Abstract
Goblet cell carcinoid (GCC) is a distinct subtype of appendiceal neoplasm that exhibits unique clinical and pathologic features. We aimed to reveal the molecular profiles of GCC compared to other appendiceal tumors, such as adenocarcinoma and neuroendocrine tumor (NET). A total of 495 appendiceal tumor samples (53 GCCs, 428 adenocarcinomas, and 14 NETs) were tested with next-generation sequencing (NGS) on a 592-gene panel and immunohistochemistry (IHC). Microsatellite instability (MSI)/mismatch repair (MMR) status were tested with a combination of NGS, IHC, and fragment analyses. Tumor mutational burden (TMB) was evaluated by NGS, and PD-L1 expression was tested by IHC (SP142). The most prevalent mutated genes within GCCs were TP53 (24.0%), ARID1A (15.4%), SMAD4 (9.4%), and KRAS (7.5%). Pathway-specific alterations were dominantly observed in cell cycle, MAPK, epigenetic, and TGF-β signaling pathways. GCCs as compared to adenocarcinomas exhibited significantly lower mutation rates in KRAS, GNAS, and APC, with significantly higher mutation rates in CDH1, CHEK2, CDC73, ERCC2, and FGFR2. GCCs as compared to NETs showed significantly lower mutation rates in KRAS, APC, BRCA2, and FANCA. In GCCs, MSI-H/dMMR, TML-high (≥17mut/Mb), and PD-L1 expression were seen in 0.0%, 0.0%, and 2.0% of tumors, respectively. No significant differences were observed in any immunotherapy-related markers examined when compared to adenocarcinomas and NETs. In conclusion, GCCs had considerably distinct mutational profiles compared to appendiceal adenocarcinomas and NETs. Understanding these molecular characteristics may be critical for a development of novel and more effective treatment strategies for GCC.
Keywords: goblet cell carcinoid, appendiceal adenocarcinoma, appendiceal neuroendocrine tumor, molecular profile
Introduction
Goblet cell carcinoid (GCC) is a very rare tumor, almost exclusively found in the appendix, with an incidence of approximately 0.01–0.05/100,000/year(1). GCC clinically behaves as a malignant disease with a tendency to spread to the surrounding bowel, lymph nodes, peritoneum, and ovaries, thus resulting in poor prognosis(2). According to a population-based analysis of appendiceal tumors, the reported 3-year overall survival rates of GCC patients were 96.6%, 91.7%, 65.3%, and 32.9% for stage I, II, III, and IV diseases, respectively, highlighting the aggressive character of GCC—particularly in the advanced stage—with similar survival rates of colorectal adenocarcinoma(3).
GCC arises from pluripotent, intestinal crypt base stem cells that are able to differentiate into both mucinous and neuroendocrine cells. The histological patterns of GCC vary and consist of a mixture of glandular and neuroendocrine components(4). Their classical pathological features include a composition of predominant goblet cells, which include intracytoplasmic mucin, with a few neuroendocrine cells(5). Recent data show the coexistence of poorly differentiated or signet-ring cell adenocarcinoma in at least half of GCC cases (i.e. “adenocarcinoma ex-GCC”), as well as rare cases with greater amounts of neuroendocrine components(6,7). A poorly defined exocrine–endocrine hybrid appearance can confuse pathologists, surgeons, and oncologists attempting to diagnose and treat patients with GCC(8).
Whether GCC should be considered as a special form of adenocarcinoma or a neuroendocrine tumor (NET) variant remains a matter of debate(9). In fact, there are some disparities between the classification system currently used and clinical guidelines. The 2010 World Health Organization classification for appendiceal tumors classifies GCC under the category of NETs(10). On the other hand, both consensus guidelines from the European Neuroendocrine Tumor Society (ENETS) and the North American Neuroendocrine Tumor Society (NANETS) recommend regarding GCC as a colorectal adenocarcinoma when managing patients, given its aggressive clinical course(1,11). Concerning treatment, both statement guidelines are based only on expert opinions following retrospective review due to a lack of any evidences from prospective clinical trials. The current situation of a lacking consensus between the classification system and treatment strategy is partly due to the unknown molecular mechanisms of GCC. There are very few studies focusing on the genetic differences between GCC and other types of appendiceal tumors(12). However, a better understanding of the molecular background of this disease could facilitate not only differential diagnoses but also facilitate better consideration of an optimal treatment strategy for GCC. To address this issue, we performed genetic and molecular profiling of GCC compared to appendiceal adenocarcinoma and NET using a comprehensive tumor profiling platform.
Materials and methods
Samples submitted to a commercial CLIA-certified laboratory (Caris Life Sciences, Phoenix, AZ) from April 2015 to September 2019 were analyzed for molecular profiles. Formalin-fixed paraffin-embedded (FFPE) samples submitted from clinical physicians around the world were sent for analysis. The tissue diagnoses were made on the basis of pathologic assessments from physicians who requested the assays, and were further verified by a board-certified oncological pathologist at the Caris laboratory. A total of 495 appendiceal tumor samples (53 GCCs, 428 adenocarcinomas and 14 NETs) were analyzed. This study was conducted in accordance with guidelines of the Declaration of Helsinki, Belmont report, and U.S. Common Rule. In keeping compliance with policy 45 CFR 46.101(b) (4), this study was performed using retrospective, de-identified clinical data. Therefore, this study is considered Institutional Review Board exempt and no patient consent was necessary.
Mutation analyses
Next-generation sequencing (NGS) was performed on genomic DNA isolated from FFPE samples using an NGS platform (Illumina, Inc., San Diego, CA). A custom-designed SureSelectXT assay was used to enrich 592 cancer-related whole-gene targets (Agilent Technologies, Santa Clara, CA). All variants were detected with >99% confidence based on allele frequency and amplicon coverage, with an average sequencing coverage depth of 750 and an analytic sensitivity of 5%. Identified genetic variants were analyzed by board-certified molecular geneticists and categorized as follows according to the American College of Medical Genetics and Genomics standards: “pathogenic,” “presumed pathogenic,” “variant of unknown significance,” “presumed benign,” or “benign.” When assessing mutation frequencies of individual genes, “pathogenic” and “presumed pathogenic” were counted as mutations, whereas “variant of unknown significance,” “presumed benign,” and “benign” were excluded.
Immunotherapy-related biomarkers
Microsatellite instability (MSI) and mismatch repair (MMR) status was tested with a combination method employing immunohistochemistry (IHC), fragment analysis and NGS, with resulting status defined as either MSI-high (MSI-H)/MMR-deficient (dMMR) or microsatellite stable/MMR-proficient. Detailed methods for assessment of MSI/MMR status are documented in the supplementary appendix.
Tumor mutational burden (TMB) was measured by counting all nonsynonymous missense mutations found per tumor (592 genes and 1.4 megabases [MB] sequenced/tumor). The threshold for a TMB-high (TMB-H) definition was ≥17 mutations/MB. This threshold was established by comparing TMB with MSI via fragment analysis in colorectal cancer cases based on reports of TMB exhibiting high concordance with MSI-H in colorectal cancer.
PD-L1 expression was tested by IHC using SP142 antibody (Spring Biosciences). The staining intensity on the tumor cells membrane was assessed on a semiquantitative scale: 0 for no staining, 1+ for weak staining, 2+ for moderate staining, and 3+ for strong staining. Tumors exhibiting ≥5% of tumor cells stained as 2+ or 3+ were regarded as being PD-L1 positive.
From February 2019 to September 2019, mRNA expression data was obtained from isolated FFPE tumor samples using Illumina NovaSeq platform (Illumina, Inc., San Diego, CA) and Agilent SureSelect Human All Exon V7 bait panel (Agilent Technologies, Santa Clara, CA). Microenvironment Cell Population-counter (MCP-counter) was used for quantification of the abundance of immune and stromal cell populations using transcriptomic data as previously described(13).
Statistical analyses
Patient and molecular characteristics of GCCs were compared with those of adenocarcinomas and NETs. Student-t test and nonparametric Kruskal-Wallis testing were used to analyze age and TMB distribution, respectively. Other categorical data were analyzed using Fisher’s exact test. Cases with any missing data information were not included in the analysis. All statistical analyses were performed with SPSS v23 (IBM SPSS Statistics), and all tests were two-sided at a significant level set to 0.05.
Results
Patient characteristics
Baseline characteristics of the 495 enrolled patients are shown in Table 1. Average age at diagnosis of GCC was significantly higher than that of NET (57.6 vs. 44.4 years, respectively, p < 0.01) and equivalent to that of adenocarcinoma (57.6 vs. 58.2 years, p = 0.75). A gender preference was not observed for GCC (47% male vs. 53% female), and the gender proportions did not differ between GCC and adenocarcinoma/NET. The information of TNM staging was available only in limited patients (N = 142). In any type of tumor, Stage IV was the most common (75% or more) (Supplementary Fig. S1).
Table 1.
Baseline characteristics
| Characteristics | GCC (N = 53) |
AC (N = 428) |
NET (N = 14) |
P-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| Age | Average | 57.6 | 58.2 | 44.4 | GCC vs AC GCC vs NET |
0.75 <0.01 |
|||
| Sex | Male (%) Female (%) |
25 28 |
(47) (53) |
193 235 |
(45) (55) |
7 7 |
(50) (50) |
GCC vs AC GCC vs NET |
0.77 0.85 |
AC, adenocarcinoma; GCC, goblet cell carcinoid; NET, neuroendocrine tumor.
Analyses of genetic alterations
In total, 50 “pathogenic” or “presumed pathogenic” mutations were detected within 25 genes in patients with GCC (Supplementary Fig. S2). Among them, pathway-specific mutations were dominantly observed within the following: cell cycle (13 mutations in TP53), MAPK (7 in KRAS, BRAF, and NF1), epigenetic (6 in ARID1A, CDC73, KDM6A, KMT2D, and SMARCA4), and TGF-β signaling (6 in SMAD2 and SMAD4) pathways. Whereas the mutations present in the WNT (2 in APC and RNF43) and PIK3 signaling (1 in PIK3CA) pathways were less frequent (Figure S1). Genes showing the highest mutation rate in GCC patients were TP53 (24.0%), ARID1A (15.4%), SMAD4 (9.4%), and KRAS (7.5%). The other 21 genes were mutated in less than 5% of patients (Fig. 1). When comparing 26 ex-GCCs and 27 pure GCCs, no differences in genetic alterations were observed (Supplementary Table S1).
Figure 1. Mutation profile of GCC.
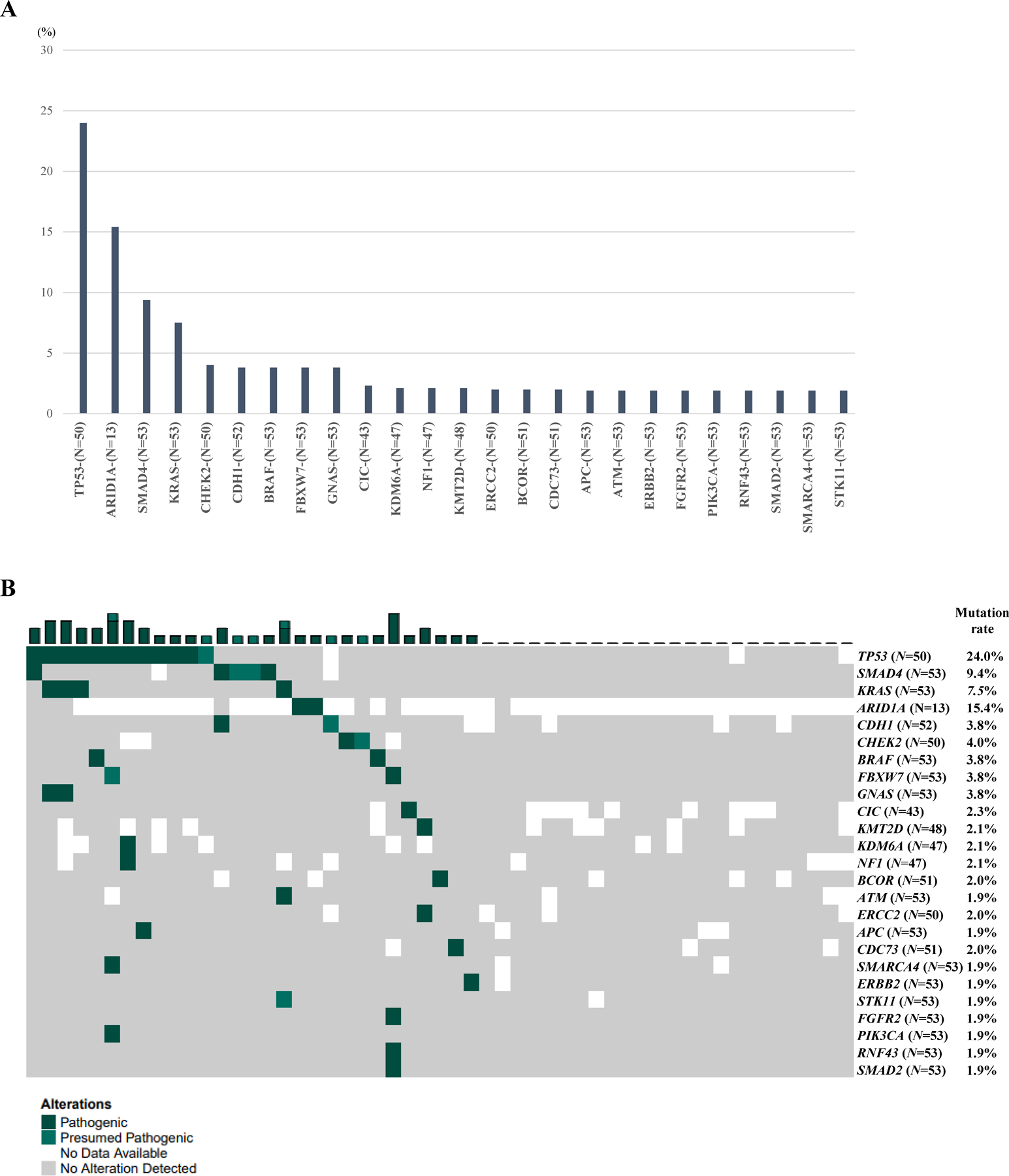
A. Most prevalent mutations within GCC. B. “Pathogenic” or “Presumed pathogenic” mutations identified within GCC. N in parentheses indicates the total number of tumors tested for the biomarker.
In the current study, a total of 71 mutated genes were identified in appendiceal adenocarcinoma (Table 2). Among them, the most frequent mutations were observed in KRAS, TP53, GNAS, ARID1A, SMAD4, and APC (mutation rate >10%). Compared to these mutation profiles of adenocarcinoma, GCC exhibited significantly lower mutation rates in KRAS (7.5% vs. 60.4% for GCC and adenocarcinoma, respectively), GNAS (3.8% vs 34.4%) and APC (1.9% vs 11.7%), and significantly higher mutation rates in CDH1 (3.8% vs 0.7%), CHEK2 (4.0% vs 0.3%), CDC73 (2.0% vs 0.0%), ERCC2 (2.0% vs 0.0%), and FGFR2 (1.9% vs 0.0%) (Fig. 2, Table 2). As for TP53—which was the second most frequently mutated gene in adenocarcinoma—GCC showed a marginally lower mutation rate as compared to adenocarcinoma (24.0% vs. 37.0%, respectively, p = 0.070).
Table 2.
Comparison of mutation frequency between appendiceal adenocarcinoma and GCC
| Gene | AC | GCC |
P-value (AC vs GCC) |
Gene | AC | GCC |
P-value (AC vs GCC) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
||||
| KRAS | 256 | 424 | 60.4 | 4 | 53 | 7.5 | <0.01 | RAD50 | 3 | 351 | 0.9 | 0 | 44 | 0.0 | 0.54 |
| TP53 | 152 | 411 | 37.0 | 12 | 50 | 24.0 | 0.07 | PRKDC | 3 | 390 | 0.8 | 0 | 49 | 0.0 | 0.54 |
| GNAS | 146 | 424 | 34.4 | 2 | 53 | 3.8 | <0.01 | CDH1 | 3 | 423 | 0.7 | 2 | 52 | 3.8 | 0.04 |
| ARID1A | 18 | 90 | 20.0 | 2 | 13 | 15.4 | 0.69 | MRE11 | 3 | 410 | 0.7 | 0 | 52 | 0.0 | 0.54 |
| SMAD4 | 78 | 426 | 18.3 | 5 | 53 | 9.4 | 0.11 | PIK3R1 | 3 | 417 | 0.7 | 0 | 52 | 0.0 | 0.54 |
| APC | 50 | 427 | 11.7 | 1 | 53 | 1.9 | 0.03 | KMT2A | 3 | 423 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| PIK3CA | 30 | 427 | 7.0 | 1 | 53 | 1.9 | 0.15 | BLM | 3 | 425 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| RNF43 | 25 | 427 | 5.9 | 1 | 53 | 1.9 | 0.23 | STK11 | 3 | 426 | 0.7 | 1 | 53 | 1.9 | 0.37 |
| ATM | 22 | 427 | 5.2 | 1 | 53 | 1.9 | 0.29 | U2AF1 | 3 | 426 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| BRAF | 17 | 427 | 4.0 | 2 | 53 | 3.8 | 0.94 | BRCA1 | 3 | 427 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| FBXW7 | 15 | 415 | 3.6 | 2 | 53 | 3.8 | 0.95 | CDK12 | 3 | 427 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| ASXL1 | 10 | 291 | 3.4 | 0 | 32 | 0.0 | 0.29 | EP300 | 3 | 427 | 0.7 | 0 | 52 | 0.0 | 0.54 |
| MED12 | 4 | 132 | 3.0 | 0 | 16 | 0.0 | 0.48 | MTOR | 3 | 427 | 0.7 | 0 | 53 | 0.0 | 0.54 |
| KDM6A | 10 | 374 | 2.7 | 1 | 47 | 2.1 | 0.82 | FANCE | 2 | 328 | 0.6 | 0 | 41 | 0.0 | 0.62 |
| BRCA2 | 11 | 427 | 2.6 | 0 | 53 | 0.0 | 0.24 | WRN | 2 | 408 | 0.5 | 0 | 53 | 0.0 | 0.61 |
| KDM5C | 4 | 158 | 2.5 | 0 | 21 | 0.0 | 0.46 | FANCC | 2 | 422 | 0.5 | 0 | 52 | 0.0 | 0.62 |
| SMAD2 | 10 | 425 | 2.4 | 1 | 53 | 1.9 | 0.83 | FH | 2 | 422 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| KMT2C | 7 | 366 | 1.9 | 0 | 45 | 0.0 | 0.35 | POT1 | 2 | 422 | 0.5 | 0 | 52 | 0.0 | 0.62 |
| CDKN1B | 8 | 425 | 1.9 | 0 | 52 | 0.0 | 0.32 | NBN | 2 | 423 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| KMT2D | 7 | 379 | 1.8 | 1 | 48 | 2.1 | 0.91 | CCND3 | 2 | 426 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| BCOR | 7 | 415 | 1.7 | 1 | 51 | 2.0 | 0.89 | CREBBP | 2 | 426 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| AMER1 | 7 | 423 | 1.7 | 0 | 53 | 0.0 | 0.35 | MAX | 2 | 426 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| ATRX | 3 | 203 | 1.5 | 0 | 22 | 0.0 | 0.57 | MITF | 2 | 426 | 0.5 | 0 | 52 | 0.0 | 0.62 |
| SMARCA4 | 6 | 420 | 1.4 | 1 | 53 | 1.9 | 0.79 | SETD2 | 2 | 426 | 0.5 | 0 | 52 | 0.0 | 0.62 |
| AKT1 | 6 | 424 | 1.4 | 0 | 53 | 0.0 | 0.38 | FLCN | 2 | 427 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| MUTYH | 6 | 425 | 1.4 | 0 | 53 | 0.0 | 0.38 | HNF1A | 2 | 427 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| ERBB2 | 6 | 427 | 1.4 | 1 | 53 | 1.9 | 0.78 | IDH1 | 2 | 427 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| PTCH1 | 4 | 317 | 1.3 | 0 | 41 | 0.0 | 0.47 | MLH1 | 2 | 427 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| PTEN | 5 | 419 | 1.2 | 0 | 53 | 0.0 | 0.42 | PALB2 | 2 | 427 | 0.5 | 0 | 53 | 0.0 | 0.62 |
| NRAS | 5 | 426 | 1.2 | 0 | 53 | 0.0 | 0.43 | CHEK2 | 1 | 399 | 0.3 | 2 | 50 | 4.0 | <0.01 |
| NF1 | 4 | 341 | 1.2 | 1 | 47 | 2.1 | 0.59 | CIC | 1 | 380 | 0.3 | 1 | 43 | 2.3 | 0.06 |
| BCL9 | 5 | 427 | 1.2 | 0 | 53 | 0.0 | 0.43 | CDC73 | 0 | 402 | 0.0 | 1 | 51 | 2.0 | <0.01 |
| MSH6 | 4 | 421 | 1.0 | 0 | 53 | 0.0 | 0.48 | ERCC2 | 0 | 418 | 0.0 | 1 | 50 | 2.0 | <0.01 |
| ARID2 | 4 | 424 | 0.9 | 0 | 53 | 0.0 | 0.48 | FGFR2 | 0 | 426 | 0.0 | 1 | 53 | 1.9 | <0.01 |
| PMS2 | 2 | 230 | 0.9 | 0 | 27 | 0.0 | 0.63 | ||||||||
The bold p-values indicate significant difference (p <0.05). AC, adenocarcinoma; GCC, goblet cell carcinoid; MT, mutant.
Figure 2. Comparison of major gene mutation rates between different appendiceal tumors.
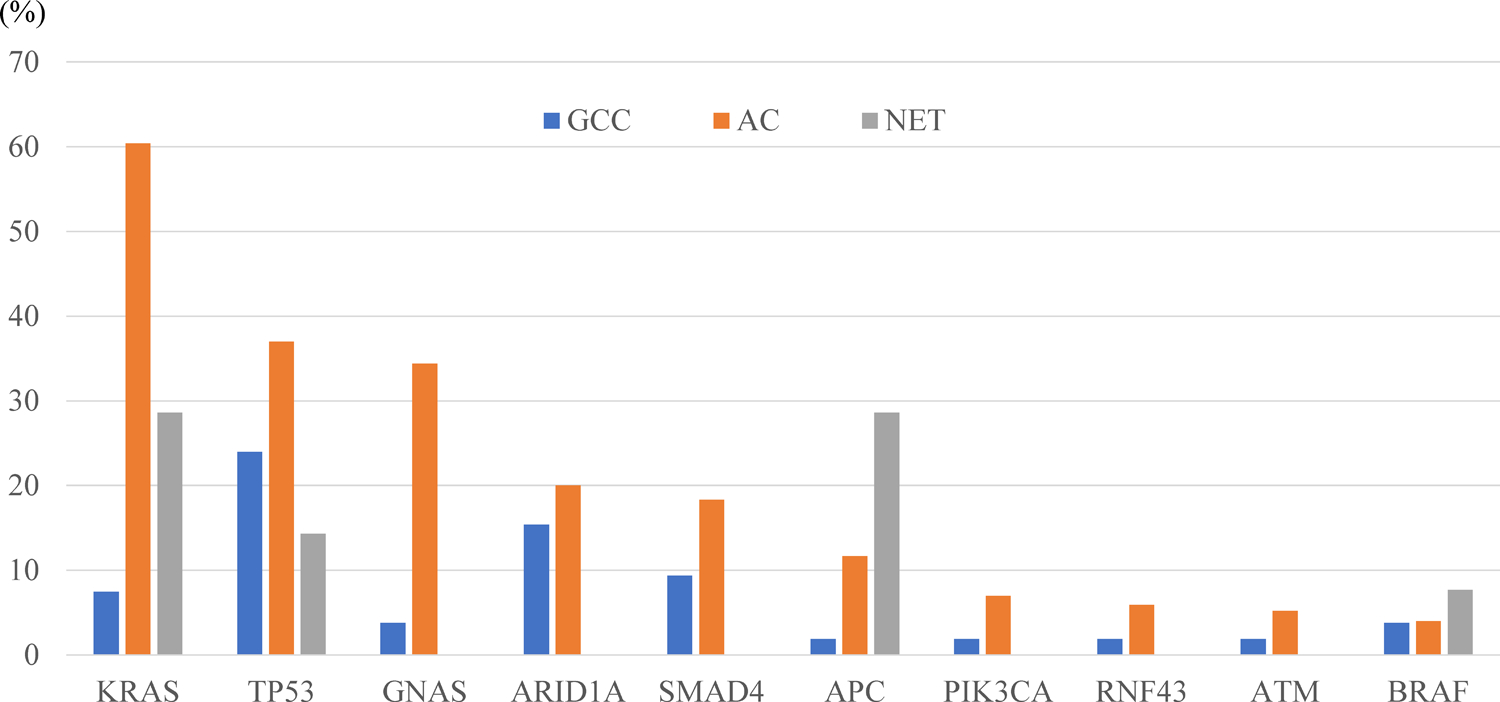
The top 10 major genes in which mutations were identified in appendiceal adenocarcinoma. Details for all data in the comparative analysis are shown in Tables 2 and 3. AC, adenocarcinoma; GCC, goblet cell carcinoid; NET, neuroendocrine tumor.
Within appendiceal NET, only nine mutated genes were observed: KRAS, APC, TP53, CDH1, BRAF, BCOR, BRCA2, FANCA, and ERBB2 (Table 3). GCC showed significantly lower mutation rates when compared to appendiceal NET in KRAS (7.5% vs. 28.6%, respectively), APC (1.9% vs. 28.6%), BRCA2 (0.0% vs. 7.1%), and FANCA (0.0% vs. 7.1%) (Fig. 2, Table 3). GCC showed a numerically higher mutation rate in TP53 (24.0% vs. 14.3%), but the difference was not statistically significant (p = 0.437). Gene amplifications in GCC were observed in MDM2 (3.8%), FUS (2.0%), SF3B1 (2.0%), and FGF23 (2.0%), while amplified MYC (2.4%), CCND1 (2.2%), FGF19 (1.7%), and FGF4 (1.5%) represented the most frequent copy number alterations observed in adenocarcinoma, and no copy number alterations were observed within NET (Supplementary Table S2). No notable gene rearrangements were detected in GCC.
Table 3.
Comparison of mutation frequency between appendiceal NET and GCC
| Gene | NET | GCC |
P-value (NET vs GCC) |
Gene | NET | GCC |
P-value (NET vs GCC) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
MT | Total | Mutation rate (%) |
||||
| KRAS | 4 | 14 | 28.6 | 4 | 53 | 7.5 | 0.03 | ATM | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 |
| APC | 4 | 14 | 28.6 | 1 | 53 | 1.9 | <0.01 | FBXW7 | 0 | 14 | 0.0 | 2 | 53 | 3.8 | 0.46 |
| TP53 | 2 | 14 | 14.3 | 12 | 50 | 24.0 | 0.44 | KDM6A | 0 | 11 | 0.0 | 1 | 47 | 2.1 | 0.63 |
| CDH1 | 1 | 13 | 7.7 | 2 | 52 | 3.8 | 0.55 | SMAD2 | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 |
| BRAF | 1 | 13 | 7.7 | 2 | 53 | 3.8 | 0.54 | KMT2D | 0 | 14 | 0.0 | 1 | 48 | 2.2 | 0.59 |
| BCOR | 1 | 13 | 7.7 | 1 | 51 | 2.0 | 0.29 | SMARCA4 | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 |
| BRCA2 | 1 | 14 | 7.1 | 0 | 53 | 0.0 | 0.05 (0.049) | NF1 | 0 | 14 | 0.0 | 1 | 47 | 2.1 | 0.58 |
| FANCA | 1 | 14 | 7.1 | 0 | 53 | 0.0 | 0.05 (0.049) | STK11 | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 |
| ERBB2 | 1 | 14 | 7.1 | 1 | 53 | 1.9 | 0.30 | CIC | 0 | 13 | 0.0 | 1 | 43 | 2.3 | 0.58 |
| GNAS | 0 | 14 | 0.0 | 2 | 53 | 3.8 | 0.46 | CHEK2 | 0 | 12 | 0.0 | 2 | 50 | 4.0 | 0.48 |
| ARID1A | 0 | 4 | 0.0 | 2 | 13 | 15.4 | 0.40 | CDC73 | 0 | 14 | 0.0 | 1 | 51 | 2.0 | 0.60 |
| SMAD4 | 0 | 14 | 0.0 | 5 | 53 | 9.4 | 0.23 | ERCC2 | 0 | 14 | 0.0 | 1 | 50 | 2.0 | 0.59 |
| PIK3CA | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 | FGFR2 | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 |
| RNF43 | 0 | 14 | 0.0 | 1 | 53 | 1.9 | 0.60 | ||||||||
The bold p-values indicate significant difference (p <0.05). GCC, goblet cell carcinoid; MT, mutant; NET, neuroendocrine tumor.
Immunotherapy-related biomarkers
Mean TMB was 5.8/Mb in GCC, which was lower than that of adenocarcinoma (7.6/Mb) and higher than that of NET (4.1/Mb). The frequency of TMB-H patients was virtually equivalent between all tumor types (GCC: 0%, adenocarcinoma: 1.7%, and NET: 0%). The frequency of MSI-H/dMMR patients was 0% for GCC, 1.9% for adenocarcinoma, and 0% for NET. PD-L1 positivity was 2.0% in GCC, 2.9% in adenocarcinoma, and 0% in NET. No significant difference was observed in these immune profiles when compared GCC and adenocarcinoma/NET (Table 4). The results of MCP-counter were obtained for 86 samples (GCC: 9, adenocarcinoma: 76, NET: 1). NET tumors only had one case with mRNA data, thus the comparative analysis was only done between GCC and adenocarcinoma. While NK cells were the only showing a trending difference, other 9 cell populations did not show any difference between GCC and adenocarcinoma (Supplementary Fig. S3).
Table 4.
Comparison of immunotherapy-related markers between GCC and appendiceal adenocarcinoma/NET
| Biomarker | GCC | AC | NET | P-value | ||
|---|---|---|---|---|---|---|
| Mean TMB | (/Mb) | 5.8 | 7.6 | 4.1 | GCC vs AC GCC vs NET |
<0.01 0.02 |
| TMB-H | (%) | 0.0 | 1.7 | 0.0 | GCC vs AC GCC vs NET |
0.34 NA |
| MSI-H/dMMR | (%) | 0.0 | 1.9 | 0.0 | GCC vs AC GCC vs NET |
0.31 NA |
| PD-L1 positive | (%) | 2.0 | 2.9 | 0.0 | GCC vs AC GCC vs NET |
0.70 0.60 |
TMB/MSI status/PD-L1 positivity were tested in 52/53/51 GCC patients, 409/427/412 AC patients and 14/14/14 NET patients, respectively. TMB-H were defined as 17 or more mutations/Mb.
ACC, adenocarcinoma; GCC, goblet cell carcinoid; MSI-H/dMMR, microsatellite instability high/deficient mismatch repair; NA, not assessed; NET, neuroendocrine tumor; TMB, tumor mutational burden; TMB-H, tumor mutational burden high.
Discussion
To the best of our knowledge, this is the largest study investigating the molecular profiles of appendiceal GCC, in which 53 patient samples were compared to other appendiceal tumors (428 adenocarcinomas and 14 NETs). We demonstrated that GCC consists of considerably different genetic alterations as compared to appendiceal adenocarcinoma and NET. Our data further increases the understanding of GCC biology, emphasizing that GCC is a molecularly distinct entity from other appendiceal tumors.
The epidemiology of GCC has been well documented, with an average age of diagnosis about 10 years higher than appendiceal NET, and no gender preference(1). In our study we confirm that the average age of diagnosis in patients with GCC was 13.2 years higher compared to NET, and no differences of distribution exist between genders.
We report here the largest studied cohort of GCC to date with comprehensive molecular profiling using a 592-gene target panel. The most prevalent mutations observed are present within TP53 (24.0%), ARID1A (15.4%), SMAD4 (9.4%), and KRAS (7.5%), and 21 minor mutant genes account for a small subset of GCC patients. In addition, the mutational spectrum reflects dominant alterations in cell cycle, MAPK, epigenetic, and TGF-β signaling pathways, indicating that these pathways are critical for GCC pathogenesis. Of note, the WNT and PIK3 signaling pathways were infrequently altered, although the well-known function of these pathways is as a key driver for tumorigenesis and progression of colorectal adenocarcinoma(14). Our findings are consistent with a previous smaller study which showed a unique distribution of altered pathways with frequent alterations in the epigenetics pathway and rare alterations of the WNT pathway within GCC(15). Our results suggest also that there is a significant overlap of molecular alterations found in pure GCC and ex-GCC, which is consistent with a previous report suggesting that both represent a single tumor type with varying differentiation grades(15).
As previously reported, the mutational profiles of appendiceal adenocarcinoma are distinct from those of colon adenocarcinoma. Specifically, appendiceal adenocarcinoma shows lower mutation rates compared to colon adenocarcinoma in TP53, APC, PIK3CA, and FBXW7, and higher mutation rates in GNAS and SMAD4(16). In the current study, the molecular profiles between 53 GCCs and 428 appendiceal adenocarcinomas are compared; we observed less frequent mutation rates in KRAS, GNAS, and APC within GCC. On the other hand, some less common mutations were more frequently detected within GCC (CDH1, CHEK2, CDC73, ERCC2, and FGFR2). In addition, the copy number alteration profiles did not overlap between GCC and appendiceal adenocarcinoma, showing more frequent amplification in MDM2, FUS, SF3B1, and FGF23 for GCC. These results suggest a variable pathogenesis of GCC with potentially different key driver alterations compared to appendiceal as well as colorectal adenocarcinoma—as observed in the previously described “adenoma-carcinoma sequence”(14).
A previous study showed loss of heterozygosity within 11q, 16q, and 18q might play a role in the pathogenesis of ileal carcinoid as well as that of GCC(17). The most frequently reported mutated gene in gastrointestinal NET (GI-NET) is CTNNB1 (18,19). However, information concerning the genetic profiles of appendiceal NET have not yet been reported. Our data are the first to show appendiceal NETs exhibit mutations in nine different genes (KRAS, APC, TP53, CDH1, BRAF, BCOR, BRCA2, FANCA, and ERBB2) and a lack of mutations in CTNNB1 (Table 3). These findings suggest that appendiceal NET may be molecularly distinct from other GI-NET. Importantly, the findings in the present study indicate that GCC contains significantly different mutation profiles compared to appendiceal NET, as well as other described GI-NET.
Certain biomarkers may become critical for patient selection for immunotherapies, including immune checkpoint inhibitors (ICI). Patients with MSI-H colorectal cancer have been shown to significantly benefit from ICI therapies(20–23). In addition to MSI status, PD-L1 expression and TMB are related to efficacy of ICI within other cancer treatments(24). In the GCC patients here, we did not detect any cases exhibiting MSI-H and/or TMB-H but we found 2% of PD-L1-positive cases. Thus, GCC is considered to be an immunologically cold tumor. The non-activated immune profiles described herein were similar to those of appendiceal adenocarcinoma and NET. Of note, MCP-counter results showed almost similar abundance of immune and stromal cell populations in tumor microenvironment between GCC and adenocarcinoma. These results indicate that ICI may not be a promising treatment for GCC nor for the other types of appendiceal tumors.
Current clinical guidelines established by the ENETS and NANETS recommend that patients with GCC are treated in accordance with colorectal cancer treatment given the aggressive clinical course(1,11). Specifically, right hemicolectomy for resectable GCC and palliative 5-fluorouracil-based chemotherapy for metastatic GCC are the recommended standard treatments. However, based on the findings of the present study, the question arises as to whether the same treatment strategy for colorectal cancer should be used for GCC, as based on the significant differences observed in molecular profiling of GCC compared to adenocarcinoma. Finding more effective and rationally based treatment strategies for patients with GCC is needed. There is no data suggesting that GCC should be treated as a NET, as significant molecular differences between these tumor types were demonstrated in this study. Our findings suggest that GCC treatment strategies should be reconsidered and instead focus on therapies targeting cell cycle, MAPK, epigenetic, and TGF-β signaling pathways. Studies of preclinical models are critical to transition new therapies into the clinic for this rare tumor.
There are some limitations within our study. First, the retrospective design could not completely exclude a selection bias. Second, we did not have certain important clinical data for the patients enrolled in this study. We just had limited information of TNM stage, but the details of treatment regimens and survival time were not available at all. Further investigations including this information would allow us to better understand the association between the genetic alterations of GCC and clinical stage, prognosis, and treatment outcome.
In conclusion, GCC has distinct genetic backgrounds compared to appendiceal adenocarcinoma and NET. These findings raise a question about reconsidering the currently used classification system and treatment strategies for this rare disease.
Supplementary Material
Acknowledgements
Funding
This work was supported by the National Cancer Institute [P30CA 014089 to H.-J.L.], Gloria Borges WunderGlo Foundation, Dhont Family Foundation, San Pedro Peninsula Cancer Guild, and Daniel Butler Research Fund.
Footnotes
Disclosure
H.-J.L. reports receiving speakers bureau honoraria from and is a consultant/advisory board member for Merck Serono, Bayer, and Genentech. A.S. received speakers bureau honoraria from Caris Life Sciences. Y.B., J.X. and W.M.K. are employers of Caris Life Sciences. All remaining authors have declared no conflicts of interest.
References
- 1.Pape UF, Perren A, Niederle B, Gross D, Gress T, Costa F, et al. ENETS Consensus Guidelines for the management of patients with neuroendocrine neoplasms from the jejuno-ileum and the appendix including goblet cell carcinomas. Neuroendocrinology 2012;95(2):135–56 doi 10.1159/000335629. [DOI] [PubMed] [Google Scholar]
- 2.Shenoy S Goblet cell carcinoids of the appendix: Tumor biology, mutations and management strategies. World journal of gastrointestinal surgery 2016;8(10):660–9 doi 10.4240/wjgs.v8.i10.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onyemkpa C, Davis A, McLeod M, Oyasiji T. Typical carcinoids, goblet cell carcinoids, mixed adenoneuroendocrine carcinomas, neuroendocrine carcinomas and adenocarcinomas of the appendix: a comparative analysis of survival profile and predictors. Journal of gastrointestinal oncology 2019;10(2):300–6 doi 10.21037/jgo.2018.11.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler JA, Houshiar A, Lin F, Wilson SE. Goblet cell carcinoid of the appendix. American journal of surgery 1994;168(6):685–7 doi 10.1016/s0002-9610(05)80145-x. [DOI] [PubMed] [Google Scholar]
- 5.Taggart MW, Abraham SC, Overman MJ, Mansfield PF, Rashid A. Goblet cell carcinoid tumor, mixed goblet cell carcinoid-adenocarcinoma, and adenocarcinoma of the appendix: comparison of clinicopathologic features and prognosis. Archives of pathology & laboratory medicine 2015;139(6):782–90 doi 10.5858/arpa.2013-0047-OA. [DOI] [PubMed] [Google Scholar]
- 6.Tang LH, Shia J, Soslow RA, Dhall D, Wong WD, O’Reilly E, et al. Pathologic classification and clinical behavior of the spectrum of goblet cell carcinoid tumors of the appendix. The American journal of surgical pathology 2008;32(10):1429–43 doi 10.1097/PAS.0b013e31817f1816. [DOI] [PubMed] [Google Scholar]
- 7.Chetty R, Klimstra DS, Henson DE, Albores-Saavedra J. Combined classical carcinoid and goblet cell carcinoid tumor: a new morphologic variant of carcinoid tumor of the appendix. The American journal of surgical pathology 2010;34(8):1163–7 doi 10.1097/PAS.0b013e3181e52916. [DOI] [PubMed] [Google Scholar]
- 8.Roy P, Chetty R. Goblet cell carcinoid tumors of the appendix: An overview. World journal of gastrointestinal oncology 2010;2(6):251–8 doi 10.4251/wjgo.v2.i6.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Meyerson C, Kassardjian A, Westbrook LM, Zheng W, Wang HL. Goblet Cell Carcinoid/Carcinoma: An Update. Advances in anatomic pathology 2019;26(2):75–83 doi 10.1097/PAP.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 10.Bosman FT, Cameiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. 2010.
- 11.Boudreaux JP, Klimstra DS, Hassan MM, Woltering EA, Jensen RT, Goldsmith SJ, et al. The NANETS consensus guideline for the diagnosis and management of neuroendocrine tumors: well-differentiated neuroendocrine tumors of the Jejunum, Ileum, Appendix, and Cecum. Pancreas 2010;39(6):753–66 doi 10.1097/MPA.0b013e3181ebb2a5. [DOI] [PubMed] [Google Scholar]
- 12.Wen KW, Grenert JP, Joseph NM, Shafizadeh N, Huang A, Hosseini M, et al. Genomic profile of appendiceal goblet cell carcinoid is distinct compared to appendiceal neuroendocrine tumor and conventional adenocarcinoma. Human pathology 2018;77:166–74 doi 10.1016/j.humpath.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 13.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome biology 2016;17(1):218 doi 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nature reviews Cancer 2017;17(2):79–92 doi 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 15.Johncilla M, Stachler M, Misdraji J, Lisovsky M, Yozu M, Lindeman N, et al. Mutational landscape of goblet cell carcinoids and adenocarcinoma ex goblet cell carcinoids of the appendix is distinct from typical carcinoids and colorectal adenocarcinomas. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2018;31(6):989–96 doi 10.1038/s41379-018-0003-0. [DOI] [PubMed] [Google Scholar]
- 16.Tokunaga R, Xiu J, Johnston C, Goldberg RM, Philip PA, Seeber A, et al. Molecular Profiling of Appendiceal Adenocarcinoma and Comparison with Right-sided and Left-sided Colorectal Cancer. Clinical Cancer Research 2019;25(10):3096–103 doi 10.1158/1078-0432.ccr-18-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stancu M, Wu TT, Wallace C, Houlihan PS, Hamilton SR, Rashid A. Genetic alterations in goblet cell carcinoids of the vermiform appendix and comparison with gastrointestinal carcinoid tumors. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc 2003;16(12):1189–98 doi 10.1097/01.MP.0000097362.10330.B1. [DOI] [PubMed] [Google Scholar]
- 18.Fujimori M, Ikeda S, Shimizu Y, Okajima M, Asahara T. Accumulation of beta-catenin protein and mutations in exon 3 of beta-catenin gene in gastrointestinal carcinoid tumor. Cancer research 2001;61(18):6656–9. [PubMed] [Google Scholar]
- 19.Oberg K. Genetics and molecular pathology of neuroendocrine gastrointestinal and pancreatic tumors (gastroenteropancreatic neuroendocrine tumors). Current opinion in endocrinology, diabetes, and obesity 2009;16(1):72–8 doi 10.1097/med.0b013e328320d845. [DOI] [PubMed] [Google Scholar]
- 20.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine 2015;372(26):2509–20 doi 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. The Lancet Oncology 2017;18(9):1182–91 doi 10.1016/s1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2018;36(8):773–9 doi 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 23.Lenz H-J, Lonardi S, Zagonel V, Cutsem EV, Limon ML, Wong KYM, et al. Nivolumab plus low-dose ipilimumab as first-line therapy in microsatellite instability-high/DNA mismatch repair deficient metastatic colorectal cancer: Clinical update. Journal of Clinical Oncology 2020;38(4_suppl):11- doi 10.1200/JCO.2020.38.4_suppl.11.31725351 [DOI] [Google Scholar]
- 24.Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. The Lancet Oncology 2016;17(12):e542–e51 doi 10.1016/s1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


