal, demonstrating shorter EFS (P < 0.01).
Table 4.
Univariate analysis of the impact of possible risk factors on overall survival and event-free survival (EFS) in critically sever and severe patients P-Value.
| Variable | No. of Patients | Mean (days ± SD) | P Value |
|---|---|---|---|
| Overall survival | |||
| Sex | |||
| male | 31 | 44.18 ± 4.17 | 0.237 |
| female | 10 | 47.80 ± 2.86 | |
| Age | |||
| Low(<median) | 19 | 43.92 ± 2.85 | 0.46 |
| High(>median) | 22 | 46.60 ± 4.38 | |
| Comorbidities | |||
| without | 15 | 35.01 ± 1.40 | 0.406 |
| with | 26 | 47.98 ± 4.17 | |
| IL-6 | |||
| Low(<median) | 20 | 37.50 ± 0.31 | 0.13 |
| High(>median) | 21 | 43.82 ± 3.60 | |
| IL-10 | |||
| Low(<median) | 20 | 42.75 ± 1.21 | 0.02 |
| High(>median) | 21 | 40.84 ± 4.17 | |
| IL-6 + IL-10 | |||
| Two positive | 20 | 39.14 ± 4.31 | 0.01 |
| other | 21 | 49.31 ± 1.63 | |
| CD3 + Cnt | |||
| Low(<median) | 14 | 52.10 ± 4.03 | 0.62 |
| High(>median) | 16 | 46.55 ± 2.68 | |
| CD3 + CD8 + Cnt | |||
| Low(<median) | 15 | 49.62 ± 4.36 | 0.25 |
| High(>median) | 15 | 48.71 ± 2.12 | |
| EFS | |||
| Sex | |||
| male | 44 | 20.05 ± 2.06 | 0.054 |
| female | 27 | 29.88 ± 3.03 | |
| Age | |||
| Low(<median) | 35 | 29.11 ± 2.63 | 0.02 |
| High(>median) | 36 | 20.12 ± 2.60 | |
| Comorbidities | |||
| without | 34 | 29.38 ± 2.70 | 0.02 |
| with | 37 | 18.91 ± 2.23 | |
| IL-6 | |||
| Low(<median) | 34 | 34.69 ± v2.13 | <0.001 |
| High(>median) | 37 | 14.54 ± 1.94 | |
| IL-10 | |||
| Low(<median) | 35 | 33.14 ± 2.25 | <0.001 |
| High(>median) | 36 | 15.41 ± 2.22 | |
| IL-6 + IL-10 | |||
| Two positive | 21 | 8.10 ± 0.77 | <0.001 |
| other | 50 | 31.11 ± 2.07 | |
| CD3 + Cnt | |||
| Low(<median) | 14 | 11.29 ± 1.81 | 0.72 |
| High(>median) | 16 | 13.19 ± 2.38 | |
| CD3 + CD8 + Cnt | |||
| Low(<median) | 15 | 10.13 ± 1.32 | 0.21 |
| High(>median) | 15 | 15.07 ± 2.93 |
P Value < 0.05 was considered statistically significant. Data were available for 71 patients. IL-6: interleukin-6; IL-6 + IL-10: interleukin-6 and interleukin-10 positive;CD3 + Cnt: absolute T lymphocyte count; CD3 + CD8 + Cnt: inhibitory/cytotoxic T lymphocyte absolute count.
4. Discussion
Due to COVID-19’s high mortality, clarifying the risk factors is urgently necessary to develop treatment strategies. WBCs, procalcitonin (PCT), and CRP, markers reflecting the severity of infection [17], were significantly higher in our critically severe patients than in the mild/moderate group. Lymphocytopenia is a unique feature of COVID-19, and low lymphocytes are suggested to be a reference index in the diagnosis [18]. T cells, particularly CD3 + CD8 + cells, play a crucial role by combating pathogens and balancing the risk of developing autoimmunity or overwhelming inflammation [19]. In this study, a substantial decrease in the total number of T lymphocytes and CD3 + CD8 + cells was observed in the severe patients, indicating that Coronavirus consumes many immune cells and inhibits the body's cellular immune function. In SARS, decreased T cells' numbers are strongly correlated with the acute disease phase [20], [21]. The consequences of low T lymphocytes and CD3 + CD8 + cells in COVID-19 are not completely clear; depletion of T cells with antiviral effects may prolong the disease and promote viral survival [22], leading to patient exacerbations [23]. Thus, CD3 + and CD3 + CD8 + T cell Abs could be considered a parameter that reflects the disease severity of COVID-19.
ARDS is associated with the induction of inflammatory cytokines, including IL-1, IL-6, IL-8, CXCL-10, and TNF-α, many of which are highly expressed in the lungs of patients [24], [25]. A high ARDS ratio was found in our COVID-19 patients, especially in the critically severe group. Therefore, elucidating the role of cytokine expression in disease severity is essential. A notable feature of SARS and MERS is that high viral replication leads to higher levels of proinflammatory cytokine production by infected epithelial cells [26], [27]. Although they may be part of a necessary initial immune response to pathogens, exacerbated expression of proinflammatory cytokines and chemokines is associated with immunopathology and ARDS [28]. In our study, too high levels of IL-6 were observed in the critically severe COVID-19 patients compared to those with mild illness. IL-6 is a major player in integrated immunity via its defense against viral infections [29]. However, in some respiratory viral infections, such as influenza and respiratory syncytial virus, elevated IL-6 has been associated with increased lung damage and worse outcomes [29], [30]. In children with pneumonia, IL-6 was correlated with disease severity [31], [32], [33], [34]. In addition to proinflammatory and anti-inflammatory functions, IL-6 enhances mucus production in the airways following allergen exposure [35]. IL-6 can directly induce Muc5AC and Muc5B mucin gene expression in human lung epithelial cell lines in vitro, and there is a possibility that IL-6 may directly affect mucus production by lung epithelial cells during allergic airway inflammation [36]. A positive correlation between IL and 6 and dyspnea was observed in childhood pneumonia, and IL-6 was the only cytokine associated with disease severity [33], [37]. The latest autopsies showed that much mucus accumulated in the pulmonary alveoli and lungs of COVID-19 patients. Excessive mucus production can lead to physical obstruction of smaller caliber airways [38]. A large amount of mucus accumulation in the airways may be the leading cause of dyspnea and respiratory distress in most COVID-19 patients. Significantly higher levels of IL-6 were present in the critically severe patients, together with a correlation between IL and 6 levels and dyspnea, supporting our hypothesis that IL-6 may contribute to mucus production in the lungs and lead to dyspnea. Elucidating the role of IL-6 in COVID-19 pathogenesis will be beneficial for therapy.
In addition to IL-6, high levels of IL-10 were also observed in the critically severe patients. The overall effects of IL-10 on antiviral immune reactions can be complicated and depend on the virus, infection site, antiviral immune response timing, and other factors. High IL-10 could be protective in early reactions to the virus but become detrimental during acute infection as they promote illness persistence [39]. In this study, a close relationship existed between Il and 10 and IL-6, and the patients with both high IL-6 and IL-10 were more likely to have dyspnea and ARDS and shorter OS and EFS, indicating that both cytokines may play a synergistic role in COVID-19 pathogenesis. Our results suggested that IL-6 and IL-10 were valuable parameters for evaluating disease prognosis.
Interferons are potent cytokines of critical importance in controlling viral infections and priming adaptive immune responses [40]. Contrary to the higher levels of IL-6 that existed in the critically severe patients, the IFN-γ level was lower in the critically severe patients than in the mild group. Although IL-6 is a negative regulator of IFN-γ [41], we did not find correlations between IFN-γ and IL-6. The mechanism of lower IFN-γ in severe disease remains to be elucidated. In viral infections, antiviral IFN acts not only to control viral infections but also to program the adaptive immune response to promote viral clearance [42]. CoV reaches high titers very early after infection and harbors multiple proteins that inhibit the IFN response, suggesting that early antagonism of the IFN response might delay or evade the innate immune response. The delayed IFN signaling further orchestrates IMM responses and sensitizes T cells to apoptosis, resulting in the dysregulated inflammatory response [26]. Except for typeⅡ (IFN-γ), typeⅠIFN (IFN-α、IFN-β、IFN-ω) was the other research focus in COVID-19. A recent study report at least 101 of 987 patients with life-threatening COVID-19 pneumonia had neutralizing immunoglobulin G (IgG) autoantibodies (auto-Abs) against type I IFNs. The auto-Abs neutralize the type I IFNs to block SARS-CoV-2 infection in vitro [43].
Despite numerous reports describing the elevation of inflammatory cytokines in primary CoV-infected patients [44], it is unknown if these cytokines are beneficial or contribute to the pathogenicity of infection. Our evidence from patients with critically severe disease suggests the role of cytokine storms in COVID-19. IL-6 and IL-10 may be involved in organ abnormalities and closely related to disease severity. Previous studies revealed that immunomodulation is an essential element in treating severe SARS-CoV diseases and avian influenza viral infections [45], [46], [47]. Unfortunately, there are few effective antiviral treatments for CoV infections, although many drugs have been tested [48]. As an independent risk factor for disease severity, IL-6 might be a potential target for treatment. Now, anti-IL-6 (anti-IL-6 monoclonal antibodies) [49] or IL-6R (tocilizumab, sarilumab, and siltuximab) [50], as well as the steroid (e.g., dexamethasone), has been used in the treatment of COVID-19 in clinical trial or animal experiment. IL-6 or IL-6R targeting inhibitors appears to offer benefits in reducing inflammation, oxygen requirements, vasopressor support, and mortality, while maintaining a good safety profile [51], [52], [53], [54]. However, There is currently no standard therapy to treat cytokine storms [14]. Conversely, how are the effects of the different treatments on cytokines and whether they could be biomarkers to monitor response to treatment. Further studies are needed to confirm this.
5. Conclusion
Significant higher IL-6 and IL-10 were observed in the critically severe COVID-19 patients than the severe and mild patients, which was closely related to dyspnea, ARDS, shorter OS, and EFS. Our results suggested that cytokines and T lymphocytes play a vital role in disease prognosis. Thus, they could be potential targets for treatment. Further studies are needed to confirm cytokines' interactions in COVID-19.
CRediT authorship contribution statement
TYT and XYJ designed and drafted the manuscript. YRL designed and revised the final manuscript. HQP, TYT and YRL collected and summarized the clinical and laboratory data. JYS, FY, YMY and TYT processed the statistical data. GMY summarized all of the data related to the virus. GY, JLF, YBP, ZWZ, SG, FLL, MZ and YWD collected the specimens and conducted the flow cytometry experiments. YRL had full access to all of the data in this study and takes responsibility for the data integrity and the accuracy of the data analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Xiao-Yue Hong, Qiao Ling Deng, Jin Li, Xiao-Juan Wu, and Xue-Ping Qiu for collecting the samples. We appreciate all of our colleagues fighting on the front lines to diagnose and treat COVID-19. We thank all of the patients involved in the study.
Funding
This study was funded by the National Key Research and Development Program of China (Grant No. 2018YFE0204500), National Natural Science Foundation of China (Grant No. 81702273), Science and Technology Key Project of Guangdong Province: Study on the Source and Epidemiology of 2019-nCoV (Grant No. 2020B111107001), Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund, Project znpy2019064 and znpy2018117, Scientific Research Project of COVID-19 Epidemic Prevention and Control at Guangdong University (Grant No. 2020KZDZX1087), and Key Project for Anti-2019 Novel Coronavirus Pneumonia from the National Key Research and Development Program of China (Grant No. 2020YFC0845500).
References
- 1.Patel A., Jernigan D.B., Abdirizak F., Abedi G., Aggarwal S., Albina D., Allen E., Andersen L., Anderson J., Anderson M., Anderson T., Anderson K., Bardossy A.C., Barry V., Beer K., Bell M., Berger S., Bertulfo J., Biggs H., Bornemann J., Bornstein J., Bower W., Bresee J., Brown C., Budd A., Buigut J., Burke S., Burke R., Burns E., Butler J., Cantrell R., Cardemil C., Cates J., Cetron M., Chatham-Stephens K., Chatham-Stevens K., Chea N., Christensen B., Chu V., Clarke K., Cleveland A., Cohen N., Cohen M., Cohn A., Collins J., Dahl R., Daley W., Dasari V., Davlantes E., Dawson P., Delaney L., Donahue M., Dowell C., Dyal J., Edens W., Eidex R., Epstein L., Evans M., Fagan R., Farris K., Feldstein L., Fox LeAnne, Frank M., Freeman B., Fry A., Fuller J., Galang R., Gerber S., Gokhale R., Goldstein S., Gorman S., Gregg W., Greim W., Grube S., Hall A., Haynes A., Hill S., Hornsby-Myers J., Hunter J., Ionta C., Isenhour C., Jacobs M., Slifka K.J., Jernigan D., Jhung M., Jones-Wormley J., Kambhampati A., Kamili S., Kennedy P., Kent C., Killerby M., Kim L., Kirking H., Koonin L., Koppaka R., Kosmos C., Kuhar D., Kuhnert-Tallman W., Kujawski S., Kumar A., Landon A., Lee L., Leung J., Lindstrom S., Link-Gelles R., Lively J., Lu X., Lynch B., Malapati L., Mandel S., Manns B., Marano N., Marlow M., Marston B., McClung N., McClure L., McDonald E., McGovern O., Messonnier N., Midgley C., Moulia D., Murray J., Noelte K., Noonan-Smith M., Nordlund K., Norton E., Oliver S., Pallansch M., Parashar U., Patel A., Patel M., Pettrone K., Pierce T., Pietz H., Pillai S., Radonovich L., Reagan-Steiner S., Reel A., Reese H., Rha B., Ricks P., Rolfes M., Roohi S., Roper L., Rotz L., Routh J., Sakthivel S.K., Sarmiento L., Schindelar J., Schneider E., Schuchat A., Scott S., Shetty V., Shockey C., Shugart J., Stenger M., Stuckey M., Sunshine B., Sykes T., Trapp J., Uyeki T., Vahey G., Valderrama A., Villanueva J., Walker T., Wallace M., Wang L., Watson J., Weber A., Weinbaum C., Weldon W., Westnedge C., Whitaker B., Whitaker M., Williams A., Williams H., Willams I., Wong K., Xie A., Yousef A. Novel Coronavirus Outbreak - United States, December 31, 2019-February 4, 2020. MMWR Morb. Mortal Wkly. Rep. 2020;69(5):140–146. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., Flemban H., Al-Nassir W.N., Balkhy H.H., Al-Hakeem R.F., Makhdoom H.Q., Zumla A.I., Memish Z.A. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet. Infect. Dis. 2013;13(9):752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Cheng V.C.C., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saad M., Omrani A.S., Baig K., Bahloul A., Elzein F., Matin M.A., Selim M.A.A., Mutairi M.A., Nakhli D.A., Aidaroos A.Y.A., Sherbeeni N.A., Al-Khashan H.I., Memish Z.A., Albarrak A.M. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int. J. Infect. Dis.: IJID: Off. Publ. Int. Soc. Infect. Dis. 2014;29:301–306. doi: 10.1016/j.ijid.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefely J.A., Christensen B.B., Gogakos T., Cone Sullivan J.K., Montgomery G.G., Barranco J.P., et al. Marked factor V activity elevation in severe COVID-19 is associated with venous thromboembolism. Am. J. Hematol. 2020;95:1522–1530. doi: 10.1002/ajh.25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meijenfeldt F.A., Havervall S., Adelmeijer J., Lundström A., Magnusson M., Mackman N., Thalin C., Lisman T. Elevated factor V activity and antigen levels in patients with Covid-19 are related to disease severity and 30-day mortality. Am. J. Hematol. 2021;96(4) doi: 10.1002/ajh.v96.410.1002/ajh.26085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin X., Duan Y., Bao T., Gu J., Chen Y., Li Y., Mao S., Chen Y., Xie W., Cox D. The values of coagulation function in COVID-19 patients. PLoS ONE. 2020;15(10):e0241329. doi: 10.1371/journal.pone.0241329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colantuoni A., Martini R., Caprari P., Ballestri M., Capecchi P.L., Gnasso A., Lo Presti R., Marcoccia A., Rossi M., Caimi G. COVID-19 Sepsis and Microcirculation Dysfunction. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Østergaard L. SARS CoV-2 related microvascular damage and symptoms during and after COVID-19: Consequences of capillary transit-time changes, tissue hypoxia and inflammation. Physiol. Reports. 2021;9(3) doi: 10.14814/phy2.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohebbi N., Abedini A., Lashgari R., Razavi F., Varahram M., Kiani A. Vascular microthrombosis associated with increased interleukin-6. A severe acute respiratory distress syndrome in COVID-19 patients treated with tocilizumab. Adv. Respiratory Med. 2020;88(5):468–469. doi: 10.5603/ARM.a2020.0123. [DOI] [PubMed] [Google Scholar]
- 13.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Seminars Immunopathol. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong J.P., Viswanathan S., Wang M., Sun L.-Q., Clark G.C., D'Elia R.V. Current and future developments in the treatment of virus-induced hypercytokinemia. Future Med. Chem. 2017;9(2):169–178. doi: 10.4155/fmc-2016-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CHIEN J.-Y., HSUEH P.-R., CHENG W.-C., YU C.-J., YANG P.-C. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11(6):715–722. doi: 10.1111/j.1440-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Li J., Zhan Y., Wu L., Yu X., Zhang W., Ye L., Xu S., Sun R., Wang Y., Lou J. Analysis of serum cytokines in patients with severe acute respiratory syndrome. Infect. Immun. 2004;72(8):4410–4415. doi: 10.1128/IAI.72.8.4410-4415.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Florin T.A., Ambroggio L. Biomarkers for community-acquired pneumonia in the emergency department. Curr Infect Dis Rep. 2014;16:451. doi: 10.1007/s11908-014-0451-8. [DOI] [PubMed] [Google Scholar]
- 18.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L.i. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cecere T.E., Todd S.M., LeRoith T. Regulatory T cells in arterivirus and coronavirus infections: do they protect against disease or enhance it? Viruses. 2012;4(5):833–846. doi: 10.3390/v4050833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T., Qiu Z., Zhang L., Han Y., He W., Liu Z., Ma X., Fan H., Lu W., Xie J., Wang H., Deng G., Wang A. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J. Infect. Dis. 2004;189(4):648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T., Qiu Z., Han Y., Wang Z., Fan H., Lu W., et al. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin. Med. J. 2003;116:985–987. [PubMed] [Google Scholar]
- 22.Mubarak A., Alturaiki W., Hemida M.G. Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Infection, Immunological Response, and Vaccine Development. J. Immunol. Res. 2019;2019:1–11. doi: 10.1155/2019/6491738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., Gao G.F. T-cell immunity of SARS-CoV: Implications for vaccine development against MERS-CoV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong S.L., Chui P., Lim B., Salto-Tellez M. Elucidating the molecular physiopathology of acute respiratory distress syndrome in severe acute respiratory syndrome patients. Virus Res. 2009;145(2):260–269. doi: 10.1016/j.virusres.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baas T., Taubenberger J.K., Chong P.Y., Chui P., Katze M.G. SARS-CoV virus-host interactions and comparative etiologies of acute respiratory distress syndrome as determined by transcriptional and cytokine profiling of formalin-fixed paraffin-embedded tissues. J. Interferon Cytokine Res.: Off. J. Int. Soc. Interferon Cytokine Res. 2006;26(5):309–317. doi: 10.1089/jir.2006.26.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Channappanavar R., Fehr A., Vijay R., Mack M., Zhao J., Meyerholz D., Perlman S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S., Lipkin W.I. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. mBio. 2015;6(3) doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugin J., Ricou B., Steinberg K.P., Suter P.M., Martin T.R. Proinflammatory activity in bronchoalveolar lavage fluids from patients with ARDS, a prominent role for interleukin-1. Am. J. Respir Crit. Care Med. 1996;153(6):1850–1856. doi: 10.1164/ajrccm.153.6.8665045. [DOI] [PubMed] [Google Scholar]
- 29.Pyle C.J., Uwadiae F.I., Swieboda D.P., Harker J.A., Lopez C.B. Early IL-6 signalling promotes IL-27 dependent maturation of regulatory T cells in the lungs and resolution of viral immunopathology. PLoS Pathog. 2017;13(9):e1006640. doi: 10.1371/journal.ppat.1006640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Gruta N.L., Kedzierska K., Stambas J., Doherty P.C. A question of self-preservation: immunopathology in influenza virus infection. Immunol. Cell Biol. 2007;85(2):85–92. doi: 10.1038/sj.icb.7100026. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes C.D., Arriaga M.B., Costa M.C.M., Costa M.C.M., Costa M.H.M., Vinhaes C.L., Silveira-Mattos P.S., Fukutani K.F., Andrade B.B. Host Inflammatory Biomarkers of Disease Severity in Pediatric Community-Acquired Pneumonia: A Systematic Review and Meta-analysis. Open Forum Infect. Dis. 2019;6(12) doi: 10.1093/ofid/ofz520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saghafian-Hedengren S., Mathew J.L., Hagel E., Singhi S., Ray P., Ygberg S., Nilsson A. Assessment of Cytokine and Chemokine Signatures as Potential Biomarkers of Childhood Community-acquired Pneumonia Severity: A Nested Cohort Study in India. Pediatr. Infect. Dis. J. 2017;36(1):102–108. doi: 10.1097/INF.0000000000001364. [DOI] [PubMed] [Google Scholar]
- 33.de Brito R.C., Lucena-Silva N., Torres L.C., Luna C.F., Correia J.B., da Silva G.A. The balance between the serum levels of IL-6 and IL-10 cytokines discriminates mild and severe acute pneumonia. BMC Pulmonary Med. 2016;16:170. doi: 10.1186/s12890-016-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haugen J., Chandyo R.K., Brokstad K.A., Mathisen M., Ulak M., Basnet S., Valentiner-Branth P., Strand T.A., Cormier S.A. Cytokine Concentrations in Plasma from Children with Severe and Non-Severe Community Acquired Pneumonia. PLoS ONE. 2015;10(9):e0138978. doi: 10.1371/journal.pone.0138978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neveu W.A., Allard J.B., Dienz O., Wargo M.J., Ciliberto G., Whittaker L.A., Rincon M. IL-6 is required for airway mucus production induced by inhaled fungal allergens. J. Immunol. 2009;183(3):1732–1738. doi: 10.4049/jimmunol.0802923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Thai P., Zhao Y.-H., Ho Y.-S., DeSouza M.M., Wu R. Stimulation of airway mucin gene expression by interleukin (IL)-17 through IL-6 paracrine/autocrine loop. J. Biol. Chem. 2003;278(19):17036–17043. doi: 10.1074/jbc.M210429200. [DOI] [PubMed] [Google Scholar]
- 37.Antunes G., Evans S.A., Lordan J.L., Frew A.J. Systemic cytokine levels in community-acquired pneumonia and their association with disease severity. Europ. Respiratory J. 2002;20(4):990–995. doi: 10.1183/09031936.02.00295102. [DOI] [PubMed] [Google Scholar]
- 38.Rogers D. Airway mucus hypersecretion in asthma: an undervalued pathology? Curr. Opin. Pharmacol. 2004;4(3):241–250. doi: 10.1016/j.coph.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Naicker D., Werner L., Kormuth E., Passmore J.-A., Mlisana K., Karim S., Ndung’u T. Interleukin-10 promoter polymorphisms influence HIV-1 susceptibility and primary HIV-1 pathogenesis. J. Infect Dis. 2009;200(3):448–452. doi: 10.1086/600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Bon A., Tough D.F. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 2002;14(4):432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 41.McLoughlin R.M., Witowski J., Robson R.L., Wilkinson T.S., Hurst S.M., Williams A.S., et al. Interplay between IFN-gamma and IL-6 signaling governs neutrophil trafficking and apoptosis during acute inflammation. J. Clin. Investig. 2003;112:598–607. doi: 10.1172/JCI17129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cervantes-Barragán L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., Ludewig B. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 2009;182(2):1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- 43.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu M. SARS Immunity and Vaccination. Cell Mol. Immunol. 2004;1:193–198. [PubMed] [Google Scholar]
- 45.Cheung C.Y., Poon L.L.M., Ng I.H.Y., Luk W., Sia S.-F., Wu M.H.S., Chan K.-H., Yuen K.-Y., Gordon S., Guan Y.i., Peiris J.S.M. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79(12):7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng B.-J., Chan K.-W., Lin Y.-P., Zhao G.-Y., Chan C., Zhang H.-J., Chen H.-L., Wong S.S.Y., Lau S.K.P., Woo P.C.Y., Chan K.-H., Jin D.-Y., Yuen K.-Y. Delayed antiviral plus immunomodulator treatment still reduces mortality in mice infected by high inoculum of influenza A/H5N1 virus. PNAS. 2008;105(23):8091–8096. doi: 10.1073/pnas.0711942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo P.CY., Lau S.KP., Li K.SM., Tsang A.KL., Yuen K.-Y. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerging Microbes Infect. 2012;1(1):1–5. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mo Y., Fisher D. A review of treatment modalities for Middle East Respiratory Syndrome. J. Antimicrob. Chemother. 2016;71(12):3340–3350. doi: 10.1093/jac/dkw338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubsamen R., Burkholz S., Massey C., Brasel T., Hodge T., Wang L.u., Herst C., Carback R., Harris P. Anti-IL-6 Versus Anti-IL-6R Blocking Antibodies to Treat Acute Ebola Infection in BALB/c Mice: Potential Implications for Treating Cytokine Release Syndrome. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.57470310.3389/fphar.2020.574703.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corominas H., Castellví I., Domingo P., Casademont J. Facing the SARS-CoV-2 (COVID-19) outbreak with IL-6R antagonists. Europ. J. Rheumatol. 2020;7(Supp2):S107–S109. doi: 10.5152/eurjrheum.2020.20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordan S.C., Zakowski P., Tran H.P., Smith E.A., Gaultier C., Marks G., Zabner R., Lowenstein H., Oft J., Bluen B., Le C., Shane R., Ammerman N., Vo A., Chen P., Kumar S., Toyoda M., Ge S., Huang E. Compassionate Use of Tocilizumab for Treatment of SARS-CoV-2 Pneumonia. Clin. Infect. Dis.: Off. Publ. Infect. Dis. Soc. Am. 2020;71(12):3168–3173. doi: 10.1093/cid/ciaa812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corominas H., Castellví I., Pomar V., Antonijoan R., Mur I., Matas L., Gich I., de Benito N., Laiz A., Castillo D., Villamarin L., Filella D., Millán A.M., Quijada M.Á., Puig M., Casademont J., Domingo P. Effectiveness and safety of intravenous tocilizumab to treat COVID-19-associated hyperinflammatory syndrome: Covizumab-6 observational cohort. Clin. Immunol. 2021;223:108631. doi: 10.1016/j.clim.2020.108631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gremese E., Cingolani A., Bosello S.L., Alivernini S., Tolusso B., Perniola S., Landi F., Pompili M., Murri R., Santoliquido A., Garcovich M., Sali M., De Pascale G., Gabrielli M., Biscetti F., Montalto M., Tosoni A., Gambassi G., Rapaccini G.L., Iaconelli A., Zileri Del Verme L., Petricca L., Fedele A.L., Lizzio M.M., Tamburrini E., Natalello G., Gigante L., Bruno D., Verardi L., Taddei E., Calabrese A., Lombardi F., Bernabei R., Cauda R., Franceschi F., Landolfi R., Richeldi L., Sanguinetti M., Fantoni M., Antonelli M., Gasbarrini A. Sarilumab use in severe SARS-CoV-2 pneumonia. EClinicalMedicine. 2020;27:100553. doi: 10.1016/j.eclinm.2020.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J.J., Zhang L.N., Hou H., Xu L., Ji K. Interleukin-6 signaling blockade treatment for cytokine release syndrome in COVID-19 (Review) Exp. Therap. Med. 2021;21:24. doi: 10.3892/etm.2020.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]