Abstract
BACKGROUND
Paracoccidioidomycosis (PCM) is a systemic mycosis with high prevalence in Latin America that is caused by thermodimorphic fungal species of the Paracoccidioides genus.
OBJECTIVES
In this study, we used quantitative polymerase chain reaction (qPCR) to investigate the expression of genes related to the virulence of Paracoccidioides brasiliensis (Pb18) and P. lutzii (Pb01) strains in their mycelial (M) and yeast (Y) forms after contact with alveolar macrophages (AMJ2-C11 cell line) and fibroblasts (MRC-5 cell line).
METHODS
The selected genes were those coding for 43 kDa glycoprotein (gp43), enolase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 14-3-3 protein (30 kDa), phospholipase, and aspartyl protease.
FINDINGS
In the Pb18 M form, the aspartyl protease gene showed the highest expression among all genes tested, both before and after infection of host cells. In the Pb18 Y form after macrophage infection, the 14-3-3 gene showed the highest expression among all genes tested, followed by the phospholipase and gp43 genes, and their expression was 50-fold, 10-fold, and 6-fold higher, respectively, than that in the M form. After fibroblast infection with the Pb18 Y form, the 14-3-3 gene showed the highest expression, followed by the phospholipase and aspartyl protease genes, and their expression was 25-fold, 10-fold, and 10-fold higher, respectively, than that in the M form. Enolase and aspartyl protease genes were expressed upon infection of both cell lines. After macrophage infection with the Pb01 Y form, the 14-3-3 gene showed the highest expression, followed by the phospholipase and aspartyl protease genes, and their expression was 18-fold, 12.5-fold, and 6-fold higher, respectively, than that in the M form.
MAIN CONCLUSIONS
In conclusion, the data show that the expression of the genes analysed may be upregulated upon fungus-host interaction. Therefore, these genes may be involved in the pathogenesis of paracoccidioidomycosis.
Key words: Paracoccidioides spp., adhesin genes, virulence factors, M form, Y form
Paracoccidioidomycosis (PCM) is a systemic granulomatous disease caused by a complex group of thermally dimorphic fungi within the genus Paracoccidioides. This disease is endemic in Latin America, and although it is not classified as a neglected tropical disease (NTD) by the World Health Organization (WHO), it meets all NTD criteria. 1 Currently, at least 10 million people are infected. 2 , 3 , 4 PCM may affect any organ in the body, predominantly the lungs, which are organs rich in cells of the mononuclear phagocyte system. 5 , 6 , 7 Even after successful antifungal treatment, most patients with a chronic form have sequelae, including pulmonary fibrosis. Therefore, these patients have respiratory problems that often hinder them from exercising their professions and activities. 8 , 9
Geographical evidence and genetic variability suggest that Paracoccidioides spp. could be composed of several genetic groups. 4 The disease may occur in areas that extend from Mexico to Argentina, and the species currently recognised are Paracoccidioides brasiliensis, P. americana, P. restrepiensis, P. venezuelensis, and P. lutzii. 10
P. brasiliensis, the most studied species, has been considered a facultative intracellular organism that can interact with epithelial cells and macrophages. 11 Macrophages constitute one of the primary defense mechanisms against infection by this fungus. 2 , 12 , 13 Phagocytes recognise the fungal cell wall, which is comprised of pathogen-associated molecular patterns (PAMPs) that are not synthesised by the host. 14 , 15 The surface molecules of the organism are involved in strategies to evade the host’s immune system and ensure survival. This is closely associated with transcriptional control involving several regulatory pathways. 14
Various components of Paracoccidioides spp., such as adhesins and others, may play a relevant role in pathogenesis. 16 , 17 , 18 , 19 , 20 , 21 de Oliveira et al. 22 evaluated the gene expression of different adhesins, including 14-3-3 protein, GP43, enolase (ENO), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), triosephosphate isomerase (TPI), and malate synthase (MLS), after mice infection with P. brasiliensis and P. lutzii. 22 The study found differences among the two species, and that 14-3-3 and ENO showed the highest expression. Other virulence factors include proteinases and phospholipases. 23 Transcriptome analysis of P. brasiliensis yeast cells (Y form) derived from infected mice 24 , 25 and after incubation in human blood and plasma 26 , 27 revealed a positively regulated serine protease transcript.
This serine proteinase (gi734682187), previously characterised during fungal growth upon nitrogen deprivation, 25 was accumulated in Y cells after lung infection, probably enabling the pathogen to enter lung tissue and spread. 28 , 29 , 30 , 31 P. brasiliensis interaction with pneumocytes and keratinocytes in vitro may be correlated with pulmonary fungal entry events and dissemination. 32 , 33 , 34 Thus, the study of the interaction of Paracoccidioides spp. with lung cells and macrophages in vitro may constitute a fundamental approach for understanding the initial stages of this fungal infection.
The majority of the studies on fungal-host interactions have been performed on the Y form of the fungus. 21 , 35 , 36 , 37 However, to study the early stage of the disease and fungal adaptation to the host’s conditions, it is best to comparatively study the M and Y forms. Thus, this study aimed to compare virulence gene expression between the M and Y forms of P. brasiliensis (Pb18) and P. lutzii (Pb01), using quantitative polymerase chain reaction (qPCR) before and after infection of alveolar macrophages and human lung fibroblasts.
MATERIALS AND METHODS
Isolates - P. brasiliensis 18 (ATCC 32069) and P. lutzii 01 (ATCC MYA-826) from the collection of the Laboratory of Clinical Mycology, Faculty of Pharmaceutical Sciences of UNESP, Araraquara (Brazil) were used in the M and Y forms. Both species were grown in Fava-Netto’s semi-solid medium at 23ºC (M form) and 37ºC (Y form).
Cell culture - AMJ2-C11 (mouse alveolar macrophages) and MRC-5 (human lung fibroblasts) cell lines were obtained from the Cell Bank of Rio de Janeiro and cultured in HAM-F12 medium at 37ºC and 5% CO2.
Culture of macrophages and lung fibroblasts - Macrophages and lung fibroblasts were grown in plastic bottles containing Dulbecco’s Modified Eagle Medium (DMEM, Sigma-Aldrich, Brazil) supplemented with foetal bovine serum (FBS), 100 µg/mL streptomycin, and 100 U/mL penicillin (Gibco BRL, Life Technologies, Rockville, MD, USA), and maintained at 36.5ºC. After three to four days, the bottles were subjected to trypsinisation. For this, the monolayer was first washed with 1.0 mL sterile phosphate-buffered saline (PBS, 0.05 M, pH 7.2), followed by addition of 1 mL ATV solution (0.2% trypsin and 0.02% EDTA - Sigma®). After 1-2 min, the cells were homogenised with varying amounts of the corresponding medium supplemented with FBS. In this step, ATV was neutralised by the FBS present in the culture medium. The obtained cell suspension was transferred to other cylinders, resulting in a cell concentration of 106 cells/mL.
Preparation and standardisation of the inocula - P. brasiliensis isolate 18 (ATCC 32069) and P. lutzii strain 01 (ATCC-MYA-826) were maintained on the solid culture medium Fava-Netto (Fava Netto) at 36ºC for the yeast form and at 23ºC for the mycelium form during five and 15 days, respectively. The cell viability was assessed by using trypan blue in a hemocytometer chamber. The preparation of the inoculum was the same as used in the M and Y form. The Paracoccidioides spp. growth was washed three times with PBS buffer after the mycelium was fragmented by glass beads using a vortex and filtered through glass wool. Besides, the cell clumps were separated by repeated passages through an 18-gauge needle (Becton Dickinson, Brazil) into a 5 mL syringe (Becton Dickinson, Brazil). The suspension was filtered through 40 µm nylon cell strainers (Corning, USA) to obtain cells of homogeneous sizes. Enough sterile PBS and fungus were employed to obtain a homogeneous turbid suspension, corresponding to 1 x 105 cells/mL, according to the MacFarland scale and adjusted by spectrophotometric reading [optical density (OD) = 0.5].
Infection of human lung fibroblasts and mouse alveolar macrophages by P. brasiliensis and P. lutzii - Y and M form of both species was employed for infection assays. After the formation of the cell monolayer, the culture medium was discarded, and 3 ml of inoculum suspension was added. The bottles containing infected lung cells were incubated with the suspension for 5 h, and bottles containing macrophages were incubated for 6 hours. A total of 1 x 105 cells/mL (Y or M form) were added to the cells to obtain a yeast/macrophage or yeast/fibroblast ratio of 1:1. After the infection period, the cells were removed with a scraper (scraper) and transferred to a Falcon®. The cells were washed 3x with PBS and stored at - 80ºC for later RNA extraction.
Differential gene expression analysis by quantitative PCR
Extraction of total RNA - In independent experiments, fungal cells were isolated and macerated after freezing with liquid nitrogen in a Falcon tube containing 3 mL Trizol and glass beads. The sample was shaken vigorously by vortexing for 15 min and, after 10 min at rest, centrifuged at 10,000 g for 10 min. The supernatant was transferred to a new tube and 200 µL chloroform was added for each 750 µL of the recovered Trizol. The tube was shaken vigorously, left to stand for 10 min, and centrifuged at 10,000 g for 15 min. Then, the supernatant was transferred to a new tube and 250 µL isopropanol, 250 µL of a 0.4 M sodium citrate solution, and 0.8 M sodium chloride were added to 750 µL Trizol. The samples were stirred gently and allowed to stand for 10 min at room temperature. Immediately after centrifugation at 10,000 g for 30 min, the supernatant was discarded, and the pellet was washed with 75% ethanol and centrifuged at 10,000 g for 5 min. The supernatant was again discarded, and the precipitate was dried in SpeedVac equipment. After drying, the pellet was re-suspended in 40 µL of a 0.01% diethyl pyrocarbonate (DEPC) solution in H2O, and the samples were stored at -80ºC. 38 , 39
Synthesis of cDNA - The synthesis of the first cDNA tape was made in a reaction containing 1 µg total RNA and 10 pmol/μL primer (5’-AAGCAGTGGTATCAACGCAGAGTACGCGGG-3’), and the mixture was heated for 2 min at 72ºC. Then, 2 μL of a mixture of dNTPs (10 mM), 2 μL of RNaseOUT (40 U/μL) (Invitrogen, Carlsbad, California, EUA), 0.4 µL DTT (100 mM), 1.2 µL magnesium chloride (50 mM), 2 μL reverse transcriptase (200 U/μL) (Superscript II RT - Invitrogen), and 4 μL buffer 5X (250 mM Tris-HCl, 375 mM KCl, 15 mM MgCl2, pH 8.3) were added. This reaction was incubated at 42ºC for 1 h and 30 min. Thereafter, 80 μL TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) was added and the mixture was heated for 10 min at 72ºC. 40
Quantitative PCR - Differential gene expression between the Y and M forms of both species after macrophage and fibroblast infection was examined using qPCR with specific primers designed using Primer 3 software (Table).
TABLE. Specific primers used for the quantitative polymerase chain reaction (qPCR) assay.
| Primers | |
| gp43 | Sense 5´-CTTGTCTGGGCCAAAAACTC-3´ |
| Antisense 5´-GCCAGGGTTTGTTTGACTGT-3´ | |
| Enolase | Sense 5´-TAGGCACCCTCACTGAATCC-3´ |
| Antisense 5´-GCTCTCAATCCCACAACGAT-3´ | |
| Phospholipases | Sense 5´-TGTTGGTGCGATCAAAAGAC-3 |
| Antisense 5´-GGATACGACGTCGCCACTAT-3 | |
| Aspartyl protease | Sense 5´-AAAGGAAACACGGAAACACG-3 |
| Antisense 5´-CGTTCCTGAGACGGTGGTAT-3 | |
| 14-3-3 (30 kDa) | Sense 5´GTTCGCTCTTGGAGACAAGC-3´ |
| Antisense 5´AGCAACCTCAGTTGCGTTCT-3´ | |
| GAPDH | Sense 5´-AAATGCTGTTGAGCACGATG-3´ |
| Antisense 5´-CTGTGCTGGATATCGCCTTT-3´ |
The reactions were performed using 1 µL cDNA, 10 µL of maximum SYBR Green/ROX qPCR Master Mix (2X) (Thermo Scientific, Waltham, Massachusetts, EUA), and 0.5 mM of each primer, to which water was added until the reaction volume was 20 µL. The reactions were performed at an initial temperature of 50ºC for 2 min, followed by 10 min at 95ºC and 40 cycles at 95ºC for 15 s, and annealing and synthesis at 60ºC for one min. PCR analysis of the melting curve was performed to confirm the presence of a single specific product. The reactions were performed in triplicate with an Applied Biosystems 7500 cycler machine. Variations in mRNA expression were calculated using the 2-ΔCT formula, where ΔCT is the difference between the targets and the reference gene β-tubulin, following previous studies performed by several authors.
Statistical analysis - All data were subjected to the Shapiro-Wilk test to check for normal distribution. Differences in gene expression between the M and Y forms of Pb18 and Pb01 after macrophage and lung fibroblast infection was assessed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparison tests and a significance level of 5%.
RESULTS
qPCR assay - The expression of the gp43 gene in the Pb18 Y form was significantly higher after macrophage infection than that after lung fibroblast infection (p < 0.05). The results were similar when the M form of the fungus was used, although the expression was lower than that in the Y form (Fig. 1). In contrast, the Pb01 M form showed higher expression of the gp43 gene after lung fibroblast infection than after macrophage infection, although with a ten-fold reduction.
Fig. 1: evaluation of gp43 gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01). Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
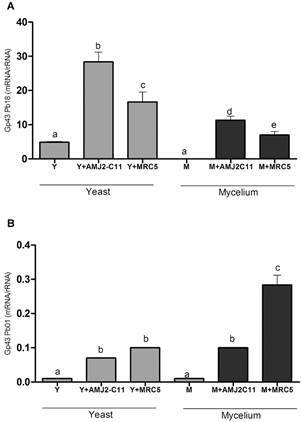
Although there was no difference in ENO gene expression between the Pb18 M and Y forms after infection of either macrophages or fibroblasts, the expression was higher after macrophage infection than after fibroblast infection (Fig. 2). ENO gene expression was lower in Pb01 than in Pb18. In Pb01, the highest expression was found after fibroblast infection with the Y form (p < 0.05).
Fig. 2: evaluation of ENO gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01) Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
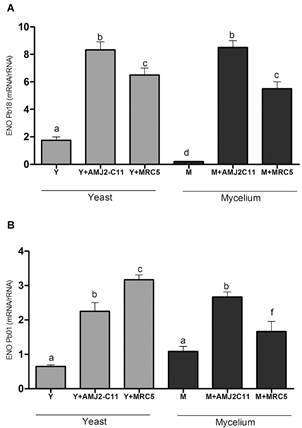
Phospholipase gene expression after macrophage infection was considerably higher in the Y form than that in the M form for both species. Phospholipase gene expression in Pb01 was similar to that in Pb18 when comparing the M forms, and lower when comparing the Y forms (Fig. 3). The highest expression was found in the Y forms of both species after macrophage infection. In contrast, the lowest expression was found in the M forms of both species after fibroblast infection.
Fig. 3: evaluation of phospholipase gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01). Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
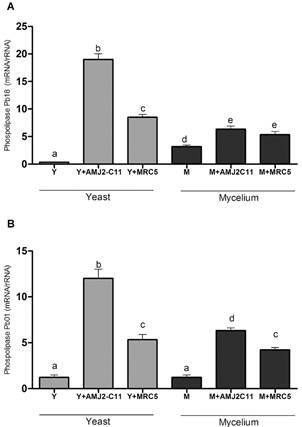
The Pb18 M form showed significantly higher expression of the aspartyl protease gene than the Y form (p < 0.05). The highest expression of this enzyme was found after macrophage infection with the Y forms (p < 0.05). We found the same level of aspartyl protease gene expression in both forms of Pb01. Fibroblasts infected with Pb01 in its Y form showed the highest expression of this enzyme, whereas, in macrophages, the levels were reduced (Fig. 4).
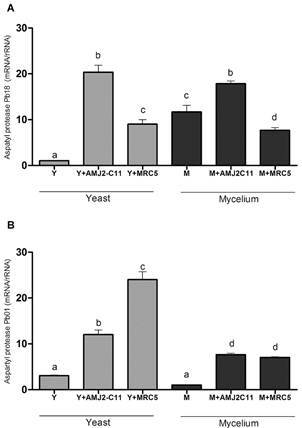
Fig. 4: evaluation of aspartyl protease gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01). Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
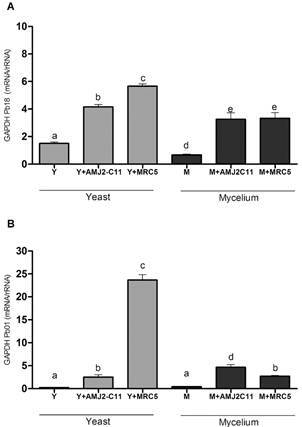
GAPDH presented the lowest expression in Pb 18 and Pb 01 in the M and Y forms, whereas this adhesin level was lower in Pb18 infected with both cells. GAPDH gene expression was higher in fibroblasts infected with the Pb01 Y form. Comparatively, the M form showed very reduced levels of GAPDH gene expression (Fig. 5).
Fig. 5: evaluation of GAPDH gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01). Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
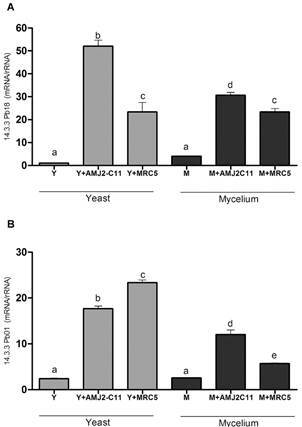
The highest expression of the 14-3-3 gene was found after macrophage infection with the Pb18 Y form, and the expression was 50% lower in the M form (Fig. 6). This gene had the highest relative level compared to the other genes evaluated. For Pb01, the highest 14-3-3 expression was found after fibroblast infection with the Y form.
Fig. 6: evaluation of 14-3-3 gene expression. Quantitative polymerase chain reaction (qPCR) for (A) Paracoccidioides brasiliensis (Pb18) and (B) P. lutzii (Pb01). Y form (Y), infected macrophages (Y + AMJ2-C11), infected lung fibroblasts (Y + MRC-5); M form (M), infected macrophages (M + AMJ2-C11), infected lung fibroblasts (M + MRC-5). Analysis of variance (ANOVA) with Tukey’s multiple comparison tests and significance at p < 0.05. *: different letters denote statistical significant.
DISCUSSION
The Paracoccidioides genus is composed of thermally dimorphic fungi, and the transition from the M to the Y form is essential for disease in humans. Nevertheless, the molecular mechanisms that control this transition still deserve attention, primarily during fungus-host interaction. In the present study, we observed different expression of genes that code for antigens, adhesins, or other potential virulence factors, after macrophage and fibroblast infection with the two forms of two Paracoccidioides species.
These experiments can help us understand the mechanisms involved in PCM and the virulence factors of these species. Virulence factors are essential for establishing an invasive fungal infection, including dimorphism, host cell adhesion, cell wall composition, and hydrolytic enzyme production. 16 , 17 , 18 , 19 , 20 , 21 , 40 Another aspect of this disease is pulmonary fibrosis, which is characterised by excessive deposition of collagen and extracellular matrix components that results in pathological remodeling of the pulmonary architecture. 40
In this study, the expression of six genes in the Y and M forms of Pb18 and Pb01 after infection of two different cell types was determined using qPCR. First, the highest expression found in the Pb18 M form corresponded to that of the aspartyl protease gene. In contrast, Pb01 expressed relatively few genes and lower levels compared with Pb18.
Among the genes expressed in the Pb18 Y form after macrophage infection, the 14-3-3 gene showed the highest expression, followed by the phospholipase and gp43 genes, and their expression was 50-fold, 10-fold, and 6-fold higher, respectively, than that in the M form. After fibroblast infection with the Y form, the 14-3-3 gene showed the highest expression, followed by the phospholipase and aspartyl protease genes, and their expression was 25-fold, 10-fold, and 10-fold higher, respectively, than that in the M form. The contact of macrophages and fibroblast cells with the Pb18 M form resulted in a similar pattern, with the 14-3-3 gene showing the highest expression, followed by the gp43 gene. ENO and aspartyl protease genes were also expressed after infection of both cell types, and the latter showed higher expression after fibroblast infection. The 14-3-3 gene was not expressed in the M and Y forms of both species.
After infection of macrophages with the Pb01 Y form, 14-3-3 showed the highest expression, followed by the phospholipase and aspartyl genes, and their expression was 18-fold, 12.5-fold, and 6-fold higher than that in the M form. The GADPH, 14-3-3, and aspartyl protease genes showed higher expression levels after fibroblast infection. In general, the 14-3-3 gene showed the highest expression under all conditions, except after fibroblast infection with the M form, where the aspartyl protease gene showed higher expression.
Based on these results, Paracoccidioides adhesins are essential for the establishment of infection. They may interact with host cells and establish the colonisation and dissemination of fungi in the host organism, 17 , 41 interacting with components of the host extracellular matrix (ECM) and lung cells. 41 Most adhesins are enzymes of the glycolytic pathway, tricarboxylic acid (TCA), and glyoxylate cycle of Paracoccidioides spp. They have adhesive properties that facilitate the interaction with the host ECM and act as ‘moonlighting’ proteins. 35
The 14-3-3 proteins are expressed in most eukaryotic organisms and play varied and crucial roles in a wide range of regulatory processes. These proteins regulate diverse signaling pathways involved in cell survival, cell cycle, and differentiation, affecting their functions via interactions with phosphorylated serine/threonine residues. In P. brasiliensis, 14-3-3 was first identified as an overexpressed 30 kDa protein after challenging mice with P. brasiliensis. 42 , 43 Furthermore, Pb 14-3-3 plays an essential role in the adhesion process and is a critical early factor in the development and subsequent progression of invasive infection. Recently, de Oliveira et al. 22 carried out an in-depth study to elucidate the importance of different adhesins in the virulence of Paracoccidioides, and 14-3-3 along with ENO were the most highly expressed adhesins during host-pathogen interactions, which partially supports the results presented in this work. Marcos et al. 35 demonstrated that downregulation of Pb 14-3-3 expression impairs cell morphology and essential features related to dimorphism, decreases the interaction with pneumocytes, and attenuates the virulence of Paracoccidioides spp. in the Galleria mellonella model.
As described earlier by our group, this gene is expressed during PCM infection. 43 Our data in this work also showed high expression of 14-3-3, especially in macrophages infected with the Pb18 Y form. Therefore, this adhesin could be extremely important as a marker of infection. In general, 14-3-3 proteins seem to promote cell survival by binding and sequestering proteins that otherwise activate signaling pathways involved in apoptosis initiation. Indeed, many studies have demonstrated the effects of 14-3-3 proteins on cell death and apoptosis. 44 , 45
Gp43 acts as an adhesin, favoring the adhesion of P. brasiliensis to host epithelial cells. 16 Studies have reported that GP43 is an immunodominant antigen in P. brasiliensis. A modified peptide derived from this molecule has been designed as a vaccine to prevent disease. 46 , 47 In our study, the gp43 gene showed the highest expression after macrophage infection with the Pb18 Y form. Considering that macrophages are fundamental in controlling fungal infections, the gp43 gene may be involved in a fungal evasion mechanism. According to Popi et al., 48 GP43 impairs the phagocytosis of P. brasiliensis and inhibits the release of reactive oxygen and nitrogen intermediates involved in the microbicidal activity of macrophages.
Our results also showed significantly lower expression levels of the gp43 gene in Pb01 alone or in contact with both cell types. Studies have demonstrated variation in the gp43 production rate of isolates of different Paracoccidioides species, and generally, P. lutzii showed low expression. 43 , 49 There are possible differences in the promoter region of this gene among the different isolates. In addition, there may be interference in post-transcriptional processes, such as RNA interference, which may influence this variability. 50
Another important and well-described enzyme in Paracoccidioides spp. is GAPDH, a key molecule of the glycolytic pathway playing a crucial role in catabolism and carbohydrate anabolism. 51 In this study, the GAPDH gene showed higher expression in macrophages infected with the Pb01 Y form. Studies performed by Sardi and collaborators. 19 demonstrated a significant increase in the expression of this gene in P. brasiliensis biofilm. Barbosa et al. 51 found that the GAPDH of P. brasiliensis is also located in the cell wall. Thus, it is probably a common receptor of proteins of the extracellular matrix, playing a crucial role in the adhesion of P. brasiliensis to host tissues. During copper (Cu) starvation of P. lutzii cultures, the expression of GADPH remains at basal levels. However, contact with the host signals the fungus to increase the expression of this protein for adherence to the host, and to obtain the necessary Cu from the host for maintenance. With these results, we can understand how the fungus uses its protein arsenal to adapt to the host and succeed during the infection process. 52
In our study, the expression of the phospholipase gene was greater after macrophage infection with the Y forms than with the M forms for both species. Phospholipase has been proposed as one of the strategies used by pathogenic bacteria, parasites, and fungi to invade host tissues and establish infection, 18 , 19 , 53 which would explain the low expression of phospholipase in situations where the fungus is not in contact with cells. Phospholipases are ubiquitous enzymes involved in a wide range of biological functions, such as membrane homeostasis, nutrient acquisition, and the generation of bioactive molecules. These enzymes are known to contribute to bacterial and fungal virulence through various interactions with eukaryotic host cells. 19 , 53
Another virulence factor addressed in this study was aspartyl protease because proteases from several families are associated with the virulence of human pathogens and play an essential role in the host invasion process of many human fungal pathogens. In our study, this enzyme was expressed in the Pb18 M form. The enzyme increase may be related to fungus pathogenesis. Parente et al. 25 studied a serine protease and demonstrated that the expression of coding transcripts is induced during the internalisation of P. brasiliensis by macrophages.
In conclusion, this study shows that the expression of the genes analysed may increase upon fungal-cell interaction. Therefore, they may be involved in the pathogenesis of paracoccidioidomycosis.
REFERENCES
- 1.Griffiths J, Colombo AL, Denning DW. The case for paracoccidioidomycosis to be accepted as a neglected tropical (fungal) disease. PLoS Negl Trop Dis. 2019;13(5):e0007195. doi: 10.1371/journal.pntd.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummer E, Castaneda E, Restrepo A. Paracoccidioidomycosis an update. Clin Microbiol Rev. 1993;6(2):89–17. doi: 10.1128/cmr.6.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez R. Epidemiology of paracoccidioidomycosis. Rev Inst Med Trop São Paulo. 2015;57(19):11–20. doi: 10.1590/S0036-46652015000700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turissini DA, Gomez OM, Teixeira MM, McEwen JG, Matute DR. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet Biol. 2017;106:9–25. doi: 10.1016/j.fgb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Almeida SM, Salvador GLO, Roza TH, Izycki LF, Dos Santos I, Aragão A. Geographical evaluation of neuroparacoccidioidomycosis and paracoccidioidomycosis in Southern Brazil. Mycoses. 2018;61(8):587–593. doi: 10.1111/myc.12782. [DOI] [PubMed] [Google Scholar]
- 6.Mendes RP, Cavalcante RS, Marques SA, Marques MEA, Venturini J, Sylvestre TF. Paracoccidioidomycosis current perspectives from Brazil. Open Microbiol J. 2017;31(11):224–282. doi: 10.2174/1874285801711010224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez R. New trends in paracoccidioidomycosis epidemiology. J Fungi (Basel) 2017;3(1):1–1. doi: 10.3390/jof3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa TA, Bazan SB, Feriotti C, Araújo EF, Bassi Ê, Loures FV. In pulmonary paracoccidioidomycosis IL-10 deficiency leads to increased immunity and regressive infection without enhancing tissue pathology. PLoS Negl Trop Dis. 2013;7(10):e2512. doi: 10.1371/journal.pntd.0002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques SA. Paracoccidioidomycosis epidemiological, clinical, diagnostic and treatment up-dating. An Bras Dermatol. 2013;88(5):700–711. doi: 10.1590/abd1806-4841.20132463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hrycyk MF, Garces HG, Bosco SMG, de Oliveira SL, Marques SA, Bagagli E. Ecology of Paracoccidioides brasiliensis, P lutzii and related species: infection in armadillos, soil occurrence and mycological aspects. Med Mycol. 2018;56(8):950–962. doi: 10.1093/mmy/myx142. [DOI] [PubMed] [Google Scholar]
- 11.Mendes-Giannini MJ, da Silva JLM, da Silva JF, Donofrio FC, Miranda ET, Andreotti PF. Interactions of Paracoccidioides brasiliensis with host cells recent advances. Mycopathologia. 2008;165(4-5):237–248. doi: 10.1007/s11046-007-9074-z. [DOI] [PubMed] [Google Scholar]
- 12.Franco M. Host-parasite relationships in paracoccidioidomycosis. J Med Vet Mycol. 1987;25(1):05–18. doi: 10.1080/02681218780000021. [DOI] [PubMed] [Google Scholar]
- 13.Benard G. An overview of the immunopathology of human paracoccidioidomycosis. Mycopathologia. 2008;165(4-5):209–221. doi: 10.1007/s11046-007-9065-0. [DOI] [PubMed] [Google Scholar]
- 14.Hernández-Chávez MJ, Pérez-García LA, Niño-Vega GA, Mora-Montes HM. Fungal strategies to evade the host immune recognition. J Fungi (Basel) 2017;3(4):51–51. doi: 10.3390/jof3040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Díaz-Jiménez DF, Pérez-García LA, Martínez-Álvarez JA, Mora-Montes HM. Role of the fungal cell wall in pathogenesis and antifungal resistance. Curr Fungal Infect Rep. 2012;6:275–282. [Google Scholar]
- 16.San-Blas G, Travassos LR, Fries BC, Goldman DL, Casadevall A, Carmona AK. Fungal morphogenesis and virulence. Med Mycol. 2000;38(1):79–86. [PubMed] [Google Scholar]
- 17.Mendes-Giannini MJ, Taylor ML, Bouchara JB, Burger E, Calich VL, Escalante ED. Pathogenesis II fungal responses to host responses: interaction of host cells with fungi. Med Mycol. 2000;38(1):113–123. [PubMed] [Google Scholar]
- 18.Soares DA, Oliveira MB, Evangelista AF, Venancio EJ, Andrade RV, Felipe MSS. Phospholipase gene expression during Paracoccidioides brasiliensis morphological transition and infection. Mem Inst Oswaldo Cruz. 2013;108(6):808–811. doi: 10.1590/0074-0276108062013021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardi JC, Pitangui NS, Voltan AR, Braz JD, Machado MP, Almeida AMF. In vitro Paracoccidioides brasiliensis biofilm and gene expression of adhesins and hydrolytic enzymes. Virulence. 2015;6(6):642–651. doi: 10.1080/21505594.2015.1031437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cattana ME, Tracogna MF, Marques I, Rojas F, Fernández M. de Los Ángeles Sosa M In Vivo Paracoccidioides sp biofilm on vascular prosthesis. Mycopathologia. 2017;182(7-8):747–749. doi: 10.1007/s11046-017-0126-8. [DOI] [PubMed] [Google Scholar]
- 21.Cezar-Dos-Santos F, Lenhard-Vidal A, Assolini JP, Marquez AS, Ono MA, Itano EN. Paracoccidioides restrepiensis B339 (PS3) and P lutzii LDR2 yeast cells and soluble components display in vitro hemolytic and hemagglutinating activities on human erythrocytes. Microbiol Immunol. 2018;doi:10–10. doi: 10.1111/1348-0421.12599. [DOI] [PubMed] [Google Scholar]
- 22.de Oliveira HC, da Silva JF, Scorzoni L, Marcos CM, Rossi SA. de Paula E Silva AC Importance of adhesins in virulence of Paracoccidioides spp. Front Microbiol. 2015;10(6):303–303. doi: 10.3389/fmicb.2015.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez A, Hernandez O. New insights into a complex fungal pathogen the case of Paracoccidioides spp. Yeast. 2016;33(4):113–128. doi: 10.1002/yea.3147. [DOI] [PubMed] [Google Scholar]
- 24.Costa M, Borges CL, Bailão AM, Meirelles GV, Mendonça YA, Dantas SFIM. Transcriptome profiling of Paracoccidioides brasiliensis yeast-phase cells recovered from infected mice brings new insights into fungal response upon host interaction. Microbiology (Reading) 2007;153(12):4194–4207. doi: 10.1099/mic.0.2007/009332-0. [DOI] [PubMed] [Google Scholar]
- 25.Parente JA, Salem-Izacc SM, Santana JM, Pereira M, Borges CL, Bailão AM. A secreted serine protease of Paracoccidioides brasiliensis and its interactions with fungal proteins. BMC Microbiol. 2010;10(292):01–10. doi: 10.1186/1471-2180-10-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailão AM, Schrank A, Borges CL, Dutra V, Molinari-Madlum EEWI, Felipe MSS. Differential gene expression by Paracoccidioides brasiliensis in host interaction conditions representational difference analysis identifies candidate genes associated with fungal pathogenesis. Microbes Infect. 2006;8(12-13):2686–2697. doi: 10.1016/j.micinf.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 27.Bailão AM, Shrank A, Borges CL, Parente JA, Dutra V, Felipe MS. The transcriptional profile of Paracoccidioides brasiliensis yeast cells is influenced by human plasma. FEMS Immunol Med Microbiol. 2007;51(1):43–57. doi: 10.1111/j.1574-695X.2007.00277.x. [DOI] [PubMed] [Google Scholar]
- 28.Pigosso LL, Parente AF, Coelho AS, Silva LP, Borges CL, Bailão AM. Comparative proteomics in the genus Paracoccidioides. Fungal Genet Biol. 2013;60:87–100. doi: 10.1016/j.fgb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Parente-Rocha JA, Tomazett MV, Pigosso LL, Bailão AM. Ferreira de Souza A.Paccez JD In vitro, ex vivo and in vivo models a comparative analysis of Paracoccidioides spp. proteomic studies. Fungal Biol. 2018;122(6):505–513. doi: 10.1016/j.funbio.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Nayak AP, Green BJ, Beezhold DH. Fungal hemolysins. Med Mycol. 2013;51(1):1–16. doi: 10.3109/13693786.2012.698025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wartenberg D, Lapp K, Jacobsen ID, Dahse HM, Kniemeyer O, Heinekamp T. Secretome analysis of Aspergillus fumigatus reveals Asp-hemolysin as a major secreted protein. Int J Med Microbiol. 2011;301(7):602–611. doi: 10.1016/j.ijmm.2011.04.016. [DOI] [PubMed] [Google Scholar]
- 32.da Silva JLM, Andreotti PF, Benard G, Soares CP, Miranda ET, Mendes-Giannini MJ. Epithelial cells treated with genistein inhibit adhesion and endocytosis of Paracoccidioides brasiliensis. Antonie Van Leeuwenhoek. 2007;92(1):129–135. doi: 10.1007/s10482-006-9129-z. [DOI] [PubMed] [Google Scholar]
- 33.Caro E, Gonzalez A, Muñoz C, Urán ME, Restrepo A, Hamilton AJ. Recognition of laminin by Paracoccidioides brasiliensis conidia a possible mechanism of adherence to human type II alveolar cells. Med Mycol. 2008;46(8):795–704. doi: 10.1080/13693780802073108. [DOI] [PubMed] [Google Scholar]
- 34.González A, Restrepo A, Cano LE. Pulmonary immune responses induced in BALB/c mice by Paracoccidioides brasiliensis conidia. Mycopathologia. 2008;165(4-5):313–330. doi: 10.1007/s11046-007-9072-1. [DOI] [PubMed] [Google Scholar]
- 35.Marcos CM, de Oliveira HC, da Silva JF, Assato PA, Yamazaki DS, da Silva RA. Identification and characterisation of elongation factor Tu, a novel protein involved in Paracoccidioides brasiliensis-host interaction. FEMS Yeast Res. 2016;16(7):01–14. doi: 10.1093/femsyr/fow079. [DOI] [PubMed] [Google Scholar]
- 36.Parente-Rocha JA, Parente AF, Baeza LC, Bonfim SM, Hernandez O, McEwen JG. Macrophage interaction with Paracoccidioides brasiliensis yeast cells modulates fungal metabolism and generates a response to oxidative stress. PLoS One. 2015;10(9):01–18. doi: 10.1371/journal.pone.0137619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castilho DG, Chaves AF, Xander P, Zelanis A, Kitano ES, Serrano SM. Exploring potential virulence regulators in Paracoccidioides brasiliensis isolates of varying virulence through quantitative proteomics. J Proteome Res. 2014;13(10):4259–4271. doi: 10.1021/pr5002274. [DOI] [PubMed] [Google Scholar]
- 38.Sardi JC, Duque C, Höfling JF, Gonçalves RB. Genetic and phenotypic evaluation of Candida albicans strains isolated from subgingival biofilm of diabetic patients with chronic periodontitis. Med Mycol. 2012;50(5):467–475. doi: 10.3109/13693786.2011.633233. [DOI] [PubMed] [Google Scholar]
- 39.Voltan AR, Sardi JC, Soares CP, Machado MP, Almeida AMF, Mendes-Giannini MJ. Early endosome antigen 1 (EEA1) decreases in macrophages infected with Paracoccidioides brasiliensis. Med Mycol. 2013;51(7):759–764. doi: 10.3109/13693786.2013.777859. [DOI] [PubMed] [Google Scholar]
- 40.Puccia R, Vallejo MC, Matsuo AL, Longo LV. The Paracoccidioides cell wall past and present layers toward understanding interaction with the host. Front Microbiol. 2011;2(257):01–07. doi: 10.3389/fmicb.2011.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arango JC, Puerta-Arias JD, Pino-Tamayo PA, Arboleda-Toro D, González A. Bone marrow-derived mesenchymal stem cells transplantation alters the course of experimental paracoccidioidomycosis by exacerbating the chronic pulmonary inflammatory response. Med Mycol. 2018;56(7):884–895. doi: 10.1093/mmy/myx128. [DOI] [PubMed] [Google Scholar]
- 42.Andreotti PF, da Silva JLM, Bailão AM, Soares CM, Benard G, Soares CP. Isolation and partial characterization of a 30 kDa adhesin from Paracoccidioides brasiliensis. Microbes Infect. 2005;7(5-6):875–881. doi: 10.1016/j.micinf.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 43.da Silva JF, de Oliveira HC, Marcos CM, da Silva RA, da Costa , TA , Calich VL. Paracoccidoides brasiliensis 30 kDa Adhesin identification as a 14-3-3 protein, cloning and subcellular localization in infection models. PLoS One. 2015;10(10):e0142028. doi: 10.1371/journal.pone.0142028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D, Janganan TK, Chen G, Marques ER, Kress MR, Goldman GH. The cAMP pathway is important for controlling the morphological switch to the pathogenic yeast form of Paracoccidioides brasiliensis. Mol Microbiol. 2007;65(3):761–779. doi: 10.1111/j.1365-2958.2007.05824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Silva JF, de Oliveira HC, Marcos CM, Assato PA, Fusco-Almeida AM, Mendes-Giannini MJS. Advances and challenges in paracoccidioidomycosis serology caused by Paracoccidioides species complex an update. Diagn Microbiol Infect Dis. 2016;84(1):87–94. doi: 10.1016/j.diagmicrobio.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 46.de Oliveira P, Juliano MA, Tanaka AS, Carmona AK, Dos Santos SM, de Barros BC. Paracoccidioides brasiliensis induces cytokine secretion in epithelial cells in a protease-activated receptor-dependent (PAR) manner. Med Microbiol Immunol. 2017;206(2):149–156. doi: 10.1007/s00430-016-0490-x. [DOI] [PubMed] [Google Scholar]
- 47.Taborda CP, Travassos LR. Peptide vaccine against paracoccidioidomycosis. Methods Mol Biol. 2017;1625:113–128. doi: 10.1007/978-1-4939-7104-6_9. [DOI] [PubMed] [Google Scholar]
- 48.Flavia Popi AF, Lopes JD, Mariano M. GP43 from Paracoccidioides brasiliensis inhibits macrophage functions An evasion mechanism of the fungus. Cell Immunol. 2002;218(1-2):87–94. doi: 10.1016/s0008-8749(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 49.Gegembauer G, Araujo LM, Pereira EF, Rodrigues AM, Paniago AM, Hahn RC. Serology of paracoccidioidomycosis due to Paracoccidioides lutzii. PLoS Negl Trop Dis. 2014;8(7):e2986. doi: 10.1371/journal.pntd.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leitão NP, Jr, Vallejo MC, Conceição PM, Camargo ZP, Hahn R, Puccia R. Paracoccidioides lutzii Plp43 is an active glucanase with partial antigenic identity with P brasiliensis gp43. PLoS Negl Trop Dis. 2014;8(8):e3111. doi: 10.1371/journal.pntd.0003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barbosa MS, Báo SN, Andreotti PF, de Faria FP, Felipe MS. dos Santos Feitosa L Glyceraldehyde-3-phosphate dehydrogenase of Paracoccidioides brasiliensis is a cell surface protein involved in fungal adhesion to extracellular matrix proteins and interaction with cells. Infect Immun. 2006;74(1):382–389. doi: 10.1128/IAI.74.1.382-389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Oliveira HC, da Silva JF, Matsumoto MT, Marcos CM, da Silva RP, da Silva RAM. Alterations of protein expression in conditions of copper-deprivation for Paracoccidioides lutzii in the presence of extracellular matrix components. BMC Microbiol. 2014;14:302–302. doi: 10.1186/s12866-014-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soares DA, de Andrade RV, Silva SS, Bocca AL, Felipe SMS, Petrofeza S. Extracellular Paracoccidioides brasiliensis phospholipase B involvement in alveolar macrophage interaction. BMC Microbiol. 2010;10:241–241. doi: 10.1186/1471-2180-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]


