Abstract
Objective:
Animal studies remain essential for understanding mechanisms of epilepsy and identifying new therapeutic targets. However, existing animal models of epilepsy do not reflect the high level of genetic diversity found in the human population. The Collaborative Cross (CC) population is a genetically diverse recombinant inbred panel of mice. The CC offers large genotypic and phenotypic diversity, inbred strains with stable genomes that allow for repeated phenotypic measurements, and genomic tools including whole genome sequence to identify candidate genes and candidate variants.
Methods:
We evaluated multiple complex epileptic traits in a sampling of 35 CC inbred strains using the flurothyl-induced seizure and kindling paradigm. We created a ~300 F2 population of extreme seizure susceptibility and performed QTL mapping to identify genomic regions associated with seizure sensitivity. We used quantitative RNA sequencing from CC hippocampal tissue to identify candidate genes and whole genome sequence to identify genetic variants likely affecting gene expression.
Results:
We identified new mouse models with extreme seizure susceptibility, seizure propagation, epileptogenesis, and SUDEP (sudden unexpected death in epilepsy). We performed QTL mapping in an F2 intercrosses of seizure susceptible and resistant strains and identified one known and seven novel loci associated with seizure sensitivity. We combined whole genome sequencing and hippocampal gene expression to pinpoint biologically plausible candidate genes (e.g. Gabra2) and variants associated with seizure sensitivity.
Significance:
New mouse models of epilepsy are needed to better understand the complex genetic architecture of seizures and to identify therapeutics. We performed a phenotypic screen utilizing a novel genetic reference population of CC mice. The data we provide enables the identification of protective/risk genes and novel molecular mechanisms linked to complex seizure traits that are currently challenging to study and treat.
Keywords: Collaborative Cross mouse, seizure susceptibility, epileptogenesis, SUDEP, Gabra2
INTRODUCTION
Epilepsies are a clinically heterogeneous group of neurological disorders with the lifetime prevalence of 0.76%1. There are no treatments to prevent epilepsy, and roughly 30% of epilepsies are intractable to current antiepileptic medications2. Uncontrolled seizures also increase the risk of sudden unexpected death in epilepsy (SUDEP), a poorly understood fatal complication of epilepsy3. New transformative treatments for epilepsy could be developed by understanding the genetic associations conferring risk or protective effects. In addition to the rapid expansion of epilepsy-associated gene lists discovered mainly with monogenic causes of seizures, human GWAS studies of generalized epilepsy identified more than a dozen novel genome-wide significant loci and biologically plausible candidate genes4–6. However, human GWAS approaches have had limited success in identifying risk loci associated with certain forms of epilepsy and specific seizure outcomes (e.g. SUDEP), partially due to complex disease etiologies and underpowered sample sizes7, 8. Animal studies remain essential for understanding the mechanisms of epilepsy and for identifying new therapeutic targets. In particular, animal models offer phenotypic repeatability and a means to study a trait in a controlled environment, as well as providing a level of experimental testing and validation that is not ethical or possible in human subjects, such as in the case of SUDEP. Most existing animal models of complex diseases such as epilepsy are limited because they do not reflect the high level of genetic diversity found in the human population. In order to identify novel genetic targets, a model research population with a high level of genetic variation is needed.
The Collaborative Cross (CC) population is a genetically diverse recombinant inbred panel derived from eight fully inbred strains that has ~42 million segregating genetic variants9. In addition, whole genome sequences have been generated for each individual CC strain, and other genomic tools have been developed to enable the identification of genetic variants from mapping studies9, 10. The CC has been used to model complex traits such as behavior11–13, cancer14, and infectious disease susceptibility15. In order for us to take advantage of the immense genetic diversity available in the CC to study epilepsy, we needed a seizure induction protocol amenable to high throughput analysis. Accordingly, we studied the CC using flurothyl seizure-induction to characterize multiple epileptic outcomes including seizure sensitivity, seizure propagation, seizure development (i.e. epileptogenesis), and SUDEP. We outlined a general approach to use the CC and its associated genetic resources to identify quantitative trait loci (QTL) and candidate genes. We demonstrated a highly reliable and repeatable seizure induction paradigm, as well as the implementation and design of a F2 genetic mapping population. We also showed the power of our approach by using the CC and transcriptomic sequence resources to identify several QTL (both novel and previously identified), candidate genes, and candidate variants associated with seizure sensitivity. These resources provide a powerful toolbox for identifying new genes, and hence new therapeutic targets, linked to seizure sensitivity, seizure propagation, epileptogenesis, and SUDEP.
MATERIALS AND METHODS
Mice
All mice (Table S1) were raised on standard mouse chow and kept on a 12:12 light/dark cycle. All mouse work was compliant with UNC Institutional Animal Care and Use Committee protocols.
Flurothyl-induced seizure and flurothyl kindling
Each mouse was placed in an enclosed chamber and 10% flurothyl in 95% ethanol was then infused at a rate of 200 μL/min onto a disk of filter paper suspended at the top of the chamber. Mice exhibit various stages of increasing seizure severity in response to flurothyl exposure, including myoclonic seizure (sudden involuntary jerk/shock-like movements involving the face, trunk, and/or limbs) and generalized seizure (also known as clonic-forebrain seizures that are characterized by clonus of the face and limbs, loss of postural control, rearing, and falling). Upon emergence of a generalized seizure, the lid of the chamber was immediately removed, allowing for rapid dissipation of the flurothyl vapors and exposure to fresh air16. C57BL/6J (B6J) mice were challenged in each round of experiments and exhibited similar seizure susceptibility, therefore ensuring lack of differences that could arise from, for example, different batches of flurothyl or different experimenter and video reviewer. For flurothyl kindling, flurothyl exposures were repeated once daily over eight consecutive days. Mouse behavior during each flurothyl exposure was video-recorded and reviewed by investigators (blind to strains) who determined latency to the onset of both myoclonic and generalized seizures. Mice that failed to survive through the eight-day kindling were excluded from analysis. The linear regression of day-1 through day-8 was used to estimate the kindling effect as previously described17.
Pentylenetetrazol (PTZ) induced acute seizure and PTZ kindling
For PTZ-induced acute seizure, each mouse was subjected to PTZ injection (40 mg/kg, i.p.) and animal behavior was monitored by video for 30 min. Video was later analyzed by investigators, blinded to strain, who measured the latency to the onset of behavioral seizures (seizure threshold) and determined seizure severity using a modified Racine’s scale. If the mouse did not exhibit any behavioral seizure within 30 min after PTZ injection, latency was recorded as 1800 s. The maximum behavioral seizure score was measured every 2 min, and the cumulative seizure score was summed. For PTZ kindling, mice received 35 mg/kg PTZ (i.p.) 10 times delivered every other day. The maximum behavioral seizure score within 30 min after each PTZ injections was assessed using modified Racine’s scale.
Repeated-low dose kainic acid (KA) induced seizures
KA (Sigma-Aldrich) was prepared fresh in sterile distilled water at a concentration of 2 mg/ml, and injected (5 mg/kg, i.p.) once every 30 min until the onset of Class 5 seizures characterized by generalized tonic-clonic convulsions with lateral recumbence or jumping and wild running followed by generalized convulsions. Latency to the onset of Class 5 seizures was determined by investigators who were blinded to mouse strain.
Genotyping
Tail biopsies were taken from mice following euthanasia, and DNA was extracted using the QIAGEN DNeasy kit as per manufacturer’s instructions. The MiniMUGA array by GeenSeek (Neogen Genomics, Lincoln, NE) was used for genotyping18. Informative markers were selected by taking markers that discriminated between B6J and CC027. Next, we identified the frequency of these markers in the F2 population and tested for expected Mendelian ratios. Markers that significantly deviated from the expected frequency distribution were removed from the initial quantitative trait loci (QTL) mapping analysis. The final list of SNP markers from MiniMUGA used for analysis was pruned to 2,440. After initial analysis, we identified an additional SNP marker on chromosome 5 at 78,300,721 bp to discriminate between the B6J haplotype in CC027 and the B6J from the other parental strain. This variant was discovered using WGS from the CC msBWT tool (http://www.csbio.unc.edu/CEGSseq/index.py)9, 10. PCR primers were designed to amplify a 431bp DNA fragment containing the SNP of interest: Forward–CTGATGTCCAGATTGCTTAGT (78,300,580); Reverse–GGATTTGAGAAGGAAGCTAGAA (78,301,010). 140 F2 samples that had ambiguous parental origin were genotyped to resolve the QTL boundary on chromosome 5.
QTL mapping
We performed QTL mapping using the R packages, Rqtl, and Rqtl219. Rqtl2 performs QTL mapping through a regression of the phenotype at each marker. Rqtl was used to perform multiple-QTL mapping (MQM), which tests for additive and interacting QTL effects. Age was included as a covariate for trait mapping. QTL significance intervals were defined by the 95% Bayesian credible interval, calculated by normalizing the area under the QTL curve20. Log of the odds ratio (LOD) was the reported mapping statistic. The significance thresholds for QTL were calculated using 10,000 permutations.
Data Availability
Genotypes for MiniMUGA are available at https://www.med.unc.edu/mmrrc/genotypes. CC sequenced genomes can be queried using the msBWT tools available at http://www.csbio.unc.edu/CEGSseq/index.py?run=MsbwtTools. Phenotype data from this study is available on the Mouse Phenome Database at https://phenome.jax.org/projects/Shorter9 with accession number MPD:660. Zenodo.org accession no. 3250238 also provides access to the pruned genotypes from MiniMUGA, FASTQ files used for RNAseq analysis, phenotypes for the CC strains, and phenotypes for the F2 mapping population (https://zenodo.org/record/3250238 ). All the QTL identified in this study were submitted to MGI (6401001–6401008).
Statistics
All experiments and analyses were performed blind to genotype and treatment, and all statistical analyses were performed using GraphPad Prism 8 software. Unless otherwise noted, comparisons were analyzed using one-way or 2-way ANOVA with Bonferroni’s or Tukey’s post hoc test. P < 0.05 was considered significant. Narrow-sense heritability was estimated using the R package “heritability” with a kinship matrix generated from RQTL221. See more methods in Supplemental Materials.
RESULTS
Extreme seizure responses in CC strains
Previous studies have reported that classical inbred strains of mice vary in their naïve responses to seizures22–24. These studies have led to the general classification of seizure resistant (e.g. B6J) and seizure susceptible (e.g. DBA/2J) mouse strains22–24. Each CC strain contains a different combination of homozygous chromosome segments derived from the 8 founder strains: A/J, B6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ and WSB. The new gene combinations in the CC lines may generate phenotypes that are more extreme than any of the founder strains9. Here, we used the flurothyl model to evaluate seizure-related phenotypes in a sampling of 35 CC inbred strains in addition to traditional seizure resistant (i.e. B6J) and susceptible (i.e. DBA/2J) inbred mice (Figure 1A). We observed wide variation (~2-fold difference) for the latency to the onset of transient myoclonic seizure/jerk (Myoclonic seizure threshold, MST; CC037: 321±16 sec vs. CC027: 165±8 sec) and generalized seizure (Generalized seizure threshold, GST; CC058: 473±24 sec vs. CC031: 191±13 sec). This variation extends beyond the traditionally resistant and particularly beyond susceptible strains (Figure 1B,C). We also estimated the heritability (h2) and observed that both traits exhibited high heritability (MST=0.63, GST=0.71), indicating a strong genetic component for flurothyl induced seizure sensitivity in this population.
Figure 1. Seizure thresholds and severity highly depend upon genetic background.
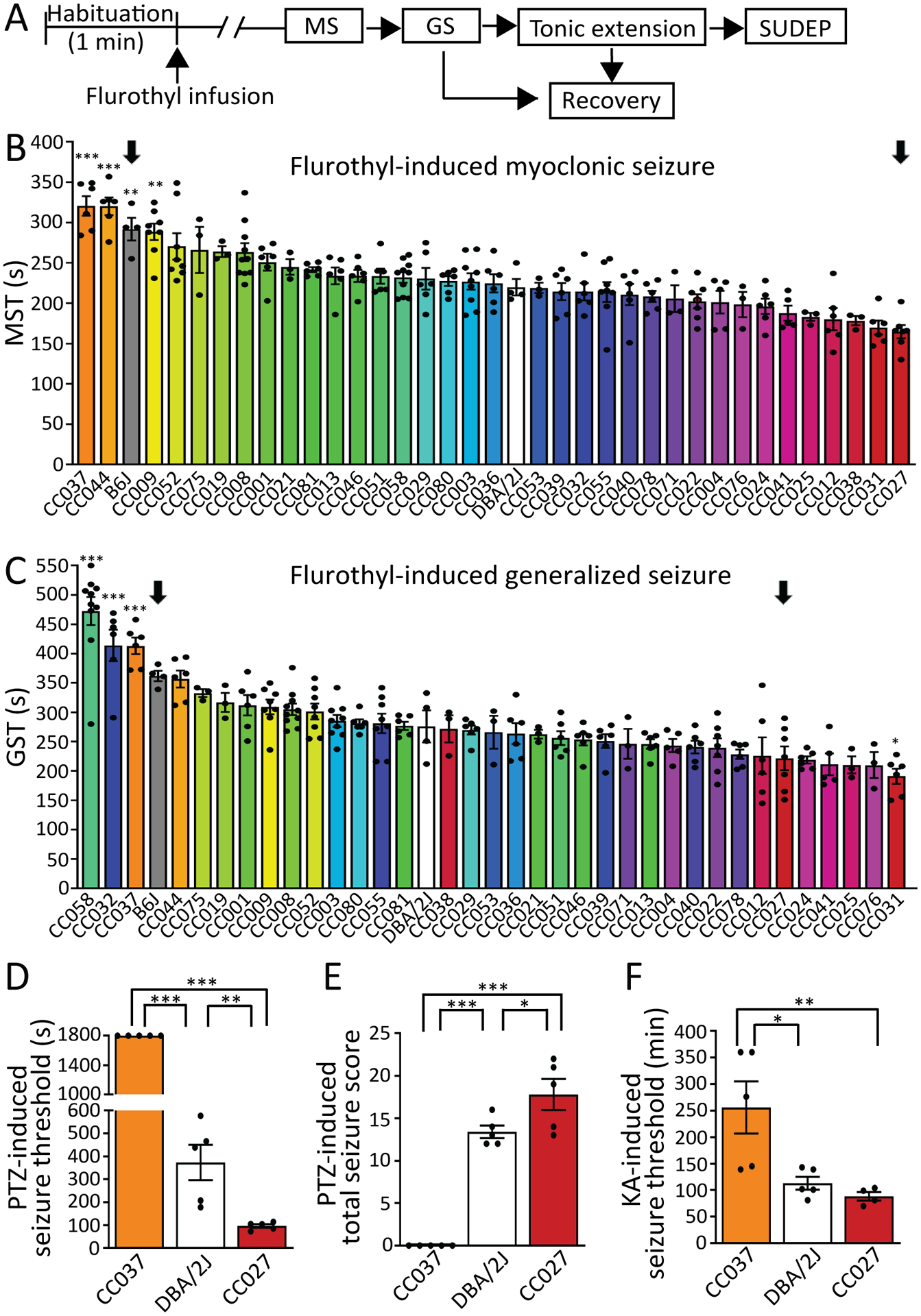
(A) Schematic of flurothyl-induced seizures. MS = myoclonic seizure; GS = generalized seizure; SUDEP = sudden unexpected death in epilepsy. (B) MS threshold (MST) and (C) GS threshold (GST) of 35 CC strains as well as B6J and DBA/2J. The order of strains was ranked from the most resistant (highest threshold) to the most susceptible (lowest threshold), n=3–10. Arrows denote parental strains used for generating F2 mapping population. Data are presented as mean ± SEM and analyzed using one-way ANOVA with post hoc Bonferroni’s multiple comparisons test. *p<0.05, **p<0.01 and ***p<0.001 compared to DBA/2J. (D) Seizure threshold and (E) sum of maximum behavioral seizure score after PTZ injection (40 mg/kg, i.p.), n=5. (F) Latency to onset of generalized seizures induced by repeated low dose of KA (5 mg/kg, i.p. every 30 min), n=4–5. Data are presented as mean ± SEM and analyzed using one-way ANOVA with post hoc Tukey’s multiple comparisons test. *p<0.05, **p<0.01 and ***p<0.001.
To test whether the flurothyl seizure susceptibility phenotypes were generalizable to other seizure models, we tested CC037 (highly resistant), DBA/2J (intermediate), and CC027 (highly susceptible) strains in two additional seizure induction paradigms — intraperitoneal injection of pentylenetetrazol (PTZ) and kainic acid (KA), as these paradigms trigger seizures via different routes and/or mechanisms25. We found that the direction of effect and the significant difference of seizure responses are consistent between flurothyl- (MST, p-value<0.001; GST, p-value<0.001), PTZ- (threshold, p-value<0.001; seizure score, p-value<0.001), and KA-induced (p-value<0.01) seizure models (Figure. 1B–F), demonstrating that the seizure responses of CC measured by flurothyl can be generalizable to other induction paradigms.
Previous research reported evidence of a significant correlation between MST and GST across multiple standard laboratory strains in their first exposure to flurothyl, suggesting an interaction between these two phenotypes26. Consistent with these observations, we also found a strong correlation of initial MST and GST (R2=0.83, p-value<0.001) across most CC as well as B6J and DBA/2J strains (Figure 2A). However, two CC strains, CC058 and CC032, exhibited a significant prolonged MS-GS difference compared to other strains (p-value<0.001) (Figure. 2B). This suggests that there are shared biological processes for sensitivity for MST and GST, but these factors can be decoupled as shown in CC058 and CC032. This also raises the possibility that target genes, and thus druggable targets, can be identified that halt seizure propagation and provide protective effects to having a generalized seizure.
Figure 2. Correlation of myoclonic and generalized seizure thresholds and identification of seizure propagation resistant CC strains.
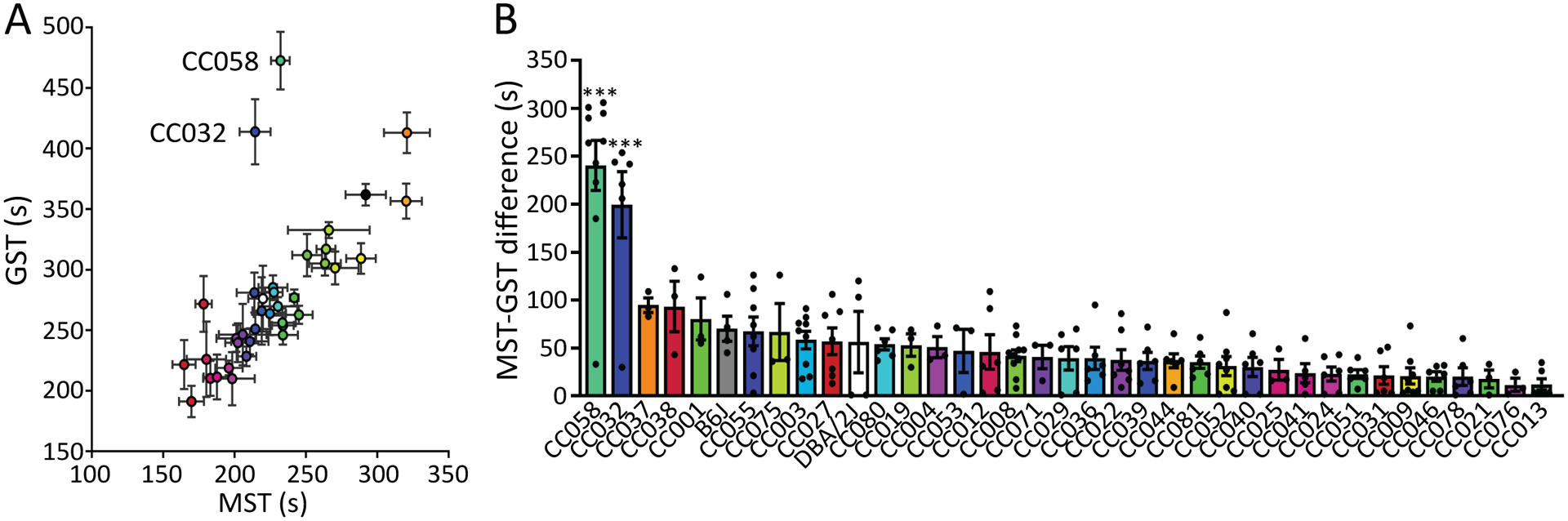
(A) Correlation of MST and GST (R2=0.824, p<0.001 without CC032 and CC058). (B) MST-GST difference of 35 CC strains as well as B6J and DBA/2J, n=3–10. Data are presented as mean ± SEM and analyzed using one-way ANOVA with post hoc Tukey’s multiple comparisons test. ***p<0.001 compared to other strains.
Variable kindling kinetics in CC strains
We measured the epileptogenic responses of the same 35 CC strains, B6J, and DBA/2J using an 8-day flurothyl kindling paradigm. We first found diverse MST and GST kindling kinetics for these strains that falls into four broad subgroups: (1) seizure threshold decreased throughout the kindling process (e.g. CC013); (2) threshold decreased and then plateaued (e.g. CC075 and CC052); (3) threshold was initially unchanged and then decreased (e.g. CC071); and (4) threshold remained similar throughout 8-days flurothyl kindling (e.g. CC031, CC055 and CC051), suggesting a remarkable resistance to epileptogenesis (Figure 3A,B and Figure S1). The kindling patterns were similar between the measurement of MST and GST, as kindling slopes of MST and GST are correlated (R2=0.33, p-value<0.001) across strains, except CC032, which exhibited the highest GST, but a more typical MST kindling slope (Figure 3C).
Figure 3. Identification of strains resistant to epileptogenesis.
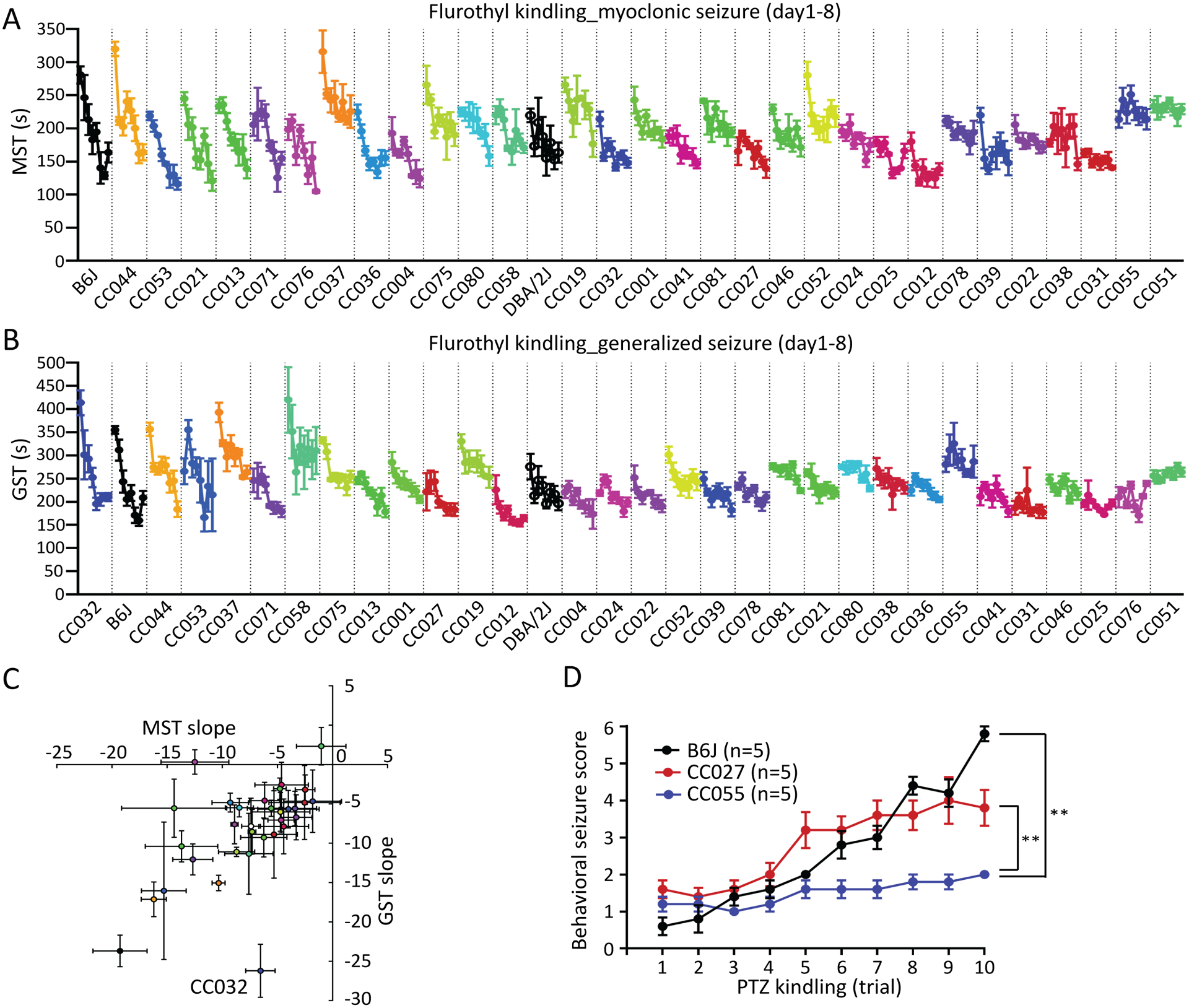
(A) MST and (B) GST during 8-day flurothyl kindling. The order of the CC strains are ranked based on the value of kindling slope of myoclonic (A) and generalized (B) seizures, respectively. (C) Kindling slopes of MST and GST (R2=0.325, p<0.001) are correlated across strains except CC032. (D) Maximum behavioral seizure score during 30 min after each injection of PTZ (35 mg/kg, i.p.) every other day over 10-trial PTZ kindling (n=5). Data are presented as mean ± SEM and analyzed using two-way ANOVA with post hoc Tukey’s multiple comparisons test, **p<0.01.
To assess whether the resistance to epileptogenesis in a particular CC strain was generalizable to another kindling paradigm, we challenged CC055 (kindling resistant), CC027 (intermediate), and B6J (kindling susceptible) mice to a PTZ kindling paradigm. PTZ kindling provides another measure of epileptogenesis with increases of behavioral seizure severity after repeated injection of PTZ at sub-convulsive doses27. Consistent with the flurothyl-kindling paradigm, behavioral seizure severity of B6J and CC027 increased over the process of PTZ kindling, whereas the seizure activity of CC055 remained sub-convulsive throughout repeated PTZ injections (p-value<0.01) (Figure 3D). Overall, these results suggest that CC055 mice are similarly resistant to kindling by repeated flurothyl or PTZ exposures. Identification of kindling resistant strains (e.g. CC055 and CC051) offers novel mouse models to study mechanistic underpinning of epileptogenesis and develop antiepileptogenic therapies.
Induction of a single seizure significantly increases the risk of death in multiple CC strains
Flurothyl induces transient convulsions that are otherwise nonfatal to naïve standard laboratory strains24. During initial screening of 211 mice, 24 mice (11%) were unable to recover from tonic extension following a single flurothyl-induced seizure and succumbed to sudden death. This susceptibility to seizure-induced sudden death provides powerful mouse model to study SUDEP in human. Twenty two of the 24 SUDEP susceptible mice (92%) were from four of the 35 CC strains: CC003 (5/9), CC008 (6/10), CC009 (7/8), and CC029 (4/6). Across the CC strains, the SUDEP phenotype did not correlate with GST or MST-GST difference, but is related to prolonged MST (Figure S2). Mice from these strains that survived the initial flurothyl challenge were still susceptible to seizure-induced sudden death during subsequent flurothyl challenges (Figure 4). We also tested whether age could be a factor for susceptibility to SUDEP. In CC009, younger mice (i.e. 2–4 months old) were significantly more susceptible to seizure-induced sudden death compared to older mice (i.e. 7–9 month old) (Figure 4, p-value<0.01, Log-rank test). Collectively, these results show that several mouse models for SUDEP exist in the CC, while the cause of the seizure-induced sudden death could be caused by brain, cardiac and/or respiratory dysfunction, which requires future investigation.
Figure 4. Mortality of SUDEP susceptible CC strains.
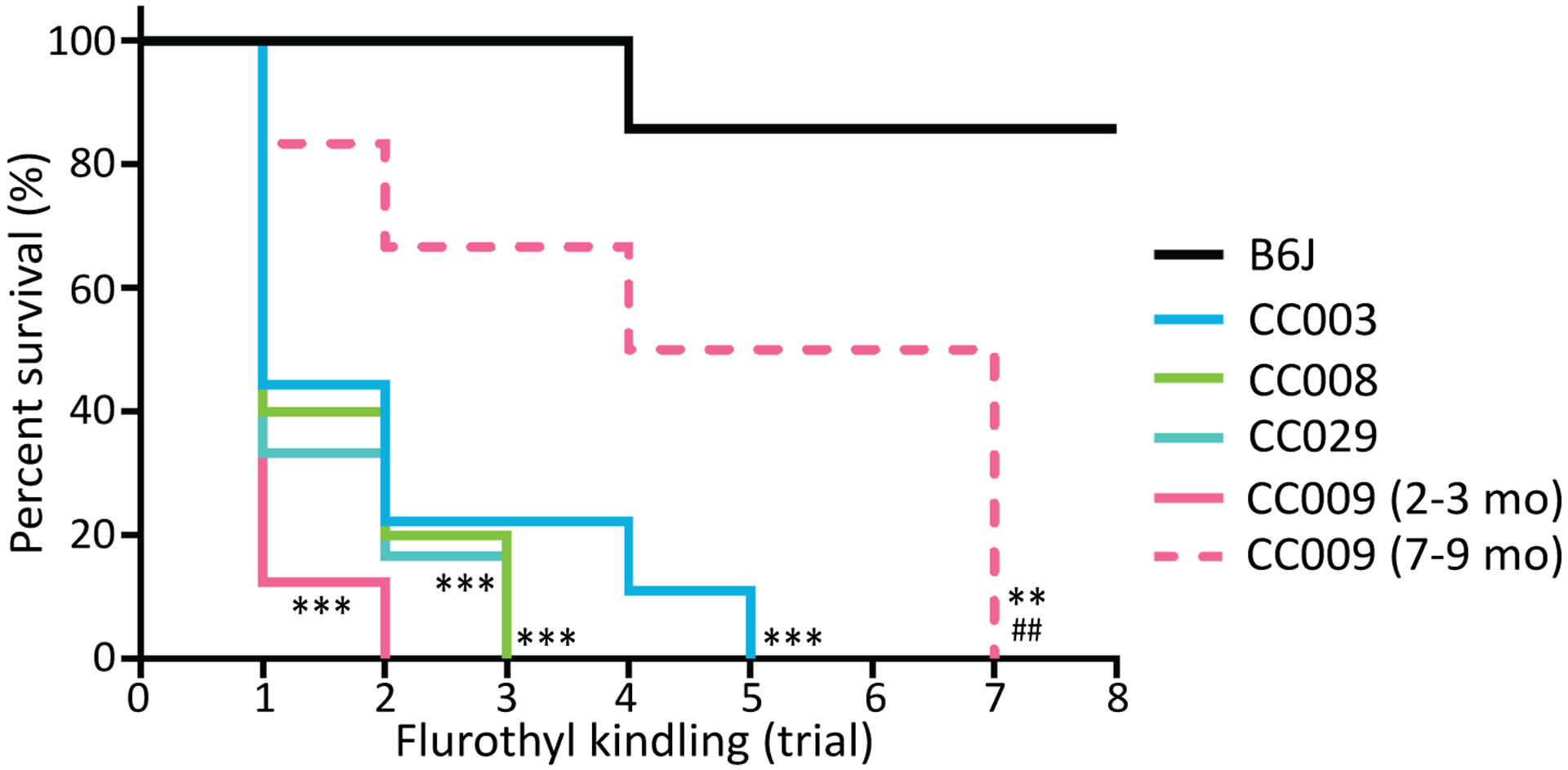
Survival curve of CC003 (n=9), CC008 (n=10), CC009 (n=8), CC029 (n=6) and B6J (n=7) during 8-day flurothyl kindling. Data are analyzed using Log-rank (Mantel-Cox) test, n=6–10, **p<0.01 and ***p<0.001 compared to B6J; and ##p<0.01 compared to young CC009 (2–3 month).
Seizure sensitivity in an F2 population
Among the four distinct seizure related traits (i.e. seizure sensitivity, seizure propagation, kindling, and SUDEP) described above, we first focused on seizure sensitivity and created an F2 population to genetically map and identify seizure sensitivity loci for MST and GST. CC027 and B6J strains were selected as the parental strains for an F2 mapping population for the following reasons. First, these two strains represent near phenotypic extremes for both myoclonic and generalized seizure thresholds (Figure 1). Second, B6J is the strain the mouse reference genome is based upon, and has been a traditional seizure resistant strain of choice in previous epilepsy research, allowing us to compare these results with previous work as proof of principle22–24. Third, CC027 exhibits similar extreme seizure sensitivities across seizure induction paradigms. We also tested F1s from CC027 and B6J mice and observed an intermediate sensitivity between the two parental strains (MST=191±5 sec; GST=265±10 sec), suggesting that additive genetic factors contribute to MST and GST (Figure S3).
We measured seizure sensitivity in 297 F2 male mice from reciprocal crosses. We only used male F2s because no significant sex differences were found in parental B6J and CC027 strains (p-value=0.201 and 0.160, respectively) (Figure S4) and female F2s were used for another experiment not related to this study. We observed a wide range of seizure thresholds for MST (mean=176, SD=39, range: 59–278 sec) and GST (mean=273, SD=57, range: 157–540 sec) (Figure S3). In the F2 population, we observed a modest but significant correlation between age of mice and seizure sensitivity for both MST (R2=0.05, p-value<0.001) and GST (R2=0.03, p-value<0.01), with younger mice being more resistant to seizure. To account for this effect, age was treated as a covariate for QTL mapping. Body weight had a non-significant effect on MST (p-value=0.125) and GST (p-value=0.09). Due to a skewed right distribution for GST and outliers with a high threshold score, GST values were normalized using a log10 transformation (Figure S3).
Genetic mapping of seizure sensitivity identifies 8 QTL
We performed QTL mapping to identify genomic regions associated with MST and GST sensitivity. Mice from the F2 cross as well as the parentals and F1s were genotyped on MiniMUGA with 2,440 genetic markers that segregate between CC027 and B6J. We performed QTL mapping with age as a covariate, and identified 5 significant QTL peaks (p-value<0.05) and 3 suggestive peaks (p-value<0.12) for MST (Myoclonic seizure susceptibility, Mss) and GST (Generalized seizure susceptibility, Gss) (Figure 5 and Table 1). There are two overlapping QTL for both seizure traits on chromosome 5 (Mss2/Gss2) and 10 (Mss3/Gss3). Other identified QTL are specific to either MST or GST. The allelic effects of these QTL are consistent with the parental strain effects, with the exception of Mss2/Gss2 on chromosome 5, which is transgressive. This means that for chromosome 5, the seizure resistant B6J strain has a seizure sensitivity allele while CC027 has a resistance allele. Gss1 on chromosome 1 overlaps the Seizure susceptibility 1 (Szs1) locus, which was previously identified using B6J and DBA/2J mice with consistent B6J allelic effects28–30. Other QTL identified in this study are partially overlapping or syntenic with previously identified seizure related loci (Table S2). Overall, the QTL mapping results demonstrate the power and utility of the CC to map both previously identified and novel genetic loci associated with two types of seizure sensitivity.
Figure 5. QTL mapping for myoclonic (slate-blue) and generalized (violet-red) seizure threshold using F2 crosses from B6J and CC027.
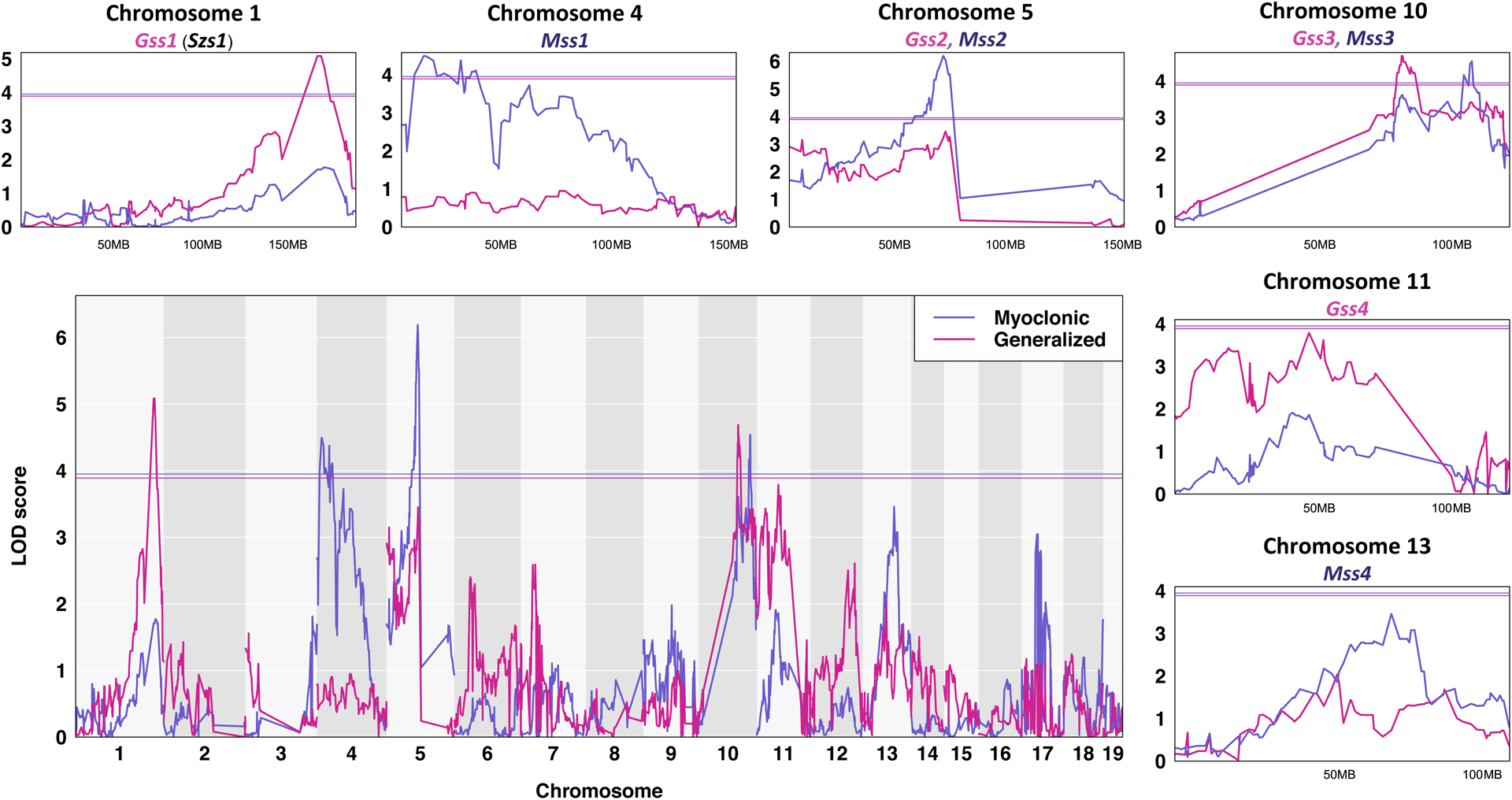
Chromosomes 1 through 19 are represented numerically on the x-axis, and the y-axis represents the LOD score. The relative width of the space allotted for each chromosome reflects the relative length of each chromosome. A magnification of each myoclonic seizure susceptibility (Mss) and generalized seizure susceptibility (Gss) locus is shown. Solid colored horizontal bar indicates the significance threshold at p = 0.05.
Table 1.
QTL of myoclonic and generalized seizure susceptibility
| Trait | Name | Chr | Position (Mb) | Size (Mb) | CI (Mb) | LOD | P-value | B6J allele | CC027 allele | Dominance |
|---|---|---|---|---|---|---|---|---|---|---|
| Myoclonic | Mss1 | 4 | 31.5 | 29.4 | 10.5–39.9 | 4.49 | 0.0111 | Resistant | Sensitive | Dominant |
| Myoclonic | Mss2 | 5 | 70.9 | 66.2 | 66.8–133 | 6.19 | 0.0001 | Sensitive | Resistant | Additive |
| Myoclonic | Mss3 | 10 | 115.8 | 76.8 | 45.2–122 | 4.54 | 0.0105 | Resistant | Sensitive | Dominant |
| Myoclonic | Mss4 | 13 | 73.3 | 48.6 | 44.6–93.2 | 3.46 | 0.1046 | Resistant | Sensitive | Dominant |
| Generalized | Gss1 | 1 | 173 | 14 | 163–177 | 5.08 | 0.0037 | Resistant | Sensitive | Additive |
| Generalized | Gss2 | 5 | 71.9 | 70.7 | 3–73.7 | 3.45 | 0.1151 | Sensitive | Resistant | Additive |
| Generalized | Gss3 | 10 | 89.6 | 77.1 | 49.9–127 | 4.69 | 0.0085 | Resistant | Sensitive | Dominant |
| Generalized | Gss4 | 11 | 51.6 | 65.6 | 10.7–76.3 | 3.79 | 0.0594 | Resistant | Sensitive | Additive |
RNA sequencing analysis
To identify candidate genes by interrogating allelic effects of genes within QTL, we performed gene expression analysis using quantitative RNA sequencing. We used CC027 and B6J mice to access the strain specific genotypic effects on the level of gene expression in the hippocampus. In total, we identified 3,069 genes that are differentially expressed in the two strains after correcting for multiple comparisons with an adjusted p-value <0.05. We identified a total of 73 differentially expressed genes within a 1.0 LOD interval drop for the 8 QTL, with 22 having been associated with epilepsy based on GeneCards ontology and score (Table S3).
As an example of a pipeline to further reduce the number of candidate genes within a single QTL, we focused on the most significant QTL, Mss2/Gss2 on chromosome 5. Within a 4 Mb region spanning the most significant marker, we identified 21 protein coding genes, of which 12 were measurably expressed in the hippocampus (Figure 6A). Notably, examination of whole genome sequence from B6J and CC027 found no protein coding variant within the genes in this region (69,000,000–73,000,000). Of these 12 genes, four (i.e. Gabra2, Gabrg1, Guf1 and Gabrb1) have genome-wide differential expression (p-value<0.05), with Gabra2 being the most differentially expressed (Figure 6B). Gabra2 in B6J has significantly decreased expression compared to CC027. In fact, Gabra2 was one of the most differentially expressed genes in the set (p-value<1.0E-100). This result is consistent with recent human and mouse studies that identified GABRA2 contributing to epilepsies5, 31–34. The B6J allele of Gabra2 was also identified as likely responsible for modifying seizure phenotype in a cross between strain B6J and 129S6 of the mouse model of Dravet syndrome35, 36.
Figure 6. Identification of genetics variants from QTL.
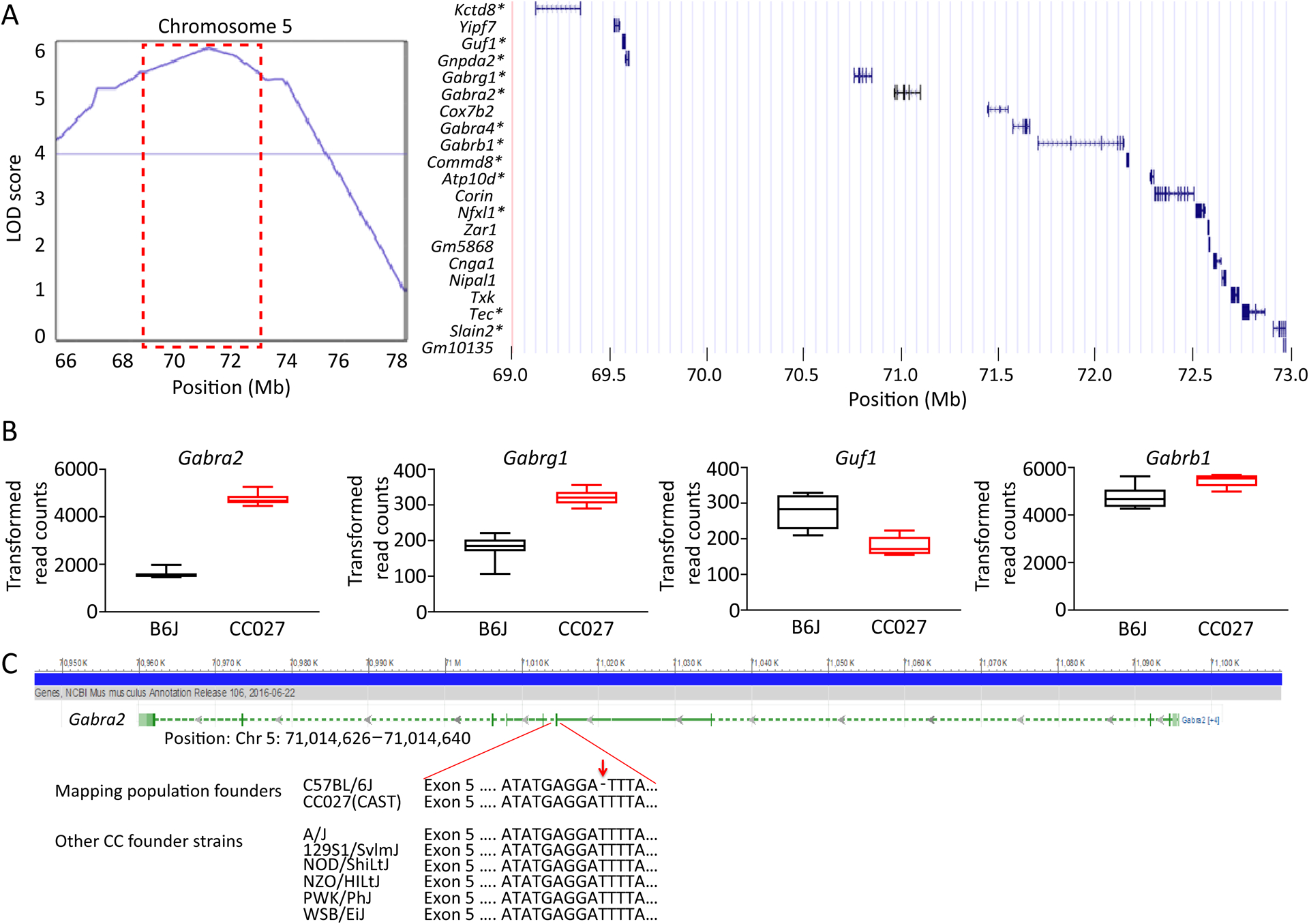
(A) Mss2 QTL region on chromosome 5 and list of 21 protein coding genes found within the interval. * denotes genes that were measurably expressed in the hippocampi. (B) Genes in the QTL region with significant differential (p<0.05, t-test) expression in the hippocampus of B6J and CC027 mice. Data are presented as median with the lower and upper hinges corresponding to 25–75 percentiles. (C) Gabra2 with an intronic base pair deletion located next to a splice acceptor site preceding exon 5. This deletion is only found in B6J mice.
Using whole genome sequence from CC027 as well as the reference B6J sequence, we were able to identify founder haplotype and genetic variation in the exons for Gabra2. The origin of the region (70.95Mb–71.10Mb) in CC027 is M. m. castaneus while in B6J this region is M. m. domesticus. As expected in regions with different subspecific origin, there are high levels of genetic diversity (at least 1,310 SNP variants, 181 short indels). Within the ten exons of Gabra2, we observed five SNPs and one indel between B6J and CC027. The indel and three of the five SNPs are UTR variants that have no reported evidence for changes in gene expression. The remaining two SNPs are synonymous variants. The most likely cause of gene expression variation in Gabra2 comes from an intronic variant. An indel, rs225241970, located at 71,031,384 bp, was recently identified as a de novo deletion in B6J that significantly reduced gene expression37. When repaired using CRISPR-Cas9 editing, it fully restored expression of Gabra2 in B6J mice37. This finding is consistent with our observations of Gabra2 expression and sequence variation. We confirmed the presence of this indel variant (71,014,638 bp) in the B6J mice used for the RNA sequencing study, and observed that B6J is the only founder strain with a deletion in the 8 founder strains in the CC (Figure 6C). We also queried this region using the msBWT tool, and determined that the variant does not create alternative splicing of the transcript. Overall, this suggests that the intronic variant in B6J is private to that founder strain and causes a reduction in expression of Gabra2, which leads to enhanced seizure sensitivity consistent with the allelic effect for Mss2/Gss2 identified in QTL mapping (Table 1).
DISCUSSION
We present new mouse model resources to guide the identification of genetic variants conferring risk/susceptibility for multiple phenotypic frontiers of epilepsy. We identified novel epileptic mouse strains with extreme epileptic responses that include but are not limited to: 1) multiple CC strains with extreme seizure susceptibility; 2) two CC strains that exhibit resistance to seizure propagation; 3) four CC strains that exhibit SUDEP; 4) CC strains with resistance to epileptogenesis as measured by kindling. Together, these CC strains provide novel animal models to explore molecular, cellular, and physiological basis of various manifestations of epilepsy. We identified a larger phenotypic range and more reliable measures of seizure susceptibility and kindling compared to the Hybrid Mouse Diversity Panel38, showing that the CC population harbors a large number of unstudied genetic variants associated with seizure sensitivity and development. The identification of new mouse models with extreme epileptic response makes the CC well suited to serve as a model for the development and deployment of targeted therapeutics for precision medicine in epilepsy. We also have the ability to create congenic mouse strains with distinct sensitivity loci and test targeted therapeutic options that are designed for a particular genetic variant associated with epilepsy. This will allow us to characterize the interaction between genetic background and candidate variants or pharmacological interventions.
Among the extreme seizure responses we identified, SUDEP is the fatal complication of epileptic individuals (~1:1,000/year). There are no current methods that effectively predict or prevent SUDEP3. The rare and unpredictable nature of SUDEP restricts traditional GWAS in humans. Respective reviews using postmortem reports have identified some candidate genes, but these studies suffer from a low number of cases and unclear causes of death7. The development of SUDEP preventions has been hindered by the limited genetic and molecular mechanistic insights, as well as the limited number of animal models of SUDEP beside, for example, DBA/1 mice39. Here we have identified four CC strains (CC003, CC008, CC009, and CC029) exhibiting high risk of death following a single episode of induced transient seizure otherwise nonfatal to most classical inbred mouse strains. The finding that younger CC009 mice were more susceptible to seizure-induced sudden death compared to older mice is consistent with clinical observations that early age is an epidemiological risk factor of SUDEP40, 41. The causes of these genetic and age effects are not yet known but are amenable to further study.
Seizure propagation is the process by which a focal seizure spreads within the brain and cause more severe behavioral manifestations. All strains we tested showed progression of seizures from transient involuntary jerks (i.e. MS) to full-blown continuous generalized clonus (i.e. GS) after flurothyl exposure. The decoupling of MS and GS in CC058 and CC032 suggests the genes segregating in these strains can control the spread of seizure activity into contiguous areas via local connections. Identification of distinct genes responsible for generalized seizure propagation may provide pharmacological targets, thereby reducing the harmful and life-threatening complications related to more severe forms of generalized seizures. Besides seizure propagation, seizure development (i.e. epileptogenesis) is another critical yet less understood domain of epilepsy. Epileptogenesis can be modeled in rodents by kindling, a process whereby repeated seizures or subconvulsive stimuli lead to increases in severity and/or susceptibility to subsequent seizures. The kindling process indicates a long-lasting plasticity of the neuronal excitability mimicking epilepsy development42. We found lack of kindling in several CC strains including some with intermediate initial seizure susceptibility, such as CC055 and CC051, suggesting that resistance to epileptogenesis is not a consequence of a floor effect of the flurothyl kindling model (Figure S5). The identification of the mouse models that exhibit resistance to epileptogenesis will permit future QTL mapping using F2 crosses between kindling resistant and kindling susceptible strains, thus providing insights into the genetic mechanism of epileptogenesis and revealing druggable targets for preventive therapies.
By combining a high throughput seizure induction model with a mapping population derived from the CC genetic reference mouse population, we were able to identify several genetic loci and predict their associations with seizure sensitivity. As a proof-of-concept, our approach validated an intronic variant exclusively found in the B6J substrain in Gabra2 as a strong candidate for the transgressive QTL on chromosome 5, notwithstanding that flurothyl screening may have biased sensitivity to phenotypes regulated by GABAR-dependent mechanisms. While this discovery may be considered to be a proof-of-concept, the resources we provide here should enable future discovery of novel genes linked to complex seizure traits, such as resistance to epileptogenesis, which have lacked appropriate animal models. Importantly, the resources and tools are publically available and supported by the Systems Genetics Core Facility at UNC (see Data Availability), making the identification of candidate risk/susceptibility genes feasible for the scientific community by generation of F2 populations from CC strains with phenotypic extremes.
Supplementary Material
KEY POINT BOX.
CC provides novel animal models of extreme epileptic responses including seizure susceptibility, seizure propagation, epileptogenesis, and SUDEP
QTL mapping in an F2 population identified one known and seven novel loci associated with seizure sensitivity
Whole genome sequencing and hippocampal gene expression pinpoint biologically plausible candidate gene and variant associated with seizure sensitivity
CC provides a powerful toolbox for studying complex features of seizures and for identifying genes associated with seizure outcomes
ACKNOWLEDGEMENTS
This work was supported in part by the following grants from the National Institutes of Health: 4U19AI100625 (FPMV), 1P01AI132130 (FPMV), 5U42OD010924 (FPMV), U24HG10100 (FPMV), R01HD093771 (BDP), T32HD040127 (JRS), Citizens United for Research in Epilepsy (BG), and the Rett Syndrome Research Trust (LHW, FPMV). The Systems Genetics Core Facility is partially supported by the University Cancer Research Fund granted to Lineberger Comprehensive Cancer Center (MCR012CCRI). MiniMUGA was developed under a service contract to FPMV from Neogen Inc., Lincoln, NE. None of the authors have a financial relationship with Neogen Inc. apart from the service contract listed above. We would like to thank Dr. Marty Ferris, Dr. Wesley Burks, and Dr. Michael Kulis (UNC) for sharing mice in the F2 study as part of the TTSAQ17P1 (FPMV).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
REFERENCES
- 1.Fiest KM, Sauro KM, Wiebe S, Patten SB, Kwon CS, Dykeman J, et al. Prevalence and incidence of epilepsy: A systematic review and meta-analysis of international studies. Neurology 2017;88:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loscher W, Klitgaard H, Twyman RE, Schmidt D. New avenues for anti-epileptic drug discovery and development. Nature reviews Drug discovery 2013;12:757–776. [DOI] [PubMed] [Google Scholar]
- 3.Sudden Devinsky O., unexpected death in epilepsy. The New England journal of medicine 2011;365:1801–1811. [DOI] [PubMed] [Google Scholar]
- 4.Steffens M, Leu C, Ruppert AK, Zara F, Striano P, Robbiano A, et al. Genome-wide association analysis of genetic generalized epilepsies implicates susceptibility loci at 1q43, 2p16.1, 2q22.3 and 17q21.32. Human molecular genetics 2012;21:5359–5372. [DOI] [PubMed] [Google Scholar]
- 5.Epilepsies ILAECoC. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nature communications 2018;9:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epilepsies ILAECoC. Genetic determinants of common epilepsies: a meta-analysis of genome-wide association studies. The Lancet Neurology 2014;13:893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman AM, Behr ER, Semsarian C, Bagnall RD, Sisodiya S, Cooper PN. Sudden unexpected death in epilepsy genetics: Molecular diagnostics and prevention. Epilepsia 2016;57 Suppl 1:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasperaviciute D, Catarino CB, Heinzen EL, Depondt C, Cavalleri GL, Caboclo LO, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain 2010;133:2136–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava A, Morgan AP, Najarian ML, Sarsani VK, Sigmon JS, Shorter JR, et al. Genomes of the Mouse Collaborative Cross. Genetics 2017;206:537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shorter JR, Najarian ML, Bell TA, Blanchard M, Ferris MT, Hock P, et al. Whole Genome Sequencing and Progress Toward Full Inbreeding of the Mouse Collaborative Cross Population. G3 2019;9:1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogue MA, Churchill GA, Chesler EJ. Collaborative Cross and Diversity Outbred data resources in the Mouse Phenome Database. Mamm Genome 2015;26:511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molenhuis RT, Bruining H, Brandt MJV, van Soldt PE, Abu-Toamih Atamni HJ, Burbach JPH, et al. Modeling the quantitative nature of neurodevelopmental disorders using Collaborative Cross mice. Molecular autism 2018;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenrock SA, Oreper D, Farrington J, McMullan RC, Ervin R, Miller DR, et al. Perinatal nutrition interacts with genetic background to alter behavior in a parent-of-origin-dependent manner in adult Collaborative Cross mice. Genes, brain, and behavior 2018;17:e12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Wang Y, Langley SA, Zhou YX, Jen KY, Sun Q, et al. Diverse tumour susceptibility in Collaborative Cross mice: identification of a new mouse model for human gastric tumourigenesis. Gut 2019;68:1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noll KE, Ferris MT, Heise MT. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell host & microbe 2019;25:484–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu B, Carstens KE, Judson MC, Dalton KA, Rougie M, Clark EP, et al. Ube3a reinstatement mitigates epileptogenesis in Angelman syndrome model mice. The Journal of clinical investigation 2019;129:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadiyala SB, Papandrea D, Herron BJ, Ferland RJ. Segregation of seizure traits in C57 black mouse substrains using the repeated-flurothyl model. PLoS One 2014;9:e90506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sigmon JS, Blanchard MW, Baric RS, Bell TA, Brennan J, Brockmann GA, et al. Content and performance of the MiniMUGA genotyping array, a new tool to improve rigor and reproducibility in mouse research. bioRxiv 2020.03.12.989400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broman KW, Gatti DM, Simecek P, Furlotte NA, Prins P, Sen S, et al. R/qtl2: Software for Mapping Quantitative Trait Loci with High-Dimensional Data and Multiparent Populations. Genetics 2019;211:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics 2001;159:371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, et al. Marker-based estimation of heritability in immortal populations. Genetics 2015;199:379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferraro TN, Golden GT, Smith GG, Berrettini WH. Differential susceptibility to seizures induced by systemic kainic acid treatment in mature DBA/2J and C57BL/6J mice. Epilepsia 1995;36:301–307. [DOI] [PubMed] [Google Scholar]
- 23.Ferraro TN, Golden GT, Smith GG, DeMuth D, Buono RJ, Berrettini WH. Mouse strain variation in maximal electroshock seizure threshold. Brain Res 2002;936:82–86. [DOI] [PubMed] [Google Scholar]
- 24.Papandrea D, Anderson TM, Herron BJ, Ferland RJ. Dissociation of seizure traits in inbred strains of mice using the flurothyl kindling model of epileptogenesis. Experimental neurology 2009;215:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandratavicius L, Balista PA, Lopes-Aguiar C, Ruggiero RN, Umeoka EH, Garcia-Cairasco N, et al. Animal models of epilepsy: use and limitations. Neuropsychiatric disease and treatment 2014;10:1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papandrea D, Kukol WS, Anderson TM, Herron BJ, Ferland RJ. Analysis of Flurothyl-induced Myoclonus in Inbred Strains of Mice. Epilepsy research 2009;87:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pentylenetetrazol Dhir A. (PTZ) kindling model of epilepsy. Current protocols in neuroscience 2012;Chapter 9:Unit9 37. [DOI] [PubMed] [Google Scholar]
- 28.Ferraro TN, Golden GT, Smith GG, Martin JF, Lohoff FW, Gieringer TA, et al. Fine mapping of a seizure susceptibility locus on mouse Chromosome 1: nomination of Kcnj10 as a causative gene. Mamm Genome 2004;15:239–251. [DOI] [PubMed] [Google Scholar]
- 29.Ferraro TN, Golden GT, Smith GG, Schork NJ, St Jean P, Ballas C, et al. Mapping murine loci for seizure response to kainic acid. Mamm Genome 1997;8:200–208. [DOI] [PubMed] [Google Scholar]
- 30.Ferraro TN, Golden GT, Smith GG, St Jean P, Schork NJ, Mulholland N, et al. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. The Journal of neuroscience 1999;19:6733–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler KM, Moody OA, Schuler E, Coryell J, Alexander JJ, Jenkins A, et al. De novo variants in GABRA2 and GABRA5 alter receptor function and contribute to early-onset epilepsy. Brain 2018;141:2392–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orenstein N, Goldberg-Stern H, Straussberg R, Bazak L, Weisz Hubshman M, Kropach N, et al. A de novo GABRA2 missense mutation in severe early-onset epileptic encephalopathy with a choreiform movement disorder. European journal of paediatric neurology 2018;22:516–524. [DOI] [PubMed] [Google Scholar]
- 33.Maljevic S, Keren B, Aung YH, Forster IC, Mignot C, Buratti J, et al. Novel GABRA2 variants in epileptic encephalopathy and intellectual disability with seizures. Brain 2019;142:e15. [DOI] [PubMed] [Google Scholar]
- 34.Nathanson AJ, Zhang Y, Smalley JL, Ollerhead TA, Rodriguez Santos MA, Andrews PM, et al. Identification of a Core Amino Acid Motif within the alpha Subunit of GABAARs that Promotes Inhibitory Synaptogenesis and Resilience to Seizures. Cell reports 2019;28:670–681.e678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hawkins NA, Zachwieja NJ, Miller AR, Anderson LL, Kearney JA. Fine Mapping of a Dravet Syndrome Modifier Locus on Mouse Chromosome 5 and Candidate Gene Analysis by RNA-Seq. PLoS genetics 2016;12:e1006398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura T, Hawkins NA, Kearney JA, George AL, Jr., Contractor A. Potentiating alpha2 subunit containing perisomatic GABAA receptors protects against seizures in a mouse model of Dravet syndrome. The Journal of physiology 2019;597:4293–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulligan MK, Abreo T, Neuner SM, Parks C, Watkins CE, Houseal MT, et al. Identification of a Functional Non-coding Variant in the GABA A Receptor alpha2 Subunit of the C57BL/6 J Mouse Reference Genome: Major Implications for Neuroscience Research. Frontiers in genetics 2019;10:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferland RJ, Smith J, Papandrea D, Gracias J, Hains L, Kadiyala SB, et al. Multidimensional Genetic Analysis of Repeated Seizures in the Hybrid Mouse Diversity Panel Reveals a Novel Epileptogenesis Susceptibility Locus. G3 2017;7:2545–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faingold CL, Randall M, Tupal S. DBA/1 mice exhibit chronic susceptibility to audiogenic seizures followed by sudden death associated with respiratory arrest. Epilepsy & behavior : E&B 2010;17:436–440. [DOI] [PubMed] [Google Scholar]
- 40.Nilsson L, Farahmand BY, Persson PG, Thiblin I, Tomson T. Risk factors for sudden unexpected death in epilepsy: a case-control study. Lancet 1999;353:888–893. [DOI] [PubMed] [Google Scholar]
- 41.Opeskin K, Berkovic SF. Risk factors for sudden unexpected death in epilepsy: a controlled prospective study based on coroners cases. Seizure 2003;12:456–464. [DOI] [PubMed] [Google Scholar]
- 42.Goldberg EM, Coulter DA. Mechanisms of epileptogenesis: a convergence on neural circuit dysfunction. Nature reviews Neuroscience 2013;14:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotypes for MiniMUGA are available at https://www.med.unc.edu/mmrrc/genotypes. CC sequenced genomes can be queried using the msBWT tools available at http://www.csbio.unc.edu/CEGSseq/index.py?run=MsbwtTools. Phenotype data from this study is available on the Mouse Phenome Database at https://phenome.jax.org/projects/Shorter9 with accession number MPD:660. Zenodo.org accession no. 3250238 also provides access to the pruned genotypes from MiniMUGA, FASTQ files used for RNAseq analysis, phenotypes for the CC strains, and phenotypes for the F2 mapping population (https://zenodo.org/record/3250238 ). All the QTL identified in this study were submitted to MGI (6401001–6401008).


