Abstract
BACKGROUND
Circulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA may represent a more reliable indicator of infection than nasal RNA, but quantitative reverse transcription PCR (RT-qPCR) lacks diagnostic sensitivity for blood samples.
METHODS
A CRISPR-augmented RT-PCR assay that sensitively detects SARS-CoV-2 RNA was employed to analyze viral RNA kinetics in longitudinal plasma samples from nonhuman primates (NHPs) after virus exposure; to evaluate the utility of blood SARS-CoV-2 RNA detection for coronavirus disease 2019 (COVID-19) diagnosis in adults cases confirmed by nasal/nasopharyngeal swab RT-PCR results; and to identify suspected COVID-19 cases in pediatric and at-risk adult populations with negative nasal swab RT-qPCR results. All blood samples were analyzed by RT-qPCR to allow direct comparisons.
RESULTS
CRISPR-augmented RT-PCR consistently detected SARS-CoV-2 RNA in the plasma of experimentally infected NHPs from 1 to 28 days after infection, and these increases preceded and correlated with rectal swab viral RNA increases. In a patient cohort (n = 159), this blood-based assay demonstrated 91.2% diagnostic sensitivity and 99.2% diagnostic specificity versus a comparator RT-qPCR nasal/nasopharyngeal test, whereas RT-qPCR exhibited 44.1% diagnostic sensitivity and 100% specificity for the same blood samples. This CRISPR-augmented RT-PCR assay also accurately identified patients with COVID-19 using one or more negative nasal swab RT-qPCR results.
CONCLUSION
Results of this study indicate that sensitive detection of SARS-CoV-2 RNA in blood by CRISPR-augmented RT-PCR permits accurate COVID-19 diagnosis, and can detect COVID-19 cases with transient or negative nasal swab RT-qPCR results, suggesting that this approach could improve COVID-19 diagnosis and the evaluation of SARS-CoV-2 infection clearance, and predict the severity of infection.
TRIAL REGISTRATION
ClinicalTrials.gov. NCT04358211.
FUNDING
Department of Defense, National Institute of Allergy and Infectious Diseases, National Institute of Child Health and Human Development, and the National Center for Research Resources.
Keywords: COVID-19
Keywords: Diagnostics, Molecular diagnosis
Introduction
The global coronavirus disease 2019 (COVID-19) pandemic, resulting from the initial outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is now responsible for more than 95 million infections and 2 million deaths in more than 200 countries (1), and has severely strained global healthcare systems (2). COVID-19 primarily manifests as a respiratory infection spread by droplet or aerosol transmission (3, 4), but mounting evidence indicates SARS-CoV-2 can infect nonrespiratory tissue (5, 6) to produce complicated extrapulmonary COVID-19 disease manifestations, which presumably arise when virus present in the respiratory tract is released into the circulation (7, 8). Quantitative reverse transcription PCR (RT-qPCR) analysis of swab specimens collected from the upper respiratory tract (e.g., nasal or nasopharyngeal swabs) is the reference standard since nasal tissue represents the most probable exposure site, expresses the SARS-CoV-2 receptor angiotensin converting enzyme-2, and is readily accessible. However, such analyses can yield false negatives due to transient viral shedding or sampling issues in these specimens (9, 10). Lower respiratory tract specimens (e.g., bronchoalveolar lavage fluid) may serve as more robust diagnostic specimens to accurately reflect virus load in the respiratory tract throughout the complete time course of a respiratory infection, but are more invasive, entail greater risk, and require additional training to safely collect, and are thus not practical for use in routine screening for, or assessment of, COVID-19 cases. Further, neither upper nor lower respiratory tract specimens are expected to accurately reflect viral load associated with extrapulmonary infections.
Sensitive detection of SARS-CoV-2 RNA in peripheral blood samples could theoretically serve as a universal diagnostic for COVID-19. SARS-CoV-2 circulation through the bloodstream appears necessary to initiate infections in the variety of tissues known to be affected by extrapulmonary SARS-CoV-2 infections (11, 12). Evidence also suggests that SARS-CoV-2 virus or subgenomic RNA may enter the circulation early in SARS-CoV-2 respiratory infection, since excessive cytokine production in SARS-CoV-2–infected pulmonary tissue can lead to pulmonary endothelial and epithelial cell injury, endothelial dysfunction, microvascular damage, and alveolar and vascular leakage (13). Similar endothelial pathology could also promote the release of viral RNA into the circulation by affected extrapulmonary tissues. Circulating SARS-CoV-2 RNA could thus serve as a potential marker for both pulmonary and extrapulmonary infection. Current blood-based COVID-19 assays, however, primarily detect virus-specific antibodies or cytokine or chemokine responses associated with COVID-19 disease severity that cannot provide direct evidence of infection (14, 15). RT-qPCR has been reported to exhibit poor and highly variable diagnostic sensitivity (1%–40%) when employed to detect SARS-CoV-2 RNA in blood samples from confirmed COVID-19 cases, with most positive samples exhibiting high Ct values indicative of low viral RNA concentration (15–17). Greater analytical sensitivity may therefore be required to reliably detect circulating SARS-CoV-2 RNA for COVID-19 diagnosis.
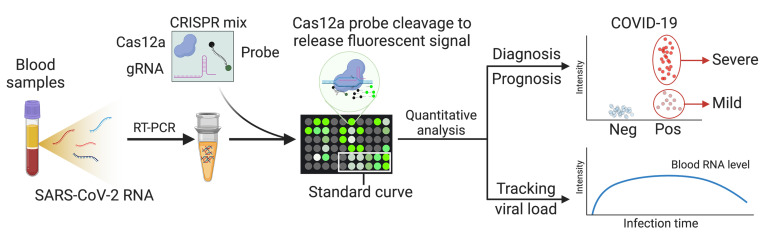
CRISPR-based nucleic acid assays have been employed to detect trace amounts of nucleic acid targets using a variety of detection methods (18, 19). RT-qPCR sensitivity for SARS-CoV-2 in nasal and nasopharyngeal swab samples can be markedly improved by using CRISPR/Cas12a activity to cleave a quenched fluorescence probe in direct correspondence with the concentration of a targeted viral amplicon following RT-PCR (20). Herein, we employed this approach to generate a CRISPR-amplified, blood-based COVID-19 (CRISPR-ABC) assay to detect SARS-CoV-2 RNA in serum and plasma from patients and a COVID-19 animal model (Figure 1). This assay detected SARS-CoV-2 RNA in the plasma of nonhuman primates (NHPs) 1 day after aerosol exposure, which increased until stabilizing at day 13 after exposure and thereafter, to precede and correlate with rectal swab viral RNA increases. Nasal swab RNA levels were much less durable, however, peaking at day 6 after exposure and then rapidly declining. CRISPR-ABC plasma results demonstrated good concordance with nasal swab RT-qPCR results, and identified COVID-19 cases in adults and children with 1 or more negative nasal swab RT-qPCR results at the time of the CRISPR-ABC–based diagnosis. Our results indicate that CRISPR-ABC provides a tractable solution for accurate COVID-19 diagnosis and infection monitoring via a plasma sample, detecting cases missed by RT-qPCR, and demonstrating durable quantification in patients who have single positive RT-qPCR results, suggesting that CRISPR-ABC analysis of plasma or serum has the potential to improve COVID-19 diagnosis and the evaluation of SARS-CoV-2 infection clearance.
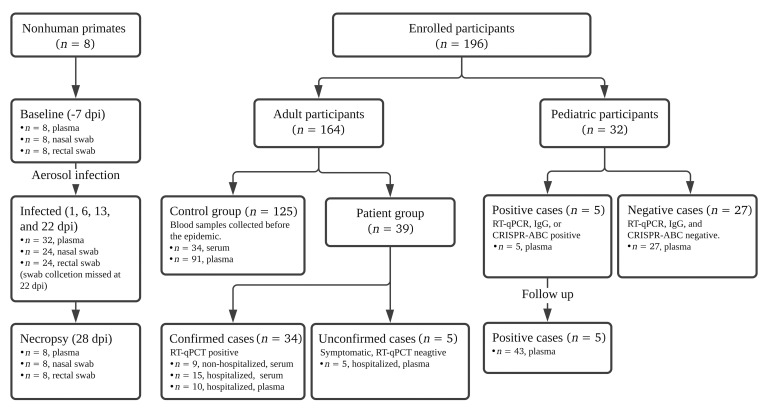
Figure 1. Flow diagram describing the numbers and disposition of the study subjects.
Results
Analytical validation of a CRISPR-enhanced assay to detect SARS-CoV-2 RNA in blood.
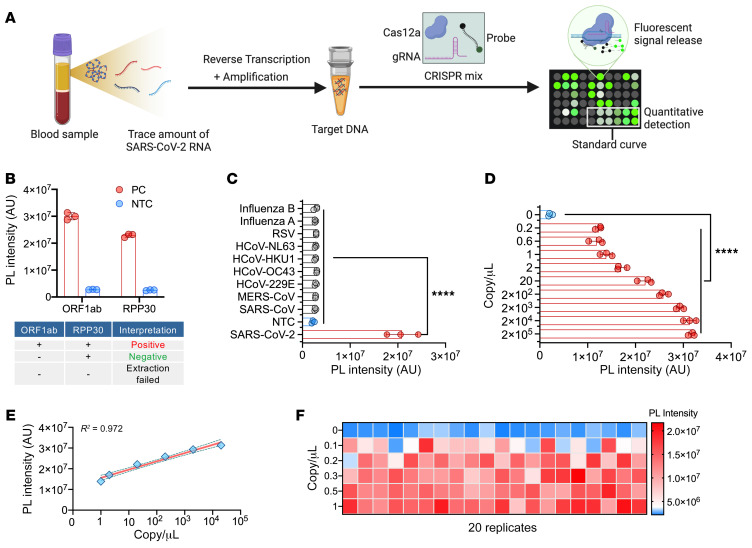
Previous studies have shown that SARS-CoV-2 RNA is detectable at highly variable rates, upon RT-qPCR analysis of peripheral blood samples from confirmed COVID-19 cases (15–17), with positive samples exhibiting low viral RNA concentrations. We therefore used a CRISPR-based signal amplification approach to enhance the detection of a RT-PCR–amplified SARS-CoV-2 gene target. In this approach, a 1-step RT-PCR reaction is employed to amplify a SARS-CoV-2 target from extracted plasma RNA, after which the guide RNA–mediated binding of Cas12a to an amplicon target activates its cleavage activity. Cas12a activity in this reaction is proportional to its binding of its target amplicon, and its cleavage of a quenched fluorescence oligonucleotide probe produces a fluorescence signal that indicates a sample’s SARS-CoV-2 RNA concentration after its comparison to a standard curve (Figure 2A). In this assay, plasma-derived RNA was analyzed to detect the SARS-CoV-2 open reading frame 1ab (ORF1ab) for COVID-19 diagnosis and the human ribonuclease P subunit p30 (RPP30) as an internal control for successful RNA extraction (Figure 2B and Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/JCI146031DS1). CRISPR-ABC exhibited robust specificity and low background when analyzing healthy human plasma spiked with RNA from viruses responsible for common human respiratory infections (Figure 2C and Supplemental Table 3). After optimizing RT-PCR and CRISPR reaction parameters (Supplemental Figures 1 and 2), CRISPR-ABC exhibited a broad linear detection range (1 × 104 – 2 × 104 copy/μL;), with an estimated limit of quantification (LoQ) of 1.1 copy/μL (Figure 2, D and E), and detected SARS-CoV-2 RNA in at least 95% of healthy plasma replicate samples spiked with at least 0.2 copy/μL heat-inactivated SARS-CoV-2 virus (Figure 2F) to yield a limit of detection (LoD) of 0.2 copy/μL. A similar result was obtained when healthy plasma replicates were directly spiked with SARS-CoV-2 RNA (Supplemental Figure 3). The CRISPR-ABC assay LoD was 5 times lower than that determined for a standard RT-qPCR assay when it was used to analyze the same samples (Supplemental Figure 4) and 5 to 100 times lower than reported for similar assays analyzing SARS-CoV-2 RNA from spiked nasal, throat, or nasopharyngeal swab RNA extract samples or standards (Supplemental Table 4).
Figure 2. Analytical validation of the CRISPR-ABC assay.
(A) CRISPR-ABC assay schematic. A SARS-CoV-2 ORF1ab target amplified from plasma RNA is quantified by comparing target- and CRISPR-mediated probe cleavage against that produced by a standard curve generated by RT-PCR of SARS-CoV-2 ORF1ab RNA samples of known concentration. (B) CRISPR-ABC signal in positive control (PC; 104 copy/μL) and no template control (NTC; nuclease-free water) samples. (C) CRISPR-ABC specificity with healthy human plasma spiked with or without indicated virus RNA or virions. (D) Limit of detection and (E) linear range of the assay. Shading denotes the 95% confidence interval of the fitted line. (F) CRISPR-ABC reproducibility for replicate plasma samples spiked with 0 to 1 copy/μL of inactivated SARS-CoV-2 virus. Graphs present the mean ± SD of 3 technical replicates for each sample. ****P < 0.0001 for a difference between the zero concentration sample and all other groups by 1-way ANOVA adjusted for multiple comparisons.
SARS-CoV-2 RNA expression in serial plasma and mucosal samples.
Given the uncertainty regarding the potential time course of detectable SARS-CoV-2 RNA in biological specimens during pulmonary and extrapulmonary infection, we employed CRISPR-ABC to evaluate viral RNA levels in nasal swab, plasma, and rectal swab samples obtained from NHPs before and after infection with aerosolized SARS-CoV-2 virus (1.4 × 104 TCID50). This group included 4 adult male African Green Monkeys aged 7.5 years and 4 adult male Indian Rhesus macaques aged 7 to 11 years (Supplemental Table 5) who had plasma and mucosal (nasal and rectal) swab samples collected 1 week prior to SARS-CoV-2 exposure and at 1, 6, 13, and 28 (necropsy) days after infection, with additional plasma samples collected at 22 days after infection (Figure 3A). Few of these NHPs exhibited overt symptoms following gross pathology at necropsy or risk factors associated with severe COVID-19, but all were found to have extended SARS-CoV-2 infections based on the detection of viral RNA in their plasma and mucosal swab samples (Figure 3, B and C) and subsequent detection of IgM specific for the SARS-CoV-2 S protein (Supplemental Figure 5), consistent with asymptomatic infection (21).
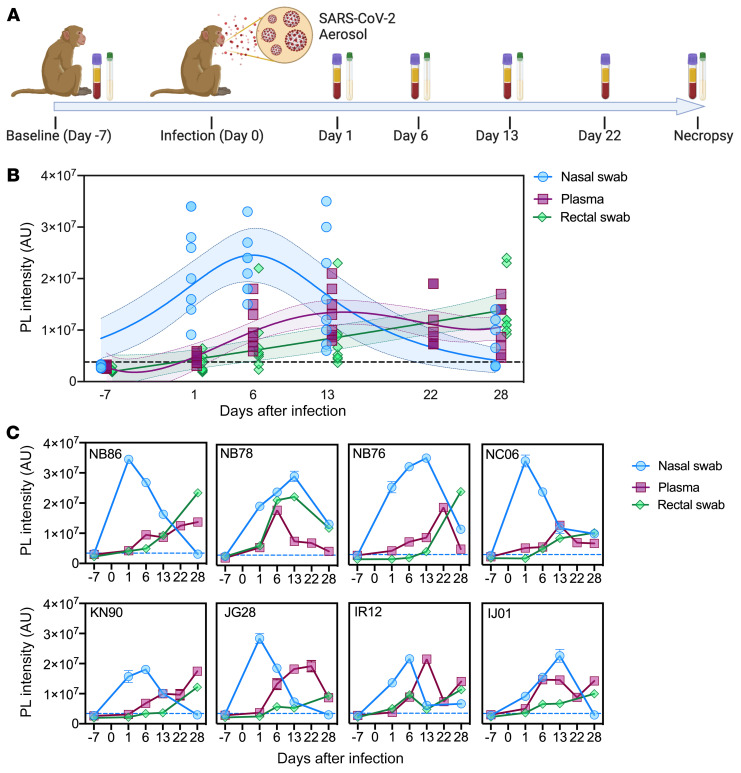
Figure 3. CRISPR-ABC analysis of samples from SARS-CoV-2-infected NHPs.
(A) Sample collection timeline (plasma and nasal and rectal swabs) versus SARS-CoV-2 infection. (B) CRISPR-ABC signal at the indicated sample time points. Shading indicates the 95% confidence interval of the fitted line. (C) CRISPR-ABC signal for samples from individual NHPs at indicated time points. SARS-CoV-2 RNA abundance is expressed as the relative photoluminescence (PL) intensity of the sample, since most samples had values below the LoQ of the CRISPR-ABC assay (Supplemental Data 1). Dotted lines indicate the positive result threshold. Data represent mean ± SD of 3 technical replicates for each sample.
All nasal swab samples were positive at day 1 after infection and tended to peak between day 1 to 13 after infection and revert to baseline by days 6 and 28 after infection (Figure 3, B and C), although individual viral peak times varied and mucosal samples were not available at day 22 after infection. Strikingly, plasma samples from most animals (5 of 8) were SARS-CoV-2 positive at day 1 after infection (Figure 3C), although virus RNA levels in plasma increased more slowly than in nasal swab samples, tending to peak at 22 to 28 days after infection (Figure 3B). SARS-CoV-2–positive expression levels observed in rectal swab samples exhibited delayed kinetics versus plasma levels, with only 3 animals demonstrating positive rectal swab results at day 1 after infection and with maximum signal not detected until day 28 after infection (Figure 3, B and C). CRISPR-ABC results for rectal swabs from most NHPs (6 of 8) exhibited gradual viral RNA increases that tended to trail but correlate with results from matching plasma (Spearman’s r = 0.9), but not nasal swab (r = 0.1) samples.
Notably, nasal swab results of 4 of these NHPs were negative at necropsy, despite continued positive plasma (and rectal swab) results (Figure 3C). Taken together, these results indicate that SARS-CoV-2 RNA circulates early after infection in NHPs that develop asymptomatic SARS-CoV-2 infections and persists after viral clearance in nasal swab samples, suggesting that changes in plasma or rectal swab results may more reliably detect unresolved infections than nasal swab results. RT-qPCR and CRISPR-ABC both detected a SARS-CoV-2 RNA signal corresponding to similar viral loads in all NHP nasal swab samples early in infection when RNA levels were high, but CRISPR-ABC detected more positive nasal swab results later in infection, and at all time points when both methods were used to analyze rectal swab and plasma samples (Supplemental Figure 6 and Supplemental Data 1), due to the greater analytical sensitivity of the CRISPR-ABC assay.
Blood-based CRISPR-ABC diagnosis of adult COVID-19 cases.
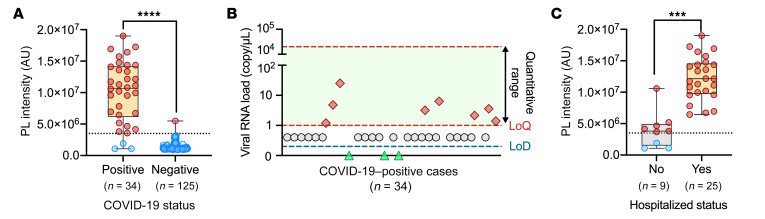
Since NHP nasal and plasma SARS-CoV-2 RNA levels demonstrated similar initial detection times following infection and overlapping expression, albeit with altered kinetics, we next evaluated the ability of CRISPR-ABC blood analysis to accurately diagnose COVID-19 cases confirmed by positive nasal or nasopharyngeal swab RT-qPCR results. Diagnostic sensitivity and specificity estimates for the CRISPR-ABC assay were determined by analyzing blood samples collected a median of 6 days after symptom onset from 34 adult symptomatic COVID-19 cases with positive nasal or nasopharyngeal RT-qPCR results (Supplemental Table 6) and archived blood samples collected from 125 individuals in 2019, prior to the first COVID-19 case reported worldwide (negative controls). The CRISPR-ABC negative response threshold defined by the negative control group (mean + 3 × standard deviation of the mean) accurately identified 31 of 34 COVID-19 cases (91.2% sensitivity) and 124 of 125 of the negative controls (99.2% specificity; Figure 4A and Supplemental Table 7). Given the current percentage of respiratory specimens testing positive in the US in late December 2020 (12%–13%) as a measure of active infections in the diagnostic population and the indicated CRISPR-ABC false- and true-positive and -negative values (22), the PPV and NPV values for the CRISPR-ABC blood assay are estimated to be 94.2% and 98.8%, respectively. Only 23.5% (8/34) of the blood samples from the COVID-19 cases revealed SARS-CoV-2 RNA concentrations above the reported 1 copy/μL LoD of RT-qPCR (23) (Figure 4B), although RT-qPCR detected SARS-CoV-2 RNA in 44.1% of these samples when a Ct less than 40 value was used as the threshold for a positive result, in agreement with the highest reported RT-qPCR sensitivity for SARS-CoV-2 RNA detection in blood (15–17). CRISPR-ABC signal intensity was significantly higher (P < 0.0002) in hospitalized versus nonhospitalized patients with COVID-19, even after employing a general linear model to adjust for age and symptom duration differences between these groups (Figure 4C and Supplemental Table 6). This agreed with results from previous studies indicating that SARS-CoV-2 RNA levels in blood were associated with disease severity (24–26). However, CRISPR-ABC signal intensity did not differ between hospitalized patients who did and did not require ventilator support or who died of COVID-19–related complications (Supplemental Figures 7 and 12). Similarly, RT-qPCR analysis of these blood samples detected SAR-CoV-2 RNA in 1 of 9 of the nonhospitalized cases and 14 of 25 of the hospitalized cases, but it was not possible to detect differences in viral RNA abundance among patients with different disease severity due to the distribution of positive results and lack of Ct variance, with most blood samples having Ct values greater than 35.
Figure 4. Blood CRISPR-ABC results of adult COVID-19 cases.
(A) CRISPR-ABC signal in baseline blood samples of 34 adults with COVID-19 diagnosed by nasal or nasopharyngeal RT-qPCR and 125 archived blood samples collected before the COVID-19 pandemic. (B) SARS-CoV-2 RNA copy number in the 34 COVID-19 subjects. (C) Comparison of CRISPR-ABC signal values of blood samples from hospitalized (n = 25) and nonhospitalized (n = 9) patients with COVID-19 by a general linear model analysis adjusted for age. A and C are box plots with maximum, Q3, median, Q1, and minimum value of PL intensity of different groups. Dotted lines indicate the positive result threshold. Dashed lines in B indicate the linear range and LoQ and LoD of the CRISPR-ABC assay. All samples were analyzed in triplicate. ****P < 0.0001 by Mann-Whitney U test; ***P < 0.001 by general linear model analysis, adjusting for age and symptom duration differences between these groups.
CRISPR-ABC diagnosis of pediatric cases with negative COVID-19 RT-qPCR results.
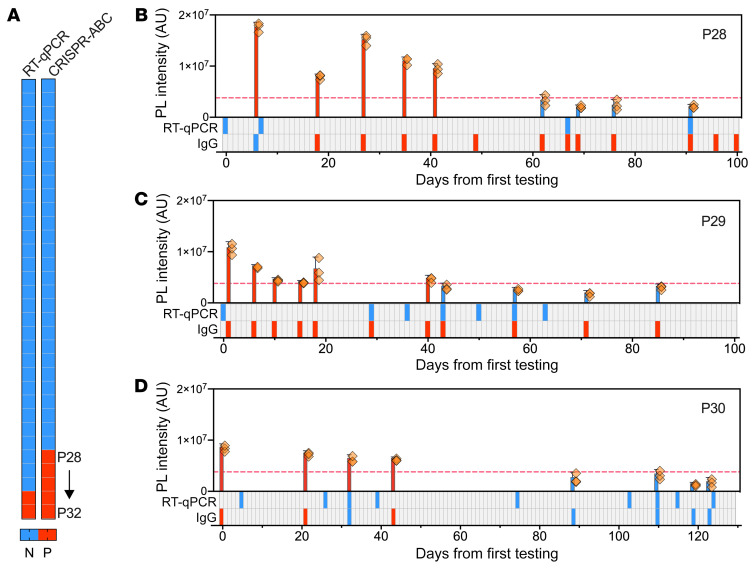
Analysis of plasma samples obtained from 32 children screened for COVID-19 during evaluation for other complaints (15 boys and 17 girls; mean age 10.3 years, range 0.2–17 years) (Supplemental Table 8) identified 27 children with negative nasal swab RT-qPCR and plasma CRISPR-ABC results, 2 children (P31 and P32) with positive results from both tests, and 3 children (P28, P29, and P30) with negative RT-qPCR results but positive CRISPR-ABC results (Figure 5A). Subsequent analysis of clinical and plasma samples obtained for the 5 children with positive plasma CRISPR-ABC results during a more than 3-month follow-up period found that none of the 3 children with negative nasal swab RT-qPCR results had a subsequent positive RT-qPCR result, although all 3 children exhibited specific antibodies at or shortly after their first evaluation (Figure 5, B–D), indicating the existence of a previous or ongoing SARS-CoV-2 infection. These children demonstrated positive plasma CRISPR-ABC results from 17 to 45 days after their initial positive result.
Figure 5. Plasma CRISPR-ABC results of pediatric cases.
(A) Positive (red) and negative (blue) results for paired nasal swab RT-qPCR and plasma CRISPR-ABC assays of 32 children screened for COVID-19. (B–D) Positive (red) and negative (blue) results for COVID-19 plasma CRISPR-ABC, nasal swab RT-qPCR, and serological results at the indicated time points after first evaluation. Dashed lines indicate the positive result threshold. Data indicate the mean ± SD of 3 technical replicates.
Both children who had positive nasal swab RT-qPCR results at or shortly after their initial evaluation had a second positive RT-qPCR test only after a sustained interval with 1 or more negative RT-qPCR tests (Supplemental Figure 8). Nasal samples collected 7 to 15 days after the first and second positive result for each child were no longer positive, although at least 1 matching and subsequent CRISPR-ABC positive samples was available for 3 of the 4 RT-qPCR positive nasal swab samples among these children. No intervening CRISPR-ABC negative sample or comparator positive plasma sample was available at the time of the second positive RT-qPCR nasal swab result for 1 of these children (Supplemental Figure 8A), preventing CRISPR-ABC confirmation. However, the second child, a 2-month-old infant at first evaluation, had both intervening negative plasma samples and positive plasma samples that matched the second positive RT-qPCR nasal swab result (Supplemental Figure 8B), suggesting this child may have contracted a second SARS-CoV-2 infection. SARS-CoV-2 IgG tests were consistently positive for this infant, although it was unclear if these results reflected maternal IgG transfer, since the infection status of the mother was not available. Finally, CRISPR-ABC results for all 5 children identified by this method demonstrated serial consistency, with no intermittent negative results aside from those observed in the single potential case of recurrent infection, and a prolonged positive interval relative to RT-qPCR, which detected no sequential positive results.
CRISPR-ABC diagnosis of at-risk patients with negative COVID-19 RT-qPCR results.
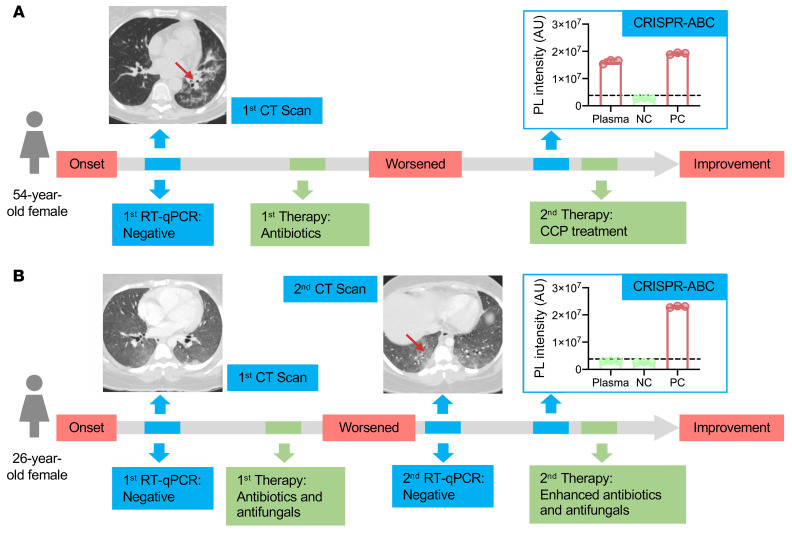
Enhanced detection of COVID-19 is necessary to improve screening and containment efforts and identify patients who are misdiagnosed due to false negative RT-qPCR results. More sensitive detection methods are also of critical importance for certain at-risk patient populations, such as individuals with chronic preexisting conditions, including cancer, where a positive diagnosis may influence available treatment options. Given that individuals with hematological cancer are reported to develop more severe disease and have higher case fatality rates (27, 28), we employed CRISPR-ABC to analyze plasma samples from a small cohort of adults with a history of leukemia who presented with symptoms consistent with COVID-19 (29, 30), including 2 cases who required supplemental oxygen during their hospitalization. RT-qPCR results for respiratory samples from all these patients were consistently negative despite concurrent clinical findings that were highly suggestive for COVID-19, but CRISPR-ABC results were positive for 4 of 5 of these patients (Figure 6 and Supplemental Figures 9–11). Two of the 4 patients with positive plasma CRISPR-ABC results improved after receiving COVID-19 convalescent plasma (CCP) therapy, 1 had milder symptoms and recovered without CCP therapy, and 1 deteriorated and died despite aggressive measures that did not include CCP treatment (Supplemental Data 2). The single patient who had a negative CRISPR-ABC result responded to enhanced antibiotic/antifungal therapy. In all cases, CRISPR-ABC results were judged to be consistent with clinical findings, as discussed in the Supplemental Results.
Figure 6. CRISPR-ABC plasma results for symptomatic adults with negative RT-qPCR results.
Case history summaries for 2 of 5 patients with 1 or more negative nasal swab RT-qPCR result. (A) Case history for a symptomatic patient with CT scan results consistent with COVID-19 who had multiple RT-qPCR negative results by nasal swab, but had a CRISPR-ABC positive plasma sample upon retroactive testing and improved after receiving COVID-19 convalescent plasma, consistent with a COVID-19 diagnosis. (B) Case history for a patient with symptoms and CT scan results consistent with COVID-19 who had negative RT-qPCR and CRISPR-ABC test results but subsequently improved after receiving enhanced antibiotic and antifungal treatment and was determined not to have had COVID-19. Red arrows on CT scan images denote COVID-19-associated “ground glass” opacity regions. The CRISPR-ABC results present the mean ± SD of 3 technical replicates for each sample.
Discussion
Nasal swab RT-qPCR results are considered the reference standard for COVID-19 diagnosis. However, mounting evidence indicates that the sensitivity of such tests varies with time since exposure, sample collection technique, and sample type. Lower respiratory tract samples tend to exhibit higher SARS-CoV-2 RNA detection rates (e.g., bronchial lavage fluid 93%; sputum 72%) than found in upper respiratory tract specimens (nasal 63%; oropharyngeal 32%), potentially due to differences in virus replication and shedding among lower and upper respiratory tract tissue, with extrapulmonary samples exhibiting even lower sensitivities (feces 29%; blood 1%) (16). RT-qPCR quantification of the amount and ratio of subgenomic to genomic SARS-CoV-2 RNA in sputum and oropharyngeal samples collected at serial time points after symptom onset has found evidence of viral replication in sputum samples until 10 to 11 days after symptom onset, the last analyzed interval, but only at 4 to 5 days after symptom onset when analyzing oropharyngeal samples (31). Nasopharyngeal swabs were not analyzed to evaluate viral replication in nasal tissue following symptom onset, but their viral genomic RNA levels correlated with those observed in oropharyngeal swabs (31).
These observations suggest that RT-qPCR analysis of nasal or nasopharyngeal swab specimens may not accurately reflect the status of lower respiratory tract infections, particularly at extended intervals after symptom onset, since oropharyngeal samples tended to decline from symptom onset, while sputum samples peaked a week after symptom development and slowly declined, in correspondence with viral RNA in stool (31).
Our results indicate that SARS-CoV-2 RNA is routinely detectable in NHP plasma 1 day after SARS-CoV-2 aerosol exposure, that viral RNA in these animals peaks by approximately 1 week after exposure in nasal samples and by 2 weeks in plasma, and that plasma SARS-CoV-2 RNA levels tend to precede and parallel rectal swab virus RNA levels. These findings are in agreement with results from human studies discussed above. Strikingly, however, SARS-CoV-2 RNA was detectable in the NHP plasma 1 day after exposure in NHPs that lacked any sign of acute respiratory infection and developed asymptomatic infections, indicating that detectable viral RNA concentrations may accumulate in plasma early after infection in patients with mild SARS-CoV-2 infections.
The emerging consensus in primate COVID-19 model development is that most species emulate asymptomatic human infection as a productive infection ensues after exposure, but that there are few clinical signs that accompany an ultimately self-limiting disease (21). Most NHP COVID-19 models develop productive infections in most mucosal and respiratory tissues, despite developing primarily asymptomatic infections in which viral RNA is detected as early as day 1 after infection in nasal and pharyngeal sites, and keep high levels of viral replication for 7 to 18 days (32–34). Clinical manifestations of human COVID-19 are dictated primarily by the presence of age and preexisting comorbidities, such as weight, that drive severe outcomes (35, 36). However, while age has been shown to increase disease severity in at least 1 NHP COVID-19 model (37), the effects of comorbidities known to promote human COVID-19 severity have not yet been evaluated in NHP disease models. Our NHP findings indicate that severe disease is not required to produce RNAemia. We observed that a lower aerosol dose than that used in previous NHP COVID-19 studies (38, 39) still induced productive infection and RNAemia in animals that developed asymptomatic disease. However, further NHP studies are required to determine the lower limit necessary to produce productive infection and/or RNAemia.
RT-qPCR exhibits poor and highly variable ability to detect SARS-CoV-2 RNA in blood samples from patients with confirmed COVID-19 (15–17). The reasons for the difference in RNAemia observed among these studies are unclear, but could reflect differences in sample collection and storage procedures. We observed that CRISPR-ABC demonstrated 91.2% diagnostic sensitivity in a small cohort of adults diagnosed with COVID-19 by their nasal/nasopharyngeal swab RT-qPCR results, whereas RT-qPCR exhibited 44.1% diagnostic sensitivity when employed to analyze the same samples. This RT-qPCR result was in agreement with the highest mean detection rate (41%) (17) reported among studies that evaluated serum or plasma SARS-CoV-2 RNA levels by standard clinical RT-qPCR (15–17). However, the reported plasma SARS-CoV-2 RNA detection rate in that study was found to be higher in severe than in mild cases (45% versus 27%), and tended to peak by the second week after admission, while the fraction of positive respiratory samples tended to peak in the first week after admission (17). A second study also reported that increased plasma SARS-CoV-2 RNA levels were associated with increased risk for progression to critical disease and death (40), although this study employed digital droplet RT-qPCR, which is not practical for use in routine high-throughput clinical applications.
CRISPR-ABC detected SARS-CoV-2 RNA in the plasma of several asymptomatic pediatric and adult patients with suspected COVID-19, but who had 1 or more negative nasal swab RT-qPCR test result, consistent with concurrent or subsequent detection of SARS-CoV-2 antibodies, clinical presentation, or response to CCP therapy. These results suggest that plasma CRISPR-ABC assays may enable detection of active SARS-CoV-2 infections in individuals not diagnosed by nasal swab RT-qPCR results. This potentially includes patients with cryptic extrapulmonary infections, as indicated by a positive CRISPR-ABC result detected for a patient with a RT-qPCR negative bronchoalveolar lavage test result. CRISPR-ABC may also be useful in evaluating or confirming disease diagnosis in patients with COVID-19 who exhibit viral clearance by nasal swab RT-qPCR results but who later exhibit evidence of disease recurrence (41, 42).
Taken together, these results support the potential for CRISPR-ABC to identify symptomatic COVID-19 cases missed by one or more nasal swab RT-qPCR tests and suggest that detection of circulating SARS-CoV-2 RNA by CRISPR-ABC may serve as a more accurate means to diagnose COVID-19 cases, judge longitudinal infection kinetics, and evaluate COVID-19 treatment responses or cures than nasal swab RT-qPCR results (Supplemental Table 9). However, one potential limitation is that this study analyzed refrigerated serum or plasma samples 3 to 7 days after their collection, and thus our results may differ from those obtained from freshly collected samples. Future studies using freshly obtained plasma and serum are required to address this question. It will also be important to determine if quantification of SARS-CoV-2 RNA level in plasma and serum by CRISPR-ABC has utility for the rapid evaluation of COVID-19 prognosis, progression, and treatment response. Finally, while the existence of secondary infection sites suggests that SARS-CoV-2 can spread through the circulation, it is unknown what fraction of SARS-CoV-2 RNA detected by our assay is present in replication-competent virions, whether this amount changes during disease development or infection severity, and how long it persists after diagnosis. This may have implications for the screening of blood donations, given rare instances of detectable viral RNA in blood from asymptomatic or presymptomatic individuals during a local outbreak but not after disease containment (43, 44). However, it is not clear if this RNA is indicative of infectious virus or if such virus might be present at levels sufficient to promote an infection, or if it would survive normal blood processing and storage procedures. Further studies are therefore necessary to address these questions and others as outlined above.
Methods
Key reagents.
SuperScript IV One-Step RT-PCR System (catalog 1235820) and nuclease-free water (catalog 4387936) were purchased from Thermo Fisher Scientific. EnGen Lba Cas12a (catalog M0653T) and NEBuffer 2.1 (catalog B7202S) were purchased from New England Biolabs. Primers, gRNA, and probes (Supplemental Table 1) were synthesized by Integrated DNA Technologies. A synthetic SARS-CoV-2 RNA reference standard (catalog NR-52358, lot 70033953) and heat inactivated 2019-nCoV/USA-WA1/2020 (catalog NR-52286, lot 70037779) were obtained from BEI Resources.
CRISPR-ABC assays.
CRISPR-ABC requires an RT-PCR–based target amplification prior to CRISPR-mediated fluorescence signal production. For RT-PCR reactions, 5 μL isolated RNA was mixed with 15 μL 1-step RT-PCR mix containing 10 μL 2X Platinum SuperFi RT-PCR Master Mix, 0.2 μL SuperScript IV RT Mix, and 2.8 μL nuclease-free water, 1 μL of 10 μM forward primer, and 1 μL of 10 μM reverse primer. RT-PCR reactions were incubated at 55°C for 10 minutes to allow cDNA synthesis then subjected to a standard PCR protocol (denaturation [5 minutes at 98°C], amplification [38 cycles: 10 seconds at 98°C, 10 seconds at 60°C, 15 seconds at 72°C], and elongation [5 minutes at 72°C]). For CRISPR reactions, 20 μL of the completed RT-PCR reaction was transferred to a 96-well half-area plate and mixed with 10 μL of the CRISPR reaction reagents (3 μL of 10 times NEBuffer 2.1, 3 μL of 300 nM gRNA, 1 μL of 1 μM EnGen Lba Cas12a, 1.5 μL of 10 μM fluorescence probe, and 1.5 μL nuclease-free water), then incubated at 37°C for 20 minutes in the dark. CRISPR-mediated fluorescence signal was then excited at 495 nm and read at 520 nm using a SpectraMax i3x Multi-Mode Microplate Reader (Molecular Devices). Refinement of assay parameters to maximize detection sensitivity by optimization of RT-PCR amplification cycles and the CRISPR cleavage reaction parameters was performed as described in Supplemental Figures 1 and 2. CRISPR-ABC specificity was evaluated in silico analysis using SnapGene software (version 5.0.8) and by triplicate CRISPR-ABC assays that analyzed 5 μL of a sample containing 1 × 104 copy/μL of a virus that represents a common cause of human respiratory infection (Supplemental Tables 2 and 3).
RT-qPCR assay.
The RT-qPCR was performed with the CDC 2019-novel coronavirus (2019-nCoV) real-time RT-qPCR diagnosis panel for target N1 gene of SARS-CoV-2. In each reaction, 5 μL RNA sample was mixed with 1.5 μL Combined Primer/Probe Mix, 5 μL TaqPath 4X 1-Step RT-PCR Master Mix (Thermo Fisher Scientific), and 8.5 μL nuclease-free water. RT-qPCR reactions were performed using a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) using the reaction conditions specified for this assay.
Standard curve LoQ, LoD, and positive result cut-off threshold.
A SARS-CoV-2 RNA standard curve was generated by serially diluting the SARS-CoV-2 RNA reference standard (1.05 × 105 RNA copy/μL) in nuclease-free water to generate 0.2, 0.6, 1, 2, 20, 2 × 102, 2 × 103, 2 × 104, and 2 × 105 copy/μL standards. The LoQ was defined as LoQ = 10 × Sy/s, where Sy is the standard deviation of the zero standard and s is the slope of the calibration curve. To assess the assay LoD, healthy donor plasma was spiked with inactivated SARS-CoV-2 and serially diluted to generate concentration standards (0.1, 0.2, 0.3, 0.5, and 1 copy/μL) that were processed for RNA, which was analyzed in 20 replicate assays. RNA was extracted from plasma samples using the Zymo Quick-DNA/RNA Viral Kit (catalog D7020). The LoD is defined as lowest concentration of SARS-CoV-2 RNA (genome copy/μL) that can be detected at least 95% of the time in replicate samples. The mean plus 3 times SD of the CRISPR-ABC value of the adult healthy control samples was used to set the threshold for positive sample results in plasma from individuals with suspected SARS-CoV-2 infections.
NHP COVID-19 models and procedures.
A total of 8 NHPs were employed in this study: 4 adult male African green monkeys aged 7.5 years and 4 adult male Indian Rhesus macaques aged 7 to 11 years (Supplemental Table 4). All animals were exposed to an inhaled dose (approximately 1.4 × 104 TCID50) of aerosolized SARS-CoV-2, and evaluated for 28 days after infection by twice daily monitoring by veterinary staff. Blood samples were drawn from all animals at 7 days prior to SARS-CoV-2 exposure and at days 1, 6, 13, 22, and 28 after infection. Nasal and rectal swab samples were not collected at day 22 after infection, but otherwise nasal and rectal swab samples were at the same time as the blood draws.
Virus information.
The SARS-CoV-2 isolate USA-WA1/2020 employed in the NHP models was acquired from BEI Resources (catalog NR-52281), and the harvested stock was determined to have a concentration of 1 × 106 TCID50/mL. The virus was passaged in Vero E6 cells in DMEM with 2% FBS sequence confirmed by PCR and/or Sanger sequencing. Plaque assays were performed in Vero E6 cells.
Clinical sample and data collection.
The human nasal swab and plasma/serum specimens analyzed in this study and demographic data were collected after obtaining prior written informed consent from adult patients or the legal guardians of pediatric patients, who also indicated their assent, or under a general research use consent, in compliance with approved IRB protocols. The samples analyzed in the adult cohort (Supplemental Table 6) were obtained from patients who had matching blood and nasal swab samples analyzed by the Weill Cornell Medicine and the Tulane Molecular Pathology Laboratories between March 17, 2020, and December 13, 2020, and whose COVID-19 status was determined based on clinical indications and current CDC guidance. Sensitivity and specificity studies were conducted using blood samples remaining after routine clinical testing at Weill Cornell Medicine and the Tulane Medical Center under a standard consent provision for research use of remnant clinical samples. Nasal swab results, demographic data, and plasma samples from indicated cases were obtained from children who were screened for COVID-19 at the Children’s Hospital New Orleans, Louisiana, USA, between March 26, 2020, and July 15, 2020, under a separate IRB (Supplemental Table 7). Eligibility criteria included any child (≤ 18 years) receiving care at the children’s hospital. Blood was drawn as part of care in the emergency room, inpatient floors, ambulatory clinics, or as part of routine preoperative studies for time-sensitive surgeries. Plasma samples corresponding to the described adult case studies were obtained from individuals who were treated at Tulane Medical Center between April 27, 2020, and July 14, 2020, under a third IRB protocol. Due to hospital regulations, refrigerated samples were released to our study team between 3 and 7 days after blood draw. All identifying data were removed and samples were coded with a unique subject identification. Clinical results for nasal swabs were determined using the CDC 2019-nCoV real-time RT-qPCR diagnostic panel.
CCP treatment of adult case studies.
ABO-compatible CCP was infused over 1 to 2 hours following premedication with 650 mg acetaminophen and 25 mg diphenhydramine. One patient was treated after obtaining individual emergency investigational new drug (eIND) approval from the FDA (Figure 4A), while a second patient (Supplemental Figure 5) was enrolled in the investigator-initiated clinical trial Expanded Access to Convalescent Plasma to Treat and Prevent Pulmonary Complications Associated With COVID-19. This clinical trial is open to enrollment at Tulane University, IND: 020073, approved by the IRB of Tulane University (IRB ref: 2020-595), and registered at the clinicaltrials.gov website under identifier NCT04358211.
Blood and swab samples collection and processing procedures.
Human and NHP blood samples were collected and rapidly processed to isolate plasma/serum. NHP plasma samples were immediately stored at –80°C until processed for RNA. Human plasma was obtained from the volume remaining in plasma stored at 4°C for potential further clinical tests. Refrigerated adult serum and pediatric plasma samples refrigerated samples were released to our study team after 3 to 7 days and 7 days after blood draw, respectively. All identifying data were removed and samples were coded with a unique subject identification. Samples were then heat inactivated for 30 minutes at 56°C, and stored at –20°C until processed for RNA. Human and NHP nasal swab samples and NHP rectal swab samples were collected in 200 μL DNA/RNA Shield (R1200, Zymo Research) and stored at –80°C until processed for RNA. NHP and clinical specimens were processed in an enhanced BL2/BL3 space in accordance with a protocol approved by the Institutional Biosafety Committee. RNA samples were isolated from 100 μL plasma or swab storage buffer using the Zymo Quick-DNA/RNA Viral Kit (D7020) following the assay protocol, and RNA was eluted in 50 μL and stored at –80°C until analysis.
COVID-19 IgG test.
Purified SARS-CoV-2 spike protein was provided by Kathryn Hastie (Scripps Research Institute, Torrey Pines, La Jolla, California, USA). The protein was used to coat wells of ELISA plates at 0.5 μg/mL in fresh 0.1M NaHCO3 for 1 hour at room temperature. Wells were washed 5 times and blocked with PBS containing 0.5% Tween, 5% dry milk, 4% whey proteins, and 10% FBS for 30 minutes at 37°C. In parallel, a set of wells not coated with antigen was incubated with blocking buffer. Sera were heat inactivated and tested at 1:100 dilution in blocking buffer. Diluted serum samples (100 μL) were incubated in wells for 1 hour at room temperature. The wells were washed and incubated with peroxidase-conjugated goat anti-human IgG-Fc (Jackson ImmunoResearch, catalog 109-035-008) diluted 1:5000 in blocking buffer. After a final wash step, color was developed by the addition of tetramethylbenzidine H2O2 as the substrate for peroxidase. Color development was stopped by the addition of 1M phosphoric acid. Color was read as absorbance (optical density) at 450 nm in a 96-well plate reader. For each sample, OD values observed with control wells were subtracted from OD values observed with S protein to calculate net OD. Positive samples had a net OD of greater than 0.4. The cut off OD value was based on preliminary screening of more than 50 pre–COVID-19 human sera in which no false positives were detected.
Statistics.
Statistical analyses were performed using GraphPad Prism 8 (version 8.4.2). Significant different of continuous characteristics between groups were determined as indicated in specific figure legends. Differences were considered statistically significant at P less than 0.05.
Study approval.
Blood samples from Weill Cornell Medicine and the Tulane Medical Center were collected under a standard consent provision for research use of remnant clinical samples. Collection of clinical samples and data from pediatric patients from the Children’s Hospital New Orleans, Louisiana, USA, was approved by Tulane University and Children’s Hospital New Orleans. Samples from adult cases treated at Tulane Medical Center were approved by that institution’s IRB. All adult studies were conducted with written informed consent in accordance with the Declaration of Helsinki. All NHP studies were performed at the Tulane National Primate Research Center, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and all animals received care that fully complied with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). The IACUC of Tulane University approved all animal procedures used in this study and the Tulane Institutional Biosafety Committee approved all procedures for sample handling, inactivation, and removal from BSL3 containment.
Author contributions
ZH, BN, CJL, and TYH conceived and designed the study and drafted manuscript. ZH, BN, BMY, AN, and MLD contributed to data collection. HSY, AN, CHM, AEM, SJB, JPL, EBN, MLD, JY, JWS, XMY, JER, NSS, KJZ, and ZZ contributed to clinical sample collection, management, and data analysis. BJB, ACF, JR, and CJR contributed to NHP model construction and data interpretation. HSY, WL, CJR, KJZ, and ZZ provided critical revisions. All authors approved the final manuscript.
Supplementary Material
Acknowledgments
This study was supported by Department of Defense grant W8IXWH1910926 and National Institute of Allergy and Infectious Diseases contract HHSN272201700033I, National Institute of Child Health and Human Development grant R01HD090927, and National Center for Research Resources and the Office of Research Infrastructure Programs grant OD011104. TYH acknowledges the generous support of the Weatherhead Presidential Endowment fund. We gratefully acknowledge BEI Resources for providing viral RNA and inactivated virus.
Version 1. 02/09/2021
In-Press Preview
Version 2. 04/01/2021
Electronic publication
Footnotes
Conflict of interest: TYH, BN, and ZH are inventors on a provisional patent application related to this work filed by Tulane University (no.63/027,530).
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(7):e146031.https://doi.org/10.1172/JCI146031.
Contributor Information
Zhen Huang, Email: zhenhuang@tulane.edu.
Bo Ning, Email: bning1@tulane.edu.
He S. Yang, Email: hey9012@med.cornell.edu.
Brady M. Youngquist, Email: byoungquist@tulane.edu.
Alex Niu, Email: aniu@tulane.edu.
Christopher J. Lyon, Email: clyon2@tulane.edu.
Brandon J. Beddingfield, Email: bbedding@tulane.edu.
Alyssa C. Fears, Email: afears@tulane.edu.
Chandler H. Monk, Email: cmonk1@tulane.edu.
Amelie E. Murrell, Email: amurrell@tulane.edu.
Samantha J. Bilton, Email: sbilton@tulane.edu.
Joshua P. Linhuber, Email: jlinhuber@tulane.edu.
Elizabeth B. Norton, Email: enorton@tulane.edu.
Monika L. Dietrich, Email: mdietri@tulane.edu.
Weihua Lai, Email: talktolaiwh@163.com.
John W. Scott, Email: jscottmd@tulane.edu.
Xiao-Ming Yin, Email: xmyin@tulane.edu.
Jay Rappaport, Email: jrappaport@tulane.edu.
James E. Robinson, Email: jrobinso@tulane.edu.
Nakhle S. Saba, Email: nsaba@tulane.edu.
Chad J. Roy, Email: croy@tulane.edu.
Kevin J. Zwezdaryk, Email: kzwezdar@tulane.edu.
Zhen Zhao, Email: zhz9010@med.cornell.edu.
Tony Y. Hu, Email: tonyhu@tulane.edu.
References
- 1. World Health Organization. WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/ Updated January 29, 2021. Accessed February 9, 2021.
- 2.Miller IF, et al. Disease and healthcare burden of COVID-19 in the United States. Nat Med. 2020;26(8):1212–1217. doi: 10.1038/s41591-020-0952-y. [DOI] [PubMed] [Google Scholar]
- 3.Anderson EL, et al. Consideration of the aerosol transmission for COVID-19 and public health. Risk Anal. 2020;40(5):902–907. doi: 10.1111/risa.13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang S, et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ Int. 2020;144:106039. doi: 10.1016/j.envint.2020.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puelles VG, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Remmelink M, et al. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit Care. 2020;24(1):495. doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu J, et al. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao AT, et al. False negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. J Med Virol. 2020;92(10):1755–1756. doi: 10.1002/jmv.25855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woloshin S, et al. False negative tests for SARS-CoV-2 infection — challenges and implications. N Engl J Med. 2020;383(6):e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, et al. Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia. Emerg Microbes Infect. 2020;9(1):727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao W, Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020;30(5):367–369. doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Q, et al. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Q-X, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 15.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broughton JP, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joung J, et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz-Fontela C, et al. Animal models for COVID-19. Nature. 2020;586(7830):509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. CDC. COVIDView: A Weekly Surveillance Summary of U.S. COVID-19 Activity. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html Updated February 5, 2021. Accessed February 9, 2021.
- 23. Center for Disease Control Prevention. In: Diseases DoV ed.: US Food and Drug Administration; 2020. [Google Scholar]
- 24.Hagman K, et al. Severe acute respiratory syndrome coronavirus 2 RNA in serum as predictor of severe outcome in coronavirus disease 2019: a retrospective cohort study. Clin Infect Dis. doi: 10.1093/cid/ciaa1285. [published online August 28, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fajnzylber J, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11(1):5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bermejo-Martin JF, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24(1):691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W, et al. COVID-19 in persons with haematological cancers. Leukemia. 2020;34(6):1637–1645. doi: 10.1038/s41375-020-0836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee LYW, et al. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: a prospective cohort study. Lancet Oncol. 2020;21(10):1309–1316. doi: 10.1016/S1470-2045(20)30442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal P, et al. Safety and efficacy of COVID-19 convalescent plasma in severe pulmonary disease: a report of 17 patients. Transfus Med. doi: 10.1111/tme.12729. [published online October 19, 2020]. [DOI] [PubMed] [Google Scholar]
- 30.Niu A, et al. COVID-19 in allogeneic stem cell transplant: high false-negative probability and role of CRISPR and convalescent plasma. Bone Marrow Transplant. 2020;55(12):2354–2356. doi: 10.1038/s41409-020-0972-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wölfel R, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 32.Lu S, et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther. 2020;5(1):157. doi: 10.1038/s41392-020-00269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockx B, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368(6494):1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.CDC COVID-19 Response Team Preliminary estimates of the prevalence of selected underlying health conditions among patients with coronavirus disease 2019 — United States, February 12-March 28, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(13):382–386. doi: 10.15585/mmwr.mm6913e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu P, et al. Age-related rhesus macaque models of COVID-19. Animal Model Exp Med. 2020;3(1):93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S, et al. Characterization of neutralizing antibody with prophylactic and therapeutic efficacy against SARS-CoV-2 in rhesus monkeys. Nat Commun. 2020;11(1):5752. doi: 10.1038/s41467-020-19568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munster VJ, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585(7824):268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veyer D, et al. Highly sensitive quantification of plasma SARS-CoV-2 RNA shelds light on its potential clinical value. Clin Infect Dis. doi: 10.1093/cid/ciaa1196. [published online August 17, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan L, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan B, et al. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10(1):11887. doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L, et al. No evidence of SARS-CoV-2 RNA among blood donors: a multicenter study in Hubei, China. Transfusion. 2020;60(9):2038–2046. doi: 10.1111/trf.15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang L, et al. Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg Infect Dis. 2020;26(7):1631–1633. doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.