Abstract
Accumulating evidence shows that extracellular vesicles (EVs) secreted by immune cells play an important role in intercellular communication. In the current report, we show that EVs released from wild-type (WT) bone marrow-derived dendritic cells (BMDCs) transfer Toll-like receptors to TLR4KO BMDCs and increase cellular responsiveness to LPS in recipient cells. The transferred EVs have exosomal characteristics and induce the activation of NFκB signaling pathways in recipient cells. We further show that BMDC-derived EVs can promote LPS-induced inflammation in TLR4KO mice in vivo. These results indicate that functional TLR4 can be transferred from WT to TLR4KO BMDCs through exosome-like EVs.
Keywords: Dendritic cells, Extracellular Vesicles, Exosome, Toll-like receptor 4
Introduction
Dendritic cells (DCs) are a class of professional antigen presenting cells (APCs) that express multiple types of Toll-like receptors (TLRs) (1). Within the family of TLRs, TLR4 is a cell membrane receptor that recognizes lipopolysaccharide (LPS), which is a major component of the outer membrane of Gram-negative bacteria (2). Upon ligand binding, TLR4 activates a set of adaptor proteins and intracellular kinases, leading to the nuclear translocation of transcription factor NFκB and the production of pro-inflammatory cytokines, such as IL-6 and TNFα (3). TLR4 activation results in enhanced inflammatory responses that are positively associated with a variety of immune diseases, such as arthritis (4) and inflammatory bowel disease (IBD) (5), and it has also been shown to be involved in mechanisms associated with immune therapy in cancer patients (6).
Recent studies have revealed that exosomes, which are extracellular vesicles (EVs) that are 30 to 100 nm in size that originate from endosomes, could be effective mediators of intercellular communication between local immune systems. Exosomes derived from dendritic cells are known to stimulate CD4-positive T cells and elicit specific immune responses (7). Additionally, it has been reported that exosomes derived from bone marrow-dendritic cells (BMDCs) incubated with recombinant IL-10 can increase the number of Treg cells in the intestinal mucosa and enhance protective effects against colitis in a mouse model (8). However, the mechanisms underlying the immune regulatory effects exerted by exosomes have not yet been completely elucidated.
EVs contain a variety of biological components, including membrane proteins, lipids, miRNA, mRNA and DNA. Previous studies demonstrated that DC-derived exosomes could exert biological effects via the functional transfer of sorted miRNAs (9). However, few studies have shown that proteins within exosomes can be transferred or induce functional changes in recipient cells (10). Here, we show for the first time that TLR4 receptors on BMDC-derived EVs that have exosome-like features can be taken up by other DCs and activate the TLR4/NFκB signaling pathway in recipient cells.
Materials and Methods
Animals
Eight-week-old C57BL/6J (WT) and C57BL/10ScNCr (TLR4KO) mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA) (https://www.jax.org/strain/003752). Animal maintenance and care procedures were conducted according to the policies the Institutional Animal Care and Use Committee (IAUCUC) at the University of Miami.
WT and TLR4KO mice received tail-vein injections of 1 mg (total protein) of exosomes in PBS. Twenty-four hours later, the mice received 500 ng/kg LPS by intraperitoneal (i.p.) injection. Two hours after this, the mice were sacrificed, and their serum, spleen and mesenteric lymph nodes (MLN) were processed for cytokine measurement.
Primary Dendritic Cell Culture
WT and TLR4KO mice were sacrificed, and their femur bones were removed for bone-marrow primary culture. The bone marrow cells were maintained in complete Iscove’s modified Dulbecco medium (IMDM; Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 20 ng/ml granulocyte macrophage-colony stimulating factor (GM-CSF), and 10 ng/ml recombinant mouse IL-4 protein (R&D Systems, MN). Bone marrow-derived dendritic cells (BMDCs) were generated by 10 to 12 days of differentiation in complete medium.
Flow Cytometric Analysis
BMDCs were stained for dendritic cell markers according to a method described in previous studies(11). Forward and side scatter gating were used to remove debris while preserving singlet events. The panel of antibodies used in these experiments included CD11b-conjugated Alexa Fluor 488 and CD11c-conjugated eFluor 660, which were obtained from eBioscience, and TLR4-BV421 and CD45-red Fluor 710, which are obtained from BD Bioscience. Flow cytometry was performed using a CytoFLEX flow cytometer (Beckman Coulter, Brea, CA, US), and the data were analyzed with CytExpert software, which is part of the CytoFLEX platform.
Electron Microscopy
Extracellular vesicles were recovered from the supernatant of BMDCs cultured in medium depleted of FBS-derived exosomes. The supernatant was centrifuged at 3000 g for 10 minutes to remove the cell debris and was then purified by filtration with 0.20 μm pore filters, followed by ultracentrifugation at 100000 g at 4°C for 2.5 hours as previously described (12) or isolation using an ExoQuick isolation kit (System Biosciences, Palo Alto, CA) based on the manufacturer’s protocol.
The isolated EVs were fixed in 2% paraformaldehyde, and 10 μl of each sample was loaded onto a Formvar/carbon-coated copper grid for 30 minutes. The grid was rinsed by floating it on top of a drop of 0.1 M PO4 buffer for 5 minutes, and then the EVs were fixed in 2% glutaraldehyde fixative for 5 minutes; the grid was washed in 0.1 M PO4 buffer for 5 minutes and then in double-distilled water for 5 minutes, after which the procedure was repeated. The grid was then floated on top of a drop of 4% uranyl acetate in double-distilled water for 5 minutes in the dark. The grid was drained and completely dried in the dark and then transferred to a grid box. Images were acquired with a JEOL JEM-1400 Transmission Electron Microscope (Peabody, MA) equipped with a Gatan Orius SC 200D digital camera. The University of Miami TEM imaging core acquired all TEM images of the EVs.
Western blot analysis
Total protein (20 μg) was extracted from BMDCs or exosome pellets lysed with radio-immunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, Waltham, MA) containing fresh protease and phosphatase inhibitors (Roche, Indianapolis, IN) and was then loaded into each lane and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis. The proteins were then transferred to a 0.45 μm pore-size nitrocellulose membrane (Bio-Rad, Hercules, CA). The antibody used in the Western blot assay is shown in Table I.
TABLE I.
Primary Abs used in Western blot assay
| Ab | Species | Dilution | Catalog No. | Brand |
|---|---|---|---|---|
| CD63 (H-193) | Rabbit | 1:200 | SC-15363 | Santa Cruz Biotechnology (Dallas, TX) |
| CD 9 (EM-04) | Rat | 1:500 | MAI-10309 | Thermo Fisher Scientific |
| TsglOl | Rabbit | 1:1000 | ab30871 | Abeam (Cambridge, U.K.) |
| β-Actin | Mouse | 1:4000 | 4967S | Cell Signaling Technology (Danvers, MA) |
| GAPDH | Rabbit | 1:2000 | PAI-987 | Thermo Fisher Scientific |
| lkBα | Rabbit | 1:500 | 9242S | Cell Signaling Technology |
| NF-kB (pl05/p50) | Rabbit | 1:1000 | ab32360 | Abeam |
| TBP | Mouse | 1:1000 | abSIS | Abeam |
lkBα, inhibitor of NF-kB kinase subunit α, also known as IKK1; TBP. TATA-binding protein; TsglOl. tumor susceptibility gene 101 protein.
Immunocytochemistry
The EVs were labeled with a lipid-associating fluorescent dye (PKH67; Sigma- Aldrich, St. Louis, MO). BMDCs were grown in a Nunc™ Lab-Tek™ II Chamber Slide™. Fluorescently-labeled exosomes or PBS-PKH67 controls were washed with PBS and incubated with the cells for 24 hours. On the following day, the cells were washed twice with PBS, fixed in 4% paraformaldehyde, and stained with Alexa Fluor™ 594 Phalloidin (Thermo Fisher Scientific, Waltham, MA) for 60 minutes. The sample slides were washed with PBS and mounted with DAPI anti-fade reagent. Images were obtained using a Leica Microscope.
Quantitative real-time PCR
Total RNA was isolated from exosomes or BMDCs with TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA). The levels of IL-6, TNFα, and TLR4 were detected by quantitative real-time PCR (qRT-PCR) using the High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA) and a LightCycler® 480 System (Roche, Indianapolis, IN). The primers used for quantitative real-time PCR are shown in Table II.
TABLE II.
Primers used in real-time PCR assays
| Genes | Orientation | Sequence |
|---|---|---|
| IL-6 F | 5’–3’ | 5’-CCGGAGAGGAGACTTCACAG-3’ |
| IL-6 R | 5’–3’ | 5’-tccacgatttcccagagaac-3’ |
| IL-1β F | 5’–3’ | 5’ -ggcaggcagtatcactcatt-3’ |
| IL-1β R | 5’–3’ | 5’-aaggtgctcatgtcctcatc-3’ |
| TNF-α F | 5’–3’ | 5’-gacgtggaactggcagaaga-3’ |
| TNF-α R | 5’–3’ | 5’ -gccacaagcaggaatgagaa-3’ |
| TLR4 F | 5’–3’ | 5’ -atatgcatgatcaacaccacag-3’ |
| TLR4 R | 5’–3’ | 5’ -TTTCCATTGCTGCCCTATAG-3’ |
| GAPDH F | 5’–3’ | 5’-cgaaggtggaagagtgggag-3’ |
| GAPDH R | 5’–3’ | 5’-tgaagcaggcatctgaggg-3’ |
| rpL32A F | 5’–3’ | 5’-gctggaggtgctgctgatgt-3’ |
| rpL32A R | 5’–3’ | 5’ -actctgatggccagctgtgc-3’ |
F, forward primer R, reverse primer; rpL32A, 60S ribosomal protein L32 A.
Enzyme-linked immunosorbent assay (ELISA)
Exosomes (100 ng total protein) were added to BMDCs and cultured in bovine exosome-free medium for 24 hours. The conditioned medium was collected and centrifuged (3000 g) to clear the supernatants, which were subsequently used for cytokine detection by ELISA. For the in vivo experiments, serum, spleen and MLN extracts from both WT and TLR4KO mice were centrifuged, and the total protein concentration in the supernatant was measured for the purpose of normalization. The cytokines in the cell culture supernatant or in animal organs were detected by TNFα and IL-6 ELISA kits (Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer’s protocols. The cytokine concentrations were determined by the measurement of absorbance in triplicate with a Spectra Max M5 plate reader and Softmax Pro 5 software.
Statistics
Data are presented as the mean ± SEM. The statistical analysis was performed with GraphPad Prism 6.0 software (GraphPad Software, San Diego, CA). Unpaired Student’s t-tests or one-way analysis of variance (ANOVA) was adopted used for the analysis. The significance levels were denoted as *p <0.05, **p < 0.01, or ***p <0.001.
Results
Dendritic cells secrete and take up extracellular vesicles with exosomal features.
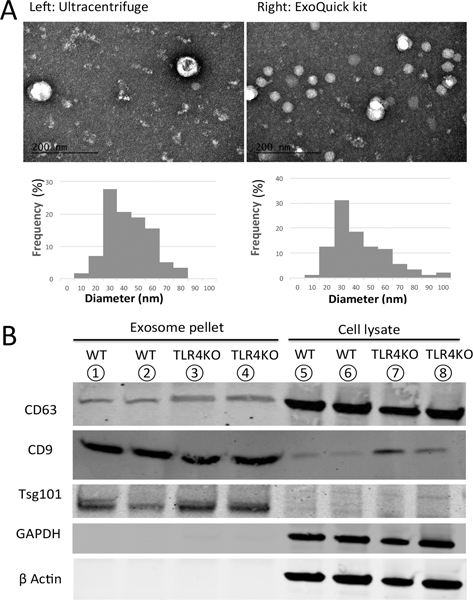
To validate the primary culture, CD45/CD11b/CD11c and TLR4 antibodies were used as cell surface markers for the flow cytometry assay. Our results show that a significant population within the bone marrow cells had differentiated into dendritic cells in 10–12 days (Supplemental Figure 1), and there was a distinct difference in TLR4 expression in WT and TLR4KO BMDCs (Supplemental Figure 2). Two methods were used to purify the EVs: ultracentrifugation or isolation with an ExoQuick Kit from System Biosciences. The EVs that were harvested from the supernatants of BMDC cultures were visualized by transmission electron microscopy (TEM). In both samples, we observed round-shaped vesicles between 20–100 nm in diameter. The particle size distribution did not show significant differences in samples that used different purification methods (Figure 1A). Exosome-specific protein markers in both of the purified EV samples were determined using Western blotting with anti-CD63, anti-CD9, and Tsg101 antibodies (Figure 1B). Overall, the EV samples isolated from BMDC culture supernatants by both methods could be characterized as exosomes.
Figure 1.
Extracellular vesicles derived from BMDC culture supernatants have exosomal features. (A) Upper panel: electron microscopy of exosome samples isolated with different methods. Lower panel: distribution of vesicle diameters (left: ultracentrifugation; right: ExoQuick kit). (B) Western blot assays revealed that both samples contained the exosomal proteins CD63, CD9, and Tsg101.
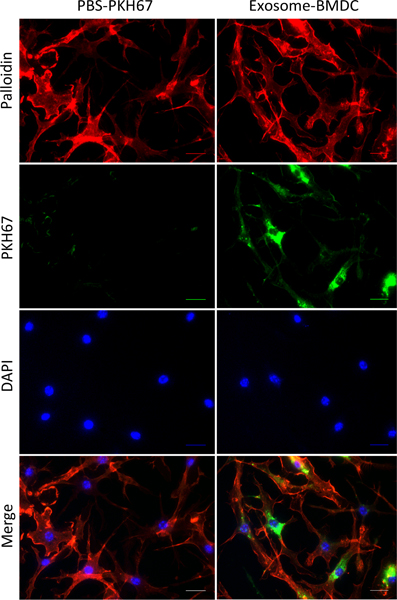
To determine whether BMDCs can take up exogenous exosomes, we labeled BMDC-secreted exosomes with PKH67 dye (green) and incubated them with BMDC cultures for 24 hours. Immunofluorescent images confirmed that recipient cells with F-actin (red) expression colocalized with PKH67-labeled exosomes (Figure 2).
Figure 2.
Incorporation of exosomes in BMDCs. Exosomes isolated from culture supernatants were dissolved in PBS, incubated with PKH67 (green fluorescent dye) for 5 minutes and then washed with 1% BSA in PBS (Exosome+ BMDC). Both exosomes and the PBS control (PBS+ PKH67) were treated with BMDCs incubated on chamber slides. After 24 hours of incubation, the PKH67-labeled exosomes were taken up by BMDCs (red: F-actin; green: PKH67; blue: DAPI; scale bar: 50 μm).
Exosomes from WT BMDCs transfer TLR4 receptors to TLR4KO BMDCs.
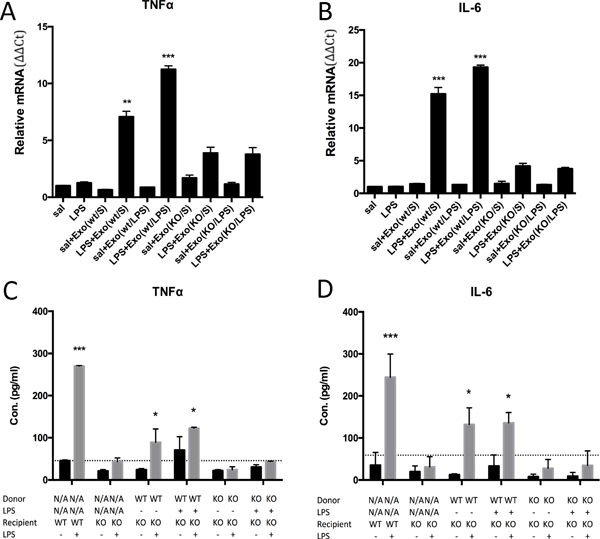
Additionally, we validated the knockout of TLR4 by real-time PCR, showing that TLR4KO BMDCs lack responsiveness to LPS upon stimulation (Supplemental Figure 3A–B). However, when we preincubated these TLR4KO BMDCs with exosomes isolated from supernatant derived from WT BMDCs, the results showed that 4 hours of LPS stimulation remarkably upregulated the mRNA (Figure 3A–B) and protein levels (Figure 3C–D) of pro-inflammatory cytokines such as TNFα and IL-6. In contrast, exosomes isolated from the supernatants of TLR4KO BMDCs did not induce any pro-inflammatory cytokine production. We tested the RNA content of exosomes using exosomes purified from the supernatants of WT or TLR4KO BMDC cultures. Quantitative PCR results showed that there were no significant differences in the levels of TNFα or IL-6 mRNA between these exosome samples (Supplemental Figure 3C). Additionally, exosomes purified from the supernatants of WT BMDCs that were pretreated with LPS alone did not stimulate TLR4KO BMDCs to produce pro-inflammatory cytokines (Figure 3 C–D). Only the addition of LPS to recipient cells in the presence of exosomes from WT BMDCs could induce the TLR4KO BMDCs to become responsive to LPS, suggesting that functional TLR4 receptors were transferred from the WT exosomes to the TLR4KO cells.
Figure 3.
BMDCs from TLR4KO mice gained responsiveness to LPS after the uptake of exosomes from WT cells. (A-B) In TLR4KO BMDCs, treatment with LPS does not stimulate cytokine production. After incubation of exosomes from WT BMDCs and LPS stimulation for 4 hours, real-time PCR showed that TNFα and IL-6 mRNA levels were significantly increased (sal: saline; Exo(wt/S): exosomes purified from the supernatant of wild-type BMDCs pretreated with saline; Exo(wt/LPS): exosomes purified from the supernatant of wild-type BMDCs pretreated with LPS). (C-D) In the supernatant of WT BMDCs, LPS stimulated a high level of TNFα and IL-6 production. In TLR4KO BMDCs, only preincubation with exosomes from WT BMDCs and treatment with LPS could stimulate the production of the pro-inflammatory cytokines TNFα and IL-6, as measured by ELISA. (The results are expressed as the mean ± SEM of triplicate measurements in each group, *p<0.05, **p<0.01, ***p<0.001).
The NF kappa B signaling pathway was activated in TLR4KO BMDCs upon exosomal uptake from WT cells.
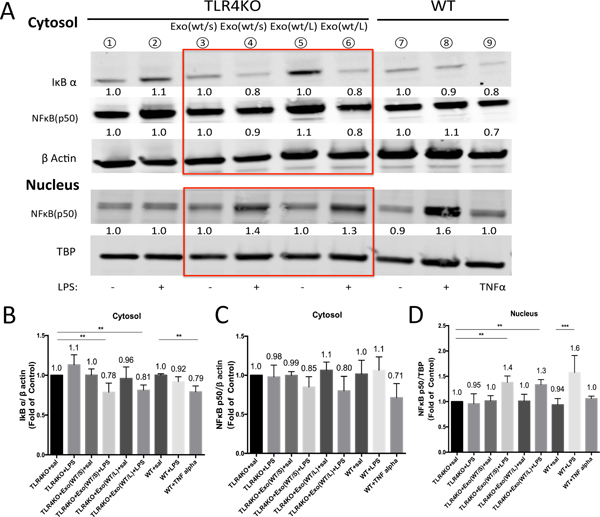
Because we observed the uptake of TLR4 receptors from WT exosomes by TLR4KO BMDCs, we next investigated the direct signaling targets of these receptors by LPS-induced activation. Western blot analysis of the cytosolic and nuclear proteins from of BMDCs showed that after the preincubation of WT exosomes and stimulation by LPS for 4 hours (Figure 4A), the IκBα protein level in the cytosol of the TLR4KO BMDCs decreased (Figure 4B) and NFκB protein translocated from the cytoplasm (Figure 4C) to the nucleus (Figure 4D), both of which indicate the activation of the NFκB signaling pathway. These results also provided evidence that exosomes transferred TLR4 between dendritic cells as cargo. The activation of these receptors led to activation of the NFκB intracellular signaling pathway and the production of inflammatory cytokines in the presence of the TLR cognate ligand LPS.
Figure 4.
The NFκB signaling pathway was activated in TLR4KO BMDCs. (A) After removing exosomes from WT cells, the levels of the signaling molecules IκBα and NFκB were altered, as shown by Western blot analysis of cytoplasmic (upper) and nuclear protein (lower) extracted from TLR4 knockout BMDCs. Exo(wt/s): exosomes purified from the supernatant of wild-type BMDCs pretreated with saline; Exo(wt/L): exosomes purified from the supernatant of wild-type BMDCs pretreated with LPS. (B-D) Quantification of the intensity of the bands shows that the preincubation of exosomes from WT BMDCs and their treatment with LPS could stimulate the translocation of NFκB protein from the cytoplasm to the nucleus and the degradation of IκBα protein (the results are expressed as the mean ± SEM from 5 independent experiments, *p<0.05, **p<0.01).
TLR4KO mice responded to LPS following i.v. injection of exosomes from WT BMDCs.
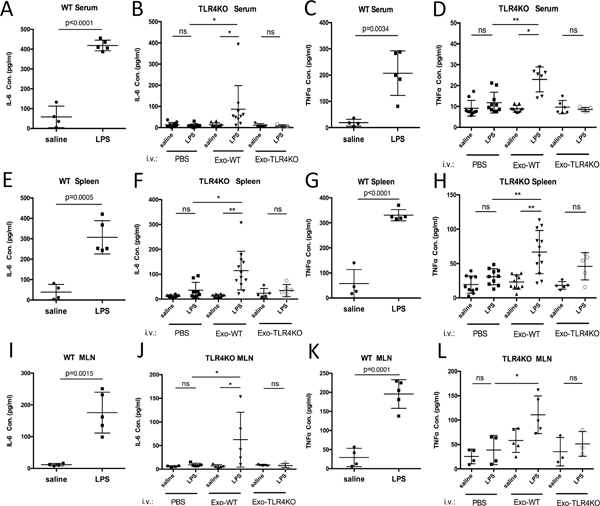
To further confirm the effect of exosomal TLR4 transportation in vivo, we administered exosomes from WT or TLR4KO BMDCs to TLR4KO mice by tail vein injection. Twenty-four hours after PBS or exosome injection, the WT or TLR4 mice were injected with LPS intraperitoneally. Two hours later, the serum, spleen and mesenteric lymph nodes (MLN) of these animals were collected for the detection of IL-6 and TNF-α by ELISA. WT mice but not TLR4KO mice responded to LPS stimulation (Figure 5). However, TLR4KO mice injected with exosomes from WT BMDCs showed significant responsiveness to LPS, as indicated by IL-6 and TNFα production in serum (Figure 5 A–D), spleen (Figure 5 E–H) and MLN (Figure 5 I–L). TLR4KO mice injected with exosomes from TLR4KO BMDCs did not show similar changes in immunomodulatory efficacy, indicating that exosomes alone do not contribute to this effect.
Figure 5.
TLR4KO mice gained responsiveness to LPS after the i.v. injection of exosomes from WT BMDCs. Measurement of TNFα and IL-6 in the serum (A-D), spleen (E-H) and mesenteric lymph nodes (MLN) (I-L) of WT and TLR4KO mice after the injection of exosomes and LPS (the results are expressed as the mean ± SEM of measurements from N=6–12 per group, and each dot represents a measurement from one animal; *p<0.05, **p<0.01, ***p<0.001).
In addition, we found that the transfer of exosomes derived from WT BMDCs to TLR2KO BMDCs induced TLR2KO BMDCs to respond to denatured Enterococcus faecalis (E.f). Interestingly, these BMDCs produced IL-6 but not TNFα after stimulation (Supplemental Figure 4). Overall, these observations support our hypothesis that functional TLR4 and TLR2 receptor proteins can be transferred via exosomes between cells.
Discussion
In this study, we used multiple approaches to confirm that exosomes derived from WT BMDCs transferred TLR4 receptors to TLR4KO cells and altered the ability of the recipient cells to respond to LPS stimulation both in vitro and in vivo. To determine whether the TLR4 membrane protein was taken up by the recipient cells and if it triggered the inflammatory responses in the TLR4KO cells, different controls were used. Exosomes from WT BMDCs alone did not induce the release of pro-inflammatory cytokines in the recipient cells, even when the donor cells were pretreated with LPS, indicating that only the TLR4 receptor but not the downstream signaling molecules in the TLR4/NFκB pathway, such as activated myd88, TRAF6 or IRAK4, were transferred via exosomes to the TLR4KO cells (Figure 3). In addition, the difference in the immunomodulatory characteristics of the recipient cells is not due to the transfer by exosomes of mRNA encoding the cytokines being investigated (Supplemental Figure 3C). The comparison between TLR4KO and WT exosomes in vivo further confirmed that functional TLR4 protein in the exosomes is required for the recipient cells to respond to LPS (Figure 5). In addition,, we are able to address the possibility that other membrane-associated proteins besides TLR4, such as TLR2, can also be transferred to other cells via exosomes (Supplemental Figure 4).
There are several mechanisms underlying the amplification of inflammatory responses to LPS in TLR4KO BMDCs. For instance, it is plausible that WT exosomes shuttle nucleic acids (mRNA or miRNA) between WT and KO cells and regulate functioning of the recipient cell at a posttranscriptional level, and many studies have demonstrated that this functional change is correlated with exosomes (12, 13). However, we believe that the most likely mechanism involves the fusion of the exosomal membrane with the recipient cell membrane to transfer the TLR4 binding moiety to the recipient cell, since most other signaling molecules are still intact in the TLR4/2 knockout dendritic cells. This mechanism of transfer is supported by studies that show that the lipid bilayer of the exosome, in addition to protecting the exosomal cargo from plasma and immune components, is also instrumental in delivering exosomal cargo and exosomal membrane components from donor cells to recipient cells by endocytosis or fusion without compromising the intrinsic functioning of the membrane or cargo (14, 15).
There is growing interest in the therapeutic uses of exosomes, both in inflammatory diseases and in cancer therapies. The small size of exosomes distributed in biofluids, such as serum and milk, allow them to travel long distances in the human body(16). Exosomes could be utilized as mediators for the exchange of bioinformation between neighboring cells and the maintenance of the homeostatic microenvironment during immune responses (9) or used as stable carriers for the delivery of RNA/protein content to a defined microenvironment (17).
Accumulating evidence has suggested that the content of exosomes can be manipulated(18), and these engineered exosomes may serve as ideal vehicles for chemoimmunotherapy involving miRNA/vaccine delivery. For instance, several clinical trials have been performed thus far that have involved immunizing patients with exosomes released in vitro by their own DCs loaded with tumor antigen-derived peptides (19). In another study, the proteomic comparative analysis of DC-derived extracellular vesicles revealed the presence of the immune regulatory molecule PD-L1 on small extracellular vesicles that were similar in size to exosomes (20). These findings provide evidence that DC-derived exosomes play a role in the mechanisms associated with immune checkpoint inhibitors in cancer treatment.
Overall, the current study significantly adds to our knowledge of the role of exosomes as important mediators of the innate immune system. The evidence that receptors can be transferred from cell to cell through exosomes to induce functional changes in recipient cells can be therapeutically exploited by the application of exosomes as valuable vectors that can be loaded with specific proteins and delivered to target cells. In addition, the corresponding signaling pathways involved in exosome biogenesis, trafficking, and exocytosis may become feasible targets for the clinical treatment of inflammatory diseases and cancer immunotherapy.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 DA043252, R01 DA037843, R01 DA044582, R01 DA047089 and R01DA034582 to Sabita Roy.
Footnotes
All named authors have read and approved this version of the manuscript and verify that there are no conflicts of interest.
REFERENCES
- 1.Visintin A, Mazzoni A, Spitzer JH, Wyllie DH, Dower SK, and Segal DM. 2001. Regulation of Toll-like receptors in human monocytes and dendritic cells. J. Immunol. 166: 249–255. [DOI] [PubMed] [Google Scholar]
- 2.Chow JC, Young DW, Golenbock DT, Christ WJ, and Gusovsky F. 1999. Toll-like receptor-4 mediates lipopolysaccharide induced signal transduction. J. Biol. Chem. 274: 10689–10692. [DOI] [PubMed] [Google Scholar]
- 3.Bamford S, Ryley H, and Jackson SK. 2007. Highly purified lipopolysaccharides from Burkholderia cepacia complex clinical isolates induce inflammatory cytokine responses via TLR4-mediated MAPK signalling pathways and activation of NFkappaB. Cell. Microbiol. 9: 532–543. [DOI] [PubMed] [Google Scholar]
- 4.Abdollahi-Roodsaz S, Joosten LAB, Roelofs MF, Radstake TRDJ, Matera G, Popa C, van der Meer JWM, Netea MG, and van den Berg WB. 2007. Inhibition of Toll-like receptor 4 breaks the inflammatory loop in autoimmune destructive arthritis. Arthritis Rheum. 56: 2957–2967. [DOI] [PubMed] [Google Scholar]
- 5.Dheer R, Santaolalla R, Davies JM, Lang JK, Phillips MC, Pastorini C, Vazquez-Pertejo MT, and Abreu MT. 2016. Intestinal Epithelial Toll-Like Receptor 4 Signaling Affects Epithelial Function and Colonic Microbiota and Promotes a Risk for Transmissible Colitis. Infect. Immun. 84: 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M, Chen J, Zhou L, Chen W, Ding G, and Cao L. 2014. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell. Immunol. 292: 65–69. [DOI] [PubMed] [Google Scholar]
- 7.Thé ry C, Duban, Segura E, Vé ron P, Lantz O, and Amigorena S. 2002. Indirect activation of näıve CD4+ T cells by dendritic cellderived exosomes. Nat. Immunol. 3: 1156–1162. [DOI] [PubMed] [Google Scholar]
- 8.Yang X, Meng S, Jiang H, Chen T, andWu. W. 2010. Exosomes derived from interleukin-10-treated dendritic cells can inhibit trinitrobenzene sulfonic acid-induced rat colitis. Scand. J. Gastroenterol. 45: 1168–1177. [DOI] [PubMed] [Google Scholar]
- 9.Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, and O’Connell RM. 2015. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 6: 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buschow SI, van Balkom BWM, Aalberts M, Heck AJR, Wauben M, and Stoorvogel W. 2010. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 88: 851–856. [DOI] [PubMed] [Google Scholar]
- 11.Yokota A, Takeuchi H, Maeda N, Ohoka Y, Kato C, Song SY, and Iwata M. 2009. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int. Immunol. 21: 361–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu G, Yao H, Chaudhuri AD, Duan M, Yelamanchili SV, Wen H, Cheney PD, Fox HS, and Buch S. 2012. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 3: e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Nardo D, De Nardo CM, Nguyen T, Hamilton JA, and Scholz GM. 2009. Signaling crosstalk during sequential TLR4 and TLR9 activation amplifies the inflammatory response of mouse macrophages. J. Immunol. 183: 8110–8118. [DOI] [PubMed] [Google Scholar]
- 14.Dalvi P, Sun B, Tang N, and Pulliam L. 2017. Immune activated monocyte exosomes alter microRNAs in brain endothelial cells and 10.4049/immunohorizons. 1900016 192 E Vs TRANSFER TLRs BETWEEN DENDRITIC CELLS ImmunoHorizons Downloaded from http://www.immunohorizons.org/ by guest on March 19, 2021 initiate an inflammatory response through the TLR4/MyD88 pathway. Sci. Rep. 7: 9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian T, Zhu Y-L, Zhou Y-Y, Liang G-F, Wang Y-Y, Hu F-H, and Xiao Z-D. 2014. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 289: 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato YT, Umezaki K, Sawada S, Mukai SA, Sasaki Y, Harada N, Shiku H, and Akiyoshi K. 2016. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 6: 21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bretz NP, Ridinger J, Rupp AK, Rimbach K, Keller S, Rupp C, Marme F´, Umansky L, Umansky V, Eigenbrod T, et al. 2013. Body fluid exosomes promote secretion of inflammatory cytokines in monocytic cells via Toll-like receptor signaling. J. Biol. Chem. 288: 36691–36702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X, Jiang X, Hou D, Chen X, Chen Y, et al. 2015. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci. Rep. 5: 17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fricke F, Lee J, Michalak M, Warnken U, Hausser I, Suarez- Carmona M, Halama N, Schnölzer M, Kopitz J, and Gebert J. 2017. TGFBR2-dependent alterations of exosomal cargo and functions in DNA mismatch repair-deficient HCT116 colorectal cancer cells. Cell Commun. Signal. 15: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitt JM, Andre F´, Amigorena S, Soria JC, Eggermont A, Kroemer G, and Zitvogel L. 2016. Dendritic cell-derived exosomes for cancer therapy. J. Clin. Invest. 126: 1224–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal- Bengtson B, Dingli F, Loew D, Tkach M, and Thé ry C. 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 113: E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.