Abstract
Background
Targeted therapy for patients with lung and colon cancer based on tumor molecular profiles is an important cancer treatment strategy, but the impact of gene mutation tests on cancer treatment and outcomes in large populations is not clear. In this study, we assessed the accuracy of an algorithm to identify tumor mutation testing in administrative claims data during a period before test-specific Current Procedural Terminology (CPT) codes were available.
Methods
We used Pennsylvania Cancer Registry data to select patients with lung or colon cancer diagnosed between 2007 and 2011 who were treated at the University of Pennsylvania Health System, and we obtained their administrative claims. A combination of CPT laboratory codes (“stacking codes”) was used to identify potential tumor mutation testing in the claims data. Patients’ electronic medical records were then searched to determine whether tumor mutation testing actually had been performed. The sensitivity, specificity, positive and negative predictive values (PPV and NPV) were calculated.
Results
An algorithm using stacking codes had moderate sensitivity (86% for lung cancer and 81% for colon cancer) and high specificity (98% for lung cancer and 96% for colon cancer). Sensitivity and specificity did not vary significantly during 2007–2011. In patients with lung cancer, PPV was 98% and NPV was 92%. In patients with colon cancer, PPV was 96% and NPV was 83%.
Conclusions
An algorithm using stacking codes can identify tumor mutation testing in administrative claims data among patients with lung and colon cancer with a high degree of accuracy.
Keywords: lung cancer, colon cancer, molecular diagnostic testing, sensitivity, specificity
INTRODUCTION
In the U.S., lung and colon cancer are leading causes of cancer-related death. Approximately 40% of lung cancer and 20% of colon cancer patients present with metastatic disease at the time of diagnosis and many will develop refractory metastatic disease after initial treatment.1 Over the last 10 years, targeted therapies have emerged as effective treatments for these two malignancies. In patients with lung adenocarcinoma, treatment with epidermal growth factor receptor (EGFR) inhibitors, such as erlotinib, leads to improvements in clinical outcomes for patients harboring specific somatic “oncogenic driver” mutations in the EGFR gene, whereas the presence of a KRAS gene mutation predicts lack of response to EGFR inhibition.2,3 Tumors from patients with metastatic lung adenocarcinoma are therefore frequently evaluated to identify EGFR and/or KRAS mutations to guide chemotherapy selection. Similarly, in patients with colon cancer, two anti-EGFR antibody-based therapies, cetuximab and panitumumab, are effective treatments for patients whose tumors do not harbor mutations in the KRAS gene (i.e., wild-type KRAS), but ineffective for patients with mutations in this gene.4,5 Thus, tumor mutation testing to identify KRAS gene mutations is necessary to restrict the use of these agents to patients with wild-type KRAS. Almost all cancer treatment guidelines now recommend determination of gene mutation status in the management of metastatic lung adenocarcinoma and colon cancer.6–8
To evaluate the impact of these tests on access to novel targeted therapies and subsequent health outcomes in large populations, researchers must first be able to reliably identify when patients did and did not receive the tests. This has been difficult using typical data sources, such as administrative claims. Until 2013, there were no unique billing codes that could be used to identify molecular testing specific to cancer treatment in administrative claims.9,10 Instead, administrative claims for molecular testing in cancer treatment used “stacking codes,” a series of Current Procedural Terminology (CPT) codes that capture the various laboratory steps involved in the performance of a molecular test, but that do not identify the specific gene target of the test.11 The codes allow for charges for procedures such as nucleic acid isolation (CPT 83891), amplification (CPT 83898), and use of sequencing to identify specific variants (CPT 83904). Code 83912 can also be used by pathologists to interpret and report the molecular findings. The use of these codes was not uniform, with considerable variability across different molecular labs in the specific codes and number of codes used. The lack of specific CPT codes for individual mutations also prevents researchers from determining which specific test was performed and is a plausible limitation in population-based research on genetic testing. In this study, we assessed the accuracy of an algorithm using stacking codes to identify tumor mutation testing in administrative claims data.
METHODS
Administrative Data
Our approach to assessing the accuracy of using stacking codes to identify clinical tumor mutation testing is shown in Figure 1. We used data from the Pennsylvania Cancer Registry (PCR), a medical-provider-mandated database of cancer cases in the Commonwealth of Pennsylvania, to identify 52,269 patients with lung cancer and 32,106 with colon cancer diagnosed between January 1, 2007 and December 31, 2011. We excluded patients who lacked clinical pathology data confirming the cancer diagnosis (n=4290), died prior to definitive diagnosis (n=2149), or lacked cancer histology information (n=3026). We further excluded lung cancer patients who had a histological subtype other than adenocarcinoma (n=22,274). There were 24,465 lung adenocarcinoma and 26,836 colon cancer patients who met these inclusion criteria.
Figure 1.
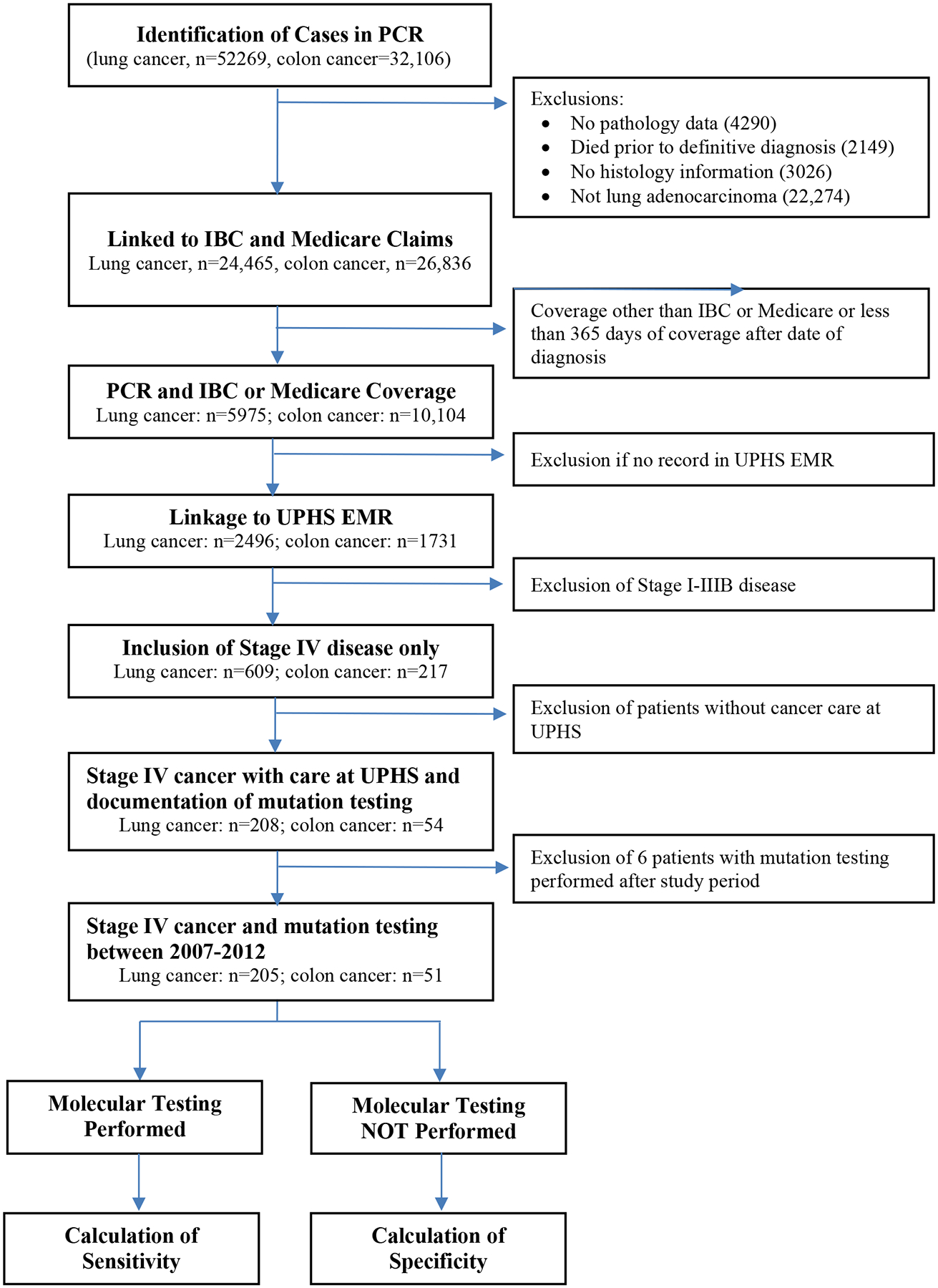
Overview of Study Design. IBC, Independence Blue Cross; UHPS, University of Pennsylvania Health System; EMR, Electronic Medical Record
Using patients’ Social Security numbers, we linked PCR patient records to administrative claims from fee-for-service Medicare, which comprised approximately two-thirds of Pennsylvania’s Medicare beneficiaries during 2007 to 2011, as well as to claims from Independence Blue Cross (Independence), one of Pennsylvania’s largest commercial health insurers. To ensure we had a complete set of claims data for each patient, we included only patients who had continuous enrollment in either Medicare or Independence for at least 365 days after diagnosis. The study cohort identified after linking to claims consisted of 5975 lung adenocarcinoma and 10,104 colon cancer patients, representing 24.4% and 37.7% respectively of the sample meeting the initial inclusion criteria. This study was approved by the University of Pennsylvania Institutional Review Board.
Analytical Cohorts
We linked patients from the claims-based cohort to administrative data from the University of Pennsylvania Health System (UPHS) using Social Security numbers to identify the subset of 2496 lung adenocarcinoma and 1731 colon cancer patients who had ever received medical care at UPHS. When compared to the broader population of lung or colon cancer patients covered by Medicare or Independence in Pennsylvania, the subset of patients ever receiving care at UPHS were slightly younger and less likely to be white, but similar in sex distribution (see Appendix).
As patients with metastatic disease at presentation (i.e., Stage IV) are much more likely to undergo mutation testing, we selected only patients with metastatic colon cancer (mCC) or lung adenocarcinoma (mLAD) by identifying those with a diagnosis of M1 disease with any T or N designation as reported in PCR enrollment data. This resulted in the inclusion of 217 mCC and 609 mLAD patients. Chart reviews of the UPHS electronic medical record (EMR) system were then performed to limit the analysis to patients who received cancer care at UPHS and to confirm each patient’s diagnosis and stage of cancer, yielding a total of 54 mCC patients and 208 mLAD patients. Diagnosis and stage were available in the EMR and matched the PCR data for all patients.) We further excluded from the calculation of diagnostic performance 6 patients (3 each with mCC or mLAD) who were diagnosed between 2007 and 2011 but underwent tumor mutation testing in 2012 or later, resulting in a final analytical cohort of 51 mCC and 205 mLAD patients for determination of performance characteristics of stacking codes.
Identification of Mutation Testing
We identified relevant CPT codes for tumor mutation testing identified from the literature and by searching the Clinical Laboratory Fee Schedule (CLFS) of the Centers for Medicare and Medicaid Services (CMS) for all potential codes that were in use for clinical molecular testing.11,12 Relevant codes for testing of somatic tumor mutations were identified and are listed in Table 1. These are generic diagnostic CPT codes that represent various steps in the molecular testing pathway, such as nucleic acid isolation (CPT 83891), amplification (CPT 83898), and use of sequencing to identify specific variants (CPT 83904). Code 83912 can also be used by pathologists to interpret and report the molecular findings. These codes have commonly been used by providers to obtain reimbursement for molecular testing from health insurers, including CMS. Because multiple codes are frequently used or “stacked” on an individual claim for a single molecular test, these are usually referred to as “stacking codes.” We searched each patient’s administrative claims for any of the CPT codes listed in Table 1 up to 1 month before and 12 months after the date of diagnosis (as identified in the PCR), as an indicator of potential tumor mutation testing. Given the variability in the number and type of codes used for molecular testing, we considered the presence of any single stack code from Table 2 in a patient’s administrative claims as evidence of mutation testing.
Table 1:
Description of CPT Codes Used for “Stacking” of Molecular Testing
| CPT Code | Description |
|---|---|
| 83890 | Molecular diagnostics; molecular isolation or extraction, each nucleic acid type |
| 83891 | Molecular diagnostics; isolation or extraction of highly purified nucleic acid, each nucleic acid type |
| 83892 | Molecular diagnostics; enzymatic digestion, each enzyme treatment |
| 83894 | Molecular diagnostics; separation by gel electrophoresis, each nucleic acid preparation |
| 83898 | Molecular diagnostics; amplification, target, each nucleic acid sequence |
| 83904 | Molecular diagnostics; mutation identification by sequencing, single segment, each segment |
| 83907 | Molecular diagnostics; lysis of cells prior to nucleic acid extraction |
| 83909 | Molecular diagnostics; separation and identification by high resolution technique |
| 83912 | Molecular diagnostics; interpretation and report |
Table 2:
Accuracy of Claims Data for Identification of Mutation Testing in Lung and Colon Cancer
| N | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | |
|---|---|---|---|---|---|
| Colon Cancer | 51 | 81 (61–90) | 96 (80–100) | 96 (77–100) | 83 (64–94) |
| Lung Cancer | 205 | 86 (77–93) | 98 (94–100) | 97 (90–100) | 92 (86–96) |
The EMR was then searched by a trained abstractor (J.I.) to identify whether molecular diagnostic testing had been performed after the lung or colon cancer diagnosis. A laboratory report documenting testing and a result for either KRAS or EGFR mutation testing in lung cancer or KRAS testing only in colon cancer was considered to be positive clinical evidence of molecular testing. All patients without evidence of mutation testing as well as a random 25% sample of patients with documented testing were evaluated by a second reviewer (A.V.).
Statistical Analysis
Molecular testing identified with the use of stacking codes was compared with testing identified via EMR chart review to determine sensitivity, specificity, positive predictive value (PPV), and negative predictive values (NPV). The sensitivity of stacking codes was calculated as the proportion of patients with documented mutational analysis via chart review who were identified correctly with any “stacking code” from Table 1. Specificity was calculated as the proportion of patients without documented mutational analysis via chart review who did not have a claim for any “stacking code” during the period of analysis. PPV was defined as the proportion of patients with a stacking code in claims data who were confirmed to have undergone molecular testing by chart review of medical records. NPV was defined as the proportion of patients without a “stacking code” in claims data who did not have evidence of molecular testing during the course of their cancer care based on the medical record. Statistical analysis was performed using Stata version 13 (Stata Corp, Texas, USA).
Temporal Trend in Mutation Testing
To provide information on temporal trends in the use of EGFR and KRAS testing, we then searched for stacking codes in the facility and provider insurance claims of a nationally representative 5% random sample of fee-for-service Medicare beneficiaries between 2005 and 2012 with any colon or lung cancer diagnosis code. The proportion of patients undergoing mutation testing in a specific year was calculated by identifying patients with lung or colon cancer (denominator), and identifying whether any stack codes were present in claims during that calendar year (numerator). Because cancer stage and date of diagnosis information are not recorded in Medicare claims, any patient with a claim with a lung or colon cancer diagnosis in a specific year was considered in the denominator for this calculation. The proportions of patients undergoing mutation testing were compared across years using χ2 tests.
RESULTS
Among UPHS patients with mCC or mLAD, molecular testing increased from 24% in 2007 to 58% in 2011 (p=0.001). The performance characteristics of stacking codes for the identification of molecular testing are summarized in Table 2. The sensitivity of stacking codes was 86% (95 percent confidence interval [95CI]: 77% to 93%) in mLAD and 81% (95CI: 61% to 90%) in mCC. The specificity of stacking codes was 98% (95CI: 94% to 100%) in mLAD and 96% (95CI: 80% to 100%) in mCC over the study period. Sensitivity and specificity did not vary significantly during the five-year study period. For patients with mLAD, the PPV and NPV of stack codes were 97% (95CI: 90% to 100%) and 92% (95CI: 86% to 96%), respectively. For patients with mCC, PPV and NPV were 96% (95CI: 77% to 100%) and 83% (95CI: 64% to 94%), respectively.
National rates of mutation testing, as identified by stacking code use, in lung and colorectal cancer patients in the 5% random sample of traditional Medicare beneficiaries increased significantly between 2005 and 2012 (Figure 2). The annual rate of mutation testing grew from 0.8% to 7.8% for patients with lung cancer (p<0.001), and from 0.7% to 5.2% for patients with colon cancer (p<0.001).
Figure 2.
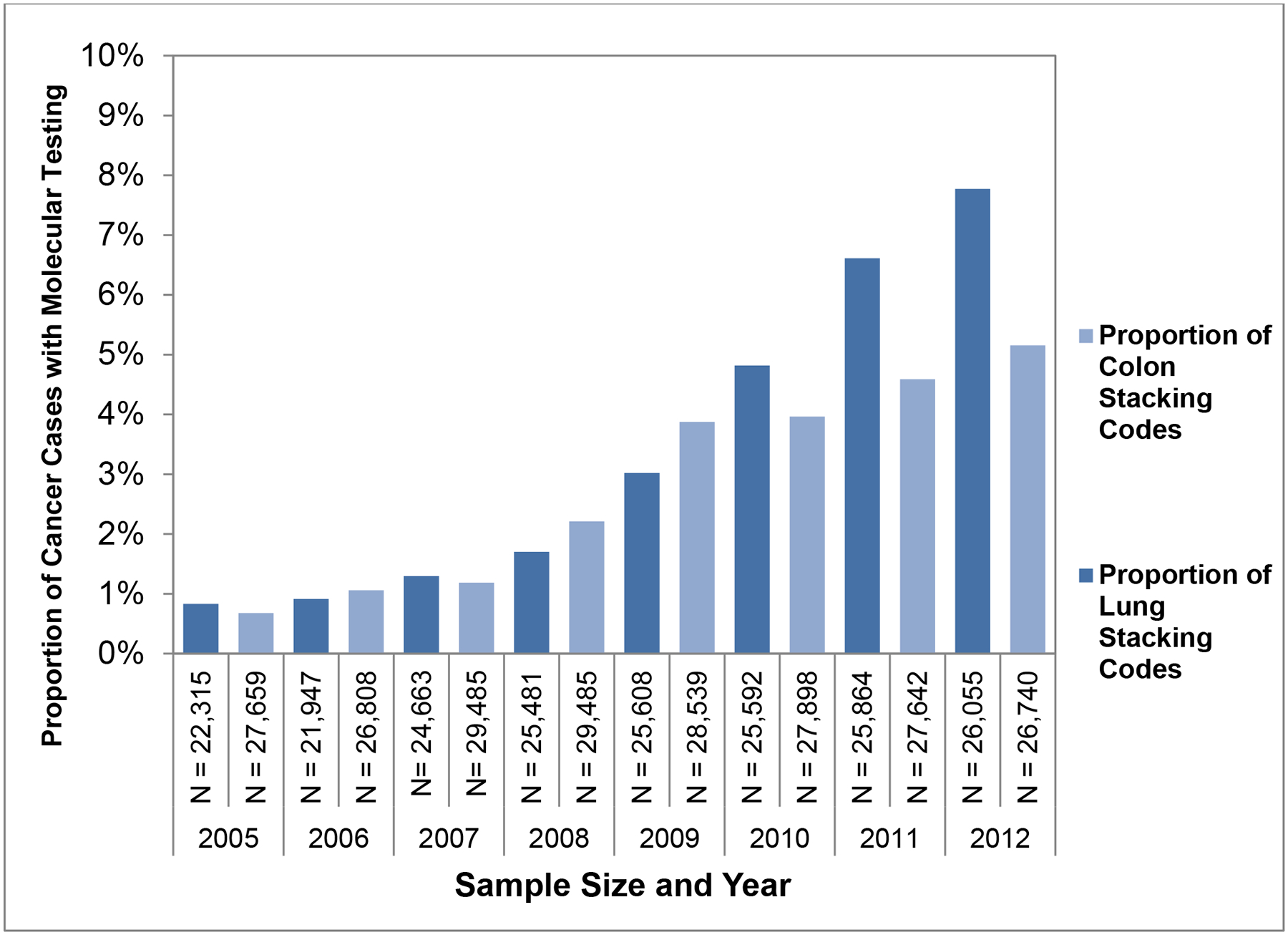
Identification of Stacking Codes in Medicare Claims
DISCUSSION
In this study, we evaluated the validity of a claims-based approach for identifying molecular testing in patients with lung and colon cancer. In patients with metastatic lung and colon cancers, the use of stacking codes enabled accurate identification of patients who have undergone tumor mutational analysis. In particular, the PPV of >95% suggests that administrative data can be used to reliably identify almost all patients undergoing mutational analysis even in the absence of specific administrative coding identifying the testing.
Although specific CPT codes for KRAS testing in lung and colon tumors were implemented in 2013, diffusion of mutation testing in lung and colon cancer started in the late 2000s.13,14 Researchers can use our algorithm to study the use of mutation testing and its effects on treatment, outcomes, and costs from its inception. As the number and diversity of tumor mutational testing continues to expand rapidly, our method of identifying mutational testing in claims data should facilitate future studies evaluating the use and clinical impact of tumor mutation testing in lung and colon cancer, and across other cancer types for similar analyses. We think such studies are critical to understanding how tumor mutational testing influences treatment selection, chemotherapy utilization, healthcare costs, and clinical outcomes. Although the use of stacking codes does not allow for identification of the specific gene evaluated or the specific mutation identified, our results suggest that this algorithm can be used in patients with a new cancer diagnosis to accurately identify the mutation testing of interest.
There are two important limitations to consider. First, our findings may not be generalizable. Our study cohort comprised patients from UPHS, a single large regional tertiary cancer center in Philadelphia, Pennsylvania. Cancer mutation testing is generally performed either internally by a hospital-based molecular pathology laboratory or by an external molecular diagnostic company. It is possible that the coding and billing practices at UPHS may differ from other medical centers or stand-alone molecular diagnostic laboratories, and thus our findings may not apply to all patients nationally. Moreover, our colon cancer cohort was small, resulting in wide confidence intervals in Table 2.
Second, although we show that stacking codes can accurately indicate that mutation testing was performed, these codes do not allow for identification of the specific mutation testing and are thus are context-specific (i.e., we assumed that the appearance of administrative claims with stacking codes in a patient with metastatic lung cancer was an indicator of lung tumor mutational testing). It is therefore likely that stacking codes will incorrectly identify the mutation testing of interest in patients with multiple simultaneous cancers, or in cases where the stacking codes occur much later than the date of cancer diagnosis, when such testing may be conducted for other clinical indications, including non-cancer-related reasons.
In conclusion, our results demonstrate that an algorithm employing generic molecular pathology billing codes can identify tumor mutation testing with high predictive value in patients with metastatic lung or colon cancer. This approach may be useful for future studies evaluating population-based use of tumor mutation testing, and its impact on clinical outcomes and healthcare utilization.
Supplementary Material
ACKNOWLEDGMENTS / FUNDING SOURCES
This study was supported by grant #4100059202 from the Pennsylvania Department of Health, which had no role in the design and conduct of the study, or in the collection, management, analysis, and interpretation of the data, or in the preparation, review, and approval of the manuscript, or in the decision to submit the manuscript for publication. Dr. Vachani had full access to all of the data and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Research support:
This research was supported by a research grant from the Commonwealth of Pennsylvania – Department of Health. None of the authors had any personal or financial conflicts of interest in regard to this study.
Footnotes
Conflicts of Interest: None of the authors have any conflicts of interest relevant to the topic, or work presented, in this manuscript.
REFERENCES
- 1.Howlader NNA, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975–2012, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted to the SEER web site, April 2015. [Google Scholar]
- 2.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958–67. [DOI] [PubMed] [Google Scholar]
- 4.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626–34. [DOI] [PubMed] [Google Scholar]
- 5.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757–65. [DOI] [PubMed] [Google Scholar]
- 6.Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091–6. [DOI] [PubMed] [Google Scholar]
- 7.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) Mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol 2011;29:2121–7. [DOI] [PubMed] [Google Scholar]
- 8.Leighl NB, Rekhtman N, Biermann WA, et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol 2014;32:3673–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Association for Molecular Pathology. Association for Molecular Pathology (AMP) proposal for CPT coding reform in molecular diagnostics. (Accessed July 21, 2015, at http://www.amp.org/committees/economics/AMPCPTReformProposal_Final.pdf.)
- 10.Medicare program: revisions to payment policies under the physician fee schedule, DME face-to-face encounters, elimination of the requirement for termination of non-random prepayment complex medical review and other revisions to part B for CY 2013; hospital outpatient prospective and ambulatory surgical center payment systems and quality reporting programs; electronic reporting pilot; inpatient rehabilitation facilities quality reporting program; quality improvement organization regulations; proposed rules. Federal Register 2012:44722–5234.
- 11.Klein RD. Reimbursement in molecular pathology: bringing genomic medicine to patients. Clin Chem 2015;61:136–8. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Medicare and Medicaid Services, Clincal Laboratory Fee Schedule (Accessed June 2014, at http://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/ClinicalLabFeeSched/clinlab.html.)
- 13.Engstrom PF, Arnoletti JP, Benson AB 3rd, et al. NCCN Clinical Practice Guidelines in Oncology: colon cancer. J Natl Compr Canc Netw 2009;7:778–831. [DOI] [PubMed] [Google Scholar]
- 14.Lynch JA, Khoury MJ, Borzecki A, et al. Utilization of epidermal growth factor receptor (EGFR) testing in the United States: a case study of T3 translational research. Genet Med 2013;15:630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


