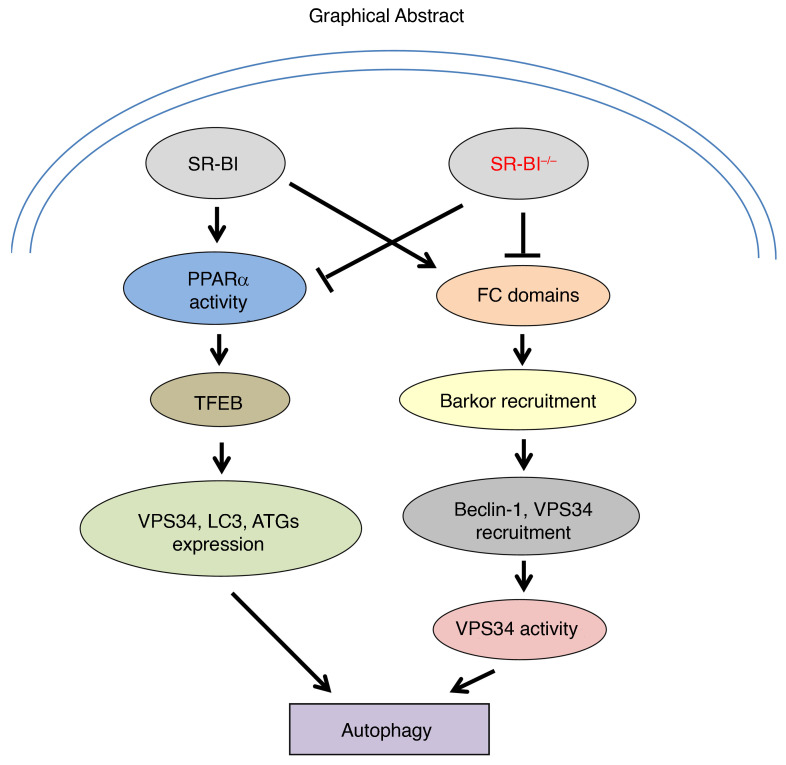
Abstract
Autophagy modulates lipid turnover, cell survival, inflammation, and atherogenesis. Scavenger receptor class B type I (SR-BI) plays a crucial role in lysosome function. Here, we demonstrate that SR-BI regulates autophagy in atherosclerosis. SR-BI deletion attenuated lipid-induced expression of autophagy mediators in macrophages and atherosclerotic aortas. Consequently, SR-BI deletion resulted in 1.8- and 2.5-fold increases in foam cell formation and apoptosis, respectively, and increased oxidized LDL–induced inflammatory cytokine expression. Pharmacological activation of autophagy failed to reduce lipid content or apoptosis in Sr-b1–/– macrophages. SR-BI deletion reduced both basal and inducible levels of transcription factor EB (TFEB), a master regulator of autophagy, causing decreased expression of autophagy genes encoding VPS34 and Beclin-1. Notably, SR-BI regulated Tfeb expression by enhancing PPARα activation. Moreover, intracellular macrophage SR-BI localized to autophagosomes, where it formed cholesterol domains resulting in enhanced association of Barkor and recruitment of the VPS34–Beclin-1 complex. Thus, SR-BI deficiency led to lower VPS34 activity in macrophages and in atherosclerotic aortic tissues. Overexpression of Tfeb or Vps34 rescued the defective autophagy in Sr-b1–/– macrophages. Taken together, our results show that macrophage SR-BI regulates autophagy via Tfeb expression and recruitment of the VPS34–Beclin-1 complex, thus identifying previously unrecognized roles for SR-BI and potentially novel targets for the treatment of atherosclerosis.
Keywords: Cardiology, Vascular Biology
Keywords: Atherosclerosis, Autophagy, Macrophages
Introduction
Autophagy is an intracellular self-digesting process that promotes degradation of damaged cellular organelles through trafficking to the lysosome (1). Three different forms of autophagy have been identified: macroautophagy, microautophagy, and chaperone-mediated autophagy. This study focuses on the role of macrophage scavenger receptor class B type I (SR-BI) in macroautophagy (hereafter, autophagy). Growing evidence demonstrates that autophagy is stimulated in human and mouse atherosclerotic lesions, and several environmental and intracellular factors have been identified as autophagy inducers in macrophages, including oxidized lipids, free cholesterol (FC), and inflammatory factors (2–4). Impairment of autophagy results in enhanced macrophage ER stress, inflammation, and death (3, 4). More recently, macrophage autophagy has been shown to be induced with cholesteryl ester (CE) loading and formation of cytoplasmic lipid inclusions (5, 6). In addition, a major portion of the FC that is made available for efflux is provided from CE being trafficked in autophagosomes to lysosomes for hydrolysis by acid lipase, as opposed to hydrolysis by neutral cholesterol esterase (6). Thus, induction of lesion cell autophagy likely reduces atherosclerosis development by reducing foam cell formation, cell death, and inflammation (2, 7). However, little is known about the exact mechanisms by which autophagy is regulated in macrophage foam cells and thus influences atherogenesis.
Autophagy processing consists of initiation (the formation of the phagophore), elongation, autophagosome closure, and autolysosome maturation, which are executed by autophagy-related genes (Atgs) and their proteins (8). During cell stress, mTOR inhibition releases the unc-51–like kinase complex (ATG1/ULK1-ATG13-ATG17) to associate with domains of damaged organelles, which leads to recruitment of the class III PI3 kinase complex (VPS34), composed of VPS15 (p150), Barkor (Beclin-1–associated autophagy-related key regulator, also known as ATG14L), and Beclin-1 (9, 10). VPS34 forms a complex with its regulator Beclin-1, and the VPS34–Beclin-1 complex associates with the forming autophagosome membrane by binding with Barkor, which preferentially interacts with highly curved membranes (11, 12). Interaction of Bif-1 with the VPS34 complex induces phosphatidylinositol-3-phosphate [PtdIns(3)P] production, which recruits other effectors critical to elongation and closure of autophagosomes (13). Formation of autophagosomes and fusion with lysosomes is accelerated by a number of factors including the immunity-related GTPase M1 (IRGM1) and the small GTPase proteins, Rab5 and Rab7 (14–17). In addition, mTOR inhibition increases nuclear translocation of transcription factor EB (TFEB), the master transcriptional regulator of genes controlling lysosome biogenesis and autophagy (10, 18, 19). Although these general mechanisms are established, the details of how specific materials are targeted to the autophagy pathway for lysosome degradation have only been identified for a few ligands.
Alterations in SR-BI influence atherosclerotic lesion development (20, 21), and SR-BI functions as a bidirectional transporter of cholesterol and lipids in the reverse cholesterol transport pathway. However, new biological functions for SR-BI are emerging, including its role as a critical factor in adaptive and innate immunity (22). Clinical studies have identified single-nucleotide polymorphisms (SNPs) in conserved regions of the extracellular domain of SR-BI in humans that are associated with reduced macrophage cholesterol efflux, impaired platelet function, and risk of coronary artery disease (23–26). Deletion of SR-BI in macrophages accelerates atherosclerosis in both low-density lipoprotein receptor–null (Ldlr–/–) and apolipoprotein E–null (Apoe–/–) mice, independent of its role as HDL’s receptor in the liver (20, 27). Sr-b1–/– versus WT macrophages in atherosclerotic lesions exhibit increased apoptosis susceptibility, and Sr-b1–/– cells are proinflammatory compared with WT macrophages (28).
Several lines of evidence suggest the possibility that SR-BI enhances autophagy. Previously, we reported that deletion of macrophage SR-BI results in FC-engorged lysosomes and extreme accumulation of cytoplasmic lipid droplets, with 97% of the cytoplasm occupied by lipid (29). In addition, SR-BI deficiency caused a failure of erythrocyte maturation in mice due to cholesterol accumulation and defective lysosome function (30). Recent studies showed that SR-BI induces autophagy in the setting of infection by forming cholesterol domains (31). Therefore, we examined the role of macrophage SR-BI in autophagy in the settings of ER stress and foam cell formation. We demonstrate that macrophage SR-BI deficiency results in impaired autophagy in response to ER stress induced by starvation, oxidized LDL (oxLDL), and FC enrichment, resulting in increased inflammation and cell death. In addition, oxLDL treatment resulted in more cytoplasmic lipid droplets in Sr-b1–/– versus WT cells. Our results demonstrate that SR-BI deletion reduces expression of key autophagy genes encoding proteins of the VPS34 complex and the master autophagy gene transcription factor, TFEB, by decreasing activation of PPARα in response to ER stress. In addition, SR-BI enhances autophagy via the VPS34 complex by increasing Barkor binding to SR-BI–associated cholesterol domains. Thus, our studies identify what we believe are novel functions of SR-BI in macrophage autophagy.
Results
Effects of SR-BI deficiency on macrophage autophagy in response to stress.
We first examined the impact of SR-BI loss of function on autophagy by quantifying the protein levels (Western blotting) of VPS34, Beclin-1, and LC3II in response to different ER stressors, including serum starvation and oxLDL (Figure 1, A and B). Studies have shown that FC enrichment of macrophages induces ER stress, and we also examined the effects of FC loading by incubating WT and Sr-b1–/– macrophages with acetylated LDL in the presence the acyl-CoA cholesterol acyltransferase (ACAT) inhibitor Sandoz 58035 (Figure 1C). Basal levels of autophagy proteins were determined in control macrophages incubated with DMEM containing 10% FBS. After serum starvation (Figure 1A) and treatment with oxLDL or FC (Figure 1, B and C), VPS34 and Beclin-1 protein levels were increased 2.1- to 2.2-fold and 1.8- to 2.5-fold, respectively, in macrophages from WT mice. In contrast, minimal changes in VPS34 and Beclin-1 protein levels were found in Sr-b1–/– macrophages in response to the 3 stress conditions. Examination of the basal levels of VPS34 and Beclin-1 in triplicate (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI94229DS1) shows that Sr-b1–/– macrophages had slightly lower levels of both proteins. In addition, we examined the effects of rapamycin, an inhibitor of the mTOR pathway, on induction of autophagy. Rapamycin treatment markedly increased VPS34 and Beclin-1 protein levels in WT compared with Sr-b1–/– macrophages (Figure 1D). Rapamycin effectively inhibited mTOR in both cell types, as demonstrated by the decreased phosphorylated mTOR (Figure 1D). Most importantly, LC3II protein levels, which can be used as a measure of autophagosome formation, were greatly increased in WT (2.1- to 3.1-fold) versus Sr-b1–/– (1.1- to 1.3-fold) macrophages after starvation, oxLDL, FC, and rapamycin treatment (Figure 1, A–D). The basal levels of LC3II on average were reduced by 30% in Sr-b1–/– versus WT macrophages (Supplemental Figure 1). The impaired autophagy in Sr-b1–/– macrophages was not the result of a decreased ER stress response, as the levels of CHOP and calnexin were increased in Sr-b1–/– versus WT macrophages during starvation, oxLDL, and FC treatments, where, under the same conditions, the WT macrophages contained much higher LC3II levels (Supplemental Figure 2). To investigate the effects of SR-BI deletion on autophagy gene expression, the mRNA levels of key autophagy factors were measured by real-time PCR. In response to FC enrichment (Figure 2), the expression levels of many autophagy/lysosome genes, including mRNAs encoding ATG5, VPS34, Beclin-1, ATG7, LC3, IRGM1, Rab5, Rab7, and Rab7 were increased in WT macrophages. Importantly, the expression of the major transcription regulator of lysosome/autophagy gene expression, TFEB, was also increased in WT macrophages. In contrast, the expression of the same genes was not enhanced in Sr-b1–/– macrophages in response to FC loading. In addition, incubation with rapamycin to inhibit mTOR, which controls TFEB phosphorylation, resulted in markedly increased mRNA levels for VPS34, Rab7, LC3, and TFEB in WT macrophages, but expression of the same genes was minimally increased in Sr-b1–/– macrophages (Supplemental Figure 3). These results suggest that SR-BI likely regulates autophagy at the transcriptional level. Consistent with this concept, transfection of both WT and Sr-b1–/– macrophages with Sr-b1, followed by enrichment in FC, resulted in increased levels of mRNAs encoding ATG5, VPS34, ATG7, Beclin-1, LC3, Rab7, and TFEB (Figure 3). Transfection with Sr-b1 resulted in similar Sr-b1 mRNA levels in WT and Sr-b1–/– macrophages, promoting similar levels of expression of these autophagy genes in both cell types (Figure 3). Formation of autophagosomes is a critical marker of autophagy. Immunofluorescence and electron microscopic analyses confirmed enhanced autophagosome formation in WT compared with Sr-b1–/– macrophages. Analysis of the number of fluorescent LC3II-positive vacuoles revealed that, compared with WT macrophages, Sr-b1–/– macrophages contained 66.7% fewer autophagosomes in response to FC enrichment (Figure 4, A and B). In addition, electron microscopic analysis showed that FC treatment resulted in 80% fewer (P < 0.05) autophagosomes in Sr-b1–/– versus WT macrophages (Figure 4, C and D). Taken together, these data demonstrate that SR-BI deletion leads to impaired autophagy in macrophages.
Figure 1. The effects of macrophage SR-BI deficiency on autophagy.
(A–D) WT and Sr-b1–/– macrophages were exposed to different levels of starvation (A), oxidized LDL (B), free cholesterol (FC) enrichment (C), or rapamycin (D). (A) For starvation, the cells were incubated for 12 or 24 hours in DMEM without serum. (B) Cells were incubated for 24 hours in DMEM containing 50 or 100 μg/mL oxidized LDL. (C) Cells were incubated for 24 hours in DMEM containing 50 or 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035 (ACAT inhibitor). (D) Cells were incubated for 24 hours with 150 or 300 nM rapamycin. In A–D, also shown are basal levels for cells that were incubated for 24 hours with DMEM containing 10% FBS. The levels of VPS34, Beclin-1, and LC3II were analyzed by Western blotting and dot quantitation. The blots are representative, and the numbers are the mean of 3 experiments, in which the values are normalized to either basal WT (green, regular font) or basal Sr-b1–/– levels (red, italic font).
Figure 2. The effects of free cholesterol (FC) enrichment on mRNA levels of autophagy players in WT versus Sr-b1–/– macrophages.
Macrophages were enriched in FC by incubation for 24 hours in DMEM containing 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. The mRNA levels for key autophagy proteins were then measured by real-time PCR in WT and Sr-b1–/– macrophages with or without FC enrichment, including ATG1, ATG5, VPS34, Beclin-1, ATG7, LC3, IRGM-1, Rab5, Rab7, Rab9, and TFEB. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05, **P < 0.01 by 1-way ANOVA with Bonferroni’s post hoc test.
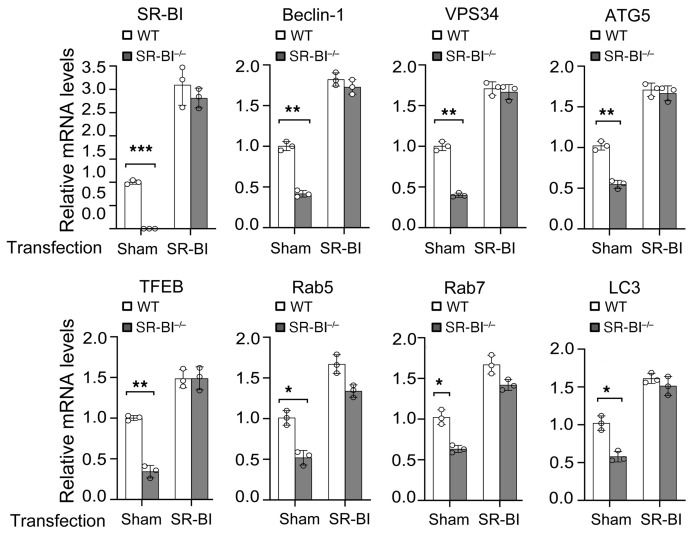
Figure 3. The effects of Sr-b1 transfection on mRNA levels of autophagy players in WT versus Sr-b1–/– macrophages.
Sr-b1–expressing or control plasmid was transfected for 48 hours into WT or Sr-b1–/– macrophages using jetPEI-Macrophage DNA Transfection reagent. Cells were enriched in FC by incubation for 24 hours with 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. The mRNA levels for key autophagy proteins were then measured by real-time PCR in cells with or without FC enrichment. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test.
Figure 4. Macrophage SR-BI deficiency causes reduced numbers of autophagosomes.
(A–D) WT or Sr-b1–/– macrophages were enriched with FC by incubation for 24 hours in DMEM containing 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. The cellular autophagosomes (APGs) were detected and quantitated by immunofluorescence (A and B) and electron microscopy (C and D). (A) Representative immunofluorescence images show LC3II-positive vacuoles. Scale bar: 20 μm. (B) LC3II dots were analyzed by ImageJ software. n = 20 per group from 3 independent experiments. The data are presented as mean ± SEM. ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test. (C) Representative electron microscopic images of APGs (arrows) are shown. Scale bars: 1 μm. (D) Quantitation of the number of APGs. Data are the mean ± SEM of 3 experiments (n = 8 per group). *P < 0.05 by Mann-Whitney test.
Effects of hematopoietic cell SR-BI deletion on autophagy in atherosclerotic lesions in Ldlr–/– mice.
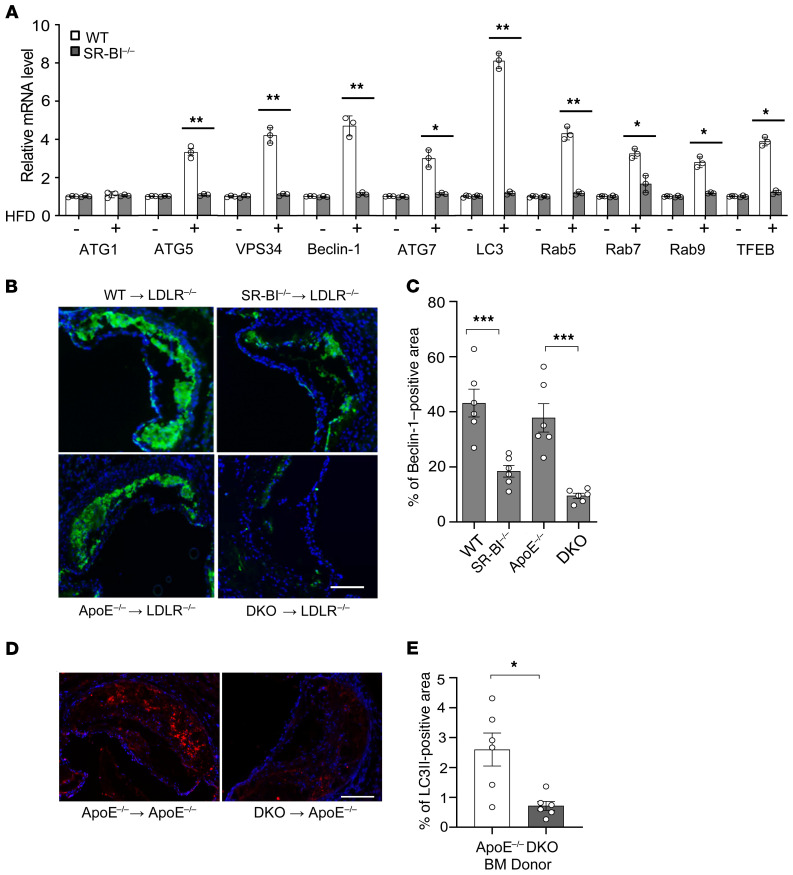
Previous studies have shown that autophagy is critical to atherosclerotic lesion formation in humans and experimental mouse models (3, 32). Therefore, we examined the effects of hematopoietic cell SR-BI deletion on atherosclerotic lesion autophagy. To examine the effects of SR-BI deletion on expression of critical autophagy players in the aortic arch, Ldlr–/– mice were transplanted with bone marrow (BM) from WT or Sr-b1–/– mice and fed either a chow or Western-type diet for 16 weeks. In Ldlr–/– mice fed a chow diet, which does not induce significant atherosclerosis development, SR-BI deletion did not cause changes in the expression levels of autophagy genes in the aortic arch tissue (Figure 5A). In Ldlr–/– mice transplanted with WT BM and fed a Western diet for 16 weeks, the expression levels of critical genes encoding proteins involved in autophagosome formation and trafficking, including ATG5, VPS34, Beclin-1, ATG7, LC3, Rab5, Rab9, and Rab7, were increased in aortic arch tissue containing atherosclerotic lesions compared with tissue devoid of atherosclerosis (Figure 5A). In contrast, atherosclerosis development failed to impact the expression of critical autophagy genes in the aortic arches of Ldlr–/– mice receiving Sr-b1–/– BM. Most interestingly, the expression of Tfeb, which is a master transcription factor of autophagy and lysosome genes, was also increased (mRNA) in atherosclerotic aortic arch tissue containing WT versus Sr-b1–/– macrophages (Figure 5A). Moreover, the expression of mRNA encoding ATG1, which is part of the unc-51–like kinase complex that is released upon mTOR inhibition, was not induced by atherosclerosis development in Ldlr–/– mice receiving WT BM. We next examined the impact of SR-BI deficiency on advanced atherosclerotic lesion Beclin-1 protein levels by immunostaining the proximal aortic sections from Ldlr–/– mice that were reconstituted with WT, Sr-b1–/–, Apoe–/–, and Apoe–/– Sr-b1–/– double-knockout (DKO) BM and fed a Western diet for 16 weeks (Figure 5, B and C). Compared with atherosclerotic lesions containing WT or Apoe–/– cells, Beclin-1 expression was reduced by 58% and 78% in lesions containing Sr-b1–/– and DKO cells, respectively. We previously published that the extent of proximal aortic atherosclerosis in these Ldlr–/– mice transplanted with WT, Sr-b1–/–, Apoe–/–, and DKO was 76.7, 192, 196, and 385 × 103 μm2, respectively, suggesting that the effects of SR-BI deficiency on autophagy impact moderate and late-stage atherosclerosis (28). Importantly, examination of the number of LC3II-positive puncta in aortic root sections of Apoe–/– mice fed a Western diet for 8 weeks showed that the number of autophagosomes was reduced by 77% in early atherosclerotic lesions containing DKO versus Apoe–/– cells (Figure 5, D and E). Taken together, these data demonstrate that hematopoietic cell SR-BI deficiency impairs autophagy in atherosclerotic lesions and suggests that the defective autophagy accelerates atherosclerosis.
Figure 5. Hematopoietic SR-BI deletion leads to attenuated autophagy in atherosclerotic lesions.
(A) Total RNA was isolated from the aortic arches of Ldlr–/– mice transplanted with WT or Sr-b1–/– bone marrow and fed either a chow or Western diet for 16 weeks. Relative gene expression was analyzed by real-time qPCR. Data are presented as mean ± SEM (n = 3 mice per group). *P < 0.05, **P < 0.01 by 1-way ANOVA with Bonferroni’s post hoc test. (B and C) Ldlr–/– mice were reconstituted with WT, Sr-b1–/–, Apoe–/–, and Sr-b1–/– Apoe–/– (DKO) bone marrow and fed a Western diet for 16 weeks. (B) Proximal aortic sections were stained using anti–Beclin-1 primary antibody and FITC-labeled secondary antibody (green). Nuclei were counterstained with Hoechst (blue). Scale bar: 100 μm. (C) Beclin-1–positive area and total lesion area were quantitated by ImageJ software. (D and E) Apoe–/– mice were reconstituted with Apoe–/– or DKO bone barrow and fed a Western diet for 8 weeks. (D) Proximal aortic sections were immunofluorescently stained with anti-LC3II primary antibody and Alexa Fluor 568–labeled secondary antibody (red). Nuclei were counterstained with Hoechst (blue). Scale bar: 100 μm. (E) LC3II-positive area and total lesion area were quantified by ImageJ software. (B–E) Data are presented as mean ± SEM (n = 6 mice per group). ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test (C) and *P < 0.05 by Mann-Whitney test (E).
SR-BI regulation of cytoplasmic lipid droplet turnover, apoptosis, and inflammation.
Recent studies demonstrated that in macrophages, cytoplasmic CE is mobilized to lysosomes via autophagy for hydrolysis to FC and release from the cell (6). Therefore, we examined the accumulation of perilipin 2–positive lipid droplets in WT and Sr-b1–/– macrophages treated with control DMEM alone, oxLDL, or oxLDL plus 300 nM rapamycin to enhance autophagy (Supplemental Figure 4, A and B). Compared with WT macrophages, Sr-b1–/– macrophages had a 1.8-fold increase in perilipin 2–positive lipid droplets (Supplemental Figure 4B). Consistent with autophagy being impaired in Sr-b1–/– cells, rapamycin treatment reduced the number of lipid droplets in WT macrophages by 51% but had no effect in Sr-b1–/– macrophages. In addition, Oil Red O staining of neutral lipids showed that Sr-b1–/– macrophages had a 1.6-fold increase in neutral lipid in the presence of oxLDL compared with WT macrophages (Supplemental Figure 4, C and D), and rapamycin treatment markedly reduced the neutral lipid accumulation in WT macrophages but had no effect in Sr-b1–/– macrophages. Measurement of cellular cholesterol confirmed that WT macrophages accumulated less cholesterol compared with Sr-b1–/– macrophages (Supplemental Figure 4E), and that induction of autophagy significantly reduced the cholesterol content of WT macrophages but did not change the cholesterol content of Sr-b1–/– macrophages. OxLDL promotes ER stress leading to apoptosis, and induction of autophagy alleviates ER stress promoting cell survival (33). Examination of oxLDL-induced apoptosis, as detected by annexin V staining, demonstrated a 36% increase of apoptosis in Sr-b1–/– compared with WT macrophages (Supplemental Figure 5, A and B). Most importantly, induction of autophagy with rapamycin reduced WT apoptosis by 68% but had no effect on apoptosis in Sr-b1–/– macrophages. Next, we tested the effects of SR-BI–mediated autophagy in regulation of the inflammatory response to oxLDL (Supplemental Figure 5, C and E). Compared with WT, Sr-b1–/– macrophages expressed 5.8-fold, 1.7-fold, and 1.9-fold more IL-1β, IL-6, and TNF-α, respectively. Induction of autophagy in WT macrophages by rapamycin reduced the expression of IL-1β, IL-6, and TNF-α by 61.3%, 52.3%, and 55.1%, respectively. Consistent with the defective autophagy observed in Sr-b1–/– macrophages, rapamycin did not affect the inflammatory response to oxLDL.
SR-BI regulates TFEB-mediated autophagy in macrophages and in atherosclerotic lesions.
TFEB is a master regulator of lysosome biogenesis and autophagy (19). Because of the observed reduction in Tfeb gene expression in Sr-b1–/– macrophages in response to FC loading, TFEB protein levels were measured in macrophages and in atherosclerotic lesions. First, SR-BI deletion in hematopoietic cells was found to result in a 65.8% decrease in TFEB protein expression in atherosclerotic lesions of Apoe–/– mice fed a Western diet for 8 weeks compared with control animals (Figure 6, A and B). Similarly, TFEB protein levels were decreased in Sr-b1–/– versus WT macrophages in response to FC enrichment (Supplemental Figure 6A). This decrease in TFEB protein was associated with reduced levels of Rab5 and Rab7 protein expression, 2 known targets of TFEB (Supplemental Figure 6A). In the absence of autophagy inducers, TFEB remains inactivated through sequestration in the cytoplasm, whereas induction of autophagy by inhibition of mTOR promotes nuclear translocation of TFEB and transcription of autophagy genes in a TFEB-dependent manner (34). Therefore, we examined the impact of loss of SR-BI on the nuclear localization of TFEB in FC-loaded macrophages. There were lower nuclear levels of TFEB in Sr-b1–/– versus WT macrophages and the nuclear to cytoplasmic ratio of TFEB was reduced by 67% (Supplemental Figure 6, B and C). Therefore, we next examined whether Tfeb overexpression could restore TFEB nuclear mobilization and autophagy in FC-enriched Sr-b1–/– macrophages. Transient expression of Tfeb increased the cellular (Supplemental Figure 7), cytoplasmic, and nuclear levels (Figure 6C) of TFEB in both WT and Sr-b1–/– macrophages. In addition, the nuclear to cytoplasmic ratio of TFEB was partially restored in Tfeb-transfected Sr-b1–/– macrophages (Figure 6D). Consistent with this result, the mRNA levels for VPS34, Rab7, and TFEB were markedly increased in Tfeb-transfected Sr-b1–/– macrophages with and without FC enrichment (Figure 6E). Importantly, transient expression of Tfeb rescued autophagy activity in Sr-b1–/– macrophages, as demonstrated by increased VPS34 and LC3II levels and by decreased p62, which is a marker of degradation via autophagy (Figure 6F and Supplemental Figure 7). These data suggest that SR-BI links the stress response elicited by lipid accumulation to autophagosome/lysosome function, TFEB expression and nuclear localization in macrophages, and atherosclerotic lesions.
Figure 6. The effects of TFEB on autophagic activity in WT versus Sr-b1–/– macrophages and in atherosclerotic lesions.
(A and B) Hematopoietic SR-BI deletion lowers the expression of TFEB in atherosclerotic lesions. Apoe–/– mice were constituted with Apoe–/– or Sr-b1–/– Apoe–/– (DKO) bone marrow and fed an atherogenic diet for 8 weeks. (A) Proximal aortic sections were stained using anti-TFEB primary antibody and FITC-labeled secondary antibody (green). Nuclei were counterstained with Hoechst (blue). Scale bar: 100 μm. (B) The TFEB-positive cells were quantitated by ImageJ software. Data are presented as mean ± SEM (n = 6 mice per group). ***P < 0.001 by unpaired Student’s t test. (C–F) Tfeb-expressing or control plasmid was transfected for 48 hours into WT or Sr-b1–/– macrophages using jetPEI-Macrophage DNA Transfection reagent. Cells were enriched with FC by incubation for 24 hours with 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. (C and D) Cytoplasmic and nuclear levels of TFEB in response to FC enrichment were analyzed by Western blotting and dot quantitation. (C) The blots are representative, and (D) the data are presented as mean ± SEM from 3 independent experiments (n = 3 per group). **P < 0.01 by 1-way ANOVA with Bonferroni’s post hoc test. (E) The mRNA levels for TFEB, VPS34, LC3, and Rab7 were measured by real-time PCR in Tfeb- or control plasmid–transfected cells with or without FC enrichment. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test. (F) Tfeb overexpression rescues defective autophagy in Sr-b1–/– macrophages as examined by Western blotting of LC3II and TFEB levels. The blots are representative, and the numbers are the mean of 3 experiments, in which the values are normalized to WT basal levels (green, regular font).
Macrophage SR-BI enhances autophagy by interacting with the VPS34 complex.
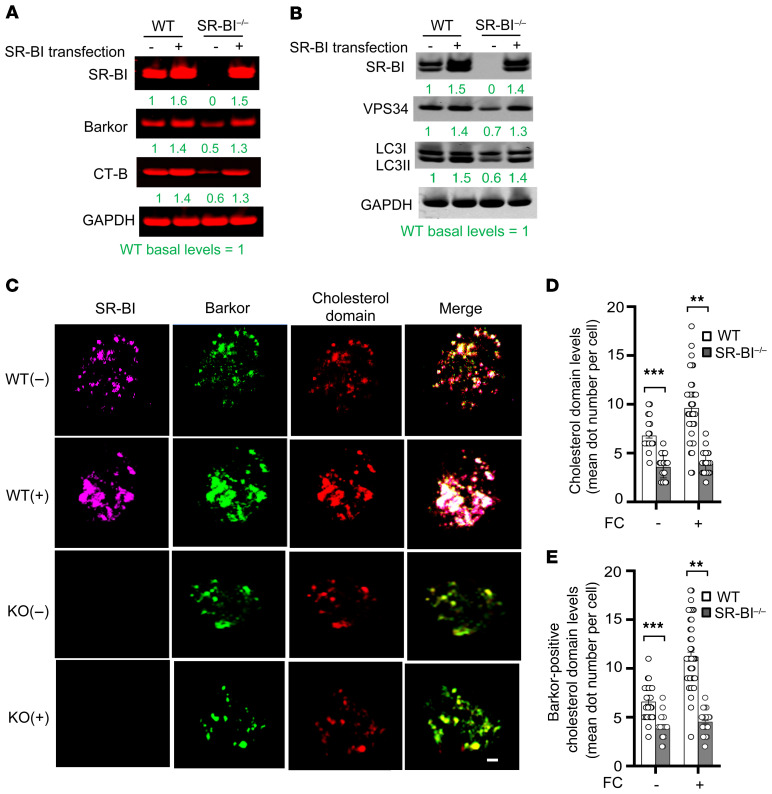
In hepatocytes and HeLa cells, SR-BI has been reported to traffic to endosomes/lysosomes (35). This observation suggests that SR-BI may play a direct role in autophagy signaling, and, to test this hypothesis, we next examined the subcellular localization of SR-BI and whether SR-BI localized to autophagosomes (Figure 7, A and B). Confocal fluorescence microscopy showed that macrophage SR-BI localizes to LC3II-positive vacuoles (Figure 7A). In addition, cellular fractionation by density-gradient ultracentrifugation (Figure 7B) showed that SR-BI was abundantly present in the plasma membrane and ER; however, macrophage SR-BI was also localized to the lysosomal fraction containing LAMP-1 (Figure 7B). Rab7 functions in the fusion of autophagosomes with lysosomes by interacting with the VPS34 complex, and studies have suggested that the trafficking of SR-BI to lysosomes in HeLa cells is Rab7 dependent (36, 37). Therefore, immunoprecipitation studies were performed to determine if macrophage SR-BI interacts with the VPS34 complex in response to FC loading stress (Figure 7C). We found that SR-BI interacts with multiple proteins in the complex, including Rab7, Barkor, VPS34, Beclin-1, and Bif-1 (Figure 7C). SR-BI specifically interacted with the VPS34 complex, as it did not coimmunoprecipitate with ATG5 or LC3. Barkor first interacts with the forming autophagosomal membrane to recruit the VPS34–Beclin-1 complex (11), and recent studies suggest that SR-BI cholesterol domains play a role in Golgi fragmentation and autophagosome formation in the setting of infection (31). Therefore, we next examined whether cholesterol domains were associated with SR-BI and Barkor (Figure 7D). FC-enriched WT macrophages were incubated with cholera toxin B (CT-B), which interacts with lipid rafts. Immunoprecipitation of the cholesterol domains with anti–CT-B antibody revealed that both SR-BI and Barkor interacted with cholesterol domains (Figure 7D). Consistent with SR-BI enhancing association of the VPS34–Beclin-1 complex to the forming autophagosomal membrane, examination of the effects of SR-BI deficiency on VPS34 activity showed that treatment with oxLDL or oxLDL plus rapamycin increased the production of PtdIns(3)P by 41% and 156% in WT macrophages, respectively, but had no effect in Sr-b1–/– macrophages (Figure 7E). In addition, VPS34 activity was 2-fold higher in the atherosclerotic aortic arch tissue in Ldlr–/– mice containing WT compared with Sr-b1–/– cells (Figure 7F). We next examined the levels of Barkor and cholesterol domains in FC-enriched Sr-b1–/– versus WT macrophages (Figure 8A). Compared with WT, Sr-b1–/– macrophages contained 50% and 40% lower levels of Barkor and cholesterol domains, respectively. In addition, transfection of Sr-b1–/– versus WT macrophages with Sr-b1 resulted in similar levels of Barkor, cholesterol domains, and VPS34 (Figure 8, A and B). Importantly, transfection of Sr-b1–/– and WT macrophages with Sr-b1 resulted in similar levels of autophagosomes, as evidenced by similar LC3II levels in FC-enriched cells (Figure 8B). In addition, transfection of Sr-b1–/– cells with increasing levels of Sr-b1 followed by FC enrichment stimulated autophagy in a dose-dependent fashion, as evidenced by the increased levels of VPS34 and LC3II (Supplemental Figure 8A). Furthermore, a comparison of WT versus Sr-b1+/– macrophages showed similar autophagic responses to FC enrichment as determined by the VPS34 and LC3II levels, which is consistent with the 2 cell types having comparable SR-BI levels (Supplemental Figure 8A). Consistent with SR-BI forming cholesterol domains, transfection of Sr-b1–/– macrophages with Sr-b1 after FC enrichment stimulated autophagy to a similar degree to that in WT macrophages, as evidenced by increased TFEB and VPS34 and decreased p62 (Supplemental Figure 8B). We next examined by confocal fluorescence microscopic analyses the number of intracellular puncta containing cholesterol domains and Barkor in WT and Sr-b1–/– macrophages (Figure 8, C–E). Basal levels of intracellular cholesterol domains were 48% higher in WT versus Sr-b1–/– macrophages and FC enrichment resulted in 64% more cholesterol domain–positive puncta in WT macrophages but did not change the number of intracellular cholesterol domains in Sr-b1–/– macrophages (Figure 8, C and D). Similar results were obtained when the cholesterol domains were measured in isolated intracellular membrane fractions from WT and Sr-b1–/– macrophages (Supplemental Figure 9A). In addition, FC-enriched WT macrophages compared with Sr-b1–/– macrophages contained 2.6-fold (P < 0.01) more intracellular puncta that were positive for both cholesterol domains and Barkor (Figure 8, C and E). Similar results were observed in colocalization of Barkor and cholesterol domains in isolated intracellular membranes (Supplemental Figure 9A). In FC-enriched WT macrophages, 68% of the puncta that were positive for cholesterol domains and Barkor were also positive for SR-BI. Consistent with SR-BI increasing VPS34 activity by enhancing association of the VPS34 complex with the forming autophagosomal membrane, FC-enriched Sr-b1–/– macrophages contained 1.6-fold more ubiquitinated VPS34 compared with WT macrophages (Supplemental Figure 9B), and inhibition of proteasomal degradation resulted in increased ubiquitinated VPS34 in both cell types.
Figure 7. Macrophage SR-BI regulates autophagy via the VPS34 complex.
(A) WT macrophages were treated with acetylated LDL and Sandoz 58035 for 24 hours and SR-BI and LC3II subcellular localization were examined by immunofluorescence confocal microscopy. Scale bar: 1 μm. (B) Plasma membrane, lysosome, ER, and nuclear fractions were isolated from WT macrophages by density-gradient ultracentrifugation, and SR-BI distribution was detected by Western blotting. LAMP-1 was used as a lysosomal marker. (C) FC-enriched WT macrophage lysates were cross-linked and immunoprecipitated with anti–SR-BI antibody or IgG and protein A/G magnetic beads. VPS34, Beclin-1, Barkor, Rab7, Bif-1, LC3, ATG5, and SR-BI were then detected in immunoprecipitated proteins by Western blotting. (D) SR-BI and Barkor are associated with cholesterol domains. FC-enriched WT macrophages were incubated with cholera toxin B (CT-B), which interacts with lipid rafts. Cell lysates were cross-linked and immunoprecipitated with anti–CT-B antibody or IgG and protein A/G magnetic beads. Barkor and SR-BI were then detected in immunoprecipitated proteins by Western blotting. In A–D, data are representative of 3 experiments. (E) VPS34 activity was measured using the Class III PI3K ELISA Kit in macrophages treated with or without 100 μg/mL oxLDL or 300 nM rapamycin for 24 hours. Data are expressed as mean ± SEM (n = 6 per group from 3 independent experiments). *P < 0.05, **P < 0.01 by 1-way ANOVA with Bonferroni’s post hoc test. (F) VPS34 activity was measured in aortic arch tissue lysates from Ldlr–/– mice reconstituted with WT or Sr-b1–/– bone marrow and fed either a chow or Western diet for 16 weeks. Data are expressed as mean ± SEM (n = 6 mice per group). *P < 0.05 by 1-way ANOVA with Bonferroni’s post hoc test.
Figure 8. SR-BI impacts autophagy by enhancing Barkor–cholesterol domain interaction.
(A and B) Sr-b1–expressing or control plasmid was transfected for 48 hours into WT or Sr-b1–/– macrophages using jetPEI-Macrophage DNA Transfection reagent. Cells were then incubated for 24 hours with 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. SR-BI, Barkor, cholesterol domains (CT-B), VPS34, and LC3II were detected by Western blotting. The blots are representative, and the numbers are the mean of 3 independent experiments in which the values are normalized to WT basal levels (green, regular font). (C–E) Cells were incubated with DMEM with 10% FBS or enriched in FC by incubation for 24 hours with 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035. The intracellular localization of cholesterol domains, Barkor, and SR-BI were examined by confocal fluorescence microscopy in cholesterol-normal (–) and FC-enriched (+) WT and Sr-b1–/– macrophages. (C) Shown are representative images of SR-BI (purple), Barkor (green), cholesterol domains (red), and merged images. Scale bar: 1 μm. (D and E) Quantitation of the average number of intracellular puncta positive for cholesterol domains alone (D) or positive for both Barkor and cholesterol domains (E) in 50 cells. **P < 0.01, ***P < 0.001 by Kruskal-Wallis test with Dunn’s multiple-comparison test. The data are presented as mean ± SEM (n = 50 per group from 3 independent experiments).
Our data suggest that SR-BI functions to enhance PtdIns(3)P production as part of the VPS34 complex; therefore, we next examined whether overexpression of Vps34 could rescue the defective autophagy observed in Sr-b1–/– macrophages (Figure 9). In response to FC loading, mRNA levels for Beclin-1, ATG5, LC3, and TFEB were increased in WT cells, but did not significantly change in Sr-b1–/– cells (Figure 9A). In contrast, transfection of Sr-b1–/– and WT macrophages with Vps34 significantly increased the mRNA levels of key autophagy proteins, including Beclin-1, ATG5, and LC3 in both cell types with and without FC enrichment (Figure 9A). Most interestingly, expression of mRNA encoding TFEB was upregulated by Vps34 overexpression in both WT and Sr-b1–/– macrophages (Figure 9A). Consistent with this observation, the whole cellular levels of TFEB protein were increased in WT and Sr-b1–/– macrophages (Supplemental Figure 10). Moreover, Vps34 overexpression resulted in increased protein levels of VPS34 and Beclin-1 in both WT and Sr-b1–/– cells with or without FC treatment (Figure 9B and Supplemental Figure 10). In addition, overexpression of Vps34 resulted in similarly increased levels of LC3II in FC-loaded WT and Sr-b1–/– macrophages, suggesting similar numbers of autophagosomes in the 2 cell types (Figure 9B). Consistent with this possibility, the levels of p62, which is a marker for degradation via autophagy, were markedly decreased in both WT and Sr-b1–/– cells with Vps34 overexpression and FC loading (Figure 9B and Supplemental Figure 10). Interestingly, overexpression of Vps34 significantly promoted TFEB nuclear localization, showing positive feedback for TFEB activity (Figure 9, C and D). Furthermore, Vps34 overexpression decreased FC-induced apoptosis by 42% and 67% in WT and Sr-b1–/– macrophages, respectively (Figure 9, E and F). Hence, overexpression of Vps34 resulted in similar numbers of apoptotic cells in cultures of WT and Sr-b1–/– macrophages (Figure 9, E and F).
Figure 9. Overexpression of Vps34 rescues defective autophagy and decreases apoptosis susceptibility in Sr-b1–/– macrophages.
(A–E) WT and Sr-b1–/– macrophages were transfected with pcDNA4-Vps34 or empty plasmid for 48 hours. The cells were then treated with or without acetylated LDL and Sandoz 58035 for 24 hours. (A) The levels of mRNA for Beclin-1, ATG5, LC3, and TFEB were measured by real-time qPCR. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). **P < 0.01, ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test. (B) VPS34, Beclin-1, p62, LC3, and GAPDH were detected by Western blotting and quantitated by using LI-COR Odyssey software. The values are normalized to WT basal levels (green, regular font). (C and D) Cytosolic and nuclear levels of TFEB were measured by Western blotting and (D) the nuclear to cytoplasmic ratio was quantitated. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05 by 1-way ANOVA with Bonferroni’s post hoc test. (E and F) Macrophage apoptosis was detected using Cy3-labeled annexin V (red). Nuclei were counterstained with Hoechst (blue). (E) Shown are representative images. Scale bar: 20 μm. (F) The percentage of apoptotic cells was quantitated, and the data are expressed as mean ± SEM (n = 9 per group from 3 independent experiments). *P < 0.05 by 1-way ANOVA with Bonferroni’s post hoc test.
SR-BI impacts autophagy via TFEB-mediated transcription of VPS34.
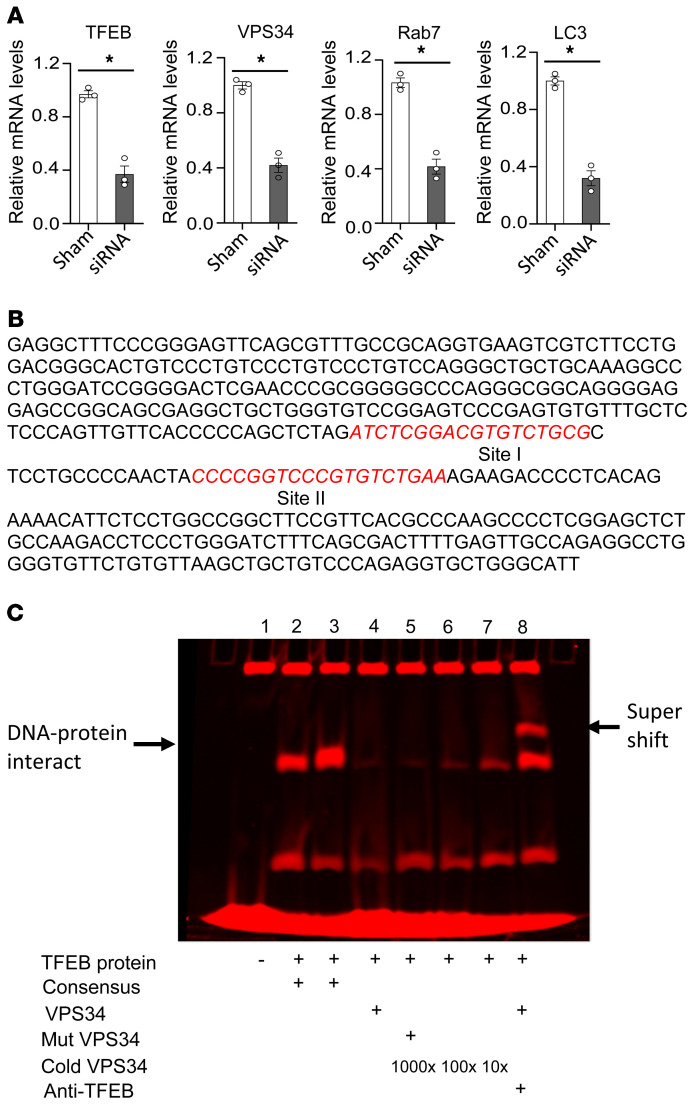
SR-BI expression increases the mRNA levels of both Tfeb and Vps34 during autophagy induction. In Sr-b1–/– macrophages — in which autophagy is impaired — the overexpression of either Vps34 or Tfeb increases expression of autophagy genes and rescues autophagy function. Therefore, we next examined whether TFEB regulates transcription of Vps34. First, we determined the effects of TFEB knockdown using siRNA in FC-enriched J774 cells. Consistent with TFEB playing a role in transcription of Vps34, knockdown of TFEB markedly reduced Vps34 mRNA levels (Figure 10A). Similarly, the mRNA levels of genes known to be transcriptionally regulated by TFEB, including mRNAs encoding Rab7 and LC3, were decreased. In addition, overexpression of Tfeb by transient transfection of cells increased VPS34 protein in FC-enriched WT and Sr-b1–/– macrophages (Figure 6F and Supplemental Figure 7). TFEB regulates expression of autophagy genes by binding to consensus DNA sequences in promoters of target genes. We next analyzed the sequence of the Vps34 gene promoter and 5′ untranslated region (5′ UTR) for the presence of potential TFEB binding sites and identified 2 sites (site I and site II) in the 5′ UTR (Figure 10B). Oligonucleotides containing these predicted TFEB consensus sites were labeled with IRDy700 fluorescent dye, and a gel shift assay revealed that TFEB protein strongly binds to DNA within site I (Figure 10C) and weakly with site II (data not shown). Importantly, mutation of site I and competition with unlabeled DNA disrupted this interaction at site I, suggesting selectivity of this effect. In addition, incubation with anti-TFEB antibody produced a clear supershift band (Figure 10C), strongly indicating that TFEB directly modulates Vps34 transcription.
Figure 10. Impact of Tfeb on transcription of VPS34.
(A) Knockdown of TFEB by siRNA attenuates Vps34 gene expression. J774A.1 macrophages were transfected with 25 nM Tfeb siRNA for 24 hours, and then treated with FC for 24 hours. The expression levels of mRNA encoding TFEB, VPS34, Rab7, and LC3 were detected by real-time PCR. Real-time PCR data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05 by unpaired Student’s t test. (B) The potential TFEB binding sites in the Vps34 gene promoter and 5′ UTR were analyzed by in silico methodology, and 2 potential sites were revealed (sites I and II) in the 5′ UTR. (C) TFEB protein interacts with Vps34 gene DNA. The specific DNA oligonucleotides (site I) were labeled with IRDy700. TFEB protein (5 μg) and 1 μL of 25 nM dsDNA were incubated in EMSA buffer for 20 minutes, and then resolved by 5% TBE polyacrylamide gel electrophoresis and the image was captured by LI-COR Odyssey.
SR-BI impacts autophagy via PPARα activation.
Our studies show that SR-BI promotes the expression of TFEB (Figures 2, 3, 5A, and 6E). Studies have shown that activation of PPARα promotes recruitment to the PPARα binding site on the Tfeb promoter, leading to enhanced expression of TFEB and autophagy (38–40). In addition, studies suggest that SR-BI modulates cellular and plasma fatty acids, which are ligands for PPARα (41). Therefore, we next examined the effects of SR-BI expression on PPARα activation by measuring nuclear PPARα levels using an ELISA. Compared with FC-enriched WT macrophages, nuclear PPARα activity was markedly decreased in Sr-b1–/– cells (Figure 11A). In contrast, PPARγ and PPARδ activity was similar in WT versus Sr-b1–/– macrophages (Supplemental Figure 11, A and B). Examination of the nuclear levels of PPARα by Western blotting also showed increased PPARα in WT versus Sr-b1–/– macrophages (Figure 11B). Incubation with the PPARα agonist CP-775146 increased the nuclear levels of PPARα in both WT and Sr-b1–/– macrophages, as determined by ELlSA (Supplemental Figure 11C) and Western blotting (Figure 11B). In addition, the PPARα antagonist GW6471 decreased nuclear PPARα levels in both cell types (Figure 11B and Supplemental Figure 11C). Consistent with PPARα promoting transcription of Tfeb, the Tfeb mRNA levels were increased in WT and Sr-b1–/– macrophages treated with CP-775146, whereas the expression of Tfeb was decreased in both cell types incubated with the antagonist, GW6471 (Figure 11D). In addition, agonism of PPARα increased the nuclear levels of TFEB in both cell types, whereas antagonism decreased nuclear TFEB (Figure 11B). Consistent with the effects of PPARα activation on TFEB, the mRNA levels for the autophagy proteins Beclin-1, Rab7, LC3, VPS34, and ATG5 were increased in WT and Sr-b1–/– macrophages treated with the PPARα agonist, CP-775146, whereas treatment with GW6471 decreased expression of the autophagy genes (Figure 11D). Similar to studies suggesting that PPARα regulates transcription of Sr-b1 (42), we found that CP-775146 increased, whereas GW6471 decreased, the Sr-b1 mRNA levels in WT cells (Figure 11D). These results demonstrate that Tfeb and Sr-b1 are coordinately regulated by PPARα. Interestingly, the mRNA levels for PPARα were increased in WT versus Sr-b1–/– macrophages, and similar to the effects of FC enrichment on Tfeb mRNA expression (Figures 2 and 6E), FC enrichment markedly increased PPARα expression in WT compared with Sr-b1–/– macrophages (Figure 11C), but did not significantly impact the expression of mRNAs for PPARγ and PPARδ in either cell type (Supplemental Figure 11D). In addition, PPARα agonist treatment of WT and Sr-b1–/– macrophages significantly increased the levels of mRNA encoding PPARα, and GW6471 decreased expression of PPARα mRNA in both cell types (Figure 11E). These results are consistent with studies showing that TFEB also regulates expression of PPARα (43, 44). Consistent with the effects of PPARα on Tfeb and autophagy genes, the PPARα agonist CP-775146 increased the protein levels of TFEB, Beclin-1, VPS34, and Rab7 in FC-enriched WT and Sr-b1–/– macrophages (Figure 11F). In addition, CP-775146 increased autophagic flux in both cell types as evidenced by decreased p62 and increased LC3II levels (Figure 11F). In contrast, the PPARα antagonist, GW6471, decreased the levels of autophagy proteins in both cell types and increased the p62 levels (Figure 11F). Furthermore, treatment of FC-enriched WT cells with the PPARγ and PPARδ agonists pioglitazone and GW501516, respectively, did not impact the levels of autophagy proteins (Supplemental Figure 11E). Importantly, PPARα protein levels were markedly increased in plaques of Apoe–/– mice with SR-BI–expressing versus Sr-b1–/– hematopoietic cells (Supplemental Figure 11, G and H). Taken together, our results show that SR-BI regulates Tfeb expression to enhance autophagy by controlling PPARα expression and activation, and that TFEB, SR-BI, and PPARα are coordinately regulated in promoting autophagy.
Figure 11. SR-BI deficiency decreases PPARα activity in macrophages.
(A) PPARα activity was measured using serial doses of nuclear extracts (NEs) from WT and Sr-b1–/– macrophages treated with 100 μg/mL acetylated LDL and 5 μg/mL Sandoz 58035 (FC enriched) for 24 hours. The data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05, **P < 0.01, ***P < 0.001 by unpaired Student’s t test. (B) WT and Sr-b1–/– cells were FC enriched and treated with or without either CP-775146 or GW6471. The levels of PPARα, TFEB, and c-Jun in nuclear extracts were then detected by Western blotting. The blots are representative, and the numbers are the mean of 2 experiments, in which the values are normalized to either basal WT (green, regular font) or basal Sr-b1–/– levels (red, italic font). (C) WT and Sr-b1–/– macrophages treated with 10% FBS or enriched with FC by incubation with acetylated LDL and Sandoz 58035 for 24 hours. The expression of mRNA encoding PPARα was then measured by real-time PCR. (D) The expression levels of mRNA for TFEB, Beclin-1, Rab7, LC3, VPS34, and ATG5 were determined by real-time PCR in FC-enriched WT and Sr-b1–/– macrophages treated with vehicle alone or with either CP-775146 or GW6471. (E) The relative PPARα mRNA levels were measured by real-time PCR in FC-enriched WT and Sr-b1–/– macrophages treated with vehicle alone or with CP-775146 or GW6471. In C–E, the data are expressed as mean ± SEM from 3 independent experiments (n = 3 per group). *P < 0.05, **P < 0.01, ***P < 0.001 by 1-way ANOVA with Bonferroni’s post hoc test. NS, not significant. (F) The expression of Beclin-1, TFEB, VPS34, LC3I, LC3II, p62, Rab7, and GAPDH was analyzed by Western blotting in FC-enriched WT and Sr-b1–/– macrophages treated with vehicle, CP-775146, or GW6471. The blots are representative, and the numbers are the mean of 2 independent experiments. In B and F, the values are normalized to either basal WT (green, regular font) or basal Sr-b1–/– levels (red, italic font).
Discussion
Autophagy is stimulated in human and experimental mouse atherosclerotic lesions by a number of ER stress factors including oxidized lipids, FC, and inflammatory cytokines (2, 3). In addition, CE loading of macrophages stimulates autophagy, with a major portion of the FC being made available for efflux via autophagic flux of cytoplasmic CE to lysosomes and hydrolysis by acid lipase (6). Defective autophagy accelerates atherosclerosis development by enhancing foam cell formation, cell death, and inflammation (2, 4, 6). In addition, SR-BI deletion accelerates atherosclerosis development in Apoe–/– and Ldlr–/– mice (21, 45). Our current study demonstrates that macrophage SR-BI enhances autophagy in foam cells of atherosclerotic lesions and thus likely influences total cellular lipid burden, inflammation, and death. Importantly, we demonstrate that SR-BI regulates autophagy by controlling expression of critical autophagy genes, including Tfeb, a major regulator of autophagy gene transcription, via PPARα activation. Moreover, SR-BI appears to have a direct effect on autophagy through interaction with Barkor as part of the VPS34 complex, where it enhances VPS34 activity. As a consequence, Sr-b1–/– macrophages exhibit increased cytoplasmic lipid accumulation and a resulting increase in cell death and a proinflammatory activation state. These findings expand on our previous work demonstrating that atherosclerotic lesions containing SR-BI–null cells have extensive cell death and massive expansion of the cellular cytoplasmic lipid volume (29). Although studies have shown that autophagy becomes impaired during late stages of atherosclerosis development (3), the defective autophagy that occurs in lesions containing SR-BI–null cells is likely not the result of increased atherosclerosis, rather the atherosclerosis is accelerated by the defective autophagy that occurs in both early and more advanced atherosclerotic lesions (Figure 5).
Tissues and cells induce autophagy in response to environmental stresses through stimulus-coupled regulation of autophagy-related genes. Precise control of this gene program is required for proper autophagosome assembly and maintenance of autophagy function. This study reveals a previously unrecognized role for SR-BI in the expression of critical autophagy effector genes induced by proatherogenic stimuli (Figure 5A) and ER stress (Figures 2, 3, and 9A). In pursuit of a candidate mechanism, we focused on the master transcription factor TFEB. TFEB coordinates induction of autophagy and lysosomal genes in response to multiple stressors (19). Here, we found that the expression (both mRNA and protein) of TFEB itself was significantly attenuated in Sr-b1–/– versus WT macrophages and in Sr-b1–/– hematopoietic cells in atherosclerotic lesions under conditions of cholesterol stress (Figures 2 and 5A). Furthermore, TFEB nuclear translocation and target gene expression were significantly reduced in Sr-b1–/– macrophages compared with WT macrophages (Supplemental Figure 6 and Figures 2 and 6). The increase in expression and nuclear localization observed in WT cells is consistent with studies identifying an autoregulatory loop involving TFEB, in which stress signals promote cytoplasmic-to-nuclear translocation of TFEB and activation of its own transcription, as well as autophagy effector genes (43). Our study suggests that this TFEB positive feedback loop is disrupted in Sr-b1–/– cells, as evidenced by the reduced TFEB levels and the defective TFEB nuclear localization. Consistent with this possibility, overexpression of Tfeb increased TFEB levels and partially restored nuclear localization, leading to rescue of targeted gene expression and of the defective autophagy observed in Sr-b1–/– macrophages (Figure 6). Importantly, our studies show that the disruption in TFEB expression and nuclear localization and autophagy gene expression in Sr-b1–/– macrophages likely results from decreased PPARα activation via SR-BI, as evidenced by decreased nuclear PPARα (Figure 11, A and B). This concept is further substantiated by our observation that agonism of PPARα increased TFEB nuclear levels and autophagy gene expression in both Sr-b1–/– and WT macrophages (Figure 11, B and D, and Supplemental Figure 11C), whereas PPARα antagonism had the opposite effects. Indeed, PPARα agonism rescued the defective autophagic flux in Sr-b1–/– macrophages (Figure 11F). This is consistent with studies demonstrating that SR-BI regulates potential PPARα ligands (46–48) and that PPARα recruitment to its binding site on the Tfeb promoter enhances Tfeb and autophagy gene expression (38–40). Interestingly, our studies suggest that PPARα, SR-BI, TFEB, and autophagy gene expression are coordinately regulated. In this regard, we found that besides enhancing Tfeb expression, PPARα agonism upregulated levels of mRNA encoding PPARα (Figure 11E). This observation suggests that TFEB also regulates PPARα expression, which is consistent with other studies (43, 44). In addition, PPARα agonism increased SR-BI expression and SR-BI deficiency reduced PPARα mRNA levels (Figure 11, C and D). Similar to the effects of SR-BI loss of function and consistent with autophagy gene expression being coordinately regulated, the overexpression of Vps34 also upregulated expression of autophagy genes (Figure 9A). The increased expression of TFEB and TFEB nuclear localization (Figure 9), resulting from VPS34 overexpression, likely contributes to the enhanced expression of autophagy genes in both WT and Sr-b1–/– macrophages. In addition, recent studies showed that VPS34 coordinates with the VPS34 complex protein, GRP78, in a positive feedback loop to regulate autophagy gene expression (49). VPS34 signaling prevents ubiquitin-mediated degradation of GRP78 and promotes its transcription via ATF6, leading to increased expression of autophagy genes by activating the unfolded protein response p-eIF2α/ATF4 pathway (50). The coordinated regulation of TFEB and VPS34 expression is further evidenced by our findings that knockdown of TFEB reduces Vps34 mRNA levels and that there is an active TFEB binding site in the Vps34 promoter (Figure 10). Taken together, these data suggest that SR-BI is an important mediator of the stress-induced signal transduction pathways that activate TFEB transcriptional responses (Figures 2, 5A, 6, 9A, and 10).
In addition to reduced expression of TFEB and its target genes, we also detected increased expression of proinflammatory genes in oxLDL-treated Sr-b1–/– macrophages. The degree and magnitude of proinflammatory signaling may also play a role in autophagy control. Indeed, prior work has shown that mycobacterium infection in macrophages impairs autophagy and inhibition of NF-κB restores its function (51–54). These data suggest that NF-κB transcriptional activity may dampen autophagy gene expression in other biological contexts. Thus, the heightened proinflammatory activation in Sr-b1–/– macrophages observed both in our work (Supplemental Figure 5) and by others (55) could interfere with induction of autophagy genes indirectly through NF-κB–dependent mechanisms.
Consistent with a direct role for SR-BI in autophagy signaling, we demonstrate that intracellular SR-BI is localized to lysosomes and LC3II-positive vacuoles in macrophages (Figure 7, A and B). This finding is consistent with other studies showing that SR-BI localizes to the late endosomal/lysosomal compartment in hepatocytes and HeLa cells (35). We observed that macrophage SR-BI interacts with members of the VPS34 complex, including VPS34, Beclin-1, Rab7, and Bif-1 (Figure 7C). The VPS34 complex is critical to the initiation and formation of autophagosomes. Formation of PtdIns(3)P by VPS34 recruits other proteins that are essential to autophagosome elongation and closure (37). Beclin-1 forms a complex with VPS34 and is recruited to the forming autophagosomal membrane by lipid-binding proteins such as Barkor and UVRAG (23). Our studies show that SR-BI increases formation of intracellular cholesterol domains that interact with Barkor (Figure 8 and Supplemental Figure 9A). Our study suggests that SR-BI formation of cholesterol domains enhances interaction with Barkor, leading to more association of the VPS34–Beclin-1 complex and increased VPS34 activity, because SR-BI–null macrophages and atherosclerotic lesions had markedly decreased VPS34 activity. Consistent with the concept of enhanced recruitment of Beclin-1 and VPS34 to the forming autophagosomal membrane, Sr-b1–/– versus WT macrophages had significantly more ubiquitinated VPS34, which is targeted for proteasomal degradation (Supplemental Figure 9B). In addition, overexpression of Vps34 rescued the defective autophagy observed in Sr-b1–/– cells and reduced apoptosis to levels similar to that of WT cells (Figure 9, B and C). However, the finding that autophagy could be rescued in the absence of SR-BI is consistent with there being multiple effectors of VPS34 in autophagy, including UVRAG, Bif-1, and Beclin-1 and with SR-BI enhancing autophagy by forming cholesterol domains that increase Barkor association (Figure 8 and Supplemental Figure 9) with the forming autophagosomal membrane (13, 56). Although the observation that forced overexpression of Vps34 or Tfeb rescues autophagy in Sr-b1–/– macrophages suggests that SR-BI is not a necessary part of the VPS34 complex, our studies show that under conditions of ER stress, macrophage SR-BI is a critical component of the VPS34 complex (Figures 7 and 8 and Supplemental Figure 9). It should also be realized that overexpression of Vps34 or Tfeb likely does not occur within the atherosclerotic lesion, whereas macrophage ER stress plays a vital role in the pathogenesis of atherosclerosis. Thus, it is likely that SR-BI is critical to enhancing VPS34 activity in the plaque, which is consistent with autophagic flux being greater in plaques containing SR-BI–expressing versus Sr-b1–/– hematopoietic cells (Figure 5D). Interestingly, studies have also shown that fusion of autophagosomes with lysosomes is dependent on Rab7 activation of VPS34. Ahras and colleagues (35) demonstrated that active Rab7 is required for trafficking of SR-BI to lysosomes in HeLa cells. The current finding that SR-BI associates with Rab7 and VPS34 in response to stress suggests that Rab7 also likely regulates VPS34 activity by interacting with SR-BI. We recently reported that macrophage SR-BI interacts with Src to activate PI3K to form the phagocytic cup in mediating the efferocytosis of apoptotic cells (28). In addition, studies have shown that the Rab7 interaction with the VPS34 complex to facilitate formation of PtdIns(3)P is essential for the fusion of phagosomes with lysosomes (57). Our data indicate that it is likely that SR-BI also functions in phagosome maturation during efferocytosis by interacting with Rab7 and VPS34.
In conclusion, our studies have uncovered a function for SR-BI in macrophage foam cell metabolism and atherosclerotic lesion development through its enhancement of autophagy, resulting in lipid clearance and in an antiinflammatory, prosurvival state. SR-BI in lipid-engorged cells regulates autophagy gene expression via PPARα-mediated transcription of Tfeb and forms cholesterol domains to enhance interaction with Barkor and the VPS34 complex to promote autophagosome formation. Hence, deletion of macrophage SR-BI resulted in increased cytoplasmic lipid accumulation, apoptosis, and inflammation, which is consistent with the extensive cytoplasmic lipid volume and cell death that occurs in SR-BI–null atherosclerotic lesions. These studies not only identify potentially novel functions for SR-BI in macrophage autophagy but also suggest therapeutic targets for the prevention and treatment of atherosclerosis.
Methods
Mice.
Sr-b1+/− mice (1:1 mixed C57BL/6 × S129 genetic background) were obtained from The Jackson Laboratory and backcrossed over 10 generations onto the C57BL/6 background. Mice were maintained on chow or a Western-type diet containing 21% milk fat and 0.15% cholesterol (Teklad).
Cell culture, autophagy induction, siRNA, and transfections.
Peritoneal macrophages were isolated from mice reconstituted with WT, Sr-b1−/−, Apoe−/−, and Sr-b1−/− Apoe−/− (DKO) BM, as previously described (28). To stimulate autophagy, an ER stress response was induced in macrophages by starvation, FC enrichment, or oxLDL treatment, as described in the Supplemental Methods. To knock down Tfeb gene expression, 25 nM siRNA for mouse Tfeb (Ambion) was transfected into J774A1 cells (ATCC) with DharmaFECT reagent (Dharmacon) for 24 hours. For transient transfections, Lipofectamine LTX with PLUS reagent (Invitrogen) or jetPEI-Macrophage DNA Transfection Reagent (Polyplus-transfection) with pcCMV6-SR-BI (Origene), pcDNA4-VPS34 (Addgene 24398; ref. 11), or pEGFP-N1-TFEB (Addgene 38119; ref. 34), or scramble control plasmids were applied to macrophages for 48 hours.
BM transplantation and atherosclerosis analyses.
Female Apoe−/− or Ldlr−/− mice were lethally irradiated and transplanted with 5 × 106 BM cells from Sr-b1+/+ Apoe−/− and DKO mice or BM from WT, Sr-b1−/−, Apoe−/−, and DKO mice, respectively. After 4 weeks, the mice were placed on a Western-type diet for 8 weeks or 16 weeks. The extent of atherosclerosis was examined both in Oil Red O–stained cross sections of the proximal aorta and by en face analysis using the KS300 imaging system (Kontron Elektronik GmbH, ref. 28). Immunofluorescence staining was used to examine the autophagosome levels in atherosclerotic lesions as described in Supplemental Methods (28).
Plasma membrane and lysosome preparation.
Plasma membranes were extracted using a plasma membrane extraction kit (BioVision). Lysosomes were prepared from macrophage lysates by density-gradient ultracentrifugation as described using the Lysosome Isolation Kit (Sigma-Aldrich) (58).
Immunoprecipitation and Western blotting.
For immunoprecipitation experiments, cross-linked whole-cell lysates were prepared as described previously (59) and immunoprecipitated with 10 μg of antibody against SR-BI (Novus) and 25 μL of magnetic beads (Invitrogen). The elicited supernatant was used for detecting mouse VPS34, Beclin-1, Rab7, LC3, Barkor, or SR-BI. For Western blotting, the cell lysates were resolved by SDS-PAGE, transferred onto nitrocellulose membranes (Amersham Bioscience), and probed with indicated primary and secondary antibodies as described in the Supplemental Methods.
Oil Red O staining of foam cells, immunofluorescence staining of lipid droplets, and measurement of cellular cholesterol.
Oil Red O staining was used to measure macrophage foam cell formation (60), as described in the Supplemental Methods. The number of cytoplasmic lipid droplets was quantitated by immunofluorescence staining of perilipin 2. Briefly, the cells were treated as indicated, fixed with fresh 4% paraformaldehyde (Sigma-Aldrich) in PBS, permeabilized with 0.2% Triton X-100, blocked with 2% BSA in PBS, and then incubated with anti–perilipin 2 antibody (Novus) and Alexa Fluor 658–labeled secondary antibody and nuclei were counterstained with Hoechst (Sigma-Aldrich). The images were processed using a fluorescence microscope (Olympus IX81) and SlideBook 6 (Intelligent-Image) software. Cellular cholesterol was measured by enzymatic cholesterol assay as described previously (61).
Electron, fluorescence, and confocal microscopic analyses.
A monolayer of cells was fixed, and then scraped and postfixed dehydrated with a series of ethanol concentrations, and gradually infiltrated with Epon resin. Thin sections were stained with uranyl acetate and lead citrate. Transmission electron microscopy was performed using an FEI Tecnai T-12 operated at 100 kiloelectron volts to ultrastructurally analyze cell autophagosome content. Macrophage autophagosome levels were also detected by immunofluorescence staining using anti-LC3II primary and fluorescently labeled secondary antibodies. The intracellular locations and expression levels of SR-BI, Barkor/ATG14L, and cholesterol domain marker CT-B were detected by immunofluorescence staining and confocal microscopy as described in Supplemental Methods.
VPS34 kinase activity assay.
A Class III PI3K ELISA Kit (Echelon) was used to measure endogenously or exogenously expressed VPS34 kinase activity in macrophages or aortic root tissue following the manufacturer’s instructions and as described in Supplemental Methods.
Aortic root RNA isolation and real-time RT-PCR.
Total RNA was isolated and purified using an Aurum Total RNA kit (Bio-Rad). Complementary DNA was synthesized with iScript reverse transcriptase (Bio-Rad). Relative quantitation of the target mRNA was performed using a Bio-Rad real-time PCR kit as described in Supplemental Methods.
Gel shift assay.
In silico methodology was applied to examine the potential existence of TFEB binding sites in the VPS34 gene promoter and 5′ UTR (62). The predicted and mutant DNA and TFEB consensus (5′-GTAGGCCACGTGACCGGG-3′) oligonucleotides were labeled with fluorescent IRDy700 dye (Invitrogen) and annealed with dsDNA. The gel shift assay was performed using the Odyssey EMSA Kit (LI-COR) as described in the Supplemental Methods.
PPAR activity assay.
Nuclear extracts were prepared from the macrophages using a nuclear extraction kit (Abcam). PPARα/γ/δ activities were measured using the PPARα/γ/δ Transcription Factor Assay Kit (Abcam). Briefly, 100 μL (5 μg protein) of nuclear extract was added to designated wells, incubated at 4°C overnight, and then primary anti-PPARα/γ/δ antibody and HRP-conjugated secondary antibody were applied to each well and incubated for 1 hour at room temperature. After washing, the wells were incubated for 45 minutes at room temperature with the supplied Transcription Factor Developing Solution, and the absorbance was read at 450 nm within 5 minutes of adding the Stop Solution.
Statistics.
Data are presented as mean ± SEM. Differences between mean values were determined by 1-way ANOVA (Bonferroni’s post hoc test), Kruskal-Wallis test (Bunn’s multiple-comparison test), Mann-Whitney test, and Student’s t test after the normality of the sample populations was examined by the Kolmogorov-Smirnov test using GraphPad Prism. P values less than 0.05 were considered significant.
Study approval.
The animal studies complied with all relevant ethical regulations for vertebrate animal research. Animal protocols were approved by and performed according to the regulations of Vanderbilt University’s Institutional Animal Care and Usage Committee.
Author contributions
HT designed and performed experiments, acquired and analyzed data, wrote the manuscript. PGY designed research, analyzed data, and wrote the manuscript. JLB, YZ, and LD conducted experiments. WGJ conducted experiments, analyzed data, and modified the manuscript. JDB and KCV analyzed data and modified the manuscript. MFL designed research, analyzed data, obtained funding, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants HL127173, HL148137, HL146134, HL116263, and DK59637. Confocal microscopy and transmission electron microscopy were performed through the use of the Vanderbilt University Cell Imaging Shared Resource (supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126).
Version 1. 03/04/2021
In-Press Preview
Version 2. 04/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(7):e94229.https://doi.org/10.1172/JCI94229.
Contributor Information
Huan Tao, Email: huan.tao@vumc.org.
Patricia G. Yancey, Email: patricia.g.yancey@vumc.org.
John L. Blakemore, Email: john.l.blakemore@vumc.org.
Youmin Zhang, Email: youmin.zhang@vumc.org.
Lei Ding, Email: lei.ding@vumc.org.
W. Gray Jerome, Email: jay.jerome@vumc.org.
Jonathan D. Brown, Email: jonathan.d.brown@vumc.org.
Kasey C. Vickers, Email: kasey.c.vickers@vumc.org.
MacRae F. Linton, Email: macrae.linton@vumc.org.
References
- 1.Behrends C, et al. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinet W, De Meyer GR. Autophagy in atherosclerosis: a cell survival and death phenomenon with therapeutic potential. Circ Res. 2009;104(3):304–317. doi: 10.1161/CIRCRESAHA.108.188318. [DOI] [PubMed] [Google Scholar]
- 3.Liao X, et al. Macrophage autophagy plays a protective role in advanced atherosclerosis. Cell Metab. 2012;15(4):545–553. doi: 10.1016/j.cmet.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razani B, et al. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012;15(4):534–544. doi: 10.1016/j.cmet.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ouimet M. Autophagy in obesity and atherosclerosis: Interrelationships between cholesterol homeostasis, lipoprotein metabolism and autophagy in macrophages and other systems. Biochim Biophys Acta. 2013;1831(6):1124–1133. doi: 10.1016/j.bbalip.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13(6):655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khaminets A, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522(7556):354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 8.Levine B, Ranganathan R. Autophagy: snapshot of the network. Nature. 2010;466(7302):38–40. doi: 10.1038/466038a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan W, et al. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci U S A. 2011;108(19):7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang CP, et al. Concanavalin A/IFN-gamma triggers autophagy-related necrotic hepatocyte death through IRGM1-mediated lysosomal membrane disruption. PLoS One. 2011;6(12):e28323. doi: 10.1371/journal.pone.0028323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng CG, et al. Interferon-inducible immunity-related GTPase Irgm1 regulates IFN gamma-dependent host defense, lymphocyte survival and autophagy. Autophagy. 2009;5(2):232–234. doi: 10.4161/auto.5.2.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh SB, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313(5792):1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 17.Bento CF, et al. The role of membrane-trafficking small GTPases in the regulation of autophagy. J Cell Sci. 2013;126(pt 5):1059–1069. doi: 10.1242/jcs.123075. [DOI] [PubMed] [Google Scholar]
- 18.Perera RM, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524(7565):361–365. doi: 10.1038/nature14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, et al. Inactivation of macrophage scavenger receptor class B type I promotes atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation. 2003;108(18):2258–2263. doi: 10.1161/01.CIR.0000093189.97429.9D. [DOI] [PubMed] [Google Scholar]
- 21.Braun A, et al. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ Res. 2002;90(3):270–276. doi: 10.1161/hh0302.104462. [DOI] [PubMed] [Google Scholar]
- 22.Zheng Z, et al. Scavenger receptor class B type I and immune dysfunctions. Curr Opin Endocrinol Diabetes Obes. 2014;21(2):121–128. doi: 10.1097/MED.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanoni P, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351(6278):1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nofer JR, van Eck M. HDL scavenger receptor class B type I and platelet function. Curr Opin Lipidol. 2011;22(4):277–282. doi: 10.1097/MOL.0b013e32834701de. [DOI] [PubMed] [Google Scholar]
- 25.Vergeer M, et al. Genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364(2):136–145. doi: 10.1056/NEJMoa0907687. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, et al. SCARB1 gene variants are associated with the phenotype of combined high high-density lipoprotein cholesterol and high lipoprotein (a) Circ Cardiovasc Genet. 2016;9(5):408–418. doi: 10.1161/CIRCGENETICS.116.001402. [DOI] [PubMed] [Google Scholar]
- 27.Covey SD, et al. Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler Thromb Vasc Biol. 2003;23(9):1589–1594. doi: 10.1161/01.ATV.0000083343.19940.A0. [DOI] [PubMed] [Google Scholar]
- 28.Tao H, et al. Macrophage SR-BI mediates efferocytosis via Src/PI3K/Rac1 signaling and reduces atherosclerotic lesion necrosis. J Lipid Res. 2015;56(8):1449–1460. doi: 10.1194/jlr.M056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yancey PG, et al. Severely altered cholesterol homeostasis in macrophages lacking apoE and SR-BI. J Lipid Res. 2007;48(5):1140–1149. doi: 10.1194/jlr.M600539-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Holm TM, et al. Failure of red blood cell maturation in mice with defects in the high-density lipoprotein receptor SR-BI. Blood. 2002;99(5):1817–1824. doi: 10.1182/blood.v99.5.1817. [DOI] [PubMed] [Google Scholar]
- 31.Pfeiler S, et al. Distinct surveillance pathway for immunopathology during acute infection via autophagy and SR-BI. Sci Rep. 2016;6:34440. doi: 10.1038/srep34440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinet W, Meyer GR. Autophagy in atherosclerosis. Curr Atheroscler Rep. 2008;10(3):216–223. doi: 10.1007/s11883-008-0034-y. [DOI] [PubMed] [Google Scholar]
- 33.Ding Z, et al. Oxidant stress in mitochondrial DNA damage, autophagy and inflammation in atherosclerosis. Sci Rep. 2013;3:1077. doi: 10.1038/srep01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roczniak-Ferguson A, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahras M, et al. Scavenger receptor class B type I localizes to a late endosomal compartment. J Lipid Res. 2008;49(7):1569–1576. doi: 10.1194/jlr.M800055-JLR200. [DOI] [PubMed] [Google Scholar]
- 36.Stein MP, et al. Interaction and functional analyses of human VPS34/p150 phosphatidylinositol 3-kinase complex with Rab7. Methods Enzymol. 2005;403:628–649. doi: 10.1016/S0076-6879(05)03055-7. [DOI] [PubMed] [Google Scholar]
- 37.Stein MP, et al. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4(11):754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 38.Kim YS, et al. PPAR-α activation mediates innate host defense through induction of TFEB and lipid catabolism. J Immunol. 2017;198(8):3283–3295. doi: 10.4049/jimmunol.1601920. [DOI] [PubMed] [Google Scholar]
- 39.Jiao M, et al. Peroxisome proliferator-activated receptor α activation attenuates the inflammatory response to protect the liver from acute failure by promoting the autophagy pathway. Cell Death Dis. 2014;5:e1397. doi: 10.1038/cddis.2014.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh A, et al. Activation of peroxisome proliferator-activated receptor α induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J Biol Chem. 2015;290(16):10309–10324. doi: 10.1074/jbc.M114.610659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mochizuki K, et al. Selectivity of fatty acid ligands for PPARalpha which correlates both with binding to cis-element and DNA binding-independent transactivity in Caco-2 cells. Life Sci. 2006;80(2):140–145. doi: 10.1016/j.lfs.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 42.Lopez D, McLean MP. Activation of the rat scavenger receptor class B type I gene by PPARalpha. Mol Cell Endocrinol. 2006;251(1–2):67–77. doi: 10.1016/j.mce.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh A, Pahan K. PPARα in lysosomal biogenesis: a perspective. Pharmacol Res. 2016;103:144–148. doi: 10.1016/j.phrs.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuller M, et al. The effects of diet on occlusive coronary artery atherosclerosis and myocardial infarction in scavenger receptor class B, type 1/low-density lipoprotein receptor double knockout mice. Arterioscler Thromb Vasc Biol. 2014;34(11):2394–2403. doi: 10.1161/ATVBAHA.114.304200. [DOI] [PubMed] [Google Scholar]
- 46.Wang W, et al. Scavenger receptor class B, type 1 facilitates cellular fatty acid uptake. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865(2):158554. doi: 10.1016/j.bbalip.2019.158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martins Cardoso R, et al. Hyperalphalipoproteinemic scavenger receptor BI knockout mice exhibit a disrupted epidermal lipid barrier. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865(3):158592. doi: 10.1016/j.bbalip.2019.158592. [DOI] [PubMed] [Google Scholar]
- 48.Manichaikul A, et al. Lp-PLA2, scavenger receptor class B type I gene (SCARB1) rs10846744 variant, and cardiovascular disease. PLoS One. 2018;13(10):e0204352. doi: 10.1371/journal.pone.0204352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, et al. A positive feedback loop between GRP78 and VPS34 is critical for GRP78-mediated autophagy in cancer cells. Exp Cell Res. 2016;351(1):24–35. doi: 10.1016/j.yexcr.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 50.B’Chir W, et al. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41(16):7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams-Bey Y, et al. Omega-3 free fatty acids suppress macrophage inflammasome activation by inhibiting NF-κB activation and enhancing autophagy. PLoS One. 2014;9(6):e97957. doi: 10.1371/journal.pone.0097957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai X, et al. Inhibition of nuclear factor-kappa B activation decreases survival of Mycobacterium tuberculosis in human macrophages. PLoS One. 2013;8(4):e61925. doi: 10.1371/journal.pone.0061925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djavaheri-Mergny M, et al. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006;281(41):30373–30382. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 54.Ouimet M, et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat Immunol. 2016;17(6):677–686. doi: 10.1038/ni.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai L, et al. Macrophage SR-BI regulates LPS-induced pro-inflammatory signaling in mice and isolated macrophages. J Lipid Res. 2012;53(8):1472–1481. doi: 10.1194/jlr.M023234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi Y, et al. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 2009;16(7):947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graham J, et al. The preparation of subcellular organelles from mouse liver in self-generated gradients of iodixanol. Anal Biochem. 1994;220(2):367–373. doi: 10.1006/abio.1994.1351. [DOI] [PubMed] [Google Scholar]
- 59.Staros JV. N-hydroxysulfosuccinimide active esters: bis(N-hydroxysulfosuccinimide) esters of two dicarboxylic acids are hydrophilic, membrane-impermeant, protein cross-linkers. Biochemistry. 1982;21(17):3950–3955. doi: 10.1021/bi00260a008. [DOI] [PubMed] [Google Scholar]
- 60.Tao H, et al. Peroxisome proliferator-activated receptor-gamma regulates the expression and function of very-low-density lipoprotein receptor. Am J Physiol Endocrinol Metab. 2010;298(1):E68–E79. doi: 10.1152/ajpendo.00367.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinet P, et al. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J Lipid Res. 2010;51(11):3364–3369. doi: 10.1194/jlr.D007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oshchepkov DY, Levitsky VG. In silico prediction of transcriptional factor-binding sites. Methods Mol Biol. 2011;760:251–267. doi: 10.1007/978-1-61779-176-5_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.