Activation of PD-1 by p53 is critical for its tumor suppression in an immunity-independent manner.
Abstract
Cancer cell–intrinsic programmed cell death protein-1 (PD-1) has emerged as a tumor regulator in an immunity-independent manner, but its precise role in modulating tumor behaviors is complex, and how PD-1 is regulated in cancer cells is largely unknown. Here, we identified PD-1 as a direct target of tumor suppressor p53. Notably, p53 acetylation at K120/164 played a critical role in p53-mediated PD-1 transcription. Acetylated p53 preferentially recruited acetyltransferase cofactors onto PD-1 promoter, selectively facilitating PD-1 transcription by enhancing local chromatin acetylation. Reexpression of PD-1 in cancer cells inhibited tumor growth, whereas depletion of cancer cell–intrinsic PD-1 compromised p53-dependent tumor suppression. Moreover, histone deacetylase inhibitor (HDACi) activated PD-1 in an acetylated p53–dependent manner, supporting a synergistic effect by HDACi and p53 on tumor suppression via stimulating cancer cell–intrinsic PD-1. Our study reveals a mechanism for activating cancer cell–intrinsic PD-1 and indicates that p53-mediated PD-1 activation is critically involved in tumor suppression in an immunity-independent manner.
INTRODUCTION
The tumor suppressor p53 plays a critical role in the prevention of malignant transformation of normal cells (1, 2). Loss of p53 in mice model results in a predisposition to spontaneous tumorigenesis (3), and more than 50% human cancers harbor p53 mutations reflecting dysfunction of p53 as a crucial event during cancerous development (4). In general, p53 serves as a sequence-specific transcription factor (1, 2), and the transactivity of p53 is required for its tumor-suppressive actions (5). In response to cellular stresses, p53 transcriptionally activates or represses a number of downstream target genes, which, in turn, participate in the modulation of multiple biological processes including cell cycle arrest, programmed cell death, cellular senescence, DNA repair, oxidative response, and metabolic regulation (6). It is considered that p53 exerts its tumor suppressive functions by eliciting distinct transcriptional profiles in a context-dependent manner, which links the selective regulation of the unique biological process(es) under a given condition to prevent tumor formation (6). Consistent with this notion, the posttranslational modifications (PTMs) such as phosphorylation and acetylation have been uncovered as a key mechanism to determine the selectivity of p53-mediated transcription (7).
The programmed cell death protein-1 (PD-1; also known as CD279), encoded by the PDCD1 gene, is a transmembrane protein that is predominantly expressed in immune cells including T cell, B cell, and macrophage, upon receiving variously initial immune stimuli by these types of cells (8, 9). PD-1 serves as an inhibitory receptor that plays a key role in peripheral immune tolerance (8). Elevated expression of PD-1 in CD8+ T cells was observed in chronic infection, which links a “T cell” functional exhaustion and, consequently, a compromised immune response (10, 11). The programmed death-ligand 1 (PD-L1; also known as B7-H1 or CD274), one major ligand of PD-1, is frequently overexpressed in cancer cells (12, 13). In local tumor microenvironment, the PD-1 signaling of immune cells can be activated upon PD-1 engagement of cancer cell–expressed PD-L1, which largely contributes to the immunological evasion of cancer cells (8, 14, 15). Using the monoclonal antibodies targeting PD-1/PD-L1 axis, the immune checkpoint therapy on several types of cancer, including advanced melanoma and lung cancer, has achieved great success during past a few years (16, 17).
In addition to the immune cells, the expression of intrinsic PD-1 was also recently found in some kind of cancer cells, suggesting a potential role of PD-1 in the regulation of cancer cell behaviors in an immunity-independent way (18–23). For example, the subpopulation of melanoma cells that express PD-1 exhibited higher ability of proliferation and tumorigenicity without need of adaptive immune response (18). Similar oncogenic effect by intrinsic PD-1 was also observed in hepatic cancer cells and pancreatic cancer cells (19, 20). On the contrary, a more recent study focused on PD-1/PD-L1 expression in lung cancer cells indicated that the intrinsic PD-1 may suppress tumor growth (23). Consistent with this view, a clinical trial also reported that the anti–PD-1 therapy promoted lung cancer progress in a patient with PD-1–positive in his/her cancer cells (21). These contradictory results implied a complex and diverse effect of cancer cell–intrinsic PD-1. Moreover, although the regulations of PD-1 have been widely studied in immune cells (24), it is still elusive how the intrinsic PD-1 is regulated in cancer cells.
In this study, we identify the tumor suppressor p53 as a key regulator that transcriptionally activates PD-1 expression in cancer cells in response to cellular stress. The acetylation of p53 at lysine-120 and lysine-164 catalyzed by acetyltransferases p300/CBP (CREB-binding protein) and TIP60 (Tat-interactive protein 60 kD), as well as the chromatin recruitment of these enzyme cofactors by p53, contributes to p53-mediated PD-1 transcription. Moreover, we confirm that the expression of intrinsic PD-1 in lung cancer cells inhibits tumor growth and remarkably participates in p53-mediated tumor suppression in vitro and in vivo. Our data on the p53-mediated PD-1 regulation in cancer cells provide a new insight into the regulation of cancer cell–intrinsic PD-1 and its immune checkpoint–independent roles in tumor suppression.
RESULTS
p53 regulates PD-1 expression in both normal and cancer cells
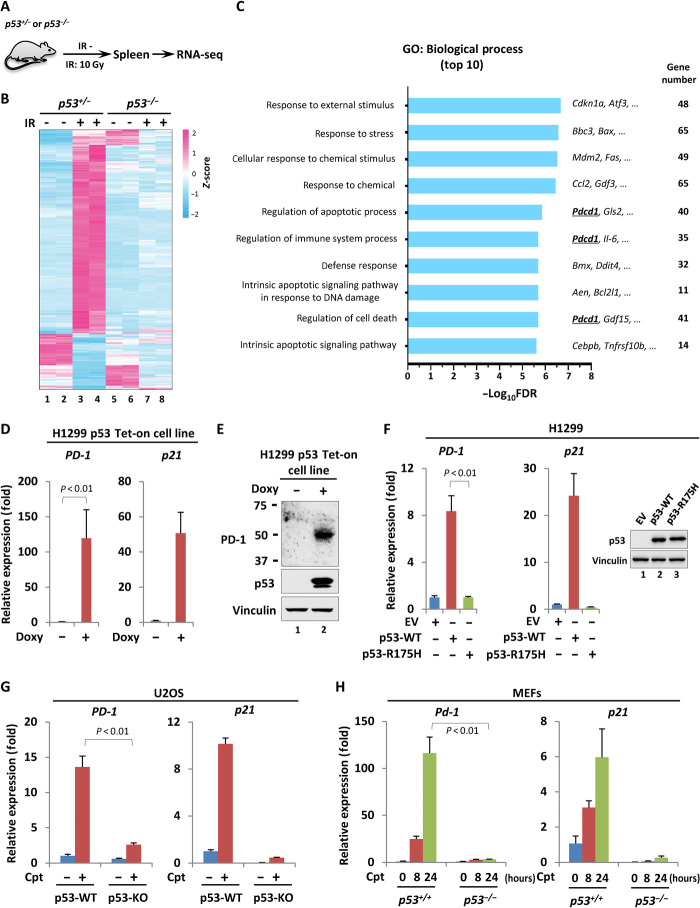
To identify novel downstream targets that contribute to p53-mediated tumor suppression, we exposed the mice of p53+/− and p53−/− littermates to the γ-radiation and screened differentially expressed genes in vivo through an RNA sequencing (RNA-seq) technique (Fig. 1A). More than 300 genes of which the expression was significantly altered by γ-radiation in splenic cells derived from p53+/−, but not p53−/−, mice were identified (Fig. 1B). Gene ontology (GO) analysis revealed that most top-ranked biological processes enriched by these genes were highly related to stimulus response and cell death, consistent with the notions regarding p53-mediated functional regulation during antitumorigenesis (Fig. 1C and table S1). Among those previously uncharacterized candidates, we paid more attentions to Pdcd1 (Pd-1), a gene that was traditionally recognized to encode an inhibitory receptor PD-1 in immune cells and was recently reported to also express in subgroup of cancer cells. In addition to the spleen, the regulation of PD-1 by p53 was also observed in thymus, a developing T cell–enriched peripheral lymphoid organ, suggesting that p53 might regulate PD-1 transcription in T cells in response to cellular stress (fig. S1, A and B). Although it is well accepted that T cell–expressed PD-1 serves as an immune checkpoint and plays a key role in the regulation of immune response, it is still obscure how PD-1 is regulated and functions in cancer cells.
Fig. 1. p53 regulates PD-1 expression in both normal and cancer cells.
(A) Schematic diagram of the working flow for the screening. p53+/− or p53−/− mice were treated with or without 10–gray (Gy) γ-irradiation (IR). After 4 hours, the splenic RNA was extracted, and the gene expression was measured by RNA-seq (n = 2). (B) Heatmap presentation of the genes regulated by γ-irradiation in a p53-dependent manner. (C) GO analysis of the top 10 biological processes enriched by the genes that were regulated in response to γ-irradiation in a p53-dependnent manner. FDR, false discovery rate. (D) Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis of the mRNA level of PD-1 and p21 in H1299 p53 Tet-on cells treated with or without doxycycline (Doxy; 1 μg/ml) for 24 hours. (E) Western blot analysis of PD-1 in H1299 p53 Tet-on cells treated with or without doxycycline (1 μg/ml) for 24 hours. (F) RT-qPCR analysis of the mRNA level of PD-1 and p21 in H1299 cells transiently transfected with empty vector (EV), wild-type, or mutant (R175H) p53-expressing construct for 24 hours. The relative expression of p53 was detected by Western blot. (G) RT-qPCR analysis of the mRNA level of PD-1 and p21 in p53 wild-type (p53-WT) or p53 knockout (p53-KO) U2OS cells treated with or without 1 μM camptothecin (Cpt) for 24 hours. (H) RT-qPCR analysis of the mRNA level of Pd-1 and p21 in p53+/+ or p53−/− MEFs treated with or without 1 μM Cpt for indicated period of time. Data were shown as means ± SD, n = 3.
To test whether p53 regulates PD-1 in cancer cells, we generated a p53 Tet-on expression system in a p53-null lung cancer cell line H1299. The basal level of PD-1 in H1299 cells was pretty low; however, doxycycline-induced expression of the ectopic p53 remarkably elevated PD-1, both in mRNA and protein level (Fig. 1, D and E). Notably, the transactivity of p53 was required to p53-mediated PD-1 induction, as the mutant p53 (R175H) that lost DNA binding ability failed to do so (Fig. 1F). To validate this modulation physiologically, we deleted the endogenous wild-type p53 by CRISPR-Cas9 technique in osteosarcoma U2OS cells and treated cells with or without camptothecin (Cpt) that serves as a DNA damage inducer to activate p53. As expected, Cpt treatment elevated PD-1 expression in parental U2OS cells; however, this up-regulation of PD-1 was largely compromised upon p53 deficiency (Fig. 1G and fig. S1C). Notably, p53 ablation had no obvious effect on the decay rate of PD-1 mRNA (fig. S1D). Similarly, the modulation of PD-1 by p53 was also observed in the melanoma cell line A375 and the pancreatic carcinoma cell line SW1990 (fig. S1, E and F), consistent with the previous report that PD-1 is regulated in other types of cancer cell line (25). These data together indicated a physiological role of p53 in the regulation of PD-1 transcription in response to DNA damage in cancer cells. In addition, a bioinformatic analysis of The Cancer Genome Atlas (TCGA), but not Cancer Cell Line Encyclopedia (CCLE), database revealed a positive correlation between the high levels of PD-1 transcript and the wild-type p53 status in multiple types of tumor samples, such as diffuse large B cell lymphoma, head-neck squamous cell carcinoma, kidney renal clear cell carcinoma, and stomach adenocarcinoma, suggesting that p53-mediated PD-1 regulation is likely widespread, to some extent, in tumors (fig. S2, A and B). Moreover, the dependency of p53 in the modulation of PD-1 was further confirmed in mouse embryonic fibroblasts (MEFs) where DNA damage–induced PD-1 up-regulation was significantly abrogated in p53−/− MEFs (Fig. 1H). Together, our data indicate that p53 regulates PD-1 expression in both normal and cancer cells.
PD-1 is a p53 direct target gene
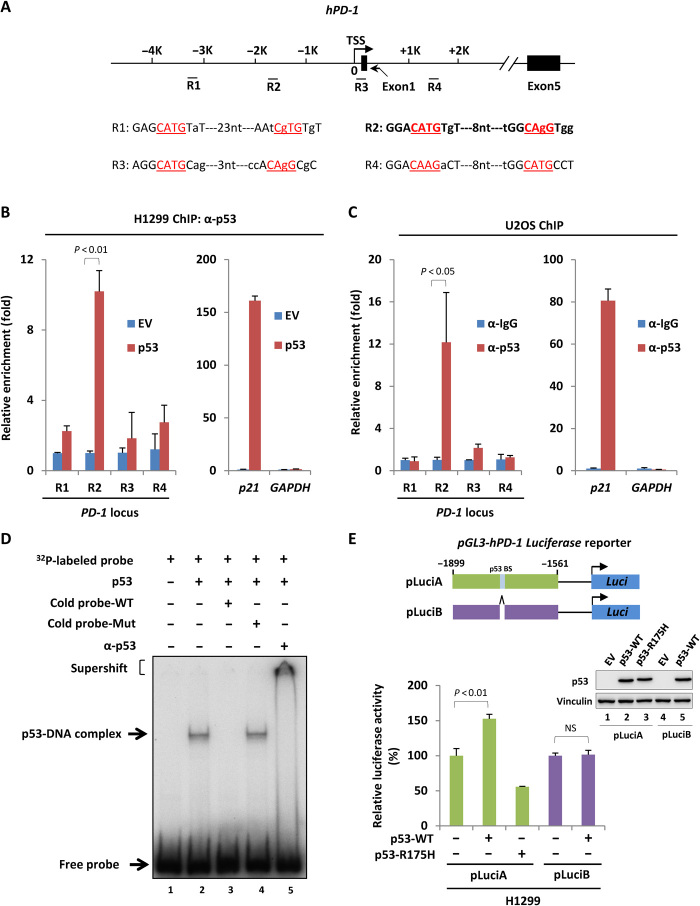
These observations above suggest that PD-1 might be a direct downstream target of p53. Therefore, we next sought the potential p53-binding loci surrounding PD-1 promoter. By comparing with the consensus binding motif of p53, we found four putative regions (R1 to R4) for p53 binding (Fig. 2A). A chromatin immunoprecipitation (ChIP) assay was performed in H1299 cells transiently transfected with empty vector or construct expressing p53. As expected, the ectopic p53 strongly bound on p21 promoter, but not on GAPDH promoter that resembles an irrelevant region without p53-binding element (Fig. 2B). Notably, under the same conditions, p53 also exhibited a binding affinity to R2 at ~−1.7-kb upstream of PD-1 transcription start site, but not to R1, R3, or R4 (Fig. 2B). ChIP assay conducted in U2OS cells revealed that the endogenous p53 bound on R2 of PD-1 promoter, further confirming the recruitment of p53 on PD-1 promoter under physiological conditions (Fig. 2C). In addition, the electrophoretic mobility shift assay (EMSA) further proved that the binding between p53 and PD-1 promoter DNA was direct and specific (Fig. 2D). Moreover, we generated a luciferase reporter construct containing wild-type or mutant p53-binding element of PD-1 promoter to evaluate whether p53 drives PD-1 expression through this binding region. Only wild-type p53 enabled to activating the luciferase reporter containing the wild type, but not the mutant, p53-binding element of PD-1 promoter (Fig. 2E), indicating that p53 directly transactivates PD-1 through binding with PD-1 promoter. Together, our data indicate that PD-1 is a direct downstream target of tumor suppressor p53.
Fig. 2. Identification of PD-1 as a p53 direct target gene.
(A) Schematic diagram of human PD-1 gene locus with four potential p53-binding regions. TSS, transcription start site. (B) ChIP-qPCR analysis of p53 occupancy on PD-1 promoter or gene body region in H1299 cells transfected with empty vector or p53-expressing construct for 24 hours. (C) ChIP-qPCR analysis of p53 occupancy on PD-1 promoter or gene body region in U2OS cells. p53-binding on p21 and GAPDH promoter in (B) and (C) was measured as positive and negative control, respectively. (D) EMSA of p53 binding with PD-1 promoter in vitro. Purified p53 was incubated with a 32P-labeled probe containing p53-binding element of PD-1 promoter. α-p53 antibody was used for supershift assay. (E) Luciferase assay of p53-driven transcription of PD-1 promoter–containing reporter expressing cassette in H1299 cells. The expression of p53 was detected by Western blot. BS, binding site. Data were shown as means ± SD, n = 3.
Acetylation contributes to p53-mediated PD-1 transcriptional activation
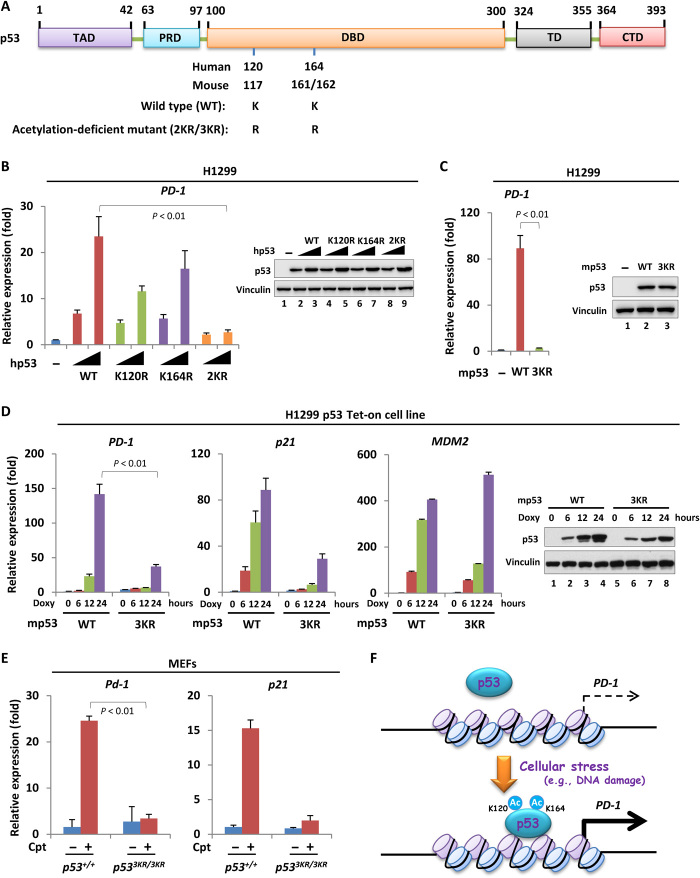
The PTMs of p53, such as phosphorylation within its transactivation domain and acetylation within its DNA binding domain (DBD) or C-terminal domain, play important roles in p53 stabilization and transactivation during response to various cellular stresses (7, 26, 27). To evaluate whether these modifications were involved in p53-mediated PD-1 transcriptional activation, we introduced a serial of mutant p53 where the serine/threonine or lysine residues were replaced with alanine or arginine, respectively, to block phosphorylation or acetylation (Fig. 3A and fig. S3A). The loss of either N-terminal phosphorylation or C-terminal acetylation had no obvious effect on p53-mediated PD-1 expression (fig. S3, B and C). In contrast, the mutant p53 harboring K120R or K164R, respectively, was compromised to transactivate PD-1 (Fig. 3B). Moreover, the alteration of both lysine residues together [K120/164R (2KR)] almost completely abolished p53-mediated PD-1 activation (Fig. 3B). These lysine residues were conserved and corresponded to K117 and K161/162 in mouse (Fig. 3A). Similarly, overexpression of mouse mutant p53 harboring K117/161/162R (3KR) in H1299 cells also failed to up-regulate PD-1 mRNA level (Fig. 3C), suggesting an evolutionary conserved regulatory mode of p53 acetylation–mediated PD-1 transcriptional activation. To further evaluate this notion, we generated a pair of H1299-inducible cells that express the wild-type p53 and the acetylation-deficient p53 mutant, respectively, upon doxycycline treatment. As expected, the induced wild-type p53 was acetylated at K120/164 (K117/161/162 in mouse) (fig. S3, D and E). In addition, the wild-type p53 promoted PD-1 expression in a time-dependent manner, whereas the acetylation-deficient p53 mutant almost completely lost the ability to induce PD-1 (Fig. 3D). More physiologically, we treated p53+/+ and p533KR/3KR MEFs, respectively, with Cpt to activate p53 and detected PD-1 expression. The acetylation of K120/164 (K117/161/162 in mouse) of endogenous p53 was easily detected in response to DNA damage (fig. S3, F and G). PD-1 expression was significantly increased in p53+/+, but not in p533KR/3KR, MEFs in response to DNA damage (Fig. 3E). Because the lysine residues could be decorated by many types of the modifications, we then examined any other potential modifications on K120/164 upon p53 activation. We performed a mass spectrometry (MS) analysis of the activated p53 purified from H1299 p53 Tet-on cells treated with doxycycline. The MS revealed 15 peptides containing K120 and 13 peptides containing K164, respectively, and no modifications other than acetylation were identified on K120/164 (fig. S3H). This result revealed that the acetylation could be the major type of the modification on K120/164 upon p53 activation. Collectively, although we cannot completely rule out the possibility of other potential modifications on K120/164 in the regulation of PD-1, our data suggested that the acetylation of K120/164 upon p53 activation is critically involved in p53-mediated PD-1 transcription (Fig. 3F).
Fig. 3. Acetylation of p53 contributes to PD-1 transcriptional activation.
(A) Schematic diagram of p53, with conserved lysine residues for acetylation in its DBD. TAD, transactivation domain; PRD, proline-rich domain; DBD, DNA-binding domain; TD, tetramerization domain; CTD, C-terminal domain. (B) RT-qPCR analysis of PD-1 expression in H1299 cells transfected with increasing amount of wild-type or acetylation-defect human p53-expressing construct for 24 hours. The p53 expression was measured by Western blot. (C) RT-qPCR analysis of PD-1 expression in H1299 cells transfected with mouse wild-type or acetylation-defect p53-expressing construct for 24 hours. The expression of p53 was detected by Western blot. (D) RT-qPCR analysis of PD-1 expression in H1299 p53-inducible cells (wild type versus acetylation defect) treated with or without doxycycline (1 μg/ml) for increasing period of time. The expression of p21 and MDM2 was also measured as the positive and negative control, respectively. The expression of p53 was detected by Western blot. (E) RT-qPCR analysis of Pd-1 and p21 expression in p53+/+ or p533KR/3KR MEFs treated with or without 1 μM Cpt for 24 hours. (F) Schematic diagram to summarize the regulatory effect of p53 acetylation (Ac) within its DBD on PD-1 transcriptional activation in response to cellular stresses (e.g., DNA damage). Data were shown as means ± SD, n = 3.
p53 links acetyltransferase cofactors to modulate PD-1 expression
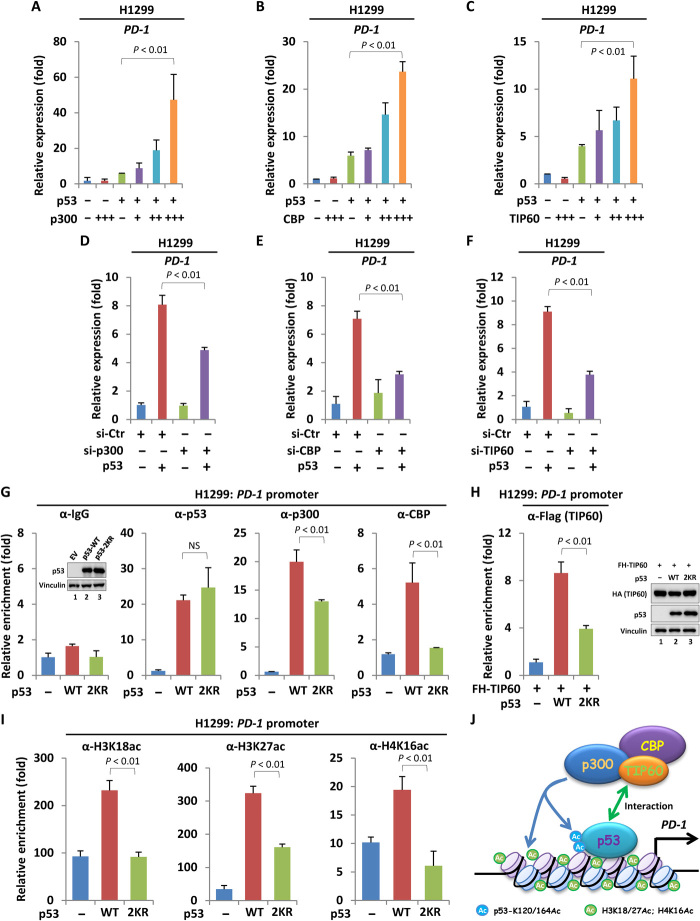
To investigate the mechanism by which the acetylation of K120/164 regulates p53-mediated PD-1 expression, we first focused on the acetyltransferase p300/CBP and TIP60. p300/CBP represents a unique type of HAT (histone acetyltransferase) domain–containing proteins that are responsible for the acetylation on K164 in human or K161/162 in mouse, whereas TIP60 is a MYST (MOZ, Ybf2/Sas3, Sas2 and TIP60) family protein that specifically catalyzes p53 acetylation on K120 in human or K117 in mouse (28–30). In addition, these enzymes also modulate histone acetylation within p53 target gene’s promoter and acts as coactivator of p53. To this end, we coexpressed p300, CBP, or TIP60, together with p53 into H1299 cell to detect PD-1 expression. All these enzymes enabled to significantly promoting p53-mediated PD-1 expression in a dose-dependent manner (Fig. 4, A to C, and fig. S4, A to C). In contrast, the acetyltransferase GCN5 (general control of amino acid synthesis 5) and PCAF (p300/CBP-associated factor) showed no obvious effect on p53-mediated PD-1 expression (fig. S4D). On the other hand, small interfering RNA (siRNA)–mediated knockdown of p300, CBP, or TIP60 attenuated p53-induced PD-1 transcription, further indicating that these acetyltransferases play key roles in p53-mediated PD-1 expression (Fig. 4, D to F, and fig. S4, E to G). Unlike the regulation of PD-1, only CBP, but not p300 or TIP60, affected p53-dependent transcription of TIGAR (TP53-inducible glycolysis and apoptosis regulator) (fig. S4H), a p53 target gene involved in cancer cell survival under certain conditions (31). To further investigate the role of these acetyltransferases in differential regulation of PD-1 controlled by wild-type p53 versus acetylation-deficient p53, we then detected the interaction between p53 and these enzymes. As expected, the wild-type p53 was easily observed to interact with p300, CBP, and TIP60. Similarly, the p53-2KR mutant bound these enzymes almost equally to that of the wild-type p53 (fig. S5, A to C), suggesting that loss of acetylation of K120/164 had no obvious effect on the overall interaction between p53 and these acetyltransferase cofactors. We then performed a ChIP assay to specifically evaluate the recruitment of p300, CBP, or TIP60 on PD-1 promoter through a p53-dependent manner. As shown in Fig. 4G, the binding between wild-type p53 and p53-2KR mutant on PD-1 promoter was comparable. However, the p53-2KR–mediated recruitment of p300, CBP, or TIP60 on PD-1 promoter was markedly reduced in comparison to that performed by wild-type p53 (Fig. 4, G and H). In contrast, the association of p300, CBP, or TIP60 on MDM2 (mouse double minute 2) promoter recruited by wild-type p53 versus the p53-2KR mutant showed no difference (fig. S5, D and E). These results implied that the enhanced chromatin recruitment of p300, CBP, or TIP60 by acetylated p53 could be promoter specific. Under cellular conditions, p300/CBP mainly catalyzes histone H3K18/27 acetylation, whereas TIP60 is predominantly responsible to H4K16 acetylation. All these histone markers were correlated to transcriptional activation. Consistent with the reduced recruitment of p300/CBP and TIP60 by p53-2KR mutant on PD-1 promoter, the p53-2KR mutant–induced elevation of H3K18/27ac and H4K16ac surrounding PD-1 promoter was significantly less than that induced by the wild-type p53 (Fig. 4I). This observation may explain, at least in part, the mechanism by which the acetylation of K120/164 facilitates p53-mediated PD-1 transcription. Together, our data indicated that acetyltransferase p300, CBP, and TIP60 serve as key cofactors and contribute to p53-mediated PD-1 transcriptional activation (Fig. 4J).
Fig. 4. p53 links acetyltransferase cofactors to modulate PD-1 expression.
(A to C) RT-qPCR analysis of PD-1 expression in H1299 cells cotransfected with p53 and increasing amount of p300-, CBP-, or TIP60-expressing construct for 24 hours. (D to F) RT-qPCR analysis of p53-mediated PD-1 expression upon depletion of p300, CBP, or TIP60. H1299 cells transfected with control siRNA or siRNA targeting p300, CBP, or TIP60 for 2 days were then transfected with empty vector or p53-expressing construct for another 24 hours, and the PD-1 mRNA level was measured. (G) ChIP-qPCR analysis of PD-1 promoter occupancy by p53, p300, or CBP in H1299 cells expressing wild-type or acetylation-defect p53. The relative expression of p53 was detected by Western blot. NS, not significant. (H) ChIP-qPCR analysis of TIP60 occupancy on PD-1 promoter in H1299 cells cotransfected with Flag–hemagglutinin (HA)–TIP60 (FH-TIP60) with wild-type or acetylation-defect p53. The relative expression of p53 and TIP60 was measured by Western blot. (I) ChIP-qPCR analysis of indicated histone modifications on PD-1 promoter in H1299 cells expressing wild-type or acetylation-defect p53. (J) Schematic diagram to summarize the mechanism by which the acetyltransferase cofactors participate in p53-mediated PD-1 expression. Data were shown as means ± SD, n = 3.
Cancer cell–intrinsic PD-1 suppresses tumor growth in an immunity-independent manner
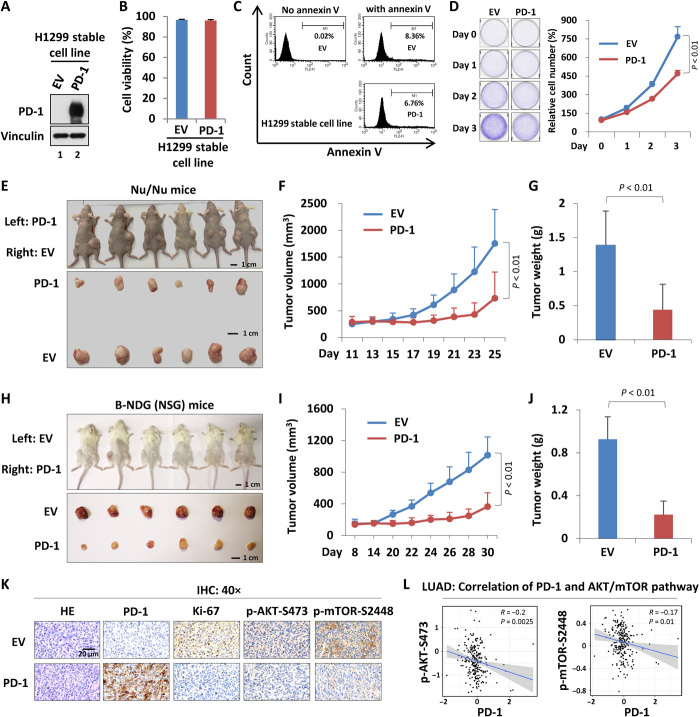
Although the inhibitory effect of PD-1 in cancer immunity has been well documented during past years, the role of intrinsic PD-1 in cancer cells is still complex (18–21, 23). To thoroughly evaluate the potential roles of cancer cell–intrinsic PD-1 in tumor behaviors, we set up our own experimental system. Because the mRNA level of PD-1 is pretty low and is without detectable protein level of PD-1 in H1299 cells (fig. S6, A and B), we then reexpressed the ectopic PD-1 in H1299 cells and generated a stable cell line (Fig. 5A). The ectopic PD-1 was localized in both cytomembrane and cytoplasm (fig. S7, A to C). Similar to the control cells, the PD-1 stable cell line exhibited negligible change on overall cell viability and the basal level of cellular apoptosis, suggesting that PD-1 reexpression has no effect on cell death (Fig. 5, B and C). Reexpression of PD-1 significantly inhibited cancer cell proliferation in vitro (Fig. 5D). Using a nude mice xenograft model, we observed a tumor-suppressive roles of cancer cell–intrinsic PD-1 in vivo (Fig. 5E), where both the volume and the weight of tumors derived from PD-1–expressed cells were markedly less than that of tumors derived from control cells (Fig. 5, F and G). The phenomenon of tumor suppression by cancer cell–intrinsic PD-1 in lung cancer cells was also recaptured using B-NDG (NSG) mice, a severe immunodeficient strain where the mature T cells, B cells, and natural killer cells are completely lost, indicating that the property of tumor suppression by cancer cell–intrinsic PD-1 is achieved through an immunity-independent manner (Fig. 5, H to J). Moreover, immunohistochemistry (IHC) of tumor sections showed a lower percentage of the nuclear Ki-67 staining in PD-1–expressing cells than that in control cells (Fig. 5K), further confirming the inhibitory roles of intrinsic PD-1 in cancer cell proliferation. Previous studies suggested that PD-1 signaling cascade may interrupt AKT–mechanistic target of rapamycin (mTOR) pathway to negatively modulate cell growth and proliferation in immune cells (32). We then next detected whether this molecular mechanism may apply to cancer cells. As shown in Fig. 5K, the PD-1–expressing tumor sections exhibited lower level of phospho-AKT/mTOR than that of tumor sections without PD-1 expression. Consistent with our observation, the RPPA (reverse phase protein array) data of lung adenocarcinoma (LUAD) samples collected from TCGA database revealed a significant negative correlation in terms of levels between phospho-AKT/mTOR and PD-1 (33) (Fig. 5L). Together, our data indicate that the cancer cell–intrinsic PD-1 negatively regulates tumor growth in an immunity-independent manner through, at least in part, inhibiting AKT/mTOR pathway.
Fig. 5. The role of cancer cell–intrinsic PD-1 in tumor growth.
(A) Western blot analysis of PD-1 expression of the H1299–PD-1 stable cell line. (B) Cell viability assay of H1299-EV and H1299–PD-1 stably expressing cells. (C) Flow cytometry analysis of the apoptotic cells in H1299-EV versus H1299–PD-1 stable cell line using annexin V. (D) Cell growth assay of H1299-EV and H1299–PD-1 cells. (E) Xenograft tumor growth of H1299-EV versus H1299–PD-1 stably expressing cells in nude mice. (F) The volume analysis of xenograft tumors based on (E). (G) The final weight of xenograft tumors based on (E). (H) Xenograft tumor growth of H1299-EV versus H1299–PD-1 stably expressing cells in B-NDG (NSG) mice. (I) The volume analysis of xenograft tumors based on (H). (J) The final weight of xenograft tumors based on (H). (K) IHC of xenograft tumor sections with indicated antibodies. (L) Correlation analysis of PD-1 expression and AKT/mTOR pathway activity in LUAD. HE, hematoxylin and eosin. Data were shown as means ± SD, n = 3 or 6. Photo credit: Zhijie Cao, Chinese Academy of Medical Sciences and Peking Union Medical College.
Activation of cancer cell–intrinsic PD-1 participates in p53-dependent tumor suppression
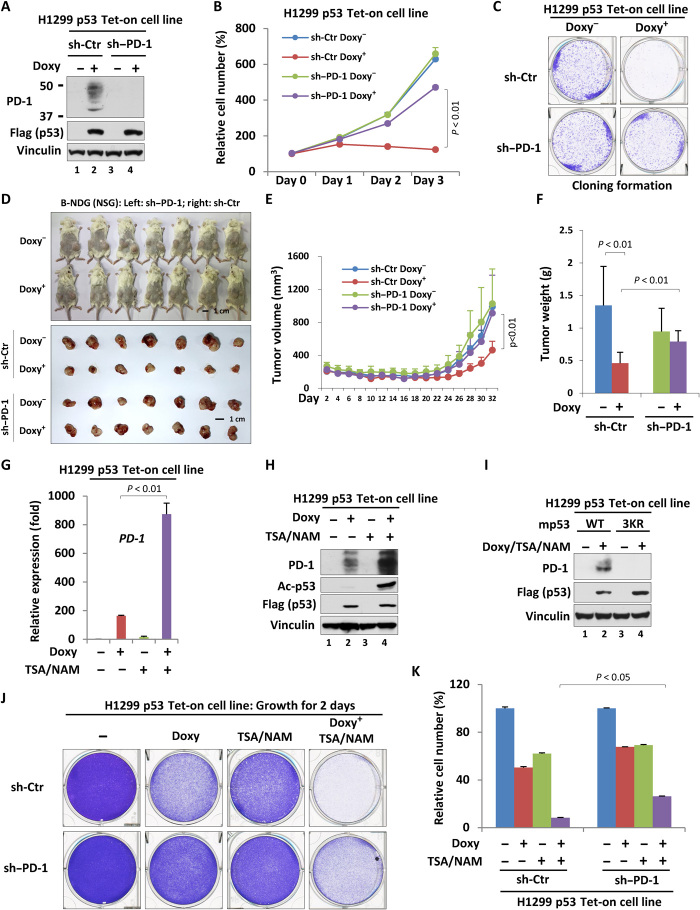
To investigate whether p53-mediated PD-1 expression is involved in the regulation of cancer cell behaviors, we generated a PD-1 stable knockdown cell line based on H1299 p53 Tet-on cells by short hairpin RNA technique. As shown in Fig. 6A, p53-induced PD-1 expression was almost completely abolished in PD-1 knockdown cells, indicating a good PD-1 knockdown efficiency. PD-1 depletion alone had no obvious effect on the growth of cells that were lack of p53 induction (Fig. 6B). In contrast, when p53 expression was induced by treating cells with doxycycline, PD-1 knockdown remarkably interrupted p53-dependent inhibition of cell proliferation (Fig. 6B and fig. S8A). Similar regulatory effect was also captured in a cloning formation assay in vitro (Fig. 6C). Next, we performed a xenograft tumor growth assay in which the control cells or PD-1 knockdown cells were subcutaneously injected into the B-NDG (NSG) mice that were then fed with or without doxycycline-containing diet. As expected, p53 expression by doxycycline resulted in the marked regression of the tumors in both tumor volume and tumor weight (Fig. 6, D to F). PD-1 knockdown significantly interrupted p53-mediated tumor shrinkage (Fig. 6, D to F), indicating that the intrinsic PD-1 transcriptional activation by p53 participates in p53-dependent tumor suppression in vivo. As our data showed that acetylation in DBD of p53 promotes p53-dependent PD-1 expression, we were then intrigued to evaluate whether histone deacetylase inhibitor (HDACi) synergizes with p53 activation to inhibit tumor growth through the regulation of PD-1. In contrast to doxycycline treatment that significantly elevated PD-1 transcription by inducing p53 expression, cells treated with HDACi [trichostatin A (TSA) and nicotinamide (NAM)] only exhibited a slightly increase in PD-1 expression (Fig. 6, G and H). However, doxycycline plus HDACi treatment further markedly promoted PD-1 expression, indicating an obvious synergistic effect of these two types of treatment on PD-1 transcription (Fig. 6, G and H). This synergistic effect was relied on the enhancement of the acetylation at K120/164 of p53 by HDACi, as the acetylation-defect mutant p53 failed to up-regulate PD-1 transcription in response to HDACi treatment (Fig. 6I). Consistent with the tumor-suppressive function of cancer cell–intrinsic PD-1, HDACi treatment further enhanced doxycycline-induced inhibition of cancer cell proliferation in control cells but less obvious in PD-1 knockdown cells (Fig. 6, J and K), suggesting that acetylated p53 may perform its tumor-suppressive function by, at least in part, regulating PD-1 expression in cells. Together, our data indicated that p53-regulated expression of cancer cell–intrinsic PD-1 significantly contributes to p53-mediated tumor suppression.
Fig. 6. Activation of cancer cell–intrinsic PD-1 participates in p53-dependent tumor suppression.
(A) Western blot analysis of PD-1 knockdown efficiency in the H1299 p53 Tet-on cell line. (B) Cell proliferation assay for three consecutive days of H1299 p53 Tet-on control or PD-1–depleted cells treated with or without doxycycline. (C) Cloning formation of H1299 p53 Tet-on control or PD-1 depleted cells treated with or without doxycycline. (D) Xenograft tumor growth analysis of PD-1 effects on p53-mediated tumor suppression in B-NDG (NSG) mice. (E) The volume analysis of xenograft tumors based on (D). (F) The final weight of xenograft tumors based on (D). (G and H) RT-qPCR (G) or Western blot (H) analysis of the mRNA or protein level of PD-1 in H1299 p53 Tet-on cells treated with doxycycline (for 24 hours) and/or TSA/NAM (for the last 6 hours). (I) Western blot analysis of PD-1 in H1299 p53 Tet-on (wild-type versus 3KR) cells treated with or without doxycycline (for 24 hours) and TSA/NAM (for the last 6 hours). (J) Cell proliferation analysis (48 hours) of H1299 p53 Tet-on control or PD-1–depleted cells treated with or without doxycycline. (K) Quantitation of cell proliferation based on (J). Data were shown as means ± SD, n = 3 or 7. Photo credit: Zhijie Cao, Chinese Academy of Medical Sciences and Peking Union Medical College.
DISCUSSION
It is conventionally thought that PD-1 is mainly expressed in immune cells. However, the emerging evidence in recent years argued that the PD-1 is also expressed in a subgroup of cancer cells, which may exert different types of functions on cancer cell behaviors, depending on the biological context (18–21, 23). Unlike the immune cell–expressed PD-1 whose regulation, especially in transcriptional level, has been extensively studied (24), how PD-1 is regulated in cancer cells is largely unknown. In this study, we demonstrated the tumor suppressor p53 as a master regulator that transcriptionally activates PD-1 in cancer cells. This regulation is largely reliant on the acetylation at K120 and K164 within DBD of p53, revealing a regulatory paradigm where the site-specific acetylation determines the selectivity of p53-driven transcriptional profile. In addition, our observations supported that the reexpression of ectopic PD-1 in lung cancer cells significantly suppresses tumor growth in an immunity-independent manner (Fig. 5, D to J), consistent with the recent study where the cancer cell–intrinsic PD-1 was reported as a tumor suppressor (23). However, unlike their observation, we did not find that depletion of PD-1 alone in H1299 cells promoted tumor growth (Fig. 6, D to F). This discrepancy might be attributed to different basal levels of endogenous PD-1 among cancer cells, as in our system, the basal protein level of endogenous PD-1 in H1299 cells is too low (fig. S6B). In support of this notion, we observed that the blockade of p53-induced expression of cancer cell–intrinsic PD-1 in H1299 cells could interrupt p53-mediated tumor regression, implying a functional importance of p53–PD-1 axis in tumor suppression (Fig. 6, D to F).
The PTMs, including phosphorylation and acetylation, represent an important layer of the mechanisms in the regulation of p53 functions (7). For example, the phosphorylation of p53 N-terminal serine or threonine residues stabilizes p53 by antagonizing p53-MDM2 interaction (5). In addition, the acetylation occurred within p53 C-terminal lysine clusters generally boosts p53 in both stability and transactivity by serving as a platform to mediate the association/dissociation of the cofactors to p53 (34–37). Moreover, accumulating evidence indicate that the acetylation within DBD of p53 may contribute to p53-dependent gene-specific transcription, which, in turn, determines the selectivity of p53-mediated biological processes, and consequently, the cell fates in a given context (28, 30, 38). In this study, we found that neither N-terminal phosphorylation nor C-terminal acetylation is necessary for p53-driven PD-1 expression. However, the acetylation at K120 and K164 on DBD largely contributes to p53-mediated PD-1 transcription, as the mutant p53 deficient in the acetylation at both K120 and K164 almost completely lost the ability to induce PD-1 expression (Fig. 3, B to D). This observation brought a new example to further emphasize the critical role of the DBD acetylation in controlling the transcriptional selectivity of p53.
In mechanism, the promoter recruitment of the acetyltransferases including p300, CBP, and TIP60 by p53 plays an important role in creating an open structure within local chromatin surrounding PD-1 promoter, thus facilitating p53-guided PD-1 transcription (Fig. 4, G to I). Because of their relatively broad spectrum of substrate adaptability, p300, CBP, and TIP60 can modify both p53 and histones effectively, which, therefore, most likely bridge the informational cross-talk between nonhistone p53 acetylation and histone acetylation in terms of the regulation of PD-1 expression. Although we cannot exclude the possibility that other cofactors, in addition to the acetyltransferase, also participate in K120/164 acetylation–dependent modulation of PD-1 transcription, our observations showed that the acetylation-defect p53 (2KR) was compromised to recruit p300, CBP, and TIP60 onto PD-1 promoter (Fig. 4, G and H), prompting us to speculate that K120/K164 acetylation contributes to the chromatin recruitment of these acetyltransferases, probably in a promoter-specific manner. Despite more study remains to dissect the detail mechanism by which K120/164 acetylation selectively regulates p53-mediated PD-1 expression, we, in current stage, would like to propose a model where the acetylated p53 at K120/164 catalyzed by p300, CBP, and TIP60 promotes its ability to recruit these acetyltransferase cofactors onto PD-1 promoter and consequently elicits PD-1 transcription by enhancing local chromatin acetylation (Fig. 4J).
The HDACis represent a group of chemical compounds that have already displayed quite potentials in clinical use for treating a range of diseases including cancer (39, 40). So far, more than 350 clinical trials have been performed by testing HDACi per se or the combination of HDACi with other types of therapeutic strategy in the treatment of various malignancies (41), and some HDACis such as vorinostat and romidepsin (also known as suberanilohydroxamic acid and depsipeptide, respectively) have been approved by the Food and Drug Administration for treatment of refractory cutaneous T cell lymphoma (42, 43). In this study, HDACi treatment under a p53-null cellular condition only induced a slightly increase in PD-1 expression (Fig. 6G). However, the PD-1 expression was significantly elevated by HDACi in the present of p53 (Fig. 6, G and H), which consequently contributed to the inhibition of cancer cell growth (Fig. 6, J and K). This observation indicated a synergistic effect by the combination of HDACi and p53 activation on PD-1 transcription. In addition, we observed that the acetylation-deficient p53 mutant failed to induce PD-1 expression in response to HDACi treatment, further indicating that HDACi up-regulated PD-1 via promoting p53 acetylation at K120 and K164 (Fig. 6I). On the basis of these observations, our study revealed a potential strategy for cancer therapy using HDACi upon p53 activation via stimulating cancer cell–intrinsic PD-1.
The recent studies by analyzing the TCGA and CCLE database revealed that the transcript of PD-1 is widespread among both tumor tissues and established cancer cell lines (23, 44). Consistent with this finding, we also successfully detected the reliable transcription of PD-1 in multiple cancer cell lines commonly used in our laboratory (fig. S6A). However, unlike the easily detectable of PD-1 transcript in these cancer lines, only U2OS and MOLT-4 cell lines exhibited PD-1 signals for its protein level, leaving others with undetectable protein level of PD-1 (fig. S6B). This discrepancy of PD-1 expression in cancer cells may reflect the unknown mechanisms by which the PD-1 was regulated by other layers including posttranscription, translation or even protein degradation. Actually, the proteasomal-dependent degradation of PD-1 in T cells has been recently reported (45, 46). It is noteworthy to evaluate whether this regulation on PD-1 stability was also applied to cancer cell–intrinsic PD-1. In this aspect, further studies may be carried on in different layers of regulation of cancer cell–intrinsic PD-1, which will be beneficial to broaden our understanding of PD-1 regulatory network in cancer cells. On the other aspect, as T cell–expressed PD-1 mainly functions as an immune checkpoint and our data suggested that p53 might regulate PD-1 in T cells (fig. S1, A and B), it could be interested in future to study whether p53 is involved in the regulation of immune response through modulating T cell–expressed PD-1. Together, our study uncovered an acetylation-involved p53–PD-1 regulatory axis in the modulation of cancer cell behaviors in an immunity-independent manner and raised a potential tumor therapy strategy by activating cancer cell–intrinsic PD-1 via the combination of HDACi and p53 induction.
MATERIALS AND METHODS
Cell culture, construct generation, transfection, and reagent treatment
H1299, U2OS, and A375 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Corning, 10-013-CVR) with 10% (v/v) fetal bovine serum (FBS; Gibco, 10099141) supplementation. The SW1990 cell line was cultured in L-15 [Cell Resource Center of Institute of Basic Medical Sciences–Chinese Academy of Medical Sciences (IBMS-CAMS)] with 10% FBS. MEFs were cultured in DMEM supplemented with 10% heat-inactivated FBS. The cell lines used in this project were originally purchased from American Type Culture Collection or Cell Resource Center of IBMS-CAMS and freshly thawed from our stock and cultured for no longer than 2 months. All cell lines were negative to mycoplasma contamination. The expressing constructs including p53, p300, CBP, TIP60, and PCAF were generated by cloning each CDS (coding sequence) into pcDNA3.1 vector (Invitrogen, K4800) or a modified pIRESneo2 vector (Clontech, 6938-1). The GCN5-expressing construct was gifted by W. Zhao (Peking University) To generate luciferase reporter, the annealed oligos containing wild-type or mutated p53-binding element of PD-1 were cloned into a pGL3-firefly luciferase vector (Promega, E1761). The construct expressing p53 with point mutation(s) was generated using a site-directed mutagenesis kit (Stratagene, 200521) according to the manufacturer’s protocol. To generate a p53-inducible cell line, the Flag-tagged p53–expressing cassette was subcloned into a modified pTRIPZ construct (GE Healthcare, RHS4750), and H1299 cells transfected with pTRIPZ-Flag-p53 plasmid were selected with puromycin (2 μg/ml; Invitrogen, A1113803) to build up the stable cell line. This inducible cell line was cultured in DMEM with 10% Tet System Approved FBS (Clontech, 631101). For stably expressing PD-1 in H1299 cells, the CDS of PD-1 was inserted into SFB-CT (S peptide-Flag-streptavidin binding peptide, C-terminal) destination construct (gifted by W. Wang, University of California Irvine) using a Gateway technique (Invitrogen, 1991274) to lastly obtain PD-1–SFB–expressing construct, or the CDS of PD-1 was inserted into pcDNA3.1/V5-His-TOPO vector (Invitrogen, K4800-01) to obtain PD-1–expressing construct (no tag) according to the manufacturer’s protocol. Empty vector or PD-1 construct was transfected into H1299 cells, and the cells were subjected to selection with puromycin/G418 (500 μg/ml, Sigma-Aldrich, H1720) to lastly obtain the stable cell lines. siRNA was synthesized from GenePharma Co. Ltd. All transfections including expressing construct and siRNA were performed using Lipofectamine 3000 (Invitrogen, L3000015) according to the manufacturer’s protocol. Doxycycline (Sigma-Aldrich, D9891) was used at 1 μg/ml, Cpt [Cell Signaling Technology (CST), 13637S] was used at 0.1 to 1 μM, TSA (Sigma-Aldrich, V900931) was used at 1 μM, and nicotinamide (Sigma-Aldrich, 72340) was used at 5 mM.
RNA extract, reverse transcription, and quantitative polymerase chain reaction
Sheared tissues or cultured cells were lysed by 1 ml of TRIzol reagent (Invitrogen, 15596018) for 5 min at room temperature and then centrifuged at 15,000 rpm for 15 min at 4°C. The supernatant (~400 μl) was carefully collected, and the equal volume of isopropanol was added into the supernatant to precipitate RNA at room temperature for 10 min. After centrifuging at 15,000 rpm for 10 min at 4°C, we got rid of the supernatant and washed the pellet (containing total RNA) once by 75% ethanol. After centrifuging at 15,000 rpm for 5 min at 4°C, we got rid of the supernatant and air-dried the pellet. We resuspended the pellet by appropriate volume of ribonuclease-free H2O. To get complementary DNA, 1 μg total RNA was reversely transcribed using the SuperScript IV VILO Master Mix (Invitrogen, 11756050) according to the manufacturer’s protocol. The relative expression of each gene was measured in Applied Biosystems 7500 Fast Real-Time PCR (polymerase chain reaction) System using the SYBR Green method (Abi, 4312704). The expression of Human or mouse β-Actin was used as an internal control.
RNA sequencing
Total RNA was first subjected to quality and integrity evaluation by a Bioanalyzer 2100 system (Agilent) to make sure that RNA integrity number is >8.0. Then, the libraries were generated using the NEBNext UltraTM RNA Library Prep Kit for Illumina [New England Biolabs (NEB), E7530] according to the manufacturer’s protocol. The library preparations were sequenced on Illumina HiSeq platform, and 150-bp paired-end reads were generated. The fastq-formatted raw data were first processed through in-house Perl scripts to obtain clean data, which were then aligned to the reference genome using Hisat2 v2.0.5. The reads number was counted by featureCounts v1.5.0-p3 and the fragments per kilobase of sequence per million mapped reads of each gene was calculated on the basis of the length of the gene and reads count mapped to each gene. Differential expression analysis was performed using DESeq2 R package (1.16.1). The heatmap was generated by pheatmap package of R software (v3.6.1). The GO enrichment analysis was performed using the STRING database (https://string-db.org). The threshold of statistical significance for GO enrichment analysis was set as false discovery rate ≤ 10%.
Coimmunoprecipitation and Western blot
Cells transfected with indicated expressing constructs for 24 hours were then lysed with a modified NP-40 buffer [50 mM tris-HCl (pH 8.0), 150 mM NaCl, 1% NP-40, 10 mM sodium butyrate, 1 mM MgCl2, benzonase (250 U/ml; Sigma-Aldrich, E1014), and 1× protease inhibitor (Sigma-Aldrich, P8340)] for 30 min on ice. After centrifuging at 15,000 rpm for 15 min at 4°C, the supernatant was carefully collected and subjected to precipitation by adding Anti-Flag Affinity Gel (Sigma-Aldrich, A2220) or Anti–HA (hemagglutinin) Agarose (Sigma-Aldrich, A2095) for further incubation at 4°C for 1 hour. After incubation, the beads were washed three times with BC100 buffer [20 mM tris-HCl (pH 7.3), 100 mM NaCl, 10% glycerol, 2 mM EDTA, and 0.1% Triton X-100], and the protein complex were then eluted by 1× Flag peptide (Sigma-Aldrich, F3290) or 1× HA peptide (Sigma-Aldrich, I2149) at room temperature for 1 hour. To assay the protein complex or crude cellular protein, the eluents or whole-cell lysates were denatured in 1× Laemmli buffer at 95°C for 5 min, separated by SDS–polyacrylamide gel electrophoresis (PAGE), and transferred into nitrocellulose membrane. The membrane was blocked with 5% nonfat milk {dissolved in TBS-T buffer [20 mM tris-HCl (pH 7.6), 137 mM NaCl, and 0.05% Triton X-100]}, then sequentially subjected to primary antibody incubation (dissolved in 1% nonfat milk), washed by TBS-T buffer, incubated with horseradish peroxidase–conjugated secondary antibody (dissolved in 1% nonfat milk), and washed by TBS-T buffer again. The membrane was lastly incubated with ECL (enhanced chemiluminescence) substrate (Pierce, 32106 or 34076) and underwent exposure.
Chromatin immunoprecipitation
Cells were fixed by 1% formaldehyde [in phosphate-buffered saline (PBS)] for 10 min at room temperature. After briefly washing by cold PBS, the fixed cells were lysed with ChIP lysis buffer [50 mM tris-HCl (pH 8.0), 5 mM EDTA, 1% SDS, and 1× protease inhibitor] for 10 min on ice. After sonication, the lysates were centrifuged at 15,000 rpm for 10 min at 4°C, and the supernatants were collected and diluted with dilution buffer [20 mM tris-HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 1× protease inhibitor) as 1:9 ratio. Preclean was performed by incubating diluted lysates with salmon sperm DNA–saturated protein A agarose (Millipore, 16-157) for 1 hour at 4°C. The precleaned lysates were aliquot and incubated with indicated antibodies overnight at 4°C after addition of saturated protein A agarose for further 2-hour incubation at 4°C. The agarose was washed with TSE I [20 mM tris-HCl (pH 8.0), 2 mM EDTA, 150 mM NaCl, 0.1% SDS, and 1% Triton X-100], TSE II [20 mM tris-HCl (pH 8.0), 2 mM EDTA, 500 mM NaCl, 0.1% SDS, and 1% Triton X-100], buffer III [10 mM tris-HCl (pH 8.0), 1 mM EDTA, 0.25 M LiCl, 1% DOC (Deoxycholate), and 1% NP-40], and buffer TE [10 mM tris-HCl (pH 8.0) and 1 mM EDTA], sequentially. The agarose-attached protein-DNA complex was eluted by elution buffer (1% SDS and 0.1 M NaHCO3) and subjected to reverse cross-link at 65°C for at least 6 hours. DNA was extracted by a PCR purification kit (QIAGEN, 28106). Real-time PCR was performed to detect relative enrichment of each protein or modification on indicated genes. Notably, the relative enrichment of histone modifications in a given genomic locus was normalized by total H3 enrichment in the same locus.
Electrophoretic mobility shift assay
The probe that containing p53-binding element of PD-1 promoter was labeled with 32P (PerkinElmer, BLU002Z250UC) through a T4 polynucleotide kinase–mediated 5′ end phosphorylation reaction (NEB, M0201), and the labeled probe was then purified using Bio-Spin column (Bio-Rad, 732-6223) according to the manufacturer’s protocol. For shift assay, the labeled probe together with or without the cold probe (unlabeled wild-type or mutated probe) was incubated with purified p53 in 1× EMSA buffer [10 mM Hepes (pH 7.6), 40 mM NaCl, 50 μM EDTA, 6.25% glycerol, 1 mM MgCl2, 1 mM spermidine, 1 mM dithiothreitol (DTT), bovine serum albumin (BSA; 50 ng/μl), and sheared single-strand salmon DNA (5 ng/μl)] for 20 min at room temperature. For supershift assay, α-p53 antibody (Santa Cruz Biotechnology, sc-126) was preincubated with purified p53 in the reaction system without probe for 30 min at room temperature, and then the labeled probe was added for further 20-min incubation. The complex was separated by 4% tris-borate EDTA–PAGE and visualized by autoradiography.
Luciferase assay
A firefly reporter containing wild-type or mutated p53-binding element of PD-1 promoter and a Renilla control reporter were cotransfected with wild-type or mutant p53-expressing constructs into H1299 cells for 48 hours, and the relative luciferase activity was measured by the Dual-Luciferase Reporter Assay System according to the manufacturer’s protocol (Promega, E1910).
Cell death analysis
Cells were digested by 0.25% trypsin (Gibco, 25200114) for a few minutes, and the digestion was stopped by adding complete DMEM medium. The cell-containing medium was thoroughly pipetted up and down several times to form single-cell suspensions. For viability evaluation, we mixed an equal volume of cell suspensions and 0.4% trypan blue solution (Gibco, 15250061) well and transferred them into chamber slides (Invitrogen, C10312). Cell viability was measured as the percentage of cells excluded by trypan blue using the Countess II Automated Cell Counter (Invitrogen, AMQAX1000). For apoptosis assay, the cells were incubated with fluorescein isothiocyanate–conjugated annexin V (BD Biosciences, 556547) and subjected to fluorescence-activated cell sorting analysis according to the manufacturer’s protocol.
Xenograft tumor growth
A total of 3 × 106 to 5 × 106 living cells were mixed with Matrigel (Corning, 354248) as 1.5:1 ratio for total 200-μl volume. The cell-Matrigel mixture was then subcutaneously injected into the nude mice (NU/NU; 6 weeks old, female; strain 088; Charles River Laboratories) or B-NDG (NSG) mice (NOD-Prkdcscid IL2rgtm1/Bcgen; 6 weeks old, female; Biocytogen; equally to NSG strain). The tumor growth was measured every 2 days, and the volume was counted as follows: volume = (width)2 × length / 2. After 3 to 4 weeks, the mice were euthanized, and the final tumor weight was measured. Maintenance and experimental procedures of mice were approved by the Institutional Animal Care and Use Committee of CAMS and Peking Union Medical College.
Immunohistochemistry
The xenograft tumor tissues were fixed in 4% paraformaldehyde (PFA) overnight after the tissues were embedded in paraffin and cut into serials of 5-μm sections. The sections were stained with indicated antibodies and visualized by DAB (3,3′-Diaminobenzidine) exposure. Hematoxylin and eosin was used for counterstaining to observe the morphology of each tissue sample. The sections were photographed using a ZEISS Axio Scope A1 microscope.
Cell proliferation and cloning formation assay
For cell proliferation, 1 × 105 living cells were seed into a six-well plate with a total of three replicates for each sample. We monitored cell growth for three consecutive days by crystal violet staining. For details, the cells were fixed with 4% PFA for 20 min at room temperature and stained with 0.1% crystal violet for 30 min at room temperature. We gently rinsed cells by double-distilled H2O three times, air-dried, and took picture. The cell-containing crystal violet was extracted by 10% acetic acid at room temperature for 30 min. The relative cell number was calculated by measuring the absorption of the extracted crystal violet at an optical density at 590 nm. For cloning formation, 500 to 1000 cells were seeded into a six-well plate and left cell growth/cloning formation under indicated treatment for 7 to 14 days, followed by staining with crystal violet as mentioned above.
RNA decay analysis
The cells were treated with 10 μM actinomycin D (Selleck, S8964) for 0, 2, 4, 6, and 8 hours, and the mRNA at each time point was extracted. The relative expression of the target gene was measured by reverse transcription quantitative PCR (RT-qPCR), and the levels of the mRNA at each time point were normalized to time zero. The half-life the target mRNA was calculated on the basis of the mRNA decay curve.
Immunofluorescence
The cells were fixed in 4% PFA and permeabilized with 0.1% Triton X-100. After blocking by 1% BSA, the cells were stained with indicated antibody, and the target molecules were visualized using confocal microscope. 4′,6-Diamidino-2-phenylindole (Solarbio, C0050) was used for counterstaining of the nucleus.
Cellular fractionation
The cells were sequentially lysed on ice by buffer A [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, 0.15% NP-40, and 1× protease inhibitor] for 10 min, buffer B [20 mM Hepes (pH 7.9), 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5% NP-40, and 1× protease inhibitor] for 15 min, and buffer C [20 mM tris-HCl (pH 8.0), 5 mM NaCl, 2.5 mM CaCl2, and micrococcal nuclease (30 U/ml)] for 15 min, to extract cytoplasm, nucleus, and chromatin fraction.
Flow cytometry
The cells were harvested by trypsin digestion and briefly centrifuging. A total of 1 × 105 cells were stained with phycoerythrin-conjugated control or target-specific antibody and analyzed by a Beckman Coulter CytoFLEX platform.
Antibodies
Antibodies used were as follows: PD-1 (CST, 86163S and 84651S; eBioscience, 12-9969-41), mouse immunoglobulin 1κ (IgG1κ) isotype control (eBioscience, 12-4714-81), p53 (Santa Cruz Biotechnology, sc-126; Leica Biosystems, P53-CM5P; CST, 2524S), p21 (Santa Cruz Biotechnology, sc-6246), vinculin (Sigma-Aldrich, V9131), normal IgG (Santa Cruz Biotechnology, sc-2025 and sc-2027), p300 (CST, 54062S), CBP (Santa Cruz Biotechnology, sc-7300-x), Flag (Sigma-Aldrich, A2220 and F7425), H3K18ac (Abcam, ab1191), H3K27ac (Abcam, ab4729), H4K16ac (Millipore, 07-329), HA (Sigma-Aldrich, A2095; Roche, 11867423001), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Santa Cruz Biotechnology, sc-32233), HDAC1 (Santa Cruz Biotechnology, sc-81598), H3 (CST, 4499T; Abcam, ab1791), Ki-67 (CST, 9449S), p-AKT-S473 (CST, 4060S), p-mTOR-S2448 (CST, 2976S), Ac-p53-K120 (homemade), and Ac-p53-K164 (homemade).
Primers
For RT-qPCR, the following primers were used: human PD-1 (forward, CCAGGATGGTTCTTAGACTCCC; reverse, TTTAGCACGAAGCTCTCCGAT), mouse Pd-1 (forward, ACCCTGGTCATTCACTTGGG; reverse, CATTTGCTCCCTCTGACACTG), human p21 (forward, CTGTCACTGTCTTGTACCCTTGT; reverse, GGTAGAAATCTGTCATGCTGGT), mouse p21 (forward, GCTGTCTTGCACTCTGGTGTCT; reverse, AATCTGCGCTTGGAGTGATAGAA), human MDM2 (forward, GGCGATTGGAGGGTAGACCT; reverse, CACATTTGCCTGGATCAGCA), human p53 (forward, CAGCACATGACGGAGGTTGT; reverse, TCATCCAAATACTCCACACGC), human p300 (forward, CTTCCCCACTGTCGCACAAT; reverse, TTTGTCGAGAAGATGCACAGTGT), human CBP (forward, TTCCAGCAGGAGGAATAACAACA; reverse, GTTGGAGCCATCGTTCATCA), human TIP60 (forward, GACGGAAGCGAAAATCGAATTG; reverse, GGTGCTGACGGTATTCCATCA), human TIGAR (forward, GACTTCGGGAAAGGAAATACG; reverse, CACTCTTCCCTGGCTGCTTTG), human β-actin (forward, GGCCAACCGCGAGAAGAT; reverse, GCCAGAGGCGTACAGGGATA), and mouse β-actin (forward, GGCTGTATTCCCCTCCATCG; reverse CCAGTTGGTAACAATGCCATGT). For ChIP-qPCR, the following primers were used: human PD-1 (forward, AGGGAAGGAGAGTGGGTGACA; reverse, GCGGGCACAGGGAGAAA), human p21 (forward, AGCAGGCTGTGGCTCTGATT; reverse, CAAAATAGCCACCAGCCTCTTCT), human GAPDH (forward, CGGGATTGTCTGCCCTAATTAT; reverse, GCACGGAAGGTCACGATGT), and human MDM2 (forward, CGGAAGTCAAGTTCAGACACGTTCCG; reverse, CCTCCAATCGCCACTGAACAC).
Oligos
Oligos used were as follows: control siRNA, UUCUCCGAACGUGUCACGU; human si-p300, AGGAGGAAGAAGAGAGAAA; human si-CBP, GCAAGAAUGCCAAGAAGAA; human si-TIP60, ACGGAAGGUGGAGGUGGUU; human si-p53, Dharmacon, L-003329-00-0005 (SMARTPool); human PD-1 wild-type probe for EMSA (forward, GTGCCCGCCCCCTACTCCAGGACATGTGTCCAAGCCCTGGCAGGTGGAATTTTGGGGGCAGGGCCTTGGTGGTGAGGAGACCTTCCAGGGGTCTGATAGCATCTCCCATCTCAGAGCCCA; reverse, TGGGCTCTGAGATGGGAGATGCTATCAGACCCCTGGAAGGTCTCCTCACCACCAAGGCCCTGCCCCCAAAATTCCACCTGCCAGGGCTTGGACACATGTCCTGGAGTAGGGGGCGGGCAC); and human PD-1 Mut probe for EMSA (forward, GTGCCCGCCCCCTACTCCAAATTTTGGGGGCAGGGCCTTGGTGGTGAGGAGACCTTCCAGGGGTCTGATAGCATCTCCCATCTCAGAGCCCA; reverse, TGGGCTCTGAGATGGGAGATGCTATCAGACCCCTGGAAGGTCTCCTCACCACCAAGGCCCTGCCCCCAAAATTTGGAGTAGGGGGCGGGCAC).
Statistical analysis
The results were presented as the means ± SD. The difference was determined using a two-tailed, unpaired Student t test and one-way or two-way analysis of variance (ANOVA) with Bonferroni post hoc test. All statistical analysis was performed using GraphPad Prism software. P < 0.05 was denoted as statistically significant.
Acknowledgments
We are grateful to W. Zhou (Analytical Instrumentation Center, Peking University) and X. Li (Westlake University) for the assistance in MS analysis. Funding: This research was supported by the National Natural Science Foundation of China (grant nos. 81872311 and 82073132), the Beijing Municipal Natural Science Foundation (grant no. 7192126), the National Key R&D Program of China (grant nos. 2019YFC1005200 and 2019YFC1005201), and the CAMS Innovation Fund for Medical Sciences (grant nos. 2017-I2M-1-008 and 2018-I2M-1-002). Author contributions: Z.C., W.G., and D.W. conceived and designed the research. Z.C. and D.W. performed the experiments. N.K., Y.L., W.X., H.Y., M.Z., J.W., Z.W., and X.Y. contributed new reagents/analytic tools. Z.C., W.-G.Z., W.G., and D.W. analyzed and interpreted the data. Z.C., W.G., and D.W. wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf4148/DC1
REFERENCES AND NOTES
- 1.Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Bieging K. T., Mello S. S., Attardi L. D., Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A. Jr., Butel J. S., Bradley A., Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B., Lane D., Levine A. J., Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Brady C. A., Jiang D., Mello S. S., Johnson T. M., Jarvis L. A., Kozak M. M., Kenzelmann Broz D., Basak S., Park E. J., McLaughlin M. E., Karnezis A. N., Attardi L. D., Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruse J. P., Gu W., Modes of p53 regulation. Cell 137, 609–622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharpe A. H., Pauken K. E., The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18, 153–167 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day C. L., Kaufmann D. E., Kiepiela P., Brown J. A., Moodley E. S., Reddy S., Mackey E. W., Miller J. D., Leslie A. J., DePierres C., Mncube Z., Duraiswamy J., Zhu B., Eichbaum Q., Altfeld M., Wherry E. J., Coovadia H. M., Goulder P. J., Klenerman P., Ahmed R., Freeman G. J., Walker B. D., PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443, 350–354 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Trautmann L., Janbazian L., Chomont N., Said E. A., Gimmig S., Bessette B., Boulassel M.-R., Delwart E., Sepulveda H., Balderas R. S., Routy J.-P., Haddad E. K., Sekaly R.-P., Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12, 1198–1202 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Zou W., Wolchok J. D., Chen L., PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 8, 328rv4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha J.-H., Chan L.-C., Li C.-W., Hsu J. L., Hung M.-C., Mechanisms controlling PD-L1 expression in cancer. Mol. Cell 76, 359–370 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalbasi A., Ribas A., Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 20, 25–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian S. L., Taube J. M., Pardoll D. M., Neoadjuvant checkpoint blockade for cancer immunotherapy. Science 367, eaax0182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robert C., Long G. V., Brady B., Dutriaux C., Maio M., Mortier L., Hassel J. C., Rutkowski P., McNeil C., Kalinka-Warzocha E., Savage K. J., Hernberg M. M., Lebbe C., Charles J., Mihalcioiu C., Chiarion-Sileni V., Mauch C., Cognetti F., Arance A., Schmidt H., Schadendorf D., Gogas H., Lundgren-Eriksson L., Horak C., Sharkey B., Waxman I. M., Atkinson V., Ascierto P. A., Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Rizvi N. A., Mazieres J., Planchard D., Stinchcombe T. E., Dy G. K., Antonia S. J., Horn L., Lena H., Minenza E., Mennecier B., Otterson G. A., Campos L. T., Gandara D. R., Levy B. P., Nair S. G., Zalcman G., Wolf J., Souquet P. J., Baldini E., Cappuzzo F., Chouaid C., Dowlati A., Sanborn R., Lopez-Chavez A., Grohe C., Huber R. M., Harbison C. T., Baudelet C., Lestini B. J., Ramalingam S. S., Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): A phase 2, single-arm trial. Lancet. Oncol. 16, 257–265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleffel S., Posch C., Barthel S. R., Mueller H., Schlapbach C., Guenova E., Elco C. P., Lee N., Juneja V. R., Zhan Q., Lian C. G., Thomi R., Hoetzenecker W., Cozzio A., Dummer R., Mihm M. C. Jr., Flaherty K. T., Frank M. H., Murphy G. F., Sharpe A. H., Kupper T. S., Schatton T., Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 162, 1242–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Li X., Liu S., Guo L., Zhang B., Zhang J., Ye Q., Programmed cell death-1 (PD-1) checkpoint blockade in combination with a mammalian target of rapamycin inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 66, 1920–1933 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Pu N., Gao S., Yin H., Li J. A., Wu W., Fang Y., Zhang L., Rong Y., Xu X., Wang D., Kuang T., Jin D., Yu J., Lou W., Cell-intrinsic PD-1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 460, 42–53 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Du S., McCall N., Park K., Guan Q., Fontina P., Ertel A., Zhan T., Dicker A. P., Lu B., Blockade of tumor-expressed PD-1 promotes lung cancer growth. Oncoimmunology 7, e1408747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Harrison D. L., Song Y., Ji J., Huang J., Hui E., Antigen-presenting cell-intrinsic PD-1 neutralizes PD-L1 in cis to attenuate PD-1 signaling in T cells. Cell Rep. 24, 379–390.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Yang X., Zhang C., Wang Y., Cheng T., Duan L., Tong Z., Tan S., Zhang H., Saw P. E., Gu Y., Wang J., Zhang Y., Shang L., Liu Y., Jiang S., Yan B., Li R., Yang Y., Yu J., Chen Y., Gao G. F., Ye Q., Gao S., Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl. Acad. Sci. U.S.A. 117, 6640–6650 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bally A. P., Austin J. W., Boss J. M., Genetic and epigenetic regulation of PD-1 expression. J. Immunol. 196, 2431–2437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon K. W., Byun S., Kwon E., Hwang S. Y., Chu K., Hiraki M., Jo S.-H., Weins A., Hakroush S., Cebulla A., Sykes D. B., Greka A., Mundel P., Fisher D. E., Mandinova A., Lee S. W., Control of signaling-mediated clearance of apoptotic cells by the tumor suppressor p53. Science 349, 1261669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner A., Bulyk M. L., Jambhekar A., Lahav G., The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 20, 199–210 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Liu Y., Tavana O., Gu W., p53 modifications: Exquisite decorations of the powerful guardian. J. Mol. Cell Biol. 11, 564–577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang Y., Luo J., Zhang W., Gu W., Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Tang Y., Zhao W., Chen Y., Zhao Y., Gu W., Acetylation is indispensable for p53 activation. Cell 133, 612–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li T., Kon N., Jiang L., Tan M., Ludwig T., Zhao Y., Baer R., Gu W., Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bensaad K., Tsuruta A., Selak M. A., Vidal M. N., Nakano K., Bartrons R., Gottlieb E., Vousden K. H., TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Parry R. V., Chemnitz J. M., Frauwirth K. A., Lanfranco A. R., Braunstein I., Kobayashi S. V., Linsley P. S., Thompson C. B., Riley J. L., CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 25, 9543–9553 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., Halazonetis T. D., Berger S. L., Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8, 1243–1254 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Mujtaba S., He Y., Zeng L., Yan S., Plotnikova O., Sachchidanand, Sanchez R., Zeleznik-Le N. J., Ronai Z., Zhou M.-M., Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell 13, 251–263 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Cai W., Su L., Liao L., Liu Z. Z., Langbein L., Dulaimi E., Testa J. R., Uzzo R. G., Zhong Z., Jiang W., Yan Q., Zhang Q., Yang H., PBRM1 acts as a p53 lysine-acetylation reader to suppress renal tumor growth. Nat. Commun. 10, 5800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., Kon N., Lasso G., Jiang L., Leng W., Zhu W.-G., Qin J., Honig B., Gu W., Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538, 118–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sykes S. M., Mellert H. S., Holbert M. A., Li K., Marmorstein R., Lane W. S., McMahon S. B., Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu W. S., Parmigiani R. B., Marks P. A., Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene 26, 5541–5552 (2007). [DOI] [PubMed] [Google Scholar]
- 40.West A. C., Johnstone R. W., New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30–39 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falkenberg K. J., Johnstone R. W., Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Mann B. S., Johnson J. R., Cohen M. H., Justice R., Pazdur R., FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist 12, 1247–1252 (2007). [DOI] [PubMed] [Google Scholar]
- 43.VanderMolen K. M., McCulloch W., Pearce C. J., Oberlies N. H., Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): A natural product recently approved for cutaneous T-cell lymphoma. J. Antibiot. 64, 525–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao H., Wang H., Li C., Fang J.-Y., Xu J., Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front. Immunol. 9, 1774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng X., Liu X., Guo X., Jiang S., Chen T., Hu Z., Liu H., Bai Y., Xue M., Hu R., Sun S.-C., Liu X., Zhou P., Huang X., Wei L., Yang W., Xu C., FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature 564, 130–135 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Zhou X. A., Zhou J., Zhao L., Yu G., Zhan J., Shi C., Yuan R., Wang Y., Chen C., Zhang W., Xu D., Ye Y., Wang W., Shen Z., Wang W., Wang J., KLHL22 maintains PD-1 homeostasis and prevents excessive T cell suppression. Proc. Natl. Acad. Sci. U.S.A. 117, 28239–28250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf4148/DC1