Abstract
Adoptive T cell therapies (ACTs) hold great promise in cancer treatment, but low overall response rates in patients with solid tumors underscore remaining challenges in realizing the potential of this cellular immunotherapy approach. Promoting CD8+ T cell adaptation to tissue residency represents an underutilized but promising strategy to improve tumor-infiltrating lymphocyte (TIL) function. Here, we report that deletion of the HIF negative regulator von Hippel-Lindau (VHL) in CD8+ T cells induced HIF-1α/HIF-2α–dependent differentiation of tissue-resident memory–like (Trm-like) TILs in mouse models of malignancy. VHL-deficient TILs accumulated in tumors and exhibited a core Trm signature despite an exhaustion-associated phenotype, which led to retained polyfunctionality and response to αPD-1 immunotherapy, resulting in tumor eradication and protective tissue-resident memory. VHL deficiency similarly facilitated enhanced accumulation of chimeric antigen receptor (CAR) T cells with a Trm-like phenotype in tumors. Thus, HIF activity in CD8+ TILs promotes accumulation and antitumor activity, providing a new strategy to enhance the efficacy of ACTs.
Keywords: Immunology, Therapeutics
Keywords: Adaptive immunity, Immunotherapy, T cells
Introduction
Immunotherapy is evolving rapidly to provide new strategies for the treatment of cancer. Current immunotherapeutic modalities, including enhancing endogenous tumor-infiltrating lymphocytes (TILs) by checkpoint blockade or adoptive transfer of chimeric antigen receptor–modified (CAR-modified) autologous T cells into patients, show evidence of sustained cancer remission or clearance in some patients, but overall response rates remain low, especially in advanced solid tumors (1, 2). One limitation of immunotherapy in solid tumors is tumor microenvironment–induced T cell exhaustion, a terminally differentiated T cell state driven by chronic T cell activation and tumor microenvironment factors (3). Exhausted T cells are defined by expression of multiple coinhibitory receptors such as PD-1, TIM-3, and LAG-3, as well as progressive loss of proliferative potential and effector cytokine production, ultimately leading to failed antitumoral responses (4, 5). Initial tumor infiltration leads to differentiation into activated, functional CD8+ T cells, more recently described as progenitor-exhausted T cells (5, 6). Phenotypically defined by high expression of T-bet and Tcf-1 transcription factors, as well as PD-1, Slamf6, and low TIM-3 expression, progenitor-exhausted T cells transition to a terminally exhausted T cell state, increasing expression of multiple coinhibitory receptors, losing Slamf6 expression and effector polyfunctionality, and altering their epigenetic and transcriptional landscape. Notably, progenitor-exhausted T cells are the target of enhanced activity and proliferation in response to checkpoint blockade immunotherapies (6–8). Although transcription factors governing exhaustion programs have been identified, such as T-box transcription factors T-bet and Eomesodermin (Eomes), the NFAT-driven transcription factors Nr4a and Tox, and the transcriptional repressor Blimp1 (5), it is clear that development of T cell exhaustion is a multifactorial process in which underlying mechanisms responsible for impaired function are not fully resolved.
A recently described subset of memory T cells residing in nonlymphoid tissues, tissue-resident memory T cells (Trm cells), is distinct from recirculating central memory (Tcm) and effector memory (Tem) cells and dynamically responds to PD-1 blockade in human tumors (9, 10). Trm cells permanently reside in peripheral tissues, where they display rapid effector functions and provide frontline protection against pathogens and malignant growth (11, 12). Trm cells are characterized by a core transcriptional signature, expression of CD69, and expression of CD103 in some tissues (13). Tumor-infiltrating CD8+ T cells expressing Trm markers found in biopsies have been associated with favorable prognosis for numerous solid tumors, including melanoma, colorectal, and lung cancer (9, 10, 14–19), and are often termed “Trm-like” (20). Single-cell sequencing and functional studies of TILs indicate that the Trm-like population is enriched in clonally expanded and tumor-reactive CD8+ T cells (15, 21, 22) with effector functions (21, 23–25). Given the emerging role of CD8+ Trm cells in antitumor immunity, exploring the mechanisms regulating the acquisition of Trm characteristics by TILs may benefit immunotherapy approaches (9, 10, 26–28).
Transcription factors controlling CD8+ Trm fate include Blimp-1, Hobit, Nr4a1, and Runx3 (12), with enforced expression of Runx3 in adoptively transferred T cells improving the formation of Trm-like TILs and boosting antitumor responses (25). Thus, genetic manipulation of Trm differentiation holds promise for clinical translation. Moreover, CD8+ Trm-like cells appear to play a role in protection against tumor recurrence (22, 29, 30), suggesting that immunotherapy strategies that promote Trm cells may provide long-term protection from disease relapse.
Our previous work highlighted a role for HIFs in regulating T cell effector function, exhaustion, and memory differentiation in infection and malignancies (31–33). Oxygen concentrations vary substantially throughout the body (5%–6% in healthy tissues to ≤0.1% in hypoxic areas of solid tumors and inflamed tissues). HIF-mediated transcription allows for cellular adaptation to areas of low oxygen tension. HIF is hydroxylated at proline residues in normoxic conditions and subsequently ubiquitinated by von Hippel-Lindau (VHL) proteins, mediating constitutive proteasomal degradation (34). At low oxygen tensions, HIF is not targeted for degradation and transcriptional activity is upregulated. Signaling through the T cell receptor (TCR) and cytokines such as TGF-β can further promote HIF transcriptional activity in T cells (34). Discrete expression of HIF-1α and HIF-2α isoforms in immune cells contributes to regulation of both overlapping and distinct target genes (34, 35). While tumor hypoxia is a hallmark of aggressive malignancies and several immunoevasion mechanisms through the recruitment of suppressive immune cell types (35), accumulating evidence indicates that direct effects of hypoxia on CD8+ T cells promotes effector functions (31, 33–35). In a mouse model of chronic lymphocytic choriomeningitis virus (LCMV) infection, VHL-deficient CD8+ T cells with constitutive HIF activity showed enhanced viral clearance and resisted “exhaustion” (31). VHL-deficient CD8+ T cells sustained cytokine production, upregulated costimulatory and cytolytic effector molecules, and paradoxically, multiple T cell exhaustion markers to a greater degree than WT T cells. In a mouse model of melanoma, HIF-1α–deficient CD8+ T cells did not accumulate and displayed reduced antitumor activity (32), while VHL-deficient CD8+ T cells showed enhanced control of tumor growth. Together, these findings suggest that HIF plays a key role in T cell regulation of malignancies; however, the basis for HIF regulation of TIL efficacy is unresolved.
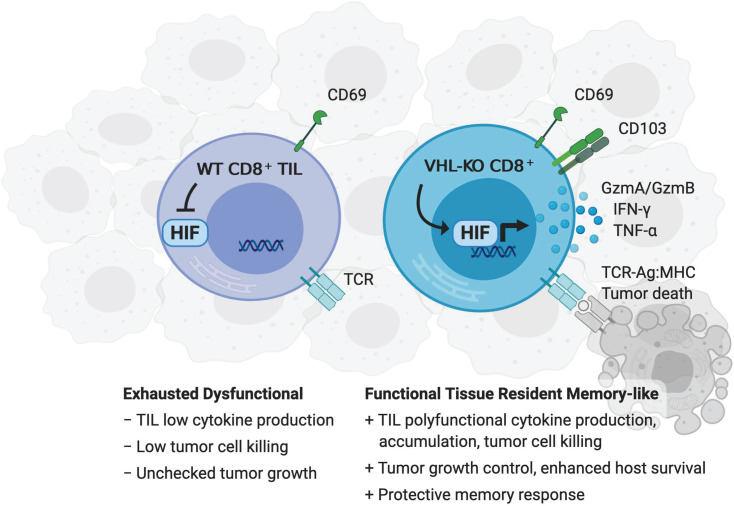
Utilizing Cre-recombinase–driven conditional deletion of Vhl, Hif1a, and Epas1 to study CD8+ T cell differentiation, localization, antitumor function, and memory formation, we demonstrated that constitutive HIF-α activity drives the formation of a prominent and highly cytotoxic CD69+CD103+ Trm–like CD8+ TIL population in solid tumors. VHL-deficient TILs expressed high levels of coinhibitory receptors yet persisted and retained polyfunctionality within the tumor microenvironment, indicating functional resistance to exhaustion and superior efficacy against primary tumors, metastatic challenge, and memory recall responses upon rechallenge. VHL-deficient Trm-like CD8+ TILs expressed a core gene-expression program associated with Trm cells that appeared closely associated with an exhaustion signature, suggesting effector function and responsiveness to checkpoint blockade by this memory subset. VHL-deficient TILs responded to anti–PD-1 therapy with elevated IFN-γ production, resulting in complete regression of large established B16 melanoma tumors. In vivo blocking studies demonstrated that HIF-α–mediated functional features were dependent on the CD103 integrin, known to enhance Trm. Finally, VHL-deficient CAR T cells exhibited the Trm-like phenotype and accumulated in solid tumors. Our results revealed a key role for the VHL/HIF axis in controlling the formation of a Trm-like CD8+ T cell subset in primary and secondary tumors that resisted functional exhaustion and mediated enhanced antitumor responses.
Results
VHL-deficient CD8+ T cells control tumor growth and improve survival.
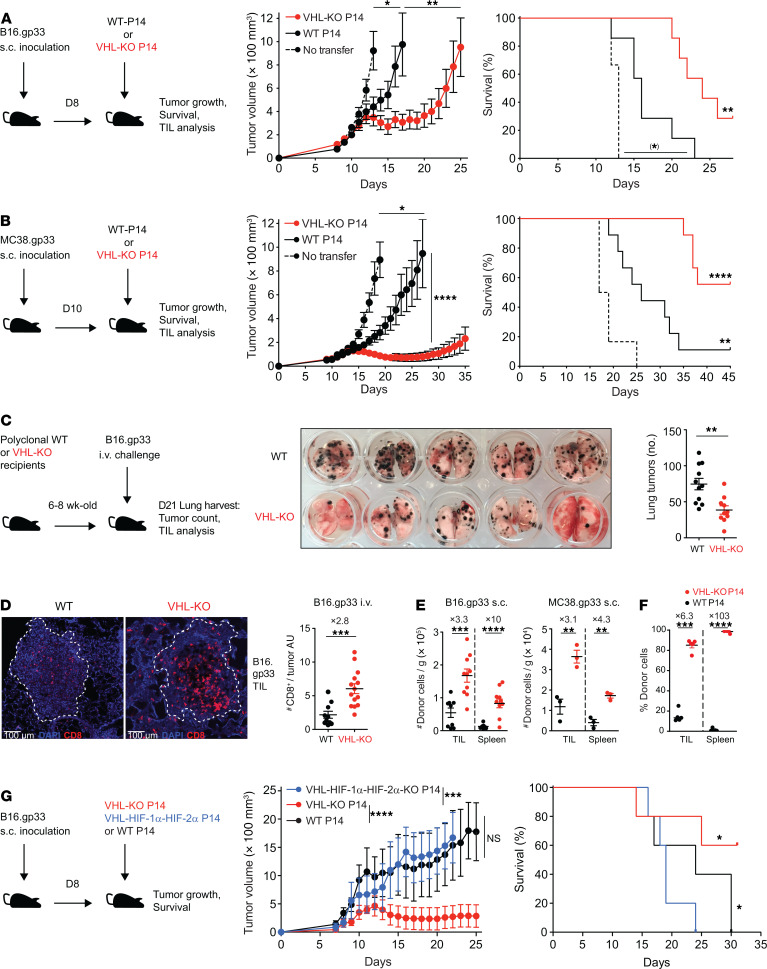
To explore the impact of VHL/HIF modulation in tumor-reactive CD8+ T cells, we utilized mice harboring loxP-flanked Vhl alleles (Vhlfl/fl) and Cre recombinase driven by the distal Lck promoter (dLck). This allowed deletion of Vhl in mature T cells, leading to elevated HIF-1α and HIF-2α protein expression (31). Crossing these mice with TCR transgenic P14 mice, which recognized LCMV-derived peptide gp33 presented by MHC class I molecule H2-Db, we assessed the ability of antigen-specific, VHL-deficient CD8+ T cells to control established tumors. WT animals were s.c. inoculated with B16.gp33 melanoma or MC38.gp33 colorectal adenocarcinoma modified to stably express the gp33 antigen, followed by adoptive transfer of ex vivo activated Vhlfl/fl dLck-Cre– (WT) or Vhlfl/fl dLck-Cre+ (VHL-KO) P14 cells (Figure 1, A and B). VHL-deficient T cells mediated superior tumor growth inhibition and survival compared with their WT counterparts in both tumor models and cured the majority of mice of MC38.gp33 tumors. Enhanced HIF activity in T cells mediated immunopathology in the lungs of animals infected with chronic virus and led to improved protection from B16 lung metastasis challenge (31, 36), suggesting enhanced lung responses. Therefore, we asked whether T cell–specific deletion of Vhl in polyclonal mice was protective against lung metastases. After i.v. challenge with B16 cells as a model for metastasis via seeding tumor lesions in the lungs, VHL-KO hosts presented significantly fewer B16 melanoma metastases (Figure 1C) compared with WT hosts. Immunofluorescence analysis of the lungs revealed increased numbers of CD8+ TILs within tumor lesions of VHL-KO compared with WT hosts (Figure 1D), without evidence of immunopathology in healthy lung parenchyma. Interestingly, VHL-KO polyclonal CD8+ TILs preferentially localized in nonhypoxic tumor areas compared with their WT counterparts (Supplemental Figure 1). VHL-KO P14 TILs also accumulated to 3–10-fold higher numbers than WT TILs in s.c. B16.gp33, MC38.gp33, and rhabdomyosarcoma KMR.gp33 (Figure 1, E and F).
Figure 1. HIF-dependent accumulation of TILs and control of tumor growth by VHL-deficient CD8+ T cells.
(A–B) Tumor growth and survival of (A) B16.gp33- or (B) MC38.gp33-bearing C57BL/6J mice after transfer of Vhlfl/fl dLck-Cre– (WT) or Vhlfl/fl dLck-Cre+ (VHL-KO) P14 cells, n = 7 in A and 9 in B (experimental) and 5 in A and 6 in B (control). (C) Images (middle) and quantification (right) of metastatic lung tumors in polyclonal WT and VHL-KO mice after i.v. challenge with B16.gp33 cells. (D) Immunofluorescence microscopy of CD8α (red) and DAPI (blue) in representative tumor lesions from C at 20× magnification; tumor regions indicated by dotted lines (left) and quantification of CD8+ TILs per tumor area unit (AU; right). Data are combined from 2 independent experiments with 6 × 105 B16.gp33 cells injected into 11 WT and 10 VHL-KO mice. Corresponding results were obtained in 2 additional experiments with 4.5 × 105 tumor cells, n = 9 mice per group. (E) Number of donor P14 cells from tumor (TILs) and spleen from mice bearing B16.gp33 (left) and MC38.gp33 (right) tumors at endpoint of efficacy experiments (A and B), n = 9 for WT and 10 for VHL-KO for B16.gp33, n = 3 per group for MC38.gp33. (F) Frequency of donor P14 cells recovered from KMR.gp33 tumors or spleen 7 days after cotransfer of WT and VHL-KO cells, n = 4. (G) Tumor growth and survival of B16.gp33 tumor–bearing mice after transfer of WT, VHL-KO, or Vhlfl/fl-Hif1afl/fl-Epas1fl/fl dLck-Cre+ (VHL-HIF-1α-HIF-2α-KO) P14 cells, n = 5 per group. Data are representative of 3 independent experiments except as stated in D. Bars and error represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 2-tailed Student’s t test or Mann-Whitney U test (tumor growth) and Bonferroni-corrected log-rank test (survival), (*) failed to reach the Bonferroni-corrected threshold although showing P < 0.05.
To assess the role of HIF in improved tumor clearance by VHL-deficient TILs, we adoptively transferred P14 cells that were Vhlfl/flHif1afl/flEpas1fl/fl dLck-Cre+ (triple knockout, TKO), VHL-KO, or WT into recipient mice bearing s.c. B16.gp33 tumors (Figure 1G). Strikingly, TKO P14 cells were unable to control tumor growth or improve survival, indicating that the enhanced antitumor activity observed by VHL-KO P14 cells was dependent on the loss of VHL-mediated degradation of HIF-α transcription factors.
VHL-deficient TILs accumulate and survive in tumors in a HIF-dependent manner.
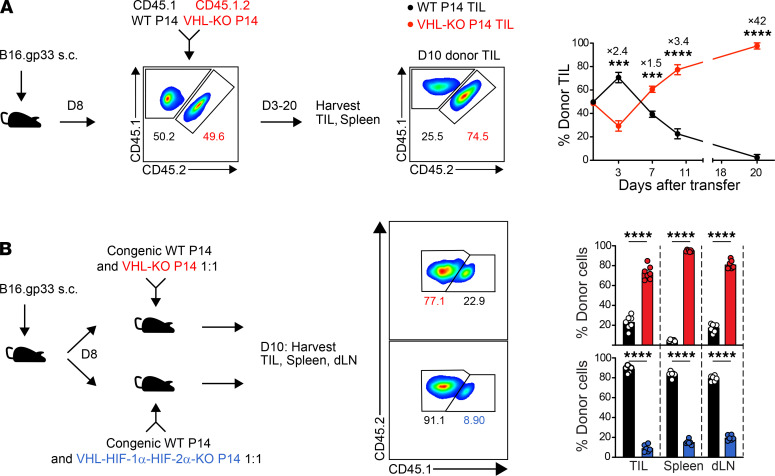
To further study VHL-KO versus WT CD8+ TILs in the same tumor microenvironment, we cotransferred equal numbers of congenically distinct VHL-KO and WT P14 cells and tracked their frequency over time in tumor-bearing hosts. Kinetic analysis of TILs in B16.gp33 tumors revealed progressive accumulation of VHL-KO TILs beginning at day 7 after transfer as assessed by both frequency (Figure 2A) and numbers (Supplemental Figure 2A). In contrast, WT P14 cell numbers progressively declined over the same time period (Supplemental Figure 2A). The increase in VHL-KO P14 cells was not due to an increase in proliferation (Supplemental Figure 2B) but likely to a survival advantage in tumors, as indicated by lower death receptor FasR (CD95) and higher antiapoptotic Bcl-2 expression compared with WT cells (Supplemental Figure 2B). Notably, VHL-KO P14 cells recovered from tumors, as well as upon prolonged culture, retained phosphorylation of the downstream signal transducer STAT5 (Supplemental Figure 2, B and C) and showed higher levels of IL-2Rα (CD25) expression (Supplemental Figure 2, B and D), consistent with regulation by HIF as also previously reported (37). Thus, VHL-KO CD8+ T cells had a cell-intrinsic survival advantage over their WT counterparts, and HIF-α was required for TIL accumulation (Figure 2B).
Figure 2. VHL-deficient P14 TILs accumulate in tumors in a HIF-dependent manner.
(A) Congenically distinct WT and VHL-KO P14 cells were cotransferred into B16.gp33 tumor–bearing mice. Flow cytometry plots show input donor cells (left) and recovered donor cells from a representative tumor on day 10 after transfer (middle). Mean recovered frequencies over time are provided (right), n = 4 at days 3–10 and n = 5 at day 20. (B) Congenically distinct donor WT and VHL-KO P14 cells (top) and WT and VHL-HIF-1α-HIF-2α-KO P14 cells (bottom) were cotransferred into B16.gp33 tumor–bearing mice (left). Representative flow cytometric analysis of donor TILs on day 10 (middle) and recovered donor P14 frequencies in indicated tissues (right), n = 6 VHL-KO/WT and 5 VHL-HIF1α-HIF-2α-KO/WT. Data are representative of 3 independent experiments. Bars, line charts, and error bars represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-tailed Student’s t test).
VHL-deficient TILs retain polyfunctionality and cytolytic capacity despite elevated exhaustion-associated markers.
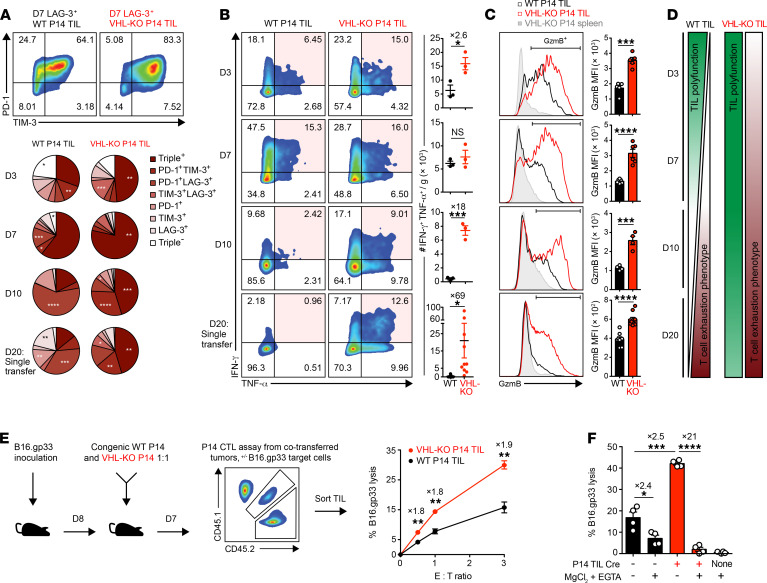
Immunotherapy of solid tumors is hampered by tumor-induced inhibitory signals that promote T cell exhaustion via multiple coinhibitory receptors on CD8+ TILs (5). Upon progressive exhaustion, CD8+ T cells lose their proliferative potential and have decreased effector cytokine production capacity (5). We sought to explore the mechanisms by which VHL-KO TILs resist tumor-induced exhaustion, sustaining their efficacy against solid tumors. Progressive accumulation of coinhibitory receptors PD-1, TIM-3, and/or LAG-3 was evident on WT and VHL-KO P14 TILs after cotransfer (Figure 3A). The VHL-KO TILs coexpressed a significantly higher frequency of these exhaustion-associated markers, yet they produced significantly more IFN-γ and TNF-α upon restimulation compared with their WT counterparts isolated from the same tumors (Figure 3B). Furthermore, the cytotoxic molecule granzyme-B was found significantly upregulated in VHL-KO over WT P14 TILs (Figure 3C). Thus, VHL-deficient TILs retain multiple effector functions in contrast to WT TILs over the course of tumor growth (Figure 3D and Supplemental Figure 2E).
Figure 3. VHL-deficient P14 TILs retain polyfunctional antitumor cell responses despite elevated exhaustion-associated markers.
(A) Coexpression of exhaustion-associated markers PD-1, TIM-3, and LAG-3 by donor P14 TILs recovered in Figure 2A. Representative flow cytometry plots of PD-1 and TIM-3 expression gated on LAG-3+ cells (top). Pie charts indicate accumulation of exhaustion markers on days 3–10 after cotransfer and on day 20 after single transfer. (B and C) IFN-γ and TNF-α coproduction (B) and granzyme-B (GzmB) expression (C) by donor TILs recovered as in A after restimulation with gp33 peptide. Representative flow cytometry plots (left) and quantification of donor cell numbers (B) and MFI (C). (D) Graphical presentation of TIL polyfunctionality (green) and tumor-induced T cell exhaustion (red) for WT and VHL-KO donor TILs. (E) Specific lysis at indicated effector-to-target ratios of B16.gp33 target cells by sorted donor P14 TIL populations recovered on day 7 after cotransfer as in A, n = 6 per condition. (F) Dependence of CTL-mediated target cell lysis on granzyme-B/perforin pathway, n = 4 per condition. Data are representative of 3 (A–D) and 2 (E and F) independent experiments. Bars, line charts, and error bars represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (2-tailed Student’s t test in A–E, 1-way ANOVA with Bonferroni correction for multiple comparisons in F).
We also cotransferred VHL-KO CD8+ T cells bearing the irrelevant OT-I transgenic TCR, and these cells did not exhibit enhanced accumulation (Supplemental Figure 2, F and G), showing that the increase in the number of TILs and elevated effector function observed with VHL deficiency was T cell intrinsic and antigen dependent. Finally, we crossed Vhlfl/fl P14 mice with those harboring estrogen receptor–controlled Cre-recombinase (ER-Cre) to induce deletion of Vhl upon in vivo tamoxifen treatment. After cotransfer of Vhlfl/fl P14 ER-Cre+ and ER-Cre– P14 cells into B16.gp33 tumor–bearing mice, mice were treated with tamoxifen and TILs were analyzed at day 9 after transfer. Even though deletion was not complete by day 9 after transfer (Supplemental Figure 2H), loss of Vhl after transfer resulted in enrichment of the ER-Cre+ P14 TIL population over their cotransferred ER-Cre–negative counterparts (Supplemental Figure 2I) and upregulation of granzyme-B (Supplemental Figure 2J).
We also investigated whether HIF activity in polyclonal CD8+ TILs correlated with the phenotype observed for VHL-deficient P14 cells. We developed a yellow fluorescence protein (YFP) reporter mouse that reported transcription of a HIF-α target gene, egl-9 family HIF 3 (Egln3), as a proxy for HIF-α transcriptional activity (Supplemental Figure 3, A–C). In s.c. B16.gp33 tumors, YFP signal was observed in the activated CD44+CD11a+ CD8+ TIL subset (Supplemental Figure 3D). The activated YFP+CD8+ TIL subset showed upregulation of multiple exhaustion-associated and costimulatory markers compared with the activated YFP– subset and had a similar pattern of expression to the VHL-KO P14 TILs (Supplemental Figure 3, E and F). Thus, the expression of cell-surface molecules associated with exhaustion by TILs reporting HIF transcriptional activity was similar to VHL-KO TILs, consistent with HIF promoting these phenotypes.
Finally, we compared cytolytic function of the P14 TIL populations (5) and found that VHL-KO P14 TILs mediated nearly 2-fold greater lysis of target cells than WT P14 TILs from the same tumors (Figure 3E), which was dependent on granzyme-B/perforin (Figure 3F). Taken together, we found that VHL deficiency promoted superior antitumor function by TILs, despite exhaustion-associated coinhibitory marker expression.
HIF transcriptional activity drives the generation of highly functional CD8+ Trm-like TILs.
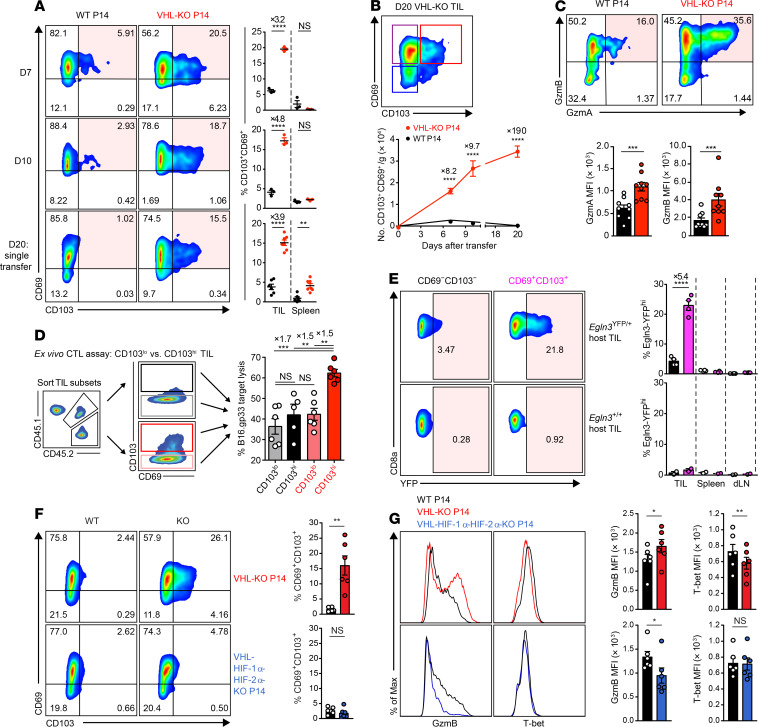
Tumor infiltration by CD8+ T cells is a positive prognostic factor in cancer, and recently the accumulation of CD8+ TILs with Trm features has been shown to predict a better clinical outcome in multiple settings (23, 24, 38–41). To understand how HIF activity affects the Trm phenotype of CD8+ TILs, we evaluated expression of the canonical tissue-residency markers CD69 and CD103 after cotransfer of WT and VHL-KO P14 cells into tumor-bearing mice. We observed a prominent CD69+CD103+ subset (15%–20% of TILs) within the VHL-KO but not the WT population (Figure 4, A and B). This CD69+CD103+ subset was observed in VHL-KO compared with WT TILs in multiple tumor settings: (a) polyclonal CD8+ TILs recovered from the lungs of VHL-KO mice after i.v. B16 challenge; (b) P14 TILs recovered from s.c. B16.gp33, MC38.gp33, and KMR.gp33 tumors; and (c) ER-Cre+ P14 TIL recovered from s.c. B16.gp33 and MC38.gp33 tumors after tamoxifen-induced in vivo deletion (Supplemental Figure 4A). Administration (i.v.) of anti-CD8α antibody before euthanization to label vascular CD8+ T cells confirmed tissue localization of the CD69+CD103+ subset within tumors (Supplemental Figure 4B).
Figure 4. HIF promotes CD69+CD103+ Trm-like CD8+ TILs with higher cytolytic capacity against target cancer cells.
(A) Representative flow cytometry (left) and frequency (right) of CD69 and CD103 expression on WT and VHL-KO P14 cells after cotransfer (days 7, 10) or single transfer (day 20) into B16.gp33-bearing mice. n = 3 (days 7–10), n = 6 WT, and n = 7 VHL-KO (day 20). (B) CD69 and CD103 expression on VHL-KO TILs on day 20 (top) and absolute number of CD69+CD103+ donor TILs over time (bottom). n = 7 (day 10), n = 6 (days 7 and 20). (C) Representative flow cytometry (top) and MFI (bottom) of granzyme-A and granzyme-B expression on WT and VHL-KO TILs after cotransfer into B16.gp33-bearing hosts, n = 9. (D) Specific target cell lysis at 1:1 effector-to-target ratio by indicated sorted CD103lo and CD103hi donor P14 TIL subsets recovered on day 7 as in A, n = 6. (E) YFP signal in polyclonal CD69–CD103– and CD69+CD103+ CD8+ Egln3-YFP-KI (Egln3YFP/+) and control (Egln3+/+) TILs stained on day 13 following B16.gp33 challenge (left) and quantification of YFP signal (right), n = 4 for Egln3YFP/+ and n = 2 for Egln3+/+. (F) Representative flow cytometry (left) and frequency (right) of CD69 and CD103 on donor TILs 7 days after cotransfer of WT and VHL-KO P14 cells (top) or WT and VHL-HIF1α-HIF-2α-KO P14 cells (bottom), n = 6 VHL-KO/WT and n = 5 VHL-HIF1α-HIF-2α-KO/WT. (G) Representative flow cytometry (left) and MFI (right) of granzyme-B and T-bet expression on P14 TIL populations recovered as in F. Data shown as mean ± SEM are representative of 4 (A), 2 (B and C), and 3 (D–F) independent experiments. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (1-way ANOVA with Bonferroni correction for multiple comparisons in D, 2-tailed Student’s t test in others).
Trm CD8+ T cells are the first-line responders of adaptive immunity within tissues and exhibit potent effector function and cytolytic capacity (42). We found increased expression of both granzyme-A and granzyme-B in VHL-KO TILs compared with WT TILs after cotransfer into B16.gp33 tumor–bearing mice (Figure 4C). Additionally, the tumor cell killing capacity of CD69+CD103+ TILs sorted into CD103hi and CD103lo subsets showed that the CD103hi VHL-KO P14 TIL was the most cytolytic subset (Figure 4D). Consistent with cytolytic function, granzyme-B was found to be upregulated by the Trm-like subset compared with other VHL-KO TIL subsets (Supplemental Figure 4, C and D). In contrast, the cytolytic advantage of VHL-KO over WT P14 TILs (as shown in Figure 3, E and F) was not observed for the CD103lo subset.
HIF–YFP-reporter activity among polyclonal CD8+ TILs showed a higher frequency of YFP+ cells among the CD69+CD103+ subset than the CD69–CD103– subset (Figure 4E). To test whether HIF-α activity supports the formation of the Trm-like CD8+ TIL subset, we transferred TKO P14 cells into B16.gp33 tumor–bearing mice, where they failed to give rise to the CD69+CD103+ TIL subset (Figure 4F). Similarly, HIF was required for the enhanced granzyme-B expression and the relative downregulation of T-bet by VHL-KO TILs (Figure 4G and Supplemental Figure 4C), which was notable because T-bet downregulation is a key characteristic of Trm differentiation (43). Considering that T-bet has been implicated in the direct suppression of CD103 expression (44), these findings support a model where HIF transcriptional activity drives the generation of CD103+ Trm in part via T-bet downregulation. Expression of additional surface molecules in a pattern consistent with reduced tissue egress and a bona fide Trm phenotype was also observed among the CD69+CD103+ VHL-KO subset; CD62L and Ly6C were downregulated, whereas CD44, which is linked to peripheral tissue retention, and CD101, which limits aberrant T cell activation, were upregulated (Supplemental Figure 4D). Consistent with these findings, analysis of the HIF reporter CD103+ TILs showed that the highest frequency of coexpressed activation/exhaustion surface receptors PD-1 and GITR was found on the YFP+ Trm-like TILs (Supplemental Figure 4E). Together, these experiments established that HIF was required for the generation of the VHL-dependent Trm-like CD8+ TIL subset.
Enrichment of Trm gene expression signature by VHL-deficient P14 TILs is associated with phenotypic hallmarks of T cell exhaustion.
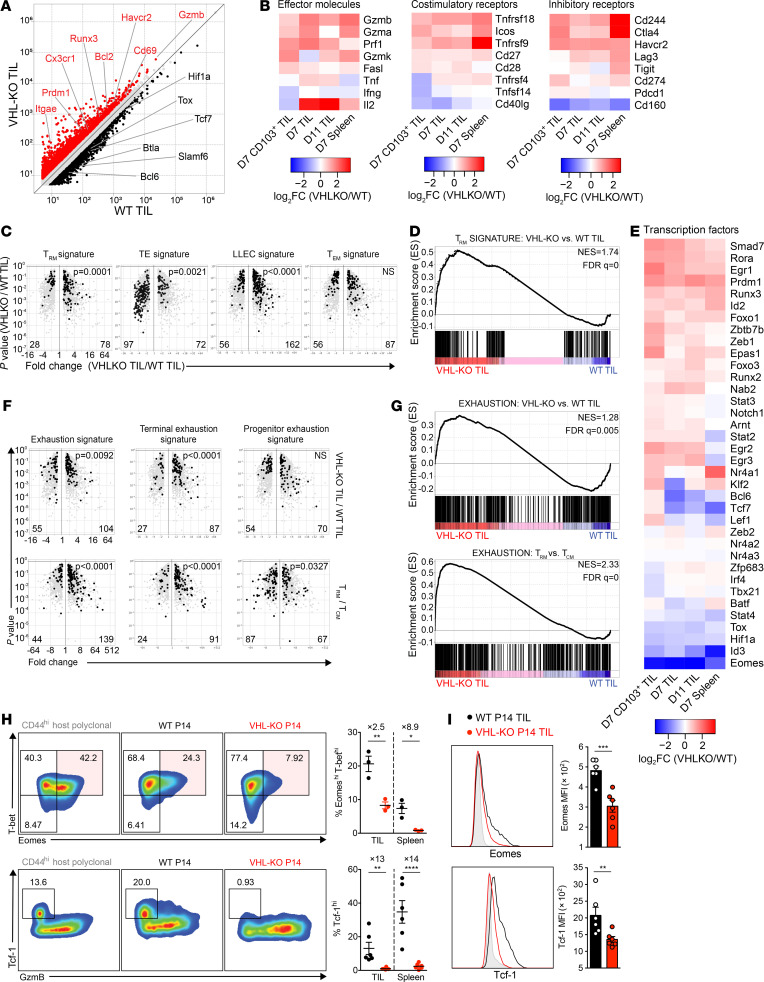
We next compared transcriptional profiles of cotransferred VHL-KO and WT P14 populations recovered from B16.gp33 tumors (GSE156342), and found differential expression of genes encoding effector, costimulatory, and coinhibitory molecules by RNA-Seq on days 7 and 11 after transfer (Figure 5, A and B). Thus, much as we observed for VHL-KO CD8+ T cells responding to chronic infection, VHL-KO TILs upregulated molecules associated with T cell activation to significantly higher levels than their WT counterparts (31). Notably, this included molecules associated with cytolytic function as well as inhibitory molecules associated with loss of T cell function, chronic stimulation, and activation. Interestingly, the VHL-KO P14 TIL population showed significant enrichment in core CD8+ Trm signature genes (Figure 5C) (25). Gene set enrichment analysis (GSEA) of the Trm signature comparing VHL-KO and WT P14 TILs corroborated this observation (Figure 5D). Furthermore, the pattern of expression for key transcription factors controlling Trm fate (12), including upregulation of Prdm1 (encodes Blimp-1), Egr2, and Runx3 and downregulation of Bcl6, Tcf7, and Eomes, was also observed for VHL-KO TILs compared with WT TILs (Figure 5E). Thus, constitutive HIF-α activity achieved by VHL deletion instructed a Trm gene expression program in tumor-specific CD8+ T cells.
Figure 5. Expression of the Trm and T cell exhaustion gene expression signatures by VHL-deficient P14 TILs.
(A) Relative gene expression by VHL-KO and WT day 11 P14 TILs. Red and black denote an increase of 1.5-fold or greater in expression by VHL-KO over WT TILs or vice versa, respectively. (B and E) Changes in gene expression between VHL-KO and WT P14 for day 7 CD103+ TILs, days 7 and 11 bulk TILs, and day 7 spleen. (C) Volcano plots showing gene expression enrichment for Trm, TE, LLEC, and Tem signature genes from published datasets (25, 45) by VHL-KO versus WT D11 TIL. Genes with less than 1.5-fold change in expression were excluded. (D) GSEA of Trm signature in VHL-KO versus WT D11 TILs. (F) Comparison of T cell exhaustion signatures for Trm, TE, LLEC, and Tem between VHL-KO and WT D11 TILs (top) and small intestine intraepithelial P14 Trm relative to spleen P14 Tcm (45) (bottom). Genes with less than 1.5-fold change in expression were excluded. (G) GSEA of T cell exhaustion signature in VHL-KO versus WT D11 TILs and P14 Trm versus P14 Tcm. (H) Representative flow cytometric analysis (left) and quantification (right) of CD8+CD44+ VHL-KO and WT P14 TILs for T-bet, Eomes, Tcf-1, and granzyme-B. Data are representative of 3 independent experiments (n = 3 for Eomes and T-bet coexpression; n = 6 for Tcf-1). (I) Representative histograms of Eomes and Tcf-1 expression (left) and MFIs (right) in cotransferred donor P14 TIL populations. Overexpressed gene counts are shown at bottom of volcano plots, and statistical P values denote comparisons between indicated groups. Error bars represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test in H and I). FC, fold change; NS, not significant; NES, normalized enrichment score; FDR q value, FDR corrected P value.
T cell exhaustion is phenotypically characterized by expression of coinhibitory receptors and activation-associated markers, many of which are also upregulated on CD8+ Trm cells (9, 18, 24). Interestingly, we found that established exhaustion and terminal-exhaustion signatures (6) were both enriched among virus-specific CD8+ Trm and VHL-KO TILs (Figure 5F), and this was further supported by equivalent exhaustion-associated gene set enrichment by CD8+ Trm compared with central memory T cells (Tcm cells) and VHL-KO TILs compared with WT TILs (Figure 5G). These data showed that the T cell exhaustion/activation phenotype observed due to VHL deletion was accompanied by transcriptional programs also observed by Trm cells. Moreover, resistance to tumor-induced exhaustion as observed for VHL-KO TILs and accumulating evidence for the functional importance of Trm-like CD8+ TILs (42) together suggest that some markers of the exhaustion phenotype in the context of Trm-like TILs do not always reflect dysfunction.
Effector cell differentiation of CD8+ T cells is derailed by tolerogenic antigen exposure, suppressive cytokine milieu, and metabolic perturbations in chronic viral infections and malignancies (20). Nevertheless, “stem-like” progenitor-exhausted CD8+ T cells arise under these conditions, possessing characteristics of classical memory subsets that give rise to terminally exhausted subsets (20). Transcription factors Tcf-1 and Eomes promote Tcm differentiation (7) and were expressed by WT TILs but were not found in VHL-KO TILs (Figure 5, H and I). Additionally, VHL-KO cells gradually downregulated expression of CD43, CD27, and CXCR3 in tumors and spleens (Supplemental Figure 4F), indicating emergence of long-lived effector cells (LLEC), a rapidly cytotoxic, cytokine-producing population that is terminally differentiated but survives to late time points. Indeed, transcriptomic analysis revealed enrichment of LLEC signature in VHL-KO TILs (Figure 5C; ref. 45), whereas effector-memory Tem and terminal-effector (TE) cell signatures were not. Together these data indicate that VHL deletion promoted sustained effector function, attributed to LLEC and Trm phenotypes, and limited terminal differentiation of TILs in tumor-bearing mice.
Combined anti–PD-1 and VHL-deficient CD8+ T cell ACT reduces tumor burden.
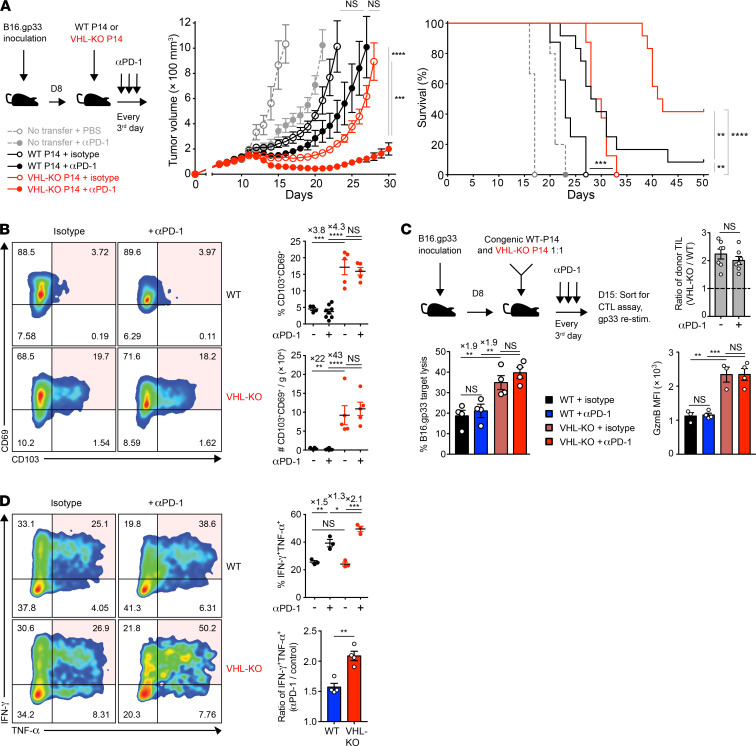
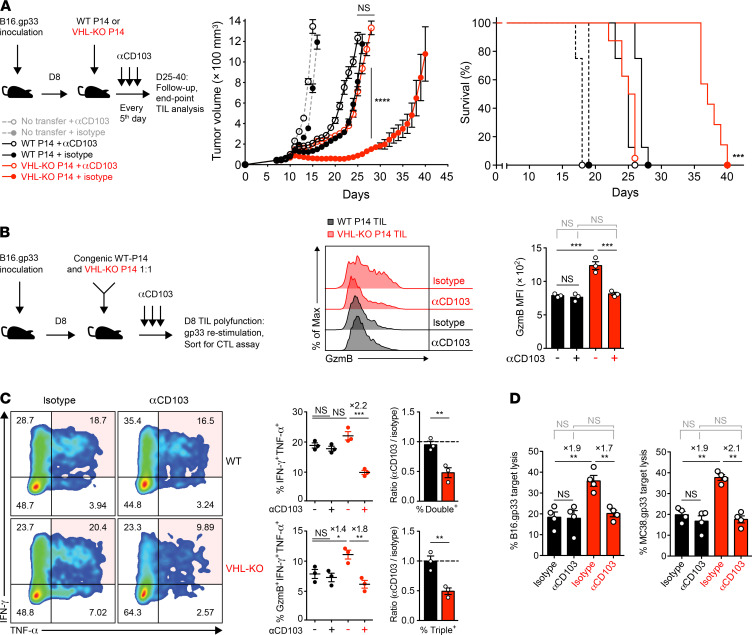
The combination of ACT and checkpoint blockade holds promise for treatment of solid tumors (46). Trm-like CD8+ TILs found in lung cancer patients have been reported to be responsive to PD-1 blockade in a CD103-dependent fashion (18), prompting us to assess VHL-KO ACT together with PD-1 blockade immunotherapy. After development of B16.gp33 tumors (>75 mm3), we transferred WT or VHL-KO P14 cells followed by treatment with PD-1 blocking or isotype control antibody. Mice receiving anti–PD-1 and VHL-KO cells showed decreased tumor burden compared with all other groups, with complete responses in 42% of mice (Figure 6A). Endpoint tumors showed effective blockade of PD-1 in both donor groups (Supplemental Figure 5A). Although anti–PD-1 immunotherapy did not further increase accumulation of donor TILs (Supplemental Figure 5A), VHL-KO cells retained the higher frequency and numbers of the CD69+CD103+ Trm-like subset over their WT counterparts (Figure 6B). Additionally, anti–PD-1 did not affect target cell killing or granzyme-B levels, which remained consistently higher for the VHL-KO population after cotransfer (Figure 6C). However, PD-1 blockade boosted the capacity of both donor TIL populations to coproduce IFN-γ and TNF-α, with a greater impact on VHL-KO TILs (Figure 6D). This is consistent with the idea that although VHL-KO cells have enhanced antitumor immunity, they are eventually overcome by exhaustion. Anti–PD-1 therapy may allow for increased proliferation of progenitor-exhausted cells and/or prevention of terminal exhaustion. These data indicate that checkpoint blockade in combination with strategies to enhance Trm-like TILs further improve responses to therapy and increase survival.
Figure 6. Immunotherapy with anti–PD-1 improves response to established tumors.
(A) Tumor growth and survival in B16.gp33-bearing mice after transfer of WT or VHL-KO P14 cells and anti–PD-1(αPD-1) or PBS treatment. Tumor growth was compared by Mann-Whitney U test (middle) and survival was assessed by Bonferroni-corrected log-rank test (right). n = 12 (αPD-1) and n = 8 (isotype) in P14 transferred mice; n = 5 (αPD-1) and n = 3 (isotype) with no transfer. (B) Representative flow cytometry (left) and quantification of frequency (top right) and weight-adjusted numbers (bottom right) of CD69+CD103+ P14 TILs recovered at endpoint in A from mice treated with anti–PD-1 or PBS. n = 8 for WT donor with αPD-1; n = 5 for rest. (C) Specific target cell lysis of sorted donor WT and VHL-KO TILs after treatment with anti–PD-1 or isotype control antibody (bottom left). Ratio of cotransferred donor VHL-KO/WT TILs recovered (top right) and MFI of granzyme-B expression on sorted populations (bottom right) are shown. n = 4 for CTL assay; n = 7 for donor cell ratio; n = 4 for αPD-1; n = 3 for isotype control in granzyme-B panel. (D) Representative flow cytometry of IFN-γ and TNF-α coproduction by donor TIL populations upon restimulation, quantitated by frequency (top right) and compared by ratio of anti–PD-1 to control treatment (bottom right), n = 3 (top right), and n = 4 pooled (bottom right). Data are representative of 2 (A–C) and 4 (D) independent experiments. Error bars represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test C top and D bottom panel, 1-way ANOVA with Bonferroni correction for multiple comparisons in others, except as specified in A).
VHL-deficient CD8+ T cells establish sustained protection from tumor challenge.
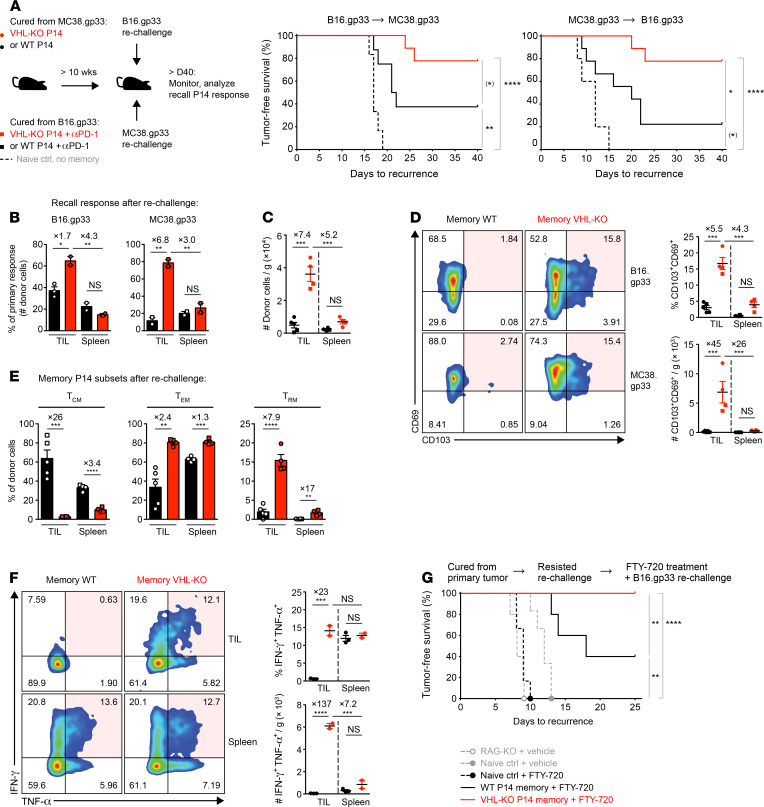
To assess antitumor memory, we rechallenged cured mice with an alternate gp33-expressing tumor cell line. Memory VHL-KO cells were significantly more effective at protecting hosts from tumor growth after rechallenge with either gp33-expressing tumor type (~78%, both) compared with the WT memory cells (MC38.gp33 [38%] and B16.gp33 [22%] rechallenge tumors) (Figure 7A). The recall response, as measured by the magnitude of TIL expansion scaled to the primary response (Figure 7B) and by recovered donor numbers (Figure 7C) were both significantly higher in mice harboring memory VHL-KO cells than their WT counterparts. The responding memory VHL-KO population included a prominent CD69+CD103+ Trm-like subset (~15%) which was absent in the WT memory population, as well as an enrichment of cells with a Tem phenotype associated with rapid effector functions (Figure 7, D and E). In contrast, animals with WT memory cells generated Tcm and Tem subsets but only a minor Trm-like component (~2%) (Supplemental Figure 5B). WT and VHL-KO memory populations recovered from spleens produced TNF-α and IFN-γ, but in the TIL compartment only the memory VHL-KO cells were capable of coproducing effector cytokines (Figure 7F). Finally, mice protected from tumor growth during rechallenge were tested for local protective tissue-resident memory by daily treatment with FTY720, which targets S1PR1, preventing T cell egress from lymph nodes (27, 47) and circulating T cells from contributing to the response, followed by further rechallenge with B16.gp33 tumor cells at the primary tumor site (Figure 7G). Control mice receiving FTY720 but without protective P14 memory succumbed to B16.gp33 challenge within 2 weeks, whereas 2 out of 5 mice with WT P14 memory and all of the mice with VHL-KO P14 memory (14 out of 14) were protected from rechallenge upon FTY720 treatment, demonstrating local protection by tissue-resident memory. Thus, tumor eradication upon VHL-deficient ACT resulted in the generation of functional antitumor T cell memory capable of protecting against local rechallenge.
Figure 7. VHL-deficient memory T cells confer superior protection from tumor rechallenge.
(A) Tumor-free survival after rechallenge with B16.gp33 (middle) or MC38.gp33 (right) in mice with complete regression of MC38.gp33 and B16.gp33 (+αPD-1) tumors, respectively (see Figure 1B and Figure 5A), n = 8 WT, n = 9 VHL-KO, n = 6 control (middle); n = 9 experimental groups, n = 5 control (right). (B) Recovered memory WT or VHL-KO P14 cell counts in animals in A were adjusted to the respective mean counts recovered in primary tumor models (see Figure 1E). (C) Weight-adjusted donor TIL and splenocyte numbers between animals receiving WT (black) and VHL-KO (red) memory cells, n = 4 VHL-KO, n = 5 WT. (D) CD69 and CD103 expression (left) and quantification (right) on recovered memory cells in C. (E) Frequency of Tcm (CD62Lhi), Tem (CD62LloCD103lo), and Trm (CD103hiCD69hiCD62Llo) phenotypes within the donor memory populations. (F) Recall function of memory cells recovered from mice bearing B16.gp33 rechallenge tumors. Representative flow cytometry (left) of IFN-γ and TNF-α coproduction by indicated memory TILs (top row) and memory splenocytes (bottom row), and quantification by frequency (top right) and weight-adjusted cell numbers (bottom right), n = 2 VHL-KO, n = 3 WT. (G) Tumor-free survival after FTY720 or vehicle and rechallenge with B16.gp33 tumor cells in mice that resisted the rechallenge tumor growth in A. RAG-KO mice were controls, n = 14 VHL-KO, n = 5 WT, n = 6 naive control, n = 5 RAG-KO. Data are representative of 2 independent (A–F) or pooled (G) experiments. Circle and square symbols in B–F indicate rechallenge with B16.gp33 and MC38.gp33, respectively, error bars represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; (A and G) Bonferroni-corrected log-rank test, (B–D, and F) 1-way ANOVA with Bonferroni correction for multiple comparisons, (E) Student’s t test. (*) failed to reach Bonferroni-corrected threshold although showing P < 0.05.
Improved antitumor activity by VHL-deficient TILs is dependent on CD103.
We next investigated the role of CD103 in VHL-KO CD8+ T cell antitumor efficacy in primary tumor models. B16.gp33-bearing mice were treated with a monoclonal anti-CD103 antibody or isotype control starting 1 day before adoptive transfer of either VHL-KO or WT P14 cells. In vivo treatment with anti-CD103 resulted in effective blockade of CD103 expression on VHL-KO TILs (Supplemental Figure 5C), which translated into loss of tumor control (Figure 8A). The mice receiving VHL-KO cells and anti-CD103 lost their survival advantage over mice receiving WT cells (Figure 8A). The blockade of CD103 resulted in partial reduction in VHL-KO cell numbers in endpoint tumors but not WT cell numbers (Supplemental Figure 5C). Although other endogenous cell types, such as DCs and CD4+ Tregs, may be affected by anti-CD103, tumor growth, survival, and TIL numbers in mice receiving WT cells were not significantly affected by the CD103 blockade (Figure 8A and Supplemental Figure 5C), indicating a dependence on the presence of VHL-KO T cells.
Figure 8. Improved antitumor efficacy and effector function by VHL-deficient TILs is dependent on CD103.
(A) Tumor growth and survival in anti-CD103 (αCD103) or isotype control treated B16.gp33 tumor–bearing mice after transfer of WT and VHL-KO P14 cells. Tumor growth was compared by Mann-Whitney U test (middle) and survival assessed by Bonferroni-corrected log-rank test (right), n = 8 for donor groups with αCD103; n = 7 for VHL-KO with isotype; n = 8 for WT with isotype; n = 4 for αCD103; n = 4 for isotype. (B) Granzyme-B expression by cotransferred P14 TIL populations for indicated antibody therapy (middle) after restimulation using cognate gp33 peptide. MFI is quantitated on the right, n = 3 per group. (C) Representative flow cytometry of IFN-γ and TNF-α coproduction by P14 TIL populations upon restimulation (left), quantitated by frequency (top middle) and compared by ratio of anti-CD103 to isotype (top right). Frequency of P14 TILs triple-positive for granzyme-B, IFN-γ, and TNF-α (bottom middle) and comparison by ratio of anti-CD103 to isotype (bottom right), n = 3 per group (middle) and pooled (right). (D) CTL assay on B16.gp33 (left) or MC38.gp33 (right) target cells of cotransferred donor P14 TIL populations sorted from the same tumors after in vivo anti-CD103 or isotype treatment at 1:1 E/T ratio. Graphs show specific target cell lysis mediated by indicated P14 TIL populations, n = 4 per group. Data are representative of 2 (A and D) and 3 (B and C) independent experiments. Mean fold increase in frequency between groups provided in C and D where appropriate. Symbols represent mean in A and individual mice in B–D, except for C (right panel) where symbols represent mean ratio of separate experiments. Error bars represent mean ± SEM. NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (Student’s t test in C right panel, 1-way ANOVA with Bonferroni correction for multiple comparisons in others, except as specified in A).
CD103 blockade resulted in a loss of the enhanced granzyme-B expression by VHL-KO over WT P14 TILs (Figure 8B). Further, coproduction of effector cytokines IFN-γ and TNF-α upon restimulation was also diminished for the CD103-treated VHL-KO TIL population compared with the isotype control (Figure 8C). In vivo treatment with anti-CD103 resulted in dramatic reduction in cytolytic advantage by the VHL-KO population in the ex vivo killing assays against B16.gp33 and MC38.gp33 target cancer cells (Figure 8D). This was accompanied by a reduction in Eomes and T-bet suppression (Supplemental Figure 5D). Thus, blockade of CD103 attenuated the improved accumulation, polycytokine production, and cytolytic capacity of VHL-KO P14 TILs, consistent with a complete loss of survival advantage over WT recipients. This observation supports the conclusion that the acquisition of the CD69+CD103+ Trm-like phenotype is a key element of enhanced tumor immunity by VHL-KO TILs.
VHL-deficient CAR T cells show enhanced accumulation and differentiation into Trm-like TILs in solid tumors.
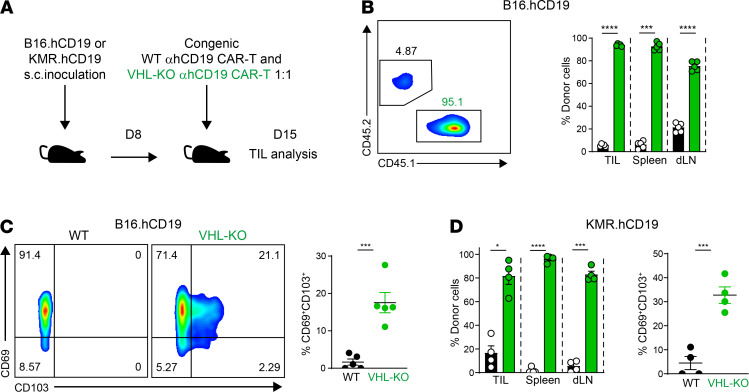
To determine whether constitutive HIF-α activity could allow for improved antitumoral responses by CAR T cells, we transduced polyclonal T cells from VHL-WT and VHL-KO donors with the second generation anti-human CD19 CAR and transferred to mice bearing B16 tumors that expressed human CD19 surface antigen (B16.hCD19) as previously described (48) (Figure 9A and Supplemental Figure 6, A–D). Eight days after T cell transfer, donor CAR T cells from tumors, spleens, and draining lymph nodes were analyzed for accumulation and expression of CD103 and CD69. As observed for VHL-KO P14 cells, VHL-KO CAR T cells again accumulated in the tumor, spleen, and draining lymph node with greater frequency than WT CAR T cells (Figure 9B). Not only was the tissue retention of CAR T cells enhanced by unregulated HIF-α expression, tumor-infiltrating CAR T cells derived from VHL-KO donors showed a significantly increased frequency of the CD69+CD103+ Trm phenotype compared with WT CAR T cells (Figure 9C). Additionally, tumor-infiltrating CAR T cells showed increased CD69 expression, and like other antigen-specific VHL-KO TILs, VHL-KO CAR T cells showed enhanced granzyme-B expression (Supplemental Figure 6E). Similar results were found with a model of rhabdomyosarcoma that we modified to express human CD19 surface antigen (KMR.hCD19, Supplemental Figure 6, A and B, and Figure 9, A and D). Collectively, these results suggest that translating the advantage of VHL deficiency in ACT to the powerful and expanding field of CAR T therapy allows for improved antitumor responses against solid tumors, and that this response correlates with the acquisition of a Trm phenotype by CAR TILs.
Figure 9. VHL-deficient CD8+ CAR T cells accumulate in tumors with an enhanced Trm-like phenotype.
(A) Schematic of experimental protocol for adoptive cotransfer of congenically distinct WT and VHL-KO anti-hCD19 (αhCD19) CAR T cells into B16.hCD19- or KMR.hCD19-bearing mice. (B) Distribution of donor WT and VHL-KO anti-hCD19 CD8+ CAR T cells 7 days after cotransfer into B16.hCD19 tumor–bearing mice as outlined in A. Representative flow cytometric analysis of donor CD8+ TILs gated on anti-hCD19 CAR+ T cells (left) and relative proportions of donor CAR T cells in tumors, spleens, and draining lymph nodes (right) are shown. (C) Expression of Trm markers CD103 and CD69 on donor CAR T cells from A. Representative flow cytometric analysis (left) and frequency of CD69+CD103+ cells (right) are shown. (D) Relative proportion of congenically distinct donor WT and VHL-KO of anti-hCD19 CAR-expressing CD8+ T cells in tumors, spleens, and draining lymph nodes, 7 days after cotransfer into KMR.hCD19 tumor–bearing mice as in A. Data are representative of 3 independent experiments with a total of 12 B16hCD19- and 12 KMRhCD19-bearing mice. Symbols represent individual mice, error bars represent mean ± SEM. *P < 0.05, ***P < 0.001, ****P < 0.0001 (Student’s t test).
Discussion
In this study, we identified a previously unknown role for the VHL/HIF axis in controlling the formation of cytotoxic CD69+CD103+ Trm-like CD8+ T cells in tumors and established the enhanced antitumor activity of this population. CD8+ Trm cells are embedded in tissue and act as sentinels, providing enhanced memory protection in peripheral tissues, which are common sites of pathogen reexposure (11). Notably, T cells with characteristics of Trm have been observed in numerous oncology studies, where they have been associated with improved clinical outcomes (19). We showed in multiple model systems that high levels of HIF-α activity in CD8+ TILs, achieved either by physiological induction in tumors (measured by Egln3YFP/+ reporter) or by VHL deletion, promoted the formation of a CD69+CD103+ Trm-like population in tumors. This subset resided within tumor tissues and expanded in numbers during the antitumor response. CD69+CD103+ Trm-like cells were phenotypically and functionally similar to bona fide virus-specific Trm cells, displaying enhanced cytolytic function and cytokine production compared with WT TILs or VHL-deficient TILs that did not display Trm characteristics. The Trm-like subset emerged within the polyclonal, TCR-transgenic, and CAR CD8+ T cell responses against melanoma, adenocarcinoma, rhabdomyosarcoma solid tumors, and a metastatic model of melanoma. VHL-deficient TILs could achieve complete tumor regression and generate long-term protection by mounting a robust protective recall response. This response was sufficient in protecting against tumor recurrence in the absence of circulating memory T cells, unlike control mice with WT memory that lacked the Trm component. Thus, HIF-α transcriptional activity drives the formation of Trm-like CD8+ TILs in a range of tumor contexts, promoting enhanced accumulation, effector function, and memory protection.
We found enrichment of the core Trm gene expression signature in VHL-deficient TILs, composed of transcription factors classically associated with differentiation toward effector and memory-precursor fates that operate in concert to program CD8+ Trm differentiation, including Egr2, Runx3, and Prdm1, and TGF-β–dependent downregulation of T-bet and Eomes required for Trm formation (43). In addition, the CD103+ subset within VHL-deficient TILs showed suppression of Klf2, which is required for downregulation of S1PR1, thus preventing egress from the tissue (49, 50). Other molecules whose upregulation is associated with tissue retention such as CD103, CD69, and CD44 were observed by VHL-deficient TILs, as well as elevated expression of prosurvival and cytolytic factors Bcl-2 and granzyme-B, consistent with the acquisition of Trm qualities. Direct effects of hypoxia can improve effector CD8+ T cell function by increasing granzyme-B and cytolytic capacity (37, 51, 52), which we found to be mediated by HIF-α factors and the granzyme-B/perforin pathway. Our findings in vivo suggest that the HIF-α–mediated functional features are dependent on the CD103 integrin, which is known to enhance Trm retention, but also functions in activation and cytotoxicity mediated via direct engagement with E-cadherin on target cells that strengthens the immune synapse and polarizes cytolytic granules toward the target cell (53, 54). These biochemical interactions support our in vivo findings demonstrating that the CD103+ Trm-like TIL subset, driven by constitutive HIF-α activity, promoted retention in the tumor, showed increased cytolytic granule composition, and mediated superior cytotoxicity and antitumor efficacy in a CD103-dependent manner.
The generation and maintenance of an exhausted T cell pool, as characterized in the context of chronic infections, is coordinated in part by transcription factors T-bet, Eomes, Tcf-1, and Tox (4, 5, 7, 55, 56). These factors were downregulated in VHL-deficient cells, potentially contributing to resistance to exhaustion. HIF-α activity in CD8+ TILs was associated with a highly activated phenotype and functional state in spite of expression of exhaustion-associated inhibitory receptors, several of which are known to be regulated by the VHL/HIF axis (31, 32, 57). Our data are in line with functional attributes of CD8+ Trm cells, both in antiviral memory and tumors, which are phenotypically reminiscent of exhausted T cells with upregulation of inhibitory receptors, yet maintain effector functions. Indeed, we found the terminal T cell exhaustion signature was also associated with Trm signatures of VHL-deficient TILs and bona fide Trm cells in our transcriptomic analysis, suggesting that these transcriptional programs are intertwined. In contrast to WT TILs that succumbed to exhaustion, VHL-deficient TILs retained their polyfunctionality within the same tumor microenvironment.
Recent advances in immunotherapy, such as checkpoint blockade and ACT, have provided lifesaving therapy for patients with malignancy. Accumulating clinical evidence indicates that Trm-like CD8+ TILs are particularly sensitive to PD-1 blockade (9, 10, 24). Notably, Trm-like CD8+ TILs isolated from lung cancer patients readily responded to PD-1 blockade in a CD103-dependent fashion (18). Indeed, we observed further enhanced effector cytokine production and complete responses in 42% of tumor-bearing mice when treated with VHL-deficient ACT and anti–PD-1 immunotherapy compared with only 8% in mice receiving WT cells and anti–PD-1. Thus, regulating HIF activity and the presence of Trm-like TILs emerges as a viable strategy to enhance checkpoint blockade efficacy.
ACT continues to evolve as an important pillar of immunotherapy for the treatment of cancer, and manipulation of the intrinsic cellular machinery of these living drugs offers opportunity for the advancement of this field. To date, use of cellular therapy for treatment of solid tumors has largely yielded disappointing results, limited in part by the inability of lymphocytes to retain functional activity and persist in the solid tumor microenvironment, as well as inadequate infiltration of tumors by CAR T cells (1, 2). Our studies demonstrated that the VHL/HIF pathway offers a single-gene target that can yield dramatically improved antitumoral responses in multiple solid tumor types. Follow-up studies are needed to connect the VHL/HIF pathway in TILs and clinical outcomes in human patients. Importantly, the CAR T model employed in our studies indicates a clear translatable tool to enhance CAR T responses. Further studies to limit VHL expression in WT donor cells using single-gene deletion techniques such as CRISPR or pharmacological ex vivo manipulation of the VHL/HIF axis in CD8+ T cells would be valuable in translating these findings to the clinic.
Methods
Additional details are provided in Supplemental Methods.
Mice
All mice were on the C57BL/6J background and bred and housed at University of California, San Diego in specific pathogen–free conditions. The following recipient mice were purchased from the Jackson Laboratory: C57BL/6J, CD45.1. (B6.SJL-Ptprca Pepcb/BoyJ) and Rag1–/– (B6.129S7-Rag1tm1Mom/J). The following mice have been described and were provided by Randall S. Johnson (Karolinska Institutet, Stockholm, Sweden; University of California, San Diego, San Diego, USA; and University of Cambridge, Cambridge, United Kingdom): Vhlfl/fl mice (58), Hif1αfl/fl mice (59), and Epas1fl/fl mice (60). Deletion of loxP-flanked genes in T cells was achieved by crossing homozygous loxP-flanked alleles to either dLck-Cre mice (61) or to Rosa26Cre-ERT2 (ER-Cre) mice (62) followed by in vivo tamoxifen treatment. For adoptive transfer experiments, the mice were further crossed to either P14 mice, which recognize LCMV glycoprotein gp33 presented by MHC class I-Db (63), or to OT-I mice that recognize SIINFEKL derived from OVA presented by MHC class I-Kb molecules (64). Egln3-YFP reporter mice were generated by inserting turboYFP after the first exon of the HIF target gene, egl-9 family HIF 3 (Egln3), via homologous recombination in C57BL/6 ES cells. Suitable clones were selected via neomycin resistance and injected into albino C57BL/6 embryos (Ozgene). Founder mice were crossed to flippase recombinase–expressing mice to remove the neomycin selection cassette. The YFP insert introduces a stop codon disrupting the Egln3 gene; thus, studies were performed with heterozygous animals (Egln3YFP/+).
For polyclonal in vivo experiments (experimental metastasis, Egln3-YFP reporter model), age-matched cohorts of littermate adult mice of each genotype were used. For adoptive transfer experiments, age-matched VHL-KO and/or VHL-HIF-1α-HIF-2α-KO and littermate WT control (Cre-negative) TCR-transgenic mice were used as donors, and age-matched or littermate C57BL/6 (CD45.2.), CD45.1., or CD45.1.2. mice were used as recipients. Donor and recipient mice were 6 to 12 weeks old, except the cured recipient mice harboring memory P14 cells that were first monitored for more than 10 weeks for remission.
Cancer cell lines
The following cancer cell lines were used: B16.gp33, MC38.gp33, KMR.gp33, B16.hCD19, and KMR.hCD19. More details can be found in Supplemental Methods.
Polyclonal in vivo tumor models
Experimental lung metastasis.
Six- to eight-week-old polyclonal Vhlfl/fl dLck-Cre– and Vhlfl/fl dLck-Cre+ littermate male mice were i.v. challenged with 6 × 105 B16.gp33 cells. Then, 21 days later, mice were euthanized and tumor lesions in lungs counted. Similar experiments were performed with injection of 4.5 × 105 B16.gp33 cells, where mice received 200 μL of pimonidazole hydrochloride solution (1.5 mg per mouse, Hypoxyprobe Red549 kit, HP7-200) or vehicle (i.v.) 90 minutes before euthanization to probe for hypoxic areas. Lungs were alveolar-fixed and expanded by intratracheal injection of 0.5% paraformaldehyde in PBS/OCT 1:1 solution (Tissue-Tek OCT compound, Sakura Finetek, 4583). Lungs were collected and fixed with 4% paraformaldehyde, 5% sucrose in PBS at 4°C on a rocker for 2.5 hours, and then dehydrated in 10% sucrose overnight and 30% sucrose for 4.5 hours, at 4°C rocking. Tissues were embedded in OCT.
To discriminate cells in the vasculature from those in the tissues, mice were injected intravenously with APC-eFluor 780–conjugated anti-CD8α antibody (eBioscience, 47-0081-82) 4 minutes before euthanization to label circulating CD8+ T cells. Tissues were immediately collected in RPMI-1640 medium with 5% bovine growth serum on ice, tumor lesions in lungs were counted, and experimental lungs bearing more than 25 lesions were prepared for flow cytometry analysis.
Egln3-YFP reporter model.
Six- to eight-week-old mice heterozygous for Egln3-YFP-KI allele (Egln3YFP/+) and littermate WT control mice (Egln3+/+) were challenged with 5 × 105 B16.gp33 melanoma cells s.c. injected into the right flank. TILs and splenocytes were isolated on days 13–14 after tumor inoculation and immediately assessed for Egln3-YFP signal and other markers by flow cytometry.
In vivo adoptive cell transfer models
Efficacy studies.
Right flanks of 6- to 12-week-old C57BL/6J (CD45.2.), CD45.1., or CD45.1.2. recipient mice were s.c. injected with 4.5 × 105 B16.gp33 or B16.hCD19 or 8 × 105 MC38.gp33 cells. B16.gp33, B16.hCD19, and MC38.gp33 tumors were inoculated for 8, 8, and 10 days, respectively,to establish palpable visible tumors. B16.gp33- and B16.hCD19-bearing mice received 3 × 106 activated and expanded (as detailed in Supplemental Methods) donor P14 cells of the appropriate genotype; MC38.gp33 tumor–bearing mice received 1 × 106 donor P14 cells of the appropriate genotype. Tumor growth and health of the animals were monitored daily and mice were euthanized by following humane endpoint and institutional guidelines. Tumors reaching tumor limit or endpoint were harvested, weighed, and processed for further analysis.
Cotransfer studies.
B16.gp33, B16.hCD19, KMR.gp33, KMR.hCD19 (4.5 × 105 cells), or MC38.gp33 (8 × 105 cells) cells were s.c. injected and tumors were established in congenically distinct recipients as in efficacy studies. Congenically distinct donor CD8+ T cells of the appropriate genotype were activated and expanded for 4 days. Cells were then collected and washed, counted and analyzed by flow cytometry, resuspended in sterile PBS, and mixed at a 1:1 ratio. Donor cells were adoptively cotransferred into recipient mice (3 × 106 total donor cells per B16.gp33-, KMR.gp33-, B16.hCD19-, and KMR.hCD19-bearing mouse, 1 × 106 total donor cells per MC38.gp33-bearing mouse). For tamoxifen-inducible ER-Cre in vivo studies, recipient mice were further treated on days 3–7 after transfer with daily 100 μL i.p. injections of either tamoxifen in sunflower oil (10 mg/mL; Cayman Chemical, 13258) or sunflower oil only (MilliporeSigma, S5007). Tamoxifen was prepared daily from frozen aliquots by shaking at 37°C immediately prior to injections. Target gene deletion in TILs upon ER-Cre induction by tamoxifen treatment was assessed on day 9 after transfer.
Antibody treatments.
All in vivo antibodies were purchased from BioXcell, stored in the dark at 4°C, and diluted in PBS immediately before i.p. injection. In anti–PD-1 efficacy experiments, 200 μg of anti-mouse PD-1 (clone RMP1-14, BE0146) or isotype control (clone 2A3, BE0089) was injected every third day, starting on the day of adoptive cell transfer. In anti-CD103 efficacy experiments, 250 μg of anti-mouse CD103 (clone M290, BE0026) or isotype control rat anti-mouse IgG2a (clone 2A3, BE0089) was injected every fifth day, starting 1 day before adoptive cell transfer, based on a previously published protocol (65).
Rechallenge experiments.
All cured mice from MC38.gp33 efficacy (VHL-KO P14 vs. WT P14) and B16.gp33 efficacy (VHL-KO P14 + anti–PD-1 vs. WT P14 + anti–PD-1) experiments were included in the rechallenge experiments. In addition, to generate more WT P14 memory mice for the MC38.gp33 rechallenge experiment, an additional experiment with a lower tumorigenic dose of 2.5 × 105 B16.gp33 cells but identical treatment protocol (WT P14 +anti–PD-1) was carried out. Mice with complete regressions of tumors and sustained remissions of over 10 weeks after the MC38.gp33 and B16.gp33 efficacy experiments were submitted for rechallenge experiments. The normal tumorigenic dose of the alternate gp33-expressing tumor cell line (4.5 × 105 B16.gp33 cells for mice cured from MC38.gp33 or 8 × 105 MC38.gp33 cells for mice cured from B16.gp33) was s.c. injected into the same location on the right flank.
For tissue-resident memory protection experiments, 1 mg/kg per mouse of FTY720 (Cayman Chemical, 10006292) or vehicle control was i.p. injected daily, starting 3 days before tumor rechallenge with 4.5 × 105 B16.gp33 cells injected into the same location on the right flank. All memory P14 mice that were protected from secondary tumor rechallenge were included in the experiment, along with 6- to 8-week-old male C57BL/6J mice without and with FTY720. FTY720 was reconstituted in DMSO (100 mg/mL; MilliporeSigma, D2650), and together with DMSO-only vehicle, were prepared daily for i.p. injections. In all rechallenge experiments, tumor growth was monitored daily and tumor recurrence was deemed when tumor volume reached 600 mm3 followed by continuous progression.
Lymphocyte isolation from tissues
Lymphocytes were isolated from harvested tumors, lungs, spleens, and tumor-draining lymph nodes. Spleen and lymph nodes were processed with the frosted ends of microscope slides and red blood cells were lysed with ACK buffer (140 mM NH4Cl and 17 mM Tris-base, pH 7.4). Tumors and lungs were cut into small pieces, digested for 30 minutes with 100 U/mL of type I collagenase (Worthington, LS004197) in RPMI 1640, 5% FBS, 2 mM MgCl2, and 2 mM CaCl2 at 37°C while shaking. Tumor and lung tissues were further dissociated using 70 μm nylon cell strainers, and single-cell suspensions were centrifugated using a 44%/67% Percoll density gradient to isolate lymphocytes.
Flow cytometry
Single-cell suspensions from isolated tumors, spleens, or tumor-draining lymph nodes were counted, stained with a fluorochrome-conjugated viability dye (eFluor 780 [65-0865-14] or eFluor 450 [65-0863-18], both from eBioscience) for 10 minutes at 4°C, washed and preincubated for 10 minutes with Fc receptor blocking medium (prepared from in-house hybridoma supernatant) followed by appropriate antibodies in PBS supplemented with 2% bovine growth serum and 0.1% sodium azide, and incubated for 30 minutes at 4°C. A list of antibodies used can be found in Supplemental Methods. After surface staining, cells were washed and prepared for analysis or further stained for intracellular molecules in the presence of Fc blocking medium for 1 hour at 4°C by using the Foxp3 Transcription Factor Staining Buffer kit (eBioscience, 00-5523) according to the manufacturer’s instructions.
Ex vivo restimulation
Freshly isolated TILs and splenocytes were counted and incubated in 200 μL of standard T cell medium containing 1× Protein Transport Inhibitor Cocktail (1:500, eBioscience, 00-4980-93) and gp33 peptide (1 μM final, KAVYNFATC; Anaspec) on a U-bottom 96-well for 4 hours. Restimulated cells were then transferred into V-bottom plates, washed twice, blocked with Fc-receptor blocking medium (prepared from in-house hybridoma supernatant), and stained for surface molecules, followed by intracellular staining to assess effector molecules and cytokine production. Aliquots of cells treated similarly but without cognate peptide were used as negative controls for cytokine production; 1× Cell Stimulation Cocktail (1:500, eBioscience, 00-4970-93) was used as a positive control.
RNA sequencing and analysis
RNA-sequencing samples were performed and analyzed independently in 2 to 3 biological replicates of 5 mice each: congenically distinct Vhlfl/fl dLck-Cre– and Vhlfl/fl dLck-Cre+ P14 cells were activated, expanded, mixed 1:1, and cotransferred into recipient mice bearing established B16.gp33 tumors as detailed above. On day 7 or 11 after transfer, TILs and splenocytes were isolated from 5 mice for each replicate. To facilitate sorting, splenocytes were further enriched for CD8+ T cells by negative selection on a MACS column as described in Supplemental Methods. FACS was performed, separating viable CD8α+ cells with donor congenic markers (bulk TILs and splenocytes), or further using CD69 and CD103 markers to separate Trm-like TIL donor subsets. Next, 1 × 103 cells of each population/subset were double-sorted into 1× TCL buffer (Qiagen, 1070498) supplemented with 1% β-mercaptoethanol (MilliporeSigma, M3148) in DNA LoBind Eppendorf tubes (MilliporeSigma, EP0030108051), centrifuged at 9,400g at 4°C for 30 seconds, incubated on ice for 5 minutes, and snap-frozen on dry ice and stored at –80°C. For library preparation, isolation of polyA+ RNA and data normalization were performed as detailed online (https://www.immgen.org/Protocols/ImmGenULI_RNAseq_methods.pdf). RNA was captured by RNAClean XP beads (Beckman Coulter) and mRNA was selected for by an anchored oligo(dT) primer (5′–AAGCAGTGGTATCAACGCAGAGTACT30VN-3′). It was converted to cDNA through reverse transcription and was PCR amplified and then fragmented with Tn5 transposon using the Nextera XT DNA Library Preparation Kit. Samples were amplified and barcoded and then pooled to be sequenced on Illumina NextSeq500 using 2 × 25 bp reads. STAR 2.5.4a (https://github.com/alexdobin/STAR/releases) was used to align to the mouse genome (GENCODE GRCm38/mm10 primary assembly and gene annotations vM16; https://www.gencodegenes.org/mouse_releases/16.html). FeatureCounts (http://subread.sourceforge.net/) was used to create a raw counts table, which was then normalized by the median of ratios method in DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). The table was converted to gene cluster text and categorical class format for use with GenePattern Multiplot Studio (66). Gene signatures were obtained from published papers as outlined in Supplemental Methods or generated by differential expression data using the multiplot module in GenePattern. Normalized enrichment scores and FDR q values were determined using the permutation test. GSEA was performed by the GSEA module in GenePattern. Sequencing data sets were deposited in the NCBI’s Gene Expression Omnibus database (GSE156342).
Statistics
Statistical parameters are reported in the figures and figure legends. Data were judged to be statistically significant when P was less than 0.05 as calculated by the following tests: tumor growth follow-up data was assessed by Mann-Whitney U test between experimental groups and versus appropriate control group(s). Survival significance was assessed by a log-rank Mantel-Cox test between experimental groups and versus appropriate control group(s), followed by post hoc Bonferroni correction of multiple comparisons. RNA sequencing was performed and analyzed independently in 2 biological replicates, and gene expression signatures were compared by Fisher’s exact tests. In all other data analyses, statistical significance was calculated by unpaired 2-tailed Student’s t test or 1-way ANOVA as indicated in figure legends. Statistical analysis was performed in GraphPad Prism software.
Study approval
All animal studies were approved by the IACUCs of the University of California, San Diego, and performed in accordance with University of California guidelines.
Author contributions
IL designed and performed experiments, analyzed the data, and wrote the manuscript. DAW and CL designed and performed experiments, analyzed the data, and wrote the manuscript. SQ analyzed the data and contributed ideas. KO, JG, and ATP designed and performed experiments and contributed ideas. NES analyzed data, contributed ideas, and wrote the manuscript. JPS and JC developed the CAR T model. AWG designed experiments, analyzed data, wrote the manuscript, and administered the project.
Supplementary Material
Acknowledgments
Funding was provided by NIH grants U19AI109976 and R01 AI067545 (to AWG); the Finnish Medical Foundation, Sigrid Jusélius Foundation, Osk. Huttunen Foundation, and Maud Kuistila Memorial Foundation (to IL); Cell and Molecular Genetics Graduate Training Program (to SQ and ATP); NIH grant K00CA222711 (to NS); NIH grants AI109842 and AI040127 (to JPS and JC). We thank Bingfei Yu and Jon Salazar and Goldrath laboratory members for discussions and experimental assistance. We thank the Immunological Genome Project (ImmGen) for reagents and sample/data processing, Alain Lamarre (INRS-Institut Armand-Frappier, Laval, Quebec, Canada) for providing B16.gp33 cell line, and Matthew Pipkin (Scripps Research Institute, Jupiter, Florida) for gp33 plasmid.
Version 1. 04/01/2021
Electronic publication
Funding Statement
Funding has been provided by NIH U19AI109976 and R01 AI067545 (A.W.G.); the Finnish Medical Foundation, Sigrid Jusélius Foundation, Osk. Huttunen Foundation and Maud Kuistila Memorial Foundation (I.L.); Cell and Molecular Genetics (CMG) Graduate Training Program (S.Q. and A.T.P); K00CA222711 (N.S.); AI109842 and AI040127 (J.P.S. and J.C.).
Footnotes
Conflict of interest: AWG is a member of the scientific advisory boards of Pandion Therapeutics and ArsenalBio.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(7):e143729.https://doi.org/10.1172/JCI143729.
Contributor Information
Ilkka Liikanen, Email: iliikanen@ucsd.edu.
Colette Lauhan, Email: clauhan@lhs.org.
Sara Quon, Email: sjquon@ucsd.edu.
Kyla Omilusik, Email: komilusik@ucsd.edu.
Anthony T. Phan, Email: atphan@vet.upenn.edu.
Amir Ferry, Email: amferry@ucsd.edu.
John Goulding, Email: jg04@cantab.net.
Joyce Chen, Email: j2chen@health.ucsd.edu.
Jason T. Yustein, Email: yustein@bcm.edu.
Deborah A. Witherden, Email: dwitherden@UCSD.EDU.
Ananda W. Goldrath, Email: agoldrath@ucsd.edu.
References
- 1.June CH, et al. CAR T cell immunotherapy for human cancer. Science. 2018;359(6382):1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 2.Newick K, et al. CAR T cell therapy for solid tumors. Annu Rev Med. 2017;68:139–152. doi: 10.1146/annurev-med-062315-120245. [DOI] [PubMed] [Google Scholar]
- 3.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blank CU, et al. Defining ‘T cell exhaustion’. Nat Rev Immunol. 2019;19(11):665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLane LM, et al. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 6.Miller BC, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20(3):326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yost KE, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25(8):1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards J, et al. CD103+ Tumor-resident CD8+ T cells are associated with improved survival in immunotherapy-naïve melanoma patients and expand significantly during anti-PD-1 treatment. Clin Cancer Res. 2018;24(13):3036–3045. doi: 10.1158/1078-0432.CCR-17-2257. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, et al. CD103+CD8+ T lymphocytes in non-small cell lung cancer are phenotypically and functionally primed to respond to PD-1 blockade. Cell Immunol. 2018;325:48–55. doi: 10.1016/j.cellimm.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Masopust D, Soerens AG. Tissue-resident T cells and other resident leukocytes. Annu Rev Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milner JJ, Goldrath AW. Transcriptional programming of tissue-resident memory CD8+ T cells. Curr Opin Immunol. 2018;51:162–169. doi: 10.1016/j.coi.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhardt T, et al. Tissue-resident memory T cells in tissue homeostasis, persistent infection, and cancer surveillance. Immunol Rev. 2018;283(1):54–76. doi: 10.1111/imr.12650. [DOI] [PubMed] [Google Scholar]
- 14.Ling KL, et al. Modulation of CD103 expression on human colon carcinoma-specific CTL. J Immunol. 2007;178(5):2908–2915. doi: 10.4049/jimmunol.178.5.2908. [DOI] [PubMed] [Google Scholar]
- 15.Boddupalli CS, et al. Interlesional diversity of T cell receptors in melanoma with immune checkpoints enriched in tissue-resident memory T cells. JCI Insight. 2016;1(21):88955. doi: 10.1172/jci.insight.88955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quinn E, et al. CD103+ intraepithelial lymphocytes--a unique population in microsatellite unstable sporadic colorectal cancer. Eur J Cancer. 2003;39(4):469–475. doi: 10.1016/S0959-8049(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 17.Hu W, et al. Prognostic significance of resident CD103+CD8+T cells in human colorectal cancer tissues. Acta Histochem. 2019;121(5):657–663. doi: 10.1016/j.acthis.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Djenidi F, et al. CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients. J Immunol. 2015;194(7):3475–3486. doi: 10.4049/jimmunol.1402711. [DOI] [PubMed] [Google Scholar]
- 19.Smazynski J, Webb JR. Resident memory-like tumor-infiltrating lymphocytes (TILRM): latest players in the immuno-oncology repertoire. Front Immunol. 2018;9:1741. doi: 10.3389/fimmu.2018.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hope JL, et al. Striking a balance-cellular and molecular drivers of memory T cell development and responses to chronic stimulation. Front Immunol. 2019;10:1595. doi: 10.3389/fimmu.2019.01595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganesan AP, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. 2017;18(8):940–950. doi: 10.1038/ni.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik BT, et al. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol. 2017;2(10):eaam6346. doi: 10.1126/sciimmunol.aam6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 24.Clarke J, et al. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J Exp Med. 2019;216(9):2128–2149. doi: 10.1084/jem.20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milner JJ, et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature. 2017;552(7684):253–257. doi: 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galvez-Cancino F, et al. Vaccination-induced skin-resident memory CD8+ T cells mediate strong protection against cutaneous melanoma. Oncoimmunology. 2018;7(7):e1442163. doi: 10.1080/2162402X.2018.1442163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nizard M, et al. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat Commun. 2017;8:15221. doi: 10.1038/ncomms15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SL, et al. Tissue-resident memory T cells in cancer immunosurveillance. Trends Immunol. 2019;40(8):735–747. doi: 10.1016/j.it.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Park SL, et al. Tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin. Nature. 2019;565(7739):366–371. doi: 10.1038/s41586-018-0812-9. [DOI] [PubMed] [Google Scholar]
- 30.Enamorado M, et al. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat Commun. 2017;8:16073. doi: 10.1038/ncomms16073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14(11):1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palazon A, et al. An HIF-1α/VEGF-A axis in cytotoxic T cells regulates tumor progression. Cancer Cell. 2017;32(5):669–683. doi: 10.1016/j.ccell.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phan AT, et al. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity. 2016;45(5):1024–1037. doi: 10.1016/j.immuni.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palazon A, et al. HIF transcription factors, inflammation, and immunity. Immunity. 2014;41(4):518–528. doi: 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor CT, Colgan SP. Regulation of immunity and inflammation by hypoxia in immunological niches. Nat Rev Immunol. 2017;17(12):774–785. doi: 10.1038/nri.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clever D, et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell. 2016;166(5):1117–1131. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caldwell CC, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J Immunol. 2001;167(11):6140–6149. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 38.Corgnac S, et al. The emerging role of CD8+ tissue resident memory T (TRM) cells in antitumor immunity: a unique functional contribution of the CD103 integrin. Front Immunol. 2018;9:1904. doi: 10.3389/fimmu.2018.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumauthioz N, et al. Tumor resident memory T cells: new players in immune surveillance and therapy. Front Immunol. 2018;9:2076. doi: 10.3389/fimmu.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egelston CA, et al. Resident memory CD8+ T cells within cancer islands mediate survival in breast cancer patients. JCI Insight. 2019;4(19):130000. doi: 10.1172/jci.insight.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molodtsov A, Turk MJ. Tissue resident CD8 memory T cell responses in cancer and autoimmunity. Front Immunol. 2018;9:2810. doi: 10.3389/fimmu.2018.02810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topham DJ, Reilly EC. Tissue-resident memory CD8+ T cells: from phenotype to function. Front Immunol. 2018;9:515. doi: 10.3389/fimmu.2018.00515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackay LK, et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity. 2015;43(6):1101–1111. doi: 10.1016/j.immuni.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Laidlaw BJ, et al. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 2014;41(4):633–645. doi: 10.1016/j.immuni.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milner JJ, et al. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc Natl Acad Sci U S A. 2020;117(41):25667–25678. doi: 10.1073/pnas.2008571117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zappasodi R, et al. Emerging concepts for immune checkpoint blockade-based combination therapies. Cancer Cell. 2018;33(4):581–598. doi: 10.1016/j.ccell.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masopust D, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207(3):553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 2019;567(7749):530–534. doi: 10.1038/s41586-019-0985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 50.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427(6972):355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 51.Gropper Y, et al. Culturing CTLs under hypoxic conditions enhances their cytolysis and improves their anti-tumor function. Cell Rep. 2017;20(11):2547–2555. doi: 10.1016/j.celrep.2017.08.071. [DOI] [PubMed] [Google Scholar]
- 52.Vuillefroy de Silly R, et al. Phenotypic switch of CD8(+) T cells reactivated under hypoxia toward IL-10 secreting, poorly proliferative effector cells. Eur J Immunol. 2015;45(8):2263–2275. doi: 10.1002/eji.201445284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franciszkiewicz K, et al. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res. 2013;73(2):617–628. doi: 10.1158/0008-5472.CAN-12-2569. [DOI] [PubMed] [Google Scholar]
- 54.Le Floc’h A, et al. Minimal engagement of CD103 on cytotoxic T lymphocytes with an E-cadherin-Fc molecule triggers lytic granule polarization via a phospholipase Cgamma-dependent pathway. Cancer Res. 2011;71(2):328–338. doi: 10.1158/0008-5472.CAN-10-2457. [DOI] [PubMed] [Google Scholar]
- 55.Buggert M, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10(7):1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann TH, Kaech SM. Tick-TOX, it’s time for T cell exhaustion. Nat Immunol. 2019;20(9):1092–1094. doi: 10.1038/s41590-019-0478-y. [DOI] [PubMed] [Google Scholar]
- 57.Palazon A, et al. The HIF-1α hypoxia response in tumor-infiltrating T lymphocytes induces functional CD137 (4-1BB) for immunotherapy. Cancer Discov. 2012;2(7):608–623. doi: 10.1158/2159-8290.CD-11-0314. [DOI] [PubMed] [Google Scholar]
- 58.Haase VH, et al. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98(4):1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryan HE, et al. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17(11):3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruber M, et al. Acute postnatal ablation of Hif-2alpha results in anemia. Proc Natl Acad Sci U S A. 2007;104(7):2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang DJ, et al. Selective expression of the Cre recombinase in late-stage thymocytes using the distal promoter of the Lck gene. J Immunol. 2005;174(11):6725–6731. doi: 10.4049/jimmunol.174.11.6725. [DOI] [PubMed] [Google Scholar]
- 62.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 63.Pircher H, et al. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342(6249):559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 64.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 65.Murray T, et al. Very late Antigen-1 marks functional tumor-resident CD8 T cells and correlates with survival of melanoma patients. Front Immunol. 2016;7:573. doi: 10.3389/fimmu.2016.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.