Restoring huntingtin-mediated intracellular trafficking rescues Huntington disease mice.
Abstract
Huntington disease (HD) damages the corticostriatal circuitry in large part by impairing transport of brain-derived neurotrophic factor (BDNF). We hypothesized that improving vesicular transport of BDNF could slow or prevent disease progression. We therefore performed selective proteomic analysis of vesicles transported within corticostriatal projecting neurons followed by in silico screening and identified palmitoylation as a pathway that could restore defective huntingtin-dependent trafficking. Using a synchronized trafficking assay and an HD network-on-a-chip, we found that increasing brain palmitoylation via ML348, which inhibits the palmitate-removing enzyme acyl-protein thioesterase 1 (APT1), restores axonal transport, synapse homeostasis, and survival signaling to wild-type levels without toxicity. In human HD induced pluripotent stem cell–derived cortical neurons, ML348 increased BDNF trafficking. In HD knock-in mice, it efficiently crossed the blood-brain barrier to restore palmitoylation levels and reverse neuropathology, locomotor deficits, and anxio-depressive behaviors. APT1 and its inhibitor ML348 thus hold therapeutic interest for HD.
INTRODUCTION
Huntington disease (HD) is caused by the abnormal expansion of a polyglutamine tract in the N-terminal domain of the huntingtin (HTT) protein (1), which interferes with its native functions (2–4). Because the mutant protein’s abnormal conformation renders it resistant to clearance, a great deal of research has been devoted to finding a safe and effective means to reduce the accumulation of mutant HTT (mHTT). Unfortunately, thus far, none of the drugs tested for reducing mutant HTT levels has yet proven useful in patients. In part, this is because loss of HTT’s wild-type (WT) functions contributes substantially to HD (5): HTT is a scaffolding protein that interacts directly with at least 400 different proteins to regulate various cellular functions from autophagy and ciliogenesis to transcription and axonal transport (2). Defects in these pathways are found in HD mutation carriers even during fetal development (6). We therefore hypothesized that restoring HTT-dependent pathways affected in HD could provide important neuroprotective effects that might delay the appearance of symptoms or slow disease progression.
HD pathogenesis centers on the corticostriatal circuitry, where early synaptic deficits precede neuronal death in both animal models and patients with HD (7–9). The corticostriatal-projecting neurons provide trophic support to striatal neurons by synthesizing and transporting brain-derived neurotrophic factor (BDNF) (2, 10–12). Studies using microfluidic devices to reconstitute the corticostriatal circuit confirm that the presynaptic cortical compartment is essential for proper functioning of the network (13). Reducing BDNF solely in corticostriatal neurons is sufficient to cause degeneration of both the striatum and the cortex in mice (14). In corticostriatal-projecting neurons affected by HD, not only is there a reduction in axonal BDNF transport, but trafficking between the endoplasmic reticulum (ER) and Golgi apparatus is also reduced, as is the secretion of vesicles from the Golgi (15–18).
Given this impairment of corticostriatal trafficking in HD, we sought to determine whether there are any HTT-dependent pathways or targets that could promote BDNF transport from the cortex to the striatum (18). We combined proteomic analyses of corticostriatal vesicles with in silico screening of the COPII pathway and HTT-specific interactors and found that palmitoylation, previously reported to be deficient in HD (19), is involved in corticostriatal BDNF trafficking. Restoring brain palmitoylation by inhibiting the acyl-protein thioesterase 1 (APT1) with the brain-permeable and selective molecule ML348 rescued trafficking in HD neurons, restored proper functioning of an HD circuit-on-a-chip, and ameliorated the behavioral phenotype and neuropathology of HD mice.
RESULTS
ER to plasma membrane trafficking is altered in HD neuronal cells
To focus on intracellular dynamics between the ER and plasma membrane in HD neuronal cells, we used the Retention Using Selective Hooks (RUSH) system (Fig. 1A) (20). This two-state assay can track the synchronous release of various cargoes from specific intracellular compartments, and it allows compounds that modify intracellular trafficking to be evaluated in real time (20). Here, we tracked the movement of glycosylphosphatidylinositol (GPI)–anchored mCherry fused to streptavidin-binding peptide (SBP). When expressed in cells, GPI-SBP-mCherry first localizes to the ER and then rapidly moves to the Golgi and then to the plasma membrane. If one expresses a hook protein composed of the KDEL motif fused to streptavidin in the same cells, GPI-SBP-mCherry remains at the ER (Fig. 1, A and B). Adding biotin to cells expressing both GPI-SBP-mCherry and KDEL-streptavidin causes the GPI-SBP-mCherry reporter to be released from the KDEL hook, traffic to the Golgi apparatus, and then traffic to the plasma membrane.
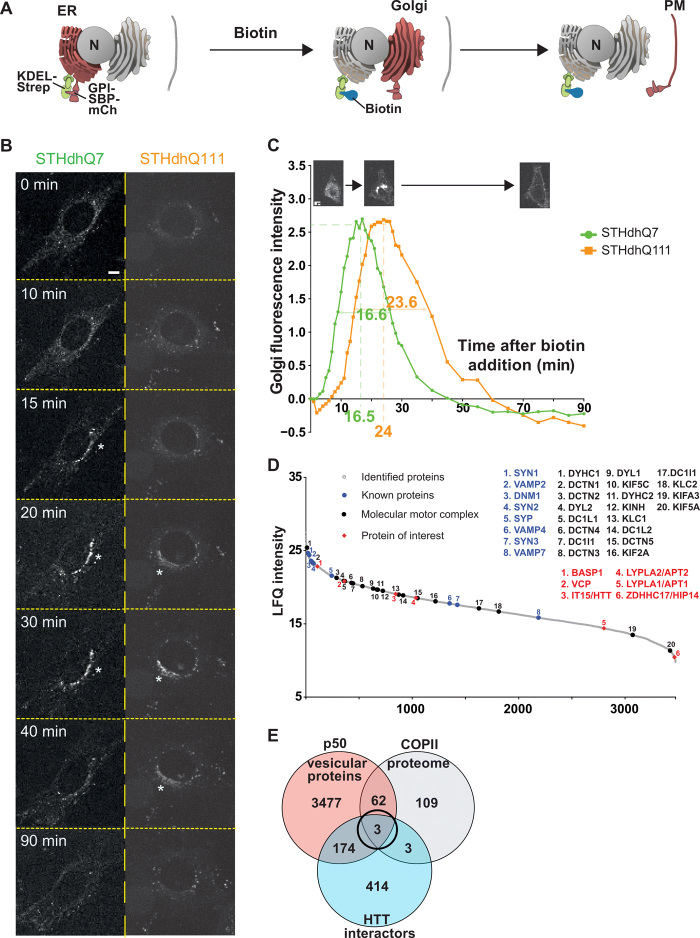
Fig. 1. Screening for modifiers of intracellular trafficking in Huntington disease.
(A) Schematic of the RUSH system KDEL-GPI-mCherry. GPI is fused with SBP and a fluorescent tag m-Cherry (mCh) (Reporter, red). The reporter is retained in the ER thanks to the Hook part (KDEL fused to streptavidin, green). The addition of biotin (blue) releases the reporter and allows trafficking of GPI-SBP-mCherry through the secretory pathway (Golgi and plasma membrane). (B) Localization of GPI-KDEL-mCherry at different time points after addition of biotin to STHdhQ7 and STHdhQ111 cells (scale bar, 10 μm). (C) Quantification of Golgi fluorescence in STHdhQ7 cells (green) and STHdhQ111 cells (orange). The green and orange numbers beneath the x axis and within the curves represent, respectively, the peak of Golgi fluorescence and the time to cross the Golgi in STHdhQ7 (N = 46) and STHdhQ111 cells (N = 54). (D) Proteins identified in the motile vesicle protein fraction were ranked by intensity and plotted according to their relative abundance (gray dots). Molecular motors and associated proteins (back dots) as well as previously identified vesicular proteins (blue dots) are among the most abundant proteins. HTT, HTT-interacting proteins present in the COPII proteome [brain abundant membrane attached signal protein 1 (BASP1), valosin-containing protein (VCP), and ZDHHC17], and APTs were identified as vesicular residents (red diamonds). (E) Venn diagram showing the overlap of proteomic (orange) and in silico screening of the HTT interactome (blue) and the COPII secretory pathway (gray) that identified three vesicular residents.
To follow these dynamics in an HD context, we transfected the GPI-SBP-mCherry and KDEL-streptavidin constructs in immortalized neurons from HdhQ111/Q111 and HdhQ7/Q7 mouse striata (hereafter referred to as STHdhQ111 and STHdhQ7 cells). These are the only available cells of neuronal origin that express endogenous levels of WT or mutant HTT (21). We tracked GPI-SBP-mCherry trafficking by real-time imaging and measured mCherry fluorescence in the Golgi apparatus over a 90-min period (Fig. 1, B and C). We found that fluorescence peaked at 16.5 min in STHdhQ7 cells, but not until 24 min in STHdhQ111 cells. This reflects delayed trafficking of GPI from the ER to Golgi and also a delay in GPI leaving the Golgi, as revealed by the longest period of GPI fluorescence in the Golgi (16.6 versus 23.6 min). These findings are in agreement with previous observations that trafficking between the ER, Golgi, and plasma membrane is hindered in HD (12, 15, 18, 22).
Proteomics and in silico screening indicate that palmitoylation is involved in HTT-mediated trafficking
To identify modifiers of ER-Golgi-plasma membrane trafficking specifically in corticostriatal neurons, we first needed to enrich vesicular fractions that are transported within corticostriatal projecting neurons. We therefore selected Thy1:p50-GFP transgenic mice (23), which express the dynactin subunit dynamitin (p50) fused to green fluorescent protein (GFP) under the neuronal promoter Thy1; this GFP construct is expressed at low enough levels that axonal transport is not affected (23). Analysis of GFP expression in these mice suggested discrete expression in some cortical regions (fig. S1A) (23). We delineated the pattern of cortical expression of p50-GFP in the Thy1:p50-GFP transgenic mice and found p50-GFP enriched in cortical layer V neurons that project to the striatum (fig. S1, B and C). These GFP-positive cells were immunopositive for Ctip2 but not Cux1, indicating their identity as layer V projecting corticostriatal neurons. We next immunostained brain sections from Thy1:p50-GFP transgenic mice for ER and Golgi markers and found p50-GFP in both compartments (fig. S2A). Last, we coexpressed p50-GFP and BDNF-mCherry in cortical neurons and observed significant cotrafficking of p50-GFP–containing organelles with BDNF-mCherry–containing vesicles (fig. S2B). This confirms the suitability of Thy1:p50-GFP transgenic mice for studying intracellular trafficking within corticostriatal projecting neurons.
We therefore dissected cortices from 2-month-old Thy1:p50-GFP mice and subjected them to subcellular fractionation and magnetic immunopurification to selectively enrich for small vesicles associated with molecular motors. We had previously used this approach to demonstrate that whole glycolysis occurs on motile vesicles (24). We subjected the purified motile vesicles to liquid chromatography–tandem mass spectrometry (LC-MS/MS) after separation by SDS–polyacrylamide gel electrophoresis (PAGE) and identified 3476 constituent proteins (table S1). Intensity ranking of the proteins revealed enrichment for synaptic proteins, molecular motors, and their associated proteins (Fig. 1D), in agreement with our previous study (24). To narrow down the list of possible targets that would be selectively altered in ER-to-Golgi trafficking, we cross-referenced this list with the recently published mammalian COPII vesicle core proteome (25) [COPII-coated vesicles promote anterograde ER-to-Golgi transport of newly synthesized secretory proteins and proteins destined for organelles other than the ER (26)]. This core proteome consists of 109 COPII-associated proteins, 74 of which appeared in our vesicular proteome (Fig. 1E and table S1). We then cross-referenced this shorter list to the HTT-interacting protein database HIPPIE (27) to identify proteins whose function might be regulated by HTT or altered in HD. This yielded only three proteins: BASP1 (brain abundant membrane attached signal protein 1), VCP (valosin-containing protein), and ZDHHC17 (Zinc Finger DHHC-Type Palmitoyltransferase 17) (Fig. 1E). BASP1 associates with organelles (28); VCP has been reported to regulate degradation processes (29, 30). ZDHHC17, also known as HTT-interacting Protein 14 (HIP14), is a palmitoyl-transferase (31) that induces palmitoylation of specific substrates.
We decided to focus on palmitoylation for several reasons. First, posttranslational protein-lipid modification by palmitoyl-transferases such as HIP14 plays an important role in regulating trafficking of various proteins in cells, including neuronal proteins such as PSD95, GAD65, synaptotagmin I, and SNAP25 (19, 32). Second, ZDHHC17/HIP14 was previously reported to be dysregulated in HD (33). Third, mice lacking HIP14 have a phenotype that overlaps with HD (cell death in the striatum, reduced numbers of corticostriatal synapses, and motor deficits) (19). Last but not least, palmitoylation is an enzymatic process that can be reversed by depalmitoylases a (dePAT) lso known as acyl-protein thioesterases or APTs—and our proteomic analysis turned up both APT1 and APT2 (Fig. 1D).
Genetic or pharmacological inhibition of APT1 restores intracellular dynamics in HD neurons
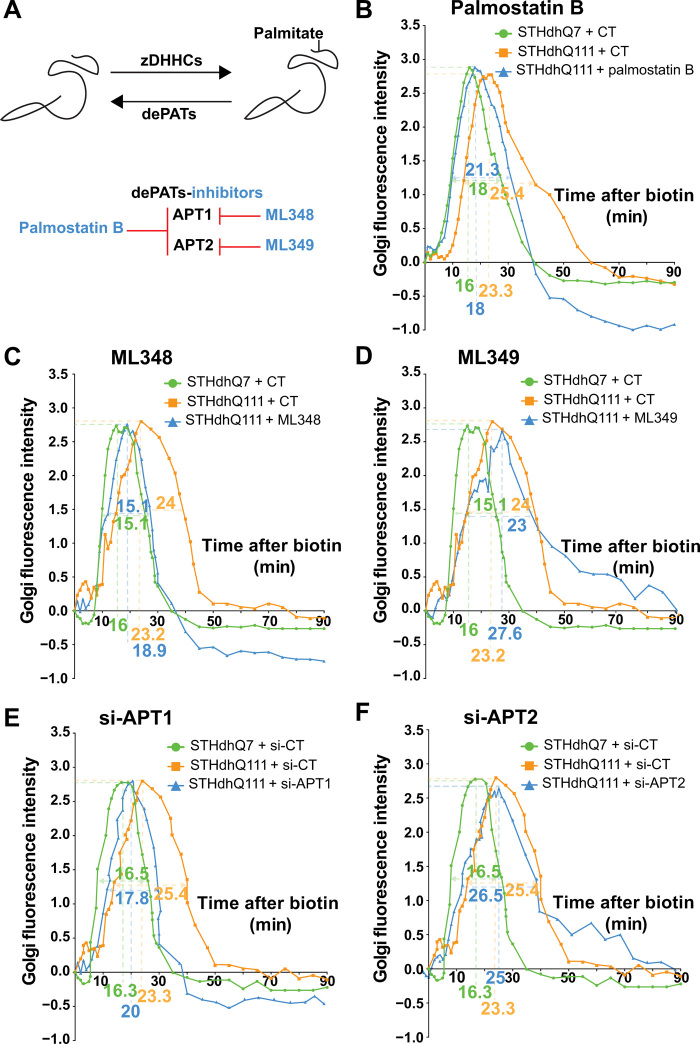
There are currently no drugs that enhance HIP14 activity. Moreover, HIP14 overexpression in neurons led to morphological changes and the formation of stress vacuoles (fig. S3A). In contrast, APT1 and APT2 can be inhibited by palmostatin B (Fig. 2A) (34, 35). We therefore tested the effect of palmostatin B on intracellular kinetics using the RUSH system. Palmostatin B restored ER-to-Golgi kinetics and decreased the time for fluorescence to exit the Golgi (Fig. 2B). Because palmostatin B targets both APT1 and APT2 with similar affinities (IC50APT1 = 5.4 nM and IC50APT2 = 37.7 nM), we investigated whether its effect was attributable to one specific dePAT by testing molecules specific for either APT1 (ML348) or APT2 (ML349) (34). Notably, treatment with ML348 rescued intracellular kinetics back to control levels, while ML349 did not. This suggested that inhibiting APT1 rather than APT2 should rescue the defective ER-to-Golgi to plasma membrane trafficking in HD cells (Fig. 2, C and D).
Fig. 2. Inhibiting APT1 restores intracellular dynamics in Huntington disease cells.
(A) Schematic representation of substrate palmitoylation and depalmitoylation induced by zDHHCs and dePATs, respectively (top). Main dePATs and their inhibitors (bottom). (B to F) Effect of pharmacological or genetic inhibition of acyl-protein thioesterases on intracellular dynamics in HD striatal cells. (B) Quantification of Golgi fluorescence in STHdhQ7 cells (green, N = 46), STHdhQ111 cells (orange, N = 46), and STHdhQ111 cells treated for 1 hour with palmostatin B at 10 μM (blue, N = 39). (C) as in (B) with STHdhQ111 cells treated for 1 hour with ML348 at 10 μM (blue, N = 27) (N = 22 STHdhQ7 CT, N = 27 STHdhQ111 CT). (D) as in (B) with STHdhQ111 cells treated for 1 hour with ML349 at 10 μM (blue, N = 23) (N = 22 STHdhQ7 CT and N = 27 STHdhQ111 CT). (E) Quantification of Golgi fluorescence in STHdhQ7 cells si-CT (green, N = 25), STHdhQ111 cells si-CT (orange, N = 26), and STHdhQ111 cells si-APT1 (blue, N = 26). (F) as in (E) with STHdhQ111 cells treated with si-APT2 (blue, N = 27) (N = 25 STHdhQ7 si-CT and N = 26 STHdhQ111 si-CT). The numbers beneath the x axis and under the curves represent, respectively, the peak of Golgi fluorescence and the time to cross the Golgi in the different conditions (represented by green, orange, and blue) N = 3 independent cultures per treatment or condition.
To further validate the target engagement of APT1 in this process, we silenced either APT1 or APT2, as shown by quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) (fig. S3B) and Western blotting for APT1 (fig. S3C; no anti-APT2 antibodies are available). Silencing APT1, but not APT2, restored intracellular trafficking in STHdhQ111 cells back to values observed in control cells (Fig. 2, E and F).
APT1 localizes with the intracellular trafficking machinery and its activity is altered in HD mice
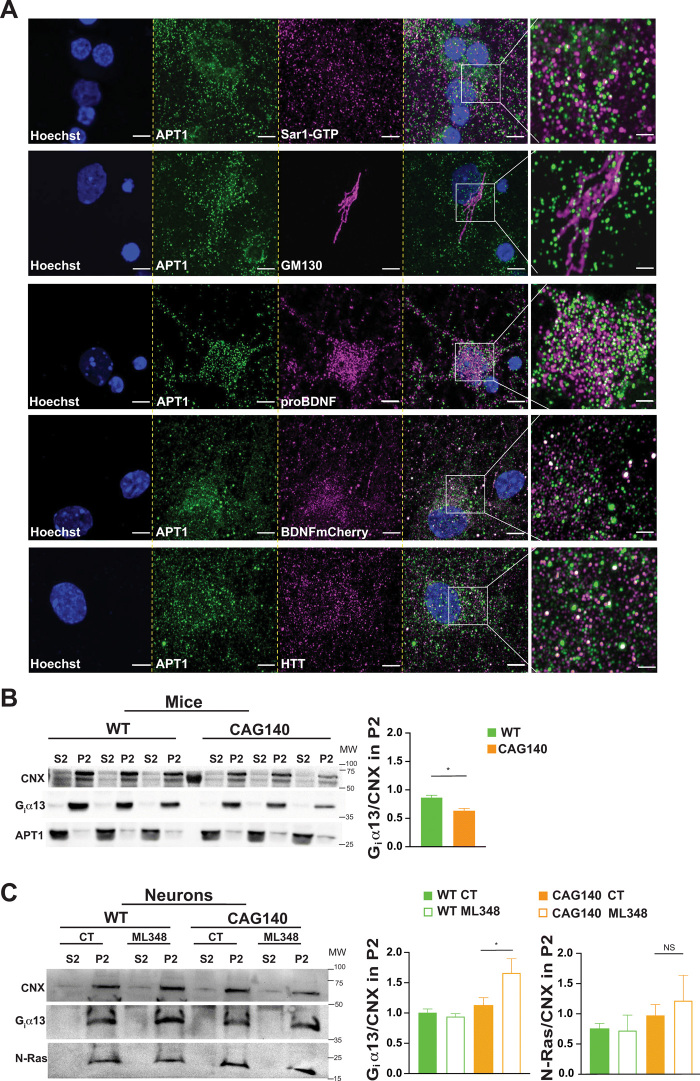
To confirm a role for APT1 in regulating intracellular trafficking in neurons in the corticostriatal circuit, we investigated the localization of endogenous and overexpressed APT1 in cortical neurons with proteins related to ER (calnexin), COPII [Sar1–guanosine 5′-triphosphate (GTP) and SEC24C], the Golgi apparatus (GM130), endogenous pro-BDNF, BDNF-containing dense core vesicles (BDNF-mCherry), and HTT [4C8 monoclonal antibody (mAb)] (Fig. 3A and fig. S4). Endogenous APT1 appeared as punctate staining in the cytosol and partially colocalized with Sar1-GTP, endogenous pro-BDNF, overexpressed BDNF-mCherry, and HTT, as well as with the GM130 as previously described (36). Ectopically expressed enhanced GFP (eGFP)–APT1 also showed partial colocalization with Sec24C and calnexin (fig. S4). These results suggest that APT1 localizes with intracellular compartments controlling trafficking.
Fig. 3. APT1 is expressed in the neuronal secretory pathway.
(A) Confocal images of endogenous APT1 in cortical neurons showing colocalization of the enzyme with proteins related to COPII (Sar1-GTP), Golgi apparatus (GM130), secreted dense core vesicles (proBDNF and BDNF-mCherry), and HTT (4C8 mAb). Nuclei were counterstained with Hoechst (scale bars, 5 μm). For each staining, the images were zoomed five times in a specific region of interest and processed by an Airyscan detector (inset scale bar, 2 μm). (B) Subcellular fractionation of three independent WT and CAG140 mouse brains showing the decrease in ER/Golgi (P2) fractions of the APT1 substrate (Giα13) (graph shows the mean of three brains per condition; *P < 0.05). (C) Primary cortical neurons from WT and CAG140 mice were treated with dimethyl sulfoxide (DMSO) or 1 μM ML348 for 4 hours and then the cytosolic (S2) and ER/Golgi (P2) were analyzed by SDS-PAGE. ML348 treatment increases Giα13 in the P2 fractions while having no effect on N-Ras (N = 3 independent cultures, *P < 0.05; one representative experiment is shown; NS, not significant).
Although brain palmitoylation is reduced in various models of HD (19, 33), APT1 levels have not been evaluated. Western blotting analyses showed that 10-week-old and 6-month-old WT mice and HdhCAG140/+ heterozygous knock-in mice (hereafter referred to as CAG140 mice) had similar levels and localization of APT1 (fig. S5, A and B) (37). We next asked whether APT1 enzymatic activity is dysregulated in the context of HD. To answer this question, we examined the localization of Giα13, which is, to our knowledge, the best-characterized substrate of APT1 (38): When APT1 activity is increased, less Giα13 localizes at the plasma membrane (38). Measurements in fractionated brain extracts revealed membrane localization of Giα13 to be lower in CAG140 than in WT mouse brains, suggesting that in HD, APT1 activity is elevated, even though its quantity remains unchanged (Fig. 3B). We then treated cortical neurons from WT and CAG140 mice with ML348 and subjected the cultured neurons to subcellular fractionation to enrich for large membranes (P2 fractions). ML348 increased Giα13 stoichiometry to P2 fractions while having no effect on N-Ras, which is not a substrate of APT1 (Fig. 3C) (38). These results indicate that the elevated enzymatic activity of APT1 in HD can be diminished by ML348 treatment. We therefore propose that APT1 is a potential target in HD and that its selective inhibitor ML348 could be of therapeutic interest.
ML348 restores BDNF axonal transport, synapse number, and postsynaptic signaling in HD corticostriatal network-on-a-chip
We next sought to determine whether inhibition of APT1 could reverse defective intracellular dynamics and restore neuronal function in a near-to-physiological HD corticostriatal circuit (13). Using primary neuron cultures from WT and CAG140 mice, we reconstituted healthy and HD corticostriatal networks in which cortical neurons project to striatal target neurons through oriented axodendritic connections (Fig. 4A). CAG140 neurons are as viable as WT neurons in the corticostriatal circuit-on-a-chip (fig. S6A), so this reconstituted neuronal circuit allows us to investigate more subtle, earlier signs of neuronal dysfunction (13) rather than waiting for late-stage events such as cell death. To determine whether ML348 is neurotoxic, we first treated WT primary cortical neuron cultures for 50 hours at increasing concentrations and found no overt toxicity for concentrations up to 10 μM (fig. S6B). We also treated cortical neurons daily for 10 days with 1 μM ML348, a concentration chosen on the basis of the ML348 IC50 for APT1 (230 nM) (34). An MTT assay did not reveal any toxic effects (fig. S6C), and we found no morphological changes or stress vacuoles in neurons treated with 1 μM ML348 up to 10 days in vitro (fig. S6D).
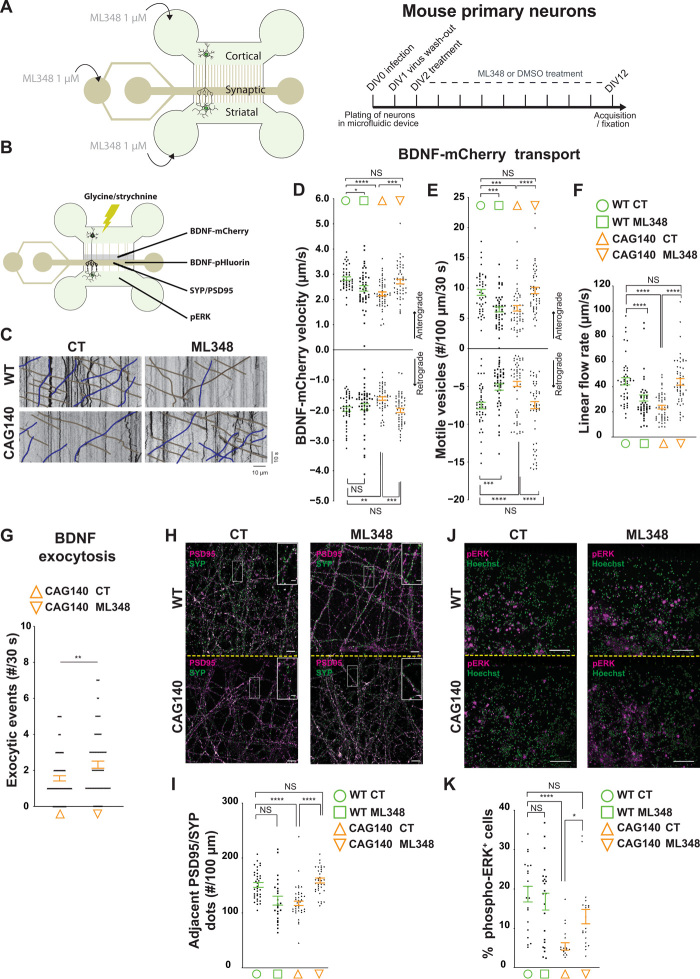
Fig. 4. Pharmacological inhibition of APT1 restores HD corticostriatal network.
(A) Schematic representation of the microfluidic device with indications of drug treatment. (B) Indications of the video-recording zone for BDNF-mCherry–containing vesicles and BDNF exocytosis and the acquisition zone for SYP/PSD95 and phospho–extracellular signal–regulated kinase (pERK) stainings. (C) Representative kymographs showing BDNF-mCherry axonal trafficking. (D) Anterograde and retrograde velocities of BDNF-mCherry vesicles (anterograde: F3,184 = 2.574, *P < 0.05, ***P < 0.001, ****P < 0.0001; retrograde: H3 = 18.94 P = 0.0003 **P < 0.01, ***P < 0.001). (E) Number of anterograde and retrograde vesicles trafficking along 100 μm of axon (anterograde: H3 = 35.57, P < 0.0001, ***P < 0.001, ****P < 0.0001; retrograde: H3 = 41.27, P < 0.0001, ***P < 0.001, ****P < 0.0001). (F) Linear flow rate (H3 = 57.61, P < 0.0001, ****P < 0.0001). (G) ML348 treatment increases BDNF exocytosis at the synapse (**P < 0.01). (H) Airyscan confocal images of PSD95 (magenta) and SYP (green) within the synaptic compartment (scale bar, 5 μm; inset scale bar, 1 μm). (I) Quantification of the number of adjacent PSD95 and SYP punctates along 100-μm neuritis (F3,128 = 2.327, ****P < 0.0001). (J) pERK (magenta) and Hoechst (green) staining within the striatal chamber (scale bar, 50 μm). (K) Quantification of pERK immunopositive cells (H3 = 26.97, P < 0.0001, *P < 0.05, ****P < 0.0001). Error bars indicate SEM.
We therefore treated neurons daily, for 10 days, with 1 μM ML348 within the three compartments of the microfluidic device (Fig. 4A). We first measured the dynamics of BDNF-containing vesicles whose transport is impaired in HD (12, 13). Cortical neurons from WT and CAG140 mice were transduced with BDNF-mCherry lentivirus and acquisitions were performed at 12 days in vitro (DIV12) in the distal part of the long microchannels to assess axonal transport of BDNF (Fig. 4, A and B). By this stage, corticostriatal neurons have established mature synapses with striatal neurons, so we are able to evaluate transport kinetics (13, 39). As previously reported, anterograde and retrograde BDNF transport took place at lower-than-normal velocities in CAG140 neurons, and there were fewer BDNF-containing vesicles traveling along the microtubules (Fig. 4, C to E); i.e., there was a reduction in linear flow (Fig. 4F). ML348 treatment restored the anterograde and retrograde velocities as well as the number of vesicles moving in both directions to control levels (Fig. 4, D and E). The linear flow rate was similar to that of controls, indicating that ML348 fully restored BDNF axonal flow to the synapse in HD neurons (Fig. 4F). In WT neurons, greater palmitoylation decreased transport, suggesting that this process is tightly regulated for proper trafficking.
There is a close correlation between anterograde transport of BDNF and its release at the synapse (12, 18). We transduced CAG140 neurons with BDNF-pHluorin lentivirus and treated the neurons with ML348 for 4 hours. We next quantified the number of BDNF exocytic events using spinning confocal video microscopy after neuronal stimulation and found a significant increase in BDNF exocytosis in mutant neurons treated with ML348 (Fig. 4G). We conclude that ML348 restores BDNF axonal flow and release at the synapse.
The HD circuit shows a significant reduction in the number of corticostriatal synapses (13). We therefore assessed synapse density using high-resolution Airyscan confocal microscopy to quantify presynaptic and postsynaptic markers (synaptophysin spots and PSD95 spots, respectively) within the synaptic chamber of the microfluidic device (Fig. 4B). As previously reported (13), there were fewer corticostriatal synapses in the HD network, but ML348 completely restored the number to that of controls (Fig. 4, H and I).
Lastly, we determined the consequences of ML348 treatment on survival signaling within the postsynaptic compartment. We used extracellular signal–regulated kinase (ERK) phosphorylation as a readout of postsynaptic signaling after cortical stimulation (13), since several studies have shown defects in phospho-ERK (pERK) signaling in the striatum in the HD context in vitro and in vivo (13, 40, 41). We stimulated cortical neurons for 15 min with glycine/strychnine and quantified the percentage of striatal neurons immunopositive for pERK (Fig. 4, B and J). ML348 treatment of CAG140 neurons completely restored postsynaptic survival signaling to values found in WT neurons (Fig. 4, J and K).
ML348 treatment increases brain palmitoylation in mice
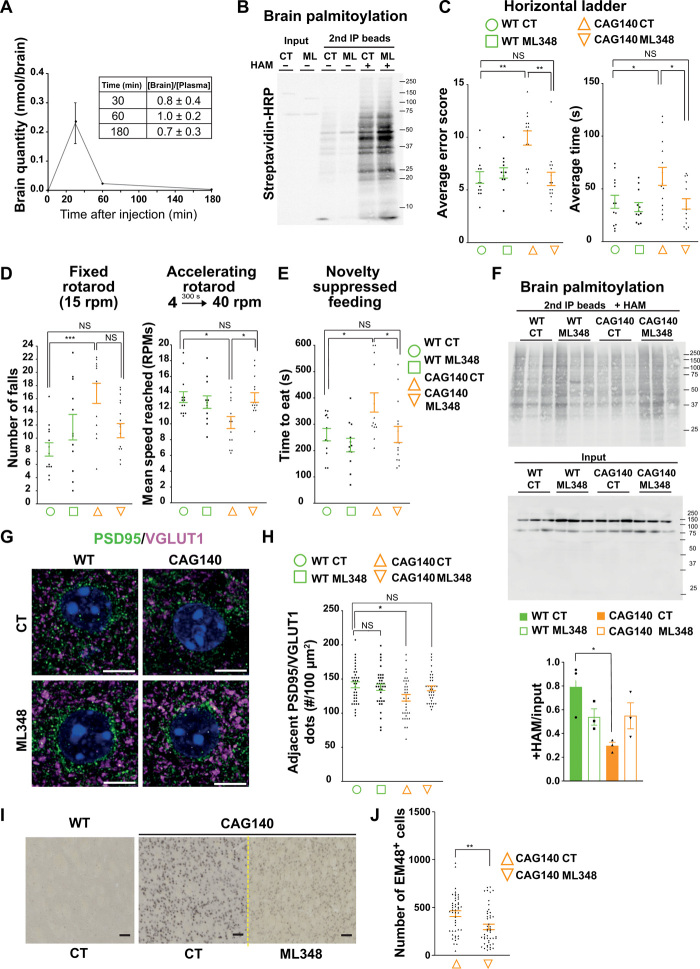
We next evaluated the pharmacokinetics of ML348 in vivo by intraperitoneally injecting WT mice with ML348 (2 mg/kg) and using LC-MS/MS to measure the brain quantity and the plasma concentration of ML348 30, 60, and 180 min after injection (Fig. 5A and fig. S7A). ML348 crossed the blood-brain barrier efficiently, as shown by the [Brain]/[Plasma] ratio value of 0.8 at 30 min after injection. We next evaluated the efficacy of the drug to increase brain palmitoylation levels 30 min after an injection of ML348 (0.3 mg/kg). We performed acyl-biotin exchange (ABE) to biotinylate selectively palmitoylated proteins before their immunoprecipitation with streptavidin-coated beads (fig. S7B). ML348 raised the level of brain palmitoylation (Fig. 5B; lane HAM+ ML+) compared to the control condition (Fig. 5B; lane HAM+ CT+).
Fig. 5. APT1 inhibition via ML348 rescues behavioral symptoms, brain palmitoylation, and neuropathology in HD mice.
(A) Pharmacokinetic analysis of brains (N = 3). (B) ABE assay for palmitoylated proteins. (C to E) Motor coordination and nonmotor behavior in HD mice. (C) Mean error score and time to cross the ladder from three trials (error score: H3 = 17.07, P = 0.0007, *P < 0.01; time: F3,50 = 4.478, *P < 0.05). (D) Mean number of falls from the fixed rotarod (F3,50 = 2.020, ***P < 0.001) and mean speed reached from the accelerating one (F3,50 = 0.4539, *P < 0.05). (E) Latency to eat in a 10-min test (F3,49 = 0.7710, *P < 0.05). (F) Immunoprecipitation of palmitoylated proteins in CAG140 treated mice (upper blot). Quantification of palmitoylated proteins relative to input (lower blot). (G) Airyscan confocal images of PSD95 (green), VGLUT1 (magenta), and Hoechst (blue) (scale bars, 5 μm). (H) Quantification of PSD95/VGLUT1 adjacent dots (F3,140 = 3.65, *P < 0.05). (I) Mutant HTT nuclear accumulation in CAG140-treated mice. EM48 immunostaining of striata from WT and CAG140 mice after ML348 or DMSO injection (scale bars, 50 μm). (J) Quantification of EM48-positive cells (t test, **P < 0.01). Error bars indicate SEM.
Chronic ML348 rescues nonmotor and motor symptoms in HD mice
Since CAG140 mice show both nonmotor and motor signs between 6 and 8 months (37), we evaluated the effects of chronic infusion of ML348 over 28 days in 7-month-old early symptomatic mice implanted with Alzet pumps (fig. S8A). Long-term treatment had no effect on APT1 protein levels (fig. S8B), body weight (fig. S8C), or exploratory behavior in the open-field test (fig. S8D).
We assessed motor coordination using the horizontal ladder test. CAG140 mice showed significant deficits in this task, with a high error rate and longer average time to cross the ladder (Fig. 5C), but ML348 treatment restored their performance to the level of WT mice (Fig. 5C and movie S1). We then explored balance and motor coordination using the rotarod test. CAG140 mice showed significant impairments in motor coordination in both the fixed-speed (15 rpm) and accelerating (4 to 40 rpm in 300 s) modes (Fig. 5D). In these three tests, ML348 significantly improved the motor coordination of CAG140 mice (Fig. 5, C and D). We also investigated anxiety- and depression-related behaviors using the novelty-suppressed feeding paradigm. The latency to feeding in a novel environment for CAG140 mice was significantly longer than for the nontreated WT mice, but ML348 treatment rescued their behavior to WT levels (Fig. 5E and fig. S8E).
Chronic ML348 increases brain palmitoylation levels and reverses neuropathology in HD mice
To determine whether improvement in brain palmitoylation accompanied these improvements in the HD mice, we performed a palmitoylation assay on brain extracts from the mice that had been subjected to the 1-month treatment. Three mice per condition were analyzed. The lower overall protein palmitoylation in CAG140 mice was restored to WT levels by ML348 treatment (Fig. 5F and fig. S8F). ML348 levels were detectable in the brains of mice that had been treated for 4 weeks (fig. S8G).
We next measured the number of synapses using VGLUT1 and PSD95 as markers. ML348 treatment restored synapse number to levels similar to WT (Fig. 5, G and H). Last, we used EM48 antibody to examine nuclear accumulation of mutant HTT, which is an early marker of HD pathogenesis (42). As expected (37), CAG140 mice showed strong nuclear localization of EM48+ mutant HTT in the striatum. Treatment with ML348 markedly reduced mutant HTT nuclear accumulation in these cells (Fig. 5, I and J).
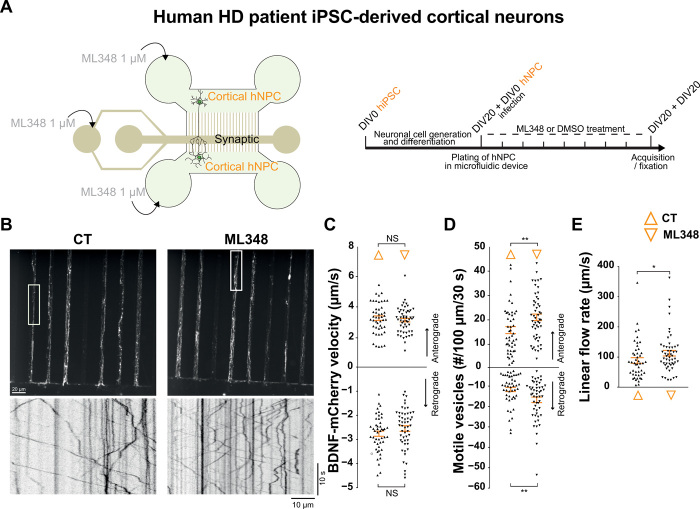
ML348 restores BDNF trafficking in human HD patient iPSC-derived cortical neurons
To investigate whether ML348 could be beneficial in human patients with HD, we plated cortical neuron precursors from human-induced pluripotent stem cells (hiPSC) into the pre- and the postsynaptic compartments of microfluidic devices. Three hours after plating, we infected cells from the presynaptic compartment with BDNF-mCherry–encoding lentivirus. Thirteen days after plating, when a mature corticocortical circuit is established, we treated the presynaptic, synaptic, and postsynaptic compartments of the microchambers (Fig. 6A) with 1 μM ML348 daily for 7 days. We then recorded the trafficking of BDNF-containing vesicles in the distal part of the presynaptic microchannels that contain only cortical axons (Fig. 6B) and generated kymographs from the single-axon recordings. We found that ML348 significantly increased the number of vesicles moving in both directions and increased the linear flow (Fig. 6, C to E).
Fig. 6. ML348 increases BDNF trafficking in hiPSC-derived cortical neurons from patients with HD.
(A) Schematic representation of the microfluidic device used to reconstruct corticocortical network with indications of drug treatment. Human HD iPSC-derived cortical neuron precursors were plated in the pre- and postsynaptic compartments. ML348 (1 μM) or DMSO was perfused within the three compartments from DIV13 to DIV20 (right). (B) z-projection and the associated kymographs showing BDNF-mCherry axonal trafficking in hiPSC-derived cortical neurons treated with DMSO (CT) or ML348 (scale bar, 20 μm). (C) Anterograde and retrograde velocities of BDNF-mCherry vesicles (N indicates the number of axons per condition in at least two independent experiments; N = 47 CT and N = 52 ML348; anterograde: P = 0.4892, retrograde: P = 0.1776 t test). (D) Number of anterograde and retrograde vesicles trafficking along 100 μm of axon (anterograde: P = 0.0068, retrograde: P = 0.0026, Mann-Whitney; **P < 0.01). (E) Linear flow rate (P = 0.0313, Mann-Whitney; *P < 0.05). Error bars indicate SEM.
DISCUSSION
The therapeutic approaches to HD currently considered the most promising are based on reducing the pathogenic accumulation of mutant HTT, such as with antisense oligonucleotides. Even if we surmount the two major challenges to this approach—delivery (intrathecal injections are invasive for the patient as well as costly) and proper targeting (only the mutant HTT and not WT protein should be reduced)—the patient will be left with insufficient WT HTT function, as we normally require expression of both alleles. The other leading approach based on HD pathogenesis, inhibiting aggregation and cell death, so far has not provided benefit in patients. We therefore sought a new approach to developing a pharmacological therapy that would be based on reversing some of the earliest corticostriatal circuit dysfunctions caused by mutant HTT.
We focused on defects in trafficking because there have been suggestions that alleviating these transport defects might prove therapeutic for several neurodegenerative disorders (43, 44). The complexity and unique features of each disease, however, necessitate creativity and precision in our approaches to developing treatments. Here, we used a combination of network-specific purification of vesicles and subsequent proteomic analysis with in silico approaches to identify targets and drugs that were then validated in cells using a medium-throughput ER-to-plasma membrane trafficking assay and a near-to-physiology HD corticostriatal circuit-on-a-chip. This bottom-up approach validated ML348 as a drug of interest for HD, which we then tested in HD mice and in iPSC-derived cortical neurons from patients with HD. The versatility of this approach and its ability to rigorously test relevant drugs based on disease mechanism before entering long and costly in vivo studies make this strategy applicable to other neurodegenerative disorders. For example, it could be used to model the defective corticohippocampal or corticocortical circuits in Alzheimer’s disease or the nigrostriatal pathway in Parkinson’s disease.
Since HIP14 is altered in HD (33), it could have been tempting to first search for factors or drugs that promote HIP14 expression or activity. However, HIP14 has been described to have oncogenic properties (31), and we found that HIP14 overexpression induced abnormal vacuolation and morphological changes in neurons. Furthermore, the defect in brain palmitoylation is likely not to be restricted to the decrease in HIP14 activity. We found that membrane association of Giα13, whose palmitoyl-dependent localization depends on APT1 activity (32), is diminished in HD mice and restored after ML348 treatment. Therefore, APT1 activity is also increased in HD, and its inhibition is sufficient to mediate neuroprotection, although ML348 does not increase palmitoylation on PSD95 (fig. S8H), a known substrate of HIP14 (35).
How does APT1 mediate neuroprotection? Giα13 is obviously not the sole APT1 substrate. Since we began this project, several additional substrates have been reported: the microtubule-associated protein 6 MAP6 (45), the CD36 receptor (46), R-RAS (47), and the Na/Ca exchanger NCX1 (48). Last, an unbiased approach recently identified 382 potential APT1 substrates, although these have yet to be validated in vivo (49). We propose that specific substrates of APT1 mediate neuroprotection in HD neurons and mice and that some could also be substrates of HIP14. Future studies investigating the contribution of particular APT1 substrates (whether shared with HIP14 or not) might lead to a better understanding of the neuroprotective mechanism of APT1 as well as additional therapeutic possibilities.
It is worth noting that ML348 reduced trafficking in WT neurons to near-HD levels, yet produced no overt toxicity. If trafficking is a fundamental aspect of HD pathogenesis—and HIP14 deficient mice, which also have trafficking defects, also develop HD-like disease (19, 32)—why did the reduced trafficking not make WT neurons and mice sick? We believe it is because they were already mature. Trafficking defects start early in embryonic development in HD, disrupting neurogenesis, neuronal migration, circuit connectivity, and other processes, setting the stage for the much later onset of overt symptoms (6). It is also the case that expressing mutant or hypomorphic HTT for only a short time during mouse development is sufficient to later produce features of HD in adult mice (50, 51). It is all the more remarkable, then, that ML348 can exert such strong benefits in HD knock-in mice, whose neurodevelopment has already been significantly altered by mutant HTT.
In addition to ZDHHC17/HIP14, our studies revealed BASP1 and VCP to be proteins of interest to restore trafficking that is defective in HD. BASP1 [also known as brain acid soluble protein, neuronal axonal membrane protein (NAP22), or CAP23] is found in neurites and growth cones, binds calmodulin and the actin cytoskeleton, and associates with synaptic vesicles (28). BASP1 expression was previously reported in the human prefrontal cortex and found increased in patients suffering from schizophrenia (52), a psychosis that can afflict patients with HD (53). The other candidate, VCP (also known as p97, Cdc48, or Ter94), plays a role in selective autophagy by directing poly-ubiquitinated substrates to proteasomes (29), a process that requires HTT and that is impaired in HD (54–57). VCP is recruited to mitochondria in HD cells and in vivo in mice and promotes abnormal mitophagy. Blocking the interaction between VCP and mHTT could be of therapeutic interest, further validating our approach (30). VCP is also found mutated in several neurodegenerative conditions, including familial amyotrophic lateral sclerosis, Charcot Marie-Tooth type 2Y, and multisystem proteinopathy (also called Inclusion body myopathy with Paget disease of bone and frontotemporal dementia) (58).
We show here the importance of brain palmitoylation homeostasis for intracellular trafficking and survival signaling in HD. Restoring palmitoylation levels by ML348, a brain-penetrant compound, represents a new, possibly viable therapeutic strategy for patients with HD. The observation that the three identified targets converge on intracellular trafficking, and more specifically on neuronal BDNF secretion, not only links HTT to trafficking defects in other neurodegenerative disorders but also suggests that modulating HTT-dependent pathways in these diseases might be of therapeutic interest as well.
MATERIALS AND METHODS
Mice
HdhCAG140/+ knock-in mice (CAG140) were generated on a C57/BL6J background to express human HTT exon 1 sequence with 140 repeats of CAG, as described previously (37). To generate the different mouse lines used in the vesicle immunopurification experiments, we crossed Thy1:p50-GFP mice, which express low levels of dynamitin fused to GFP (23), with HdhCAG111/CAG111 (59) mice expressing human HTT exon 1 with 111 repeats of CAG, to obtain Thy1:p50-GFP HdhCAG111/+ mice that were then bred with HdhCAG111/+ mice to produce the desired genotypes.
All mice were maintained with access to food and water ad libitum and kept at a constant temperature (19° to 22°C) and humidity (40 to 50%) on a 12-hour light/12-hour dark cycle. All experimental procedures were performed in an authorized establishment (Grenoble Institut Neurosciences, INSERM U1216, license #B3851610008) in strict accordance with the directive of the European Community (63/2010/EU). The project was approved by the French Ethical Committee (Authorization number: APAFIS#18126-2018103018299125 v2) for care and use of laboratory animals and performed under the supervision of authorized investigators.
RUSH system
STHdhQ7/Q7 and STHdhQ111/Q111 were electroporated with the KDEL-GPI-mCherry plasmid (gift of F. Perez) and when indicated with si-CT (Sigma-Aldrich, SIC001), si-APT1 (Sigma-Aldrich, SASI_Mm01_00077477), and si-APT2 (Sigma-Aldrich, SASI_Mm01_00051133) using the cell line nucleofactor kit L (Lonza) according to the manufacturer’s instructions. STHdh cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 1% of nonessential amino acids, 2 mM l-glutamine, and at 33°C in a humidified 5% CO2 atmosphere. After adding 40 μM biotin, images were taken every 30 s over a 90-min period using an inverted microscope (Axio Observer, Zeiss) coupled to a spinning-disk confocal system (CSU-W1-T3, Yokogawa) connected to a wide-field electron-multiplying charge-coupled device (CCD) camera (ProEM+1024, Princeton Instrument) [63× oil-immersion objective, 1.46 numerical aperture (NA)] and maintained at 33°C and 5% CO2. Integrated fluorescence intensity was measured in the Golgi region, corrected for background and normalized to the maximum value.
Vesicle purification and MS analysis
Thy1:p50-GFP Hdh+/+ and Thy1:p50-GFP HdhQ111/Q111 mice were euthanized by cervical dislocation. Brains were carefully removed from the skulls and placed in a petri dish on ice. We then separated cortical hemispheres along the middle of the sagittal plane and isolated the cortex from the remaining midbrain and hippocampus. The cortices were then homogenized in lysis buffer (4 mM Hepes and 320 mM sucrose, pH 7.4) containing protease inhibitor cocktail (Sigma-Aldrich). The homogenate was first spun for 10 min at 800g to remove unlysed cells and nuclei. The supernatant was centrifuged for 40 min at 12,000g to pellet mitochondria. The resulting supernatant was then further centrifuged for 90 min at 100,000g (Beckman, TLA 100.1). The high-speed pellet enriched in small vesicles (Vesicle Fraction) was resuspended in lysis buffer. To selectively purify small vesicles bound to the molecular motor complex, we immunopurified dynamitin-GFP–positive vesicles by incubating the Vesicle Fraction with anti-GFP iron μBeads or iron μBeads not bound to antibodies as control (Miltenyi Biotec) for 30 min at 4°C. Separation μColumns were placed in a magnetic field, and the incubated solution was applied onto the column. The flow-through containing unbound material was collected, and then the column was rinsed three times with lysis buffer. The Motile Vesicle fraction (IP) was eluted from the column by applying 150 μl of elution buffer [0.1 M triethylamine (pH 11.8) and 0.1% Triton X-100] in a tube containing 9 μl of 1 M MES (pH 3) for neutralization.
For MS, proteins were precipitated using 10% trichloroacetic acid (TCA) and resuspended in Laemmli buffer. Twenty micrograms of the sample and the equivalent volume of control immunopurifications were loaded in a 10% acrylamide gel and further stained with Coomassie Brilliant Blue.
MS sample preparation
In-gel digestion was performed on SDS-PAGE bands containing proteins with sequencing grade trypsin (12.5 μg/μl; Promega) after reduction with dithiothreitol (10 mM; Acros Organics) and alkylation with iodoacétamide (55 mM; Acros Organics).
LC-MS/MS acquisition
Peptide analyses were performed by an LTQ Velos Orbitrap (Thermo Fisher Scientific) coupled to an Easy-nLC Proxeon 1000 chromatographic system (Thermo Fisher Scientific). Chromatographic separation of peptides was performed with the following parameters: Acclaim Pepmap100 precolumn [5 mm, 300-μm inner diameter (i.d.), C18, 5 μm, and 100 Å] and Acclaim PepMap-RSLC Proxeon column (50 cm, 75-μm i.d., C18, 2 μm, and 100 Å), 300 nl/min flow, and gradient rising from 90% solvent A (2% acetonitrile and 0.1% formic acid) to 40% solvent B (100% acetonitrile and 0.1% formic acid) in 100 min and then rising to 80% B in 5 min.
The peptides were analyzed in the Orbitrap in full ion scan mode at a resolution of 30,000 [at mass/charge ratio (m/z) 400], a mass range of 400 to 1800 m/z, and with an MS full scan maximum ion time of 100 ms. Fragments were obtained with collision-induced dissociation activation with a collisional energy of 40% and an activation collisional endothermicity of 0.250 for 10 ms and were analyzed in the LTQ in a second scan event. The ion-trap MS/MS maximum ion time was 50 ms. MS/MS data were acquired in a data-dependent mode in which the 20 most intense precursor ions were isolated, with a dynamic exclusion of 20 s and an exclusion mass width of 10 parts per million (ppm).
LC-MS/MS data processing
Data were processed with Proteome Discoverer 1.4 software (Thermo Fisher Scientific) coupled to an in-house Mascot search server (Matrix Science; version 2.4). The mass tolerance of fragment ions was set to 7 ppm for precursor ions and 0.5 Da for fragments. Identification of peptides related to proteins was performed on Mus musculus taxonomy with an in-house modified database from Swiss-Prot. Posttranslational modifications were searched with carbamidomethylation (C) in constant parameters. The following modifications were used in dynamics parameters: oxidation (M), phosphorylation (S/T/Y), acetylation (K/N-terminal), deamidation (N/Q), and palmitoylation (C). The maximum number of missed cleavages was limited to two for trypsin digestion. Q values of peptides were calculated using the percolator algorithm, and a 1% filter was applied as a false discovery rate threshold.
To quantify ML348 brain concentration, frozen mouse brains were ground into 400 μl of H2O before adding 800 μl of acetonitrile. Samples were spun to remove insoluble residues at 15,000g for 5 min at 16°C. Supernatant was analyzed by LC-MS/MS. LC-MS/MS was assessed by Ultra-high performance liquid chromatography (UHPLC) coupled with a triple quadrupole (Shimadzu LC-MS 8030). Analyses were performed from three different mice for each time point.
Microfluidic chambers, neuronal plating, and transport analysis
Microfluidic devices were generated as previously described (13). Briefly, striatal and cortical primary culture were performed from E15.5 CAG140 knock-in and WT embryos. Dissociated cortical and striatal neurons were resuspended in growing medium and plated in the chamber with a final density of ~7000 cells/mm2 using growing medium (Neurobasal medium supplemented with 2% B27, 2 mM Glutamax, and 1% penicillin/streptomycin) and then placed at 37°C and 5% CO2. For p50-GFP overexpression {DCTN2 (GFP-tagged) [Origene catalog #RG214771]}, neurons were electroporated with 4 μg of DNA + 80 μl of Mouse Neuron Nucleofector Kit (Lonza, catalog #VPG-1001). Before the treatment, at DIV0, the cortical neurons were infected with LV.CMV.BDNF-mCh or LV.BDNF-pHluorin for 24 hours.
p50-GFP and BDNF-mCherry trafficking were observed at DIV6. At DIV12, for the BDNF-mCh experiments, we used an inverted microscope (Axio Observer, Zeiss) coupled to a spinning-disk confocal system (CSU-W1-T3, Yokogawa) connected to wide-field electron-multiplying CCD camera (ProEM+1024, Princeton Instrument) and maintained at 37°C and 5% CO2. We took images every 200 ms for 30-s BDNF-mCh trafficking (63× oil-immersion objective, 1.46 NA). The following number of axons from at least three independent neuronal cultures were analyzed with the KymoToolBox plugin for ImageJ (60) (N = 43 WT CT, N = 45 WT ML348, N = 50 CAG140 CT, and N = 50 CAG140 ML348). The parameters were calculated as follows:
Anterograde velocity
Retrograde velocity
Number of anterograde vesicles per 100 μm
Number of retrograde vesicles per 100 μm
Linear flow rate
Net flux
For BDNF-pHluorin experiments, we stimulated cortical neurons with chemical glycine/strychnine solution [25 mM Hepes (pH 7.5), 124 mM NaCl, 3 mM KCl, 2 mM CaCl2, 10 mM glucose, 200 μM glycine, and 1 μM strychnine] and then acquired BDNF-pHluorin signal at DIV13 to DIV14 using a total internal reflection fluorescence microscope (Nikon/Roper, Eclipse Ti) equipped with a camera Prime 95B scientific complementary metal-oxide semiconductor (sCMOS) (Telelyne Photometrics). Images were taken every 200 ms for 30 s (×100 oil-immersion objective, 1.49 NA).
For the PSD95/SYP experiments, the microchambers were fixed with a 4% paraformaldehyde (PFA) + 4% sucrose solution in phosphate-buffered saline (PBS) for 20 min at room temperature, and the synaptic chamber was incubated overnight at 4°C with anti-PSD95 (Millipore, #MAB1598; 1:1000) and anti-Synaptophysin (Abcam, no. AB14692; 1:200) antibodies. Immunostaining was imaged with a 63× oil-immersion objective (1.4 NA) using an inverted confocal microscope (LSM 710, Zeiss) coupled to an Airyscan detector. Images were analyzed with the free software ImageJ. N indicates the number of field analyzed in at least two microchambers from three independent experiments; N = 36 WT CT, N = 24 WT ML348, N = 36 CAG140 CT, and N = 36 CAG140 ML348.
For the pERK experiments, cortical neurons were stimulated for 15 min at 37°C either with a glycine/strychnine or with a control solution [25 mM Hepes (pH 7.5), 124 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM glucose, and 1 μM strychnine]. Cells within the devices were then fixed as previously described, and the striatal compartment was incubated with pERK antibody overnight at 4°C (Cell Signaling, no. 9106; 1:100). After incubation with secondary antibody, samples were incubated with Hoechst (Sigma-Aldrich no. B2261; 1:5000). Images were acquired no more than 1 week before the fixation and analyzed with the software ImageJ. N indicates the number of fields analyzed in at least two microchambers from three independent experiments; N = 20 WT CT, N = 23 WT ML348, N = 20 CAG140 CT, and N = 20 CAG140 ML348.
Drug treatments
ML348 (gift of B. Martin or Tocris-5345), palmostatin B (Merck Millipore-178501), and ML349 (gift of B. Martin) were diluted in 100% dimethyl sulfoxide (DMSO) for primary concentration. For STHdh cells and immunofluorescence of neurons plated on coverslips, we administered acute treatment with 10 μM for 1 hour before and during the acquisition time. For the fractionation experiments, we plated cortical neurons on 10-cm dishes and we treated cells for 4 hours with 1 μM ML348 before lysis. For neurons within microchambers, 1 μM ML348 was administered within each compartment of the microfluidic device beginning on DIV2 and continuing daily until DIV12, while for BDNF pHluorin analysis, compartments were treated for 4 hours with 1 μM ML348 before acquisition. For in vivo experiments, we infused 0.3 mg/kg per day of either ML348 or control solution (saline + DMSO) for 28 days using an Alzet mini-osmotic pump (2004) implanted subcutaneously on the back of the mouse.
Behavioral assays
There were four experimental groups for behavioral assays: WT CT (13 mice: 7 male and 6 female), WT ML348 (12 mice: 7 male and 5 female), CAG140 CT (14 mice: 6 male and 8 female), and CAG140 ML348 (15 mice: 6 male and 9 female). Mouse littermates were group-housed as mixed treatment groups.
Males and females were 7 months old at the beginning of the treatment. For each specific behavioral assay, all experimental groups were tested on the same day, during the light phase, and between days 16 and 28 of treatment. Before testing, the mice were acclimatized to the test room for 30 min. Treatments were assigned randomly, and behavioral tests were performed 12 days before the end of the delivery of the treatment. Body weight was measured regularly from before the start of treatment until the end.
Mice were excluded from analysis if their performance was calculated to be a significant outlier (Grubb’s test). During the novelty-suppressed feeding test, one mouse (WT CT male) jumped out from the cage and was excluded from analysis.
Open field
Spontaneous locomotion was measured as total distance traveled and general anxiety was estimated from the number of entries in the internal zone. The open-field test was conducted once in an opaque Plexiglas square chamber (50 cm by 50 cm) and the internal chamber was defined with a quadrangular form (12.5 cm by 12.5 cm). The mice were filmed for 60 min. Distance traveled and the number of entries into the internal chamber in each 5-min interval was measured using ViewPoint tracking software.
Rotarod
The rotarod apparatus was used to assess motor coordination and balance. The Rotarod test was performed over six consecutive days, with 3 days of accelerating rotarod and then 3 days of fixed rotarod, with either rate of acceleration or speed changing each day. For the accelerating rotarod, on the first day, the mouse was subjected to three trials at speeds increasing from 4 to 40 rpm over 600 s; on the second day, there were three trials at speeds increasing from 4 to 40 rpm over 300 s; on the third day, there were three trials at speeds increasing from 4 to 40 rpm over 120 s. Each trial was separated by 5-min resting periods. The latency and the speed to fall from the rotarod were recorded up to 300 s.
For the fixed rotarod test, mice were evaluated for three trials at 10, 15, and 20 rpm for days 4, 5, and 6, respectively. For a 300-s period, the animals were put back on the rotarod each time they fell. Each trial was separated by a 15-min resting period. Both the latency to the first fall and the number of falls were reported. The data were expressed as the average of the three trials.
Horizontal ladder
A horizontal ladder is used to assess forelimb and hindlimb coordination. The horizontal ladder is composed of two clear Plexiglas walls (69.5 cm by 15 cm) containing metal rungs (0.2 cm in diameter) irregularly spaced (0.5 to 2 cm apart). Mice were habituated to walk on a horizontal ladder during two consecutive days (three trials per day). Then, the mice were tested (three trials) with a different pattern from that which they learned during the 2 days of habituation. Test trials were video-recorded, and the motor performance using the foot fault scoring system (61) and the latency to complete the task were quantified. The data were expressed as the average of the three trials of the third day.
Novelty-suppressed feeding test
The test was performed as in (62). Briefly, animals were food-deprived for 24 hours to increase motivation to consume food. On the day of testing, mice were placed within a plastic box (50 cm by 30 cm by 15 cm), the floor of which was covered with wooden bedding. Mice were video-recorded and allowed to freely explore until they ate the food pellet. The experiment was stopped after a maximum of 10 min.
ABE assay
Brains were mechanically lysed in ice-cold buffer [1% IGEPAL CA-630, 50 mM tris-HCl (pH 7.5), 150 mM NaCl, and 10% glycerol] containing 1:100 of protease inhibitor cocktail (Sigma-Aldrich, P8340), 1 mM phenylmethylsulfonyl fluoride, 1:200 of phosphatase inhibitor (Sigma-Aldrich, P5726), and 50 mM N-ethylmaleimide (Sigma-Aldrich, E3876) to block free cystein. Brain lysates were rotated at 4°C for 30 min before the insoluble material was removed by centrifugation at 15,000 rpm for 10 min. Lysates were precleared by incubation with Dynabeads M-280 Streptavidin (Invitrogen-112.05D) for 30 min at 4°C on the wheel. Proteins from precleared samples were then precipitated using chloroform-methanol assay. Each pellet was divided and resuspended in +HAM (hydroxylamine) (2:3 of the pellet) [lysis buffer containing 1 mM HAM (Sigma-Aldrich, 46780-4)] or −HAM (1:3 of the pellet) for 1 hour at room temperature on the wheel. This step is a specific cleavage and unmasking of the palmitoylated cysteine’s thiol group by HAM. Proteins were precipitated with the chloroform-methanol method and the pellets were resuspended in Biotin-BMCC buffer [lysis buffer with 5 μM EZ-Link Biotin-BMCC (Thermo Fisher Scientific 21900)] for 1 hour at room temperature on the wheel. Then, proteins were again precipitated by the chloroform-methanol method and the pellets were resuspended in lysis buffer. Palmitoylated proteins were immunoprecipitated with Dynabeads M-280 Streptavidin (Invitrogen-112.05D) for 30 min at 4°C with rocking. Proteins were eluted by boiling the samples with eluent buffer [2.5% SDS, 2.5% glycerol, 62.5 mM tris-HCl (pH 6.8), 0.005% bromophenol blue, and 5 mM dithiothreitol]. Samples were loaded into a stain-free gel 12% acrylamide followed by SDS-PAGE and blotting to nitrocellulose. Stain-free gel revealed the loading control and palmitoylation was detected using horseradish peroxidase–conjugated streptavidin 1:10,000 (Thermo Fisher Scientific 21126). To detect APT1 level, the primary antibodies used were APT1 1:1000 (Abcam-ab91606) and α-tubulin 1:1000 (Sigma-Aldrich, T9026).
Toxicity assay
Toxicity was assessed using the MTT assay, which depends on mitochondrial respiration and indirectly assesses the energy capacity of the cell. Briefly, cortical free culture was treated every day as previously described. After treatment, MTT (Life-M6494) solution was added to the culture medium at 1.2 mM and incubated for 3 hours. The reaction was terminated adding the solvent solution (4 mM HCl and 0.1% NP-40 in isopropanol). The absorbance value was measured at 595 and 690 nm.
Western blotting
For analysis of APT1 expression in WT and CAG140 mice, whole forebrains were lysed in Hepes/sucrose buffer [320 mM sucrose and 4 mM Hepes (pH 7.4)]. For subcellular fractionation, frozen forebrains were triturated using a Dounce homogenizer containing 1 ml of homogenization buffer (320 mM sucrose) and then samples were transferred to ultracentrifuge tubes and centrifuged twice at 47,000 rpm for 10 min (obtaining S1 and P1). The supernatants (S1) were combined into new ultracentrifuge tubes, and a new ultracentrifuge was performed at 120,000 rcf for 40 min (S2 and P2). The entire pellet was resuspended in resuspension buffer [10 mM Hepes (pH 7.3) and 320 mM]. Subcellular fractionations in WT and CAG140 cortical neurons were performed in Hepes/sucrose buffer [320 mM sucrose and 4 mM Hepes (pH 7.4)]. Briefly, after a first centrifugation at 1200 rpm, the total fraction was collected and then the S1 and P1 were separated after a centrifugation at 3000 rpm for 10 min. The S1 was then subjected to an additional centrifugation (12,000 rcf for 40 min) to obtain S2 and P2 fractions. To analyze down-regulation of APT1 by small interfering RNA, cells were lysed in Net120 buffer after the RUSH experiments. Protein extracts were denatured at 95°C for 5 min and then subjected to SDS-PAGE. Primary antibodies were used as follows: anti-HTT (Merck-Millipore #MAB2166; 1:1000), anti-tubulin (Sigma-Aldrich #T7816; 1:10,000), anti-APT1 (Abcam #ab91606; 1:1000), anti-GM130 (Abcam #ab52649; 1:1000; BD Transduction Laboratories #610822; 1:1000), anti-calnexin (Sigma-Aldrich #C4731; 1:1000), anti-vinculin (Sigma-Aldrich #V9131; 1:1000), anti-Giα13 (Santa Cruz #sc-293424; 1:500), anti-N-Ras (Santa Cruz #sc-31; 1:500).
Immunocytochemistry
Primary cortical neurons were transfected with eGFP or HIP14-GFP or APT1-turboGFP plasmid at DIV0, infected at DIV3 with BDNF-mCherry, and then fixed at DIV10 with 4% PFA 20 min at room temperature and then, after one wash in PBS, treated for 15 min with 50 mM NH4Cl. After permeabilization with 0.1% Triton X-100, neurons were incubated for 1 hour at room temperature with blocking solution (0.1% Triton X-100 and 5% NGS in PBS). The incubation with primary antibodies was made overnight at 4°C in blocking solution: anti-HTT (Merck-Millipore #MAB2166; 1:100), anti-APT1 (Abcam #ab91606; 1:100), anti-GM130 (Abcam #ab52649; 1:500), anti-mCherry (Institute Curie Cat #A-P-R#13; 1:200), anti-calnexin (Sigma-Aldrich #C4731; 1:500), anti-turboGFP (CliniSciences #TA150041; 1:100), anti-Sec24C (Sigma-Aldrich #HPA040196; 1:100), anti-Active Sar1-GTP (Biomol-NewEast Biosciences #NB-26916; 1:100), anti-GFP (Millipore #AB16901; 1:200), anti-proBDNF (Santa Cruz #sc-65514; 1:100), and anti-GRASP65 (Thermo Fisher Scientific #PA3-910; 1:300). After 2 hours of incubation at room temperature with II antibodies, the coverslips were incubated with Hoechst (Sigma-Aldrich #B2261; 1:5000) and mounted on glass slides with ProLong Diamond Antifade Mounting (Life Technologies #P36961). Images were acquired with a 63× oil-immersion objective using an inverted confocal microscope (LSM 710, Zeiss) coupled to an Airyscan detector.
For immunofluorescence on mouse Thy1-p50 GFP brain slices, samples were fixed after mouse perfusion for 2 hours in 4% PFA, washed three times with PBS, and then incubated for 16 hours in 20% sucrose in PBS. Brains were then incubated in 30% sucrose in PBS. After inclusion in Tissue-Tek Optimal Cutting Temperature (OCT), samples were frozen at −20°C and kept at −80°C for long storage. Slices (35 μm) were prepared at the cryostat and mounted on Thermo Fisher Scientific SuperFrost Ultra Plus GOLD Adhesion Slides. At this step, slices were acquired with a 20× objective (0.45 NA) using a slide scanner (AxioScan Z1, Zeiss) or processed for immunostaining as follows. An unmasking step was performed with Citrate buffer (Sigma-Aldrich #C9999; 1:1000 in H2O) for 20 min at 95°C and then slices were incubated with 5% Normal Donkey Serum in PBS–0.3% Triton X-100 for 1 hour. Primary antibodies were incubated overnight at 4°C in blocking solution with the following concentrations: anti-CTIP2 (Abcam #Ab18465; 1:300), anti-CUX1 (Novus Biologicals #NBP2-13883; 1:100), anti-TBR1 (Abcam #ab31940; 1:200), anti-GFP (Millipore #AB16901; 1:500), anti-GRASP65 (Thermo Fisher Scientific #PA3-910; 1:300), and anti-calnexin (Sigma-Aldrich #C4731; 1:500). After incubation with secondary antibodies, samples were incubated with Hoechst (Sigma-Aldrich #B2261; 1:5000) and then coverslips were mounted with Dako Fluorescent Mounting Medium (Agilent). Acquisitions were made using a 20× or 40× oil-immersion objective using an inverted confocal microscope (LSM 710, Zeiss) and z-stacks were made with five slices of 5 μm each for a total of 20 μm. For calnexin and GRASP65 staining, images were acquired with a 63× oil-immersion objective (1.4 NA) using an inverted confocal microscope (LSM 710, Zeiss) coupled to an Airyscan detector to improve signal-to-noise ratio and increase resolution; z-stacks were made with 20 to 25 slices of 0.3 μm each for a total of 6 to 7 μm.
For PSD95/VGLUT1, palmitoylated PSD95, and EM48 experiments, animals were anesthetized with ketamine (40 mg/kg) and xylazine (10 mg/kg) and were perfused transcardially with cold PBS followed by 4% PFA. Brains were removed, fixed overnight in 4% PFA, and then cryoprotected in 30% (w/v) sucrose in PBS before embedding in OCT. Coronal sections (30 μm) were made on a cryostat. For PSD95/VGLUT1 and palmitoylated PSD95 immunostainings, epitopes were unmasked with L.A.B. solutions (PolySciences, #24310) for 20 min at room temperature. After blocking for 2 hours at room temperature in 5% normal goat serum, 1% bovine serum albumin, and 0.1% Triton X-100 in PBS, sections were incubated overnight at 4°C with anti-PSD95 (Millipore, #MAB1598; 1:400), anti-VGLUT1 (Millipore, #AB5905; 1:1000), or anti–palmitoylated PSD95 (gift of F. Perez; 1:200). After three washes, sections were incubated at room temperature for 2 hours with appropriate fluorescent secondary antibodies, washed again three times, then incubated with Hoechst (Sigma-Aldrich #B2261; 1:5000), and lastly mounted on slides in DAKO. Immunostaining was acquired with a 63× oil-immersion objective (1.4 NA) using an inverted confocal microscope (LSM 710, Zeiss) coupled to an Airyscan detector to improve signal-to-noise ratio and to increase resolution. For PSD95/VGLUT1, the following number of fields were analyzed with the free software ImageJ in at least three sections from three different mice: N = 36 WT CT, N = 36 WT ML348, N = 36 CAG140 CT, and N = 36 CAG140 ML348. For PSD95/palm-PSD95, the following number of fields were analyzed with the free software ImageJ in at least three sections from three different mice: N = 36 WT CT, N = 36 WT ML348, N = 36 CAG140 CT, and N = 36 CAG140 ML348.
For EM48 immunostaining, sections were mounted on Thermo Fisher Scientific SuperFrost Plus Adhesion Slides and kept at room temperature overnight. Epitopes were unmasked by placing the slides in a pressure cooker containing Citrate Buffer (Zytomed Systems, #ZUC028) at 95°C for 15 min and for another 15 min with the cooker off. Sections were washed three times with PBS, incubated in 3% H2O2 in PBS for 30 min at room temperature to inhibit endogenous peroxidase, and then washed again three times with PBS. Sections were then incubated with blocking solution (3% normal donkey serum in PBS 0.1% Triton X-100) for 2 hours before overnight incubation with anti EM48 antibodies (Sigma-Aldrich, #MAB 5374; 1:100) at 4°C in blocking solution. Sections were then washed three times with PBS and incubated at room temperature for 2 hours with biotinylated secondary antibodies Biotin-SP-Conjugated affiniPure Donkey-anti-Mouse immunoglobulin G (IgG) (H + L) (Jackson ImmunoResearch #715-065-150; 1:250). After three washes with PBS, sections were incubated with ABC Kit (VectaStain, #PK6100) for 1 hour at room temperature and washed again three times with PBS. Sections were developed using the DAB Kit (Vector, #SK4100) for 2 to 5 min, at room temperature under swirling. Sections were washed three times in PBS dehydrated using different baths of ethanol (50°, 75°, 95°, and 100°) before covering the section with mounting medium (Pertex, HistoLab, #871-0500). The scoring of EM48+ neurons was performed as follows. Immunostaining was acquired with a 20× objective (0.45 NA) using a slide scanner (AxioScan Z1, Zeiss). Sections were analyzed using ImageJ with the threshold set from CAG140 control brain sections incubated with mouse IgG only and revealed with DAB. The threshold was then applied to WT control, CAG140 control, and CAG140 ML348 sections. After watershed-image treatment, cells were counted automatically with the ImageJ plugin Analyze Particles with the minimum size for particles set at 0.2 μm2. Forty-seven fields for the CAG140 CT condition and 48 fields for the CAG140 ML348 condition were analyzed in two independent sections from three different mice.
RT-qPCR
Total RNA was isolated using miRNeasy (Qiagen, #74104) and monitored using Qubit RNA HS assay kit (Life, #32852). RNA samples were retrotranscribed using the iScript room temperature Supermix for RT-qPCR kit (Bio-Rad, #170-8840). Five hundred nanograms of cDNAs were submitted to RT-qPCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, #172-5270) with the following: APT1 (5′-AACAGGCAGCAGAAACCGTA-3′, 5′-CTGTGTGGTGAGAGCAGTGT-3′) and APT2 (5′-CATCCGGCTTCCTCATGTCA-3′, 5′-GCCTTCTTGATGCCAGCTTC-3′) oligonucleotides. CycloG (5′-AAGAATCGTCGCTGGTATG-3′, 5′-AAAGCCGATGACAAGGAG-3′), MTBP (5′-GGGGTCATAGGAGTCATTGG-3′, 5′-ACATCTCAGCAACCCACACA-3′), GAPDH (5′-CACCACCCTGTTGCTGTA-3′, 5′-AACTCCCACTCTTCCACCT-3′), and Actin (5′-GGAAGGTGGACAGTGAGG-3′, 5′-CCTAGCACCATGAAGATCAA-3′) genes were used as internal controls. Fold changes were calculated using the ddCT method.
Generation of cortical neuron precursors from hiPSCs
The hiPSC “109Q” line ND42222 (RRID:CVCL_Y844 passage 42) was obtained from Coriell repository. This line is heterozygous for HTT p.Gln18[109] and thus has 109 CAG repeats in one of the two alleles for HTT. hiPSC amplification, neuronal cell generation, and terminal differentiation were performed as previously described (63). 109Q iPS cells were maintained on vitronectin-coated (Life Technology) plates in mTeSRplus medium (STEMCELL Technologies). Cultures were fed every other day and passaged via manual dissociation using 0.02% EDTA (pH 7.2) (Merck Sigma-Aldrich) every 4 to 5 days. For neural differentiation, hiPSC colonies were treated (DIV0) as previously described (64) in N2B27 media consisting of 50% DMEMF-12 Glutamax, 50% Neurobasal medium, 2% B27 supplement 50× minus vitamin A, 1% N2 supplement, 0.1% penicillin/streptomycin, and 50 μM β-mercaptoethanol (Thermo Fisher Scientific). Neural differentiation was initiated, passaging the hiPSC in N2B27 media supplemented with SB431542 (20 μM; Tocris), LDN-193189 (100 nM; Sigma-Aldrich), XAV-939 (1 μM; Tocris), and 10 μM ROCK inhibitor (Y27632, Calbiochem) in low-adherence culture plate (Greiner) for 6 hours. Media were changed every day from DIV0 to DIV20. At DIV1, hiPSC aggregates were transferred on poly-ornithine laminin-coated dishes without Y27632. From DIV5 to DIV9, SB431542 was removed and FGF2 (10 ng/ml) and cyclopamine 1 μM (Merck) were added. From DIV10 to DIV20, LDN-193189 and XAV-939 were removed and CHIR99021 0.4 μM (Stemgent) was added. At DIV20, cortical neuron precursor cells were enzymatically dissociated using Accutase (Invitrogen), resuspended at 5 × 106 cells/ml in Cryostor (Merck) cell cryopreservation media, frozen, and stored in liquid nitrogen vapor at −150°C.
Neuronal differentiation of hiPSC-derived cortical neuron precursors in microfluidic devices
Microfluidic devices were coated with poly-d-lysine (Merck) in the proximal and synaptic compartments and poly-d-lysine/laminin (Thermo Fisher Scientific) in the distal compartment. Cortical neuron precursors were suspended in N2B27 medium supplemented with BDNF (20 ng/ml; PeproTech), cAMP (100 μM; Merck), DAPT (10 μM; Tocris), Cdk4i (1 μM; Merck), and ROCK inhibitor (Y-27632; STEMCELL Technologies) and plated to a final density of ~7000 cells/mm2 in the distal compartment of the microfluidic chamber. Three hours after plating, cells were infected with BDNF-mCherry lentivirus. The day after seeding, the medium was replaced by fresh N2B27-supplemented medium without ROCK inhibitor. Medium was then changed every 7 days. Cells were exposed from DIV13 to DIV20 (postseeding in microfluidic device) every day with 1 μM ML348 or DMSO added in both the proximal synaptic and distal compartment of the microfluidic device.
Statistical analysis
GraphPad Prism (GraphPad Software Inc.) software was used for statistical analysis. All in vitro experiments consisted of at least three independent replicates. Data are expressed as means ± SEM. The normality of the data distribution was tested by performing D’Agostino and Pearson test with threshold set at α = 0.05. If the data were normally distributed, we compared the groups using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis; if not, we compared the groups using a Kruskal-Wallis test followed by a Dunn’s post hoc analysis. When comparing two groups, we used the unpaired two-tailed Student’s t test when the data were normally distributed or the Mann-Whitney test when the data were not normally distributed. Toxicity of ML348 and the weight of the mice were analyzed using two-way ANOVA followed by Tukey post hoc test when comparing more than two variables (concentration and time, or genotype and day after treatment). When the n was too small to be analyzed by a normality test (e.g., Western blotting analyses), the normality was assumed. The criterion for statistical significance was set at P < 0.05.
Acknowledgments
We thank D. Zala for pilot experiments; M. Barnat and M. Cazorla for advice; B. Martin for his gift of ML348 and ML349 molecules; G. Boncompain and F. Perez for sharing constructs, antibodies, and help with experiments; C. Benstaali for mouse colony handling; V. Brandt for critical editing of the manuscript; members of the Humbert and Saudou laboratories for helpful discussions; the GIN imaging facility (PIC-GIN) for help with image acquisitions; T. Léger and C. Garcia from the Plateforme Protéomique Structurale et Fonctionnelle from the Institut Jacques Monod, CNRS Université Paris-Diderot for mass spectrometry experiments; G. Froment, D. Nègre, and C. Costa from SFR Biosciences (UMS3444/CNRS, US8/Inserm, ENS de Lyon, UCBL) for lentivirus production; and P. Gizzi from the PCBIS “Plate-forme de Chimie Biologique Intégrative de Strasbourg” (UMS3286, ESBS, Strasbourg) for pharmacokinetic analyses. Funding: This work was supported by grants from Agence Nationale de la Recherche: ANR-14-CE35-0027-01 PASSAGE (F.S.); ANR-11-INBS-011 NeurARTRIS (A.L.P. and P.L.); ANR-15-IDEX-02 NeuroCoG and PalmHunt (F.S. and S.H.) in the framework of the “Investissements d’avenir” program; ANR-10-LABX-73 REVIVE (A.L.P.); European Commission H2020 Project Joint Programme – Neurodegenerative Disease Research (JPND) ModelPolyQ grant 643417 (M.L. and A.L.P.); Fondation pour la Recherche Médicale [FRM, DEI20151234418 (F.S.) and équipe labellisée, DEQ20170336752 (S.H.)]; Fondation pour la Recherche sur le Cerveau (FRC) (F.S.); Fondation Bettencourt Schueller (F.S.); INSERM (F.S., S.H., and A.L.P.); and AGEMED program from INSERM (F.S. and S.H.). The Saudou and Humbert laboratories are part of the Grenoble Center of Excellence in Neurodegeneration (GREEN). I-Stem is supported by the Association Française contre les Myopathies (AFM). A.V. was supported by a PhD fellowship from DIM “Cerveau et Pensée” and FRM (FDT2016043504); C.S. was supported by a Postdoctoral fellowship from FRM (SPF20140129405) and by EMBO LTF (ALTF 693-2015). Author contributions: A.V., S.H., and F.S. provided the conceptual framework for the study. A.V., C.S., S.L., M.L., M.-V.H., A.L.P., S.H., and F.S. designed the experiments. A.V. performed the trafficking assays and drug treatment on RUSH and microfluidic devices and all experiments performed on mice (treatment, behavior, biochemistry, and immunostaining). C.S. performed trafficking of p50GFP, immunostaining in neurons and mouse brains, and BDNF pHluorin experiments. C.S. analyzed the protein levels in total brain lysates and subcellular fractions in neurons and mouse brains. R.C. did bioinformatic analysis of the proteomic data. S.L. did the trafficking experiments in human iPSC–derived cortical neurons and brain immunohistochemistry analyses. M.L. and P.L. did the culture and differentiation of human cells. A.G. produced the microfluidic devices and assisted in brain immunohistochemistry. M.-V.H. performed the purification of vesicles for proteomic analysis. A.L.P. supervised the experiments in human iPSC–derived cortical neurons. A.V., C.S., S.H., and F.S. wrote the manuscript. Competing interests: A.V. and F.S. have the awarded patent number WO 2020/170208; FR3092992: Inhibitors of acyl-thioesterases for the treatment and/or prevention of Huntington disease. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabb0799/DC1
REFERENCES AND NOTES
- 1.Ross C. A., Tabrizi S. J., Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Saudou F., Humbert S., The biology of huntingtin. Neuron 89, 910–926 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Langfelder P., Cantle J. P., Chatzopoulou D., Wang N., Gao F., Al-Ramahi I., Lu X. H., Ramos E. M., El-Zein K., Zhao Y., Deverasetty S., Tebbe A., Schaab C., Lavery D. J., Howland D., Kwak S., Botas J., Aaronson J. S., Rosinski J., Coppola G., Horvath S., Yang X. W., Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Ramahi I., Lu B., Di Paola S., Pang K., de Haro M., Peluso I., Gallego-Flores T., Malik N. T., Erikson K., Bleiberg B. A., Avalos M., Fan G., Rivers L. E., Laitman A. M., Diaz-Garcia J. R., Hild M., Palacino J., Liu Z., Medina D. L., Botas J., High-throughput functional analysis distinguishes pathogenic, nonpathogenic, and compensatory transcriptional changes in neurodegeneration. Cell Syst. 7, 28–40.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becanovic K., Norremolle A., Neal S. J., Kay C., Collins J. A., Arenillas D., Lilja T., Gaudenzi G., Manoharan S., Doty C. N., Beck J., Lahiri N., Portales-Casamar E., Warby S. C., Connolly C., De Souza R. A.; REGISTRY Investigators of the European Huntington’s Disease Network, Tabrizi S. J., Hermanson O., Langbehn D. R., Hayden M. R., Wasserman W. W., Leavitt B. R., A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 18, 807–816 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Barnat M., Capizzi M., Aparicio E., Boluda S., Wennagel D., Kacher R., Kassem R., Lenoir S., Agasse F., Braz B. Y., Liu J. P., Ighil J., Tessier A., Zeitlin S. O., Duyckaerts C., Dommergues M., Durr A., Humbert S., Huntington’s disease alters human neurodevelopment. Science 369, 787–793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cepeda C., Hurst R. S., Calvert C. R., Hernandez-Echeagaray E., Nguyen O. K., Jocoy E., Christian L. J., Ariano M. A., Levine M. S., Transient and progressive electrophysiological alterations in the corticostriatal pathway in a mouse model of Huntington’s disease. J. Neurosci. 23, 961–969 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosas H. D., Lee S. Y., Bender A. C., Zaleta A. K., Vangel M., Yu P., Fischl B., Pappu V., Onorato C., Cha J. H., Salat D. H., Hersch S. M., Altered white matter microstructure in the corpus callosum in Huntington’s disease: Implications for cortical “disconnection”. Neuroimage 49, 2995–3004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabrizi S. J., Scahill R. I., Durr A., Roos R. A., Leavitt B. R., Jones R., Landwehrmeyer G. B., Fox N. C., Johnson H., Hicks S. L., Kennard C., Craufurd D., Frost C., Langbehn D. R., Reilmann R., Stout J. C.; TRACK-HD Investigators , Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: The 12-month longitudinal analysis. Lancet Neurol. 10, 31–42 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Cepeda C., Wu N., Andre V. M., Cummings D. M., Levine M. S., The corticostriatal pathway in Huntington’s disease. Prog. Neurobiol. 81, 253–271 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccato C., Valenza M., Cattaneo E., Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 90, 905–981 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Gauthier L. R., Charrin B. C., Borrell-Pages M., Dompierre J. P., Rangone H., Cordelieres F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., Saudou F., Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–138 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Virlogeux A., Moutaux E., Christaller W., Genoux A., Bruyere J., Fino E., Charlot B., Cazorla M., Saudou F., Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington’s disease. Cell Rep. 22, 110–122 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Baquet Z. C., Gorski J. A., Jones K. R., Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 24, 4250–4258 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandstaetter H., Kruppa A. J., Buss F., Huntingtin is required for ER-to-Golgi transport and for secretory vesicle fusion at the plasma membrane. Dis. Model. Mech. 7, 1335–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.del Toro D., Alberch J., Lazaro-Dieguez F., Martin-Ibanez R., Xifro X., Egea G., Canals J. M., Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell 20, 1478–1492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strehlow A. N., Li J. Z., Myers R. M., Wild-type huntingtin participates in protein trafficking between the Golgi and the extracellular space. Hum. Mol. Genet. 16, 391–409 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Borrell-Pages M., Canals J. M., Cordelieres F. P., Parker J. A., Pineda J. R., Grange G., Bryson E. A., Guillermier M., Hirsch E., Hantraye P., Cheetham M. E., Neri C., Alberch J., Brouillet E., Saudou F., Humbert S., Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J. Clin. Invest. 116, 1410–1424 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singaraja R. R., Huang K., Sanders S. S., Milnerwood A. J., Hines R., Lerch J. P., Franciosi S., Drisdel R. C., Vaid K., Young F. B., Doty C., Wan J., Bissada N., Henkelman R. M., Green W. N., Davis N. G., Raymond L. A., Hayden M. R., Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum. Mol. Genet. 20, 3899–3909 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., Perez F., Synchronization of secretory protein traffic in populations of cells. Nat. Methods 9, 493–498 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V. C., Sharp A. H., Persichetti F., Cattaneo E., MacDonald M. E., Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet. 9, 2799–2809 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Pardo R., Molina-Calavita M., Poizat G., Keryer G., Humbert S., Saudou F., pARIS-htt: An optimised expression platform to study huntingtin reveals functional domains required for vesicular trafficking. Mol. Brain 3, 17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross J. L., Wallace K., Shuman H., Goldman Y. E., Holzbaur E. L., Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat. Cell Biol. 8, 562–570 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Hinckelmann M. V., Virlogeux A., Niehage C., Poujol C., Choquet D., Hoflack B., Zala D., Saudou F., Self-propelling vesicles define glycolysis as the minimal energy machinery for neuronal transport. Nat. Commun. 7, 13233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adolf F., Rhiel M., Hessling B., Gao Q., Hellwig A., Bethune J., Wieland F. T., Proteomic profiling of mammalian COPII and COPI vesicles. Cell Rep. 26, 250–265.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Miller E. A., Schekman R., COPII—A flexible vesicle formation system. Curr. Opin. Cell Biol. 25, 420–427 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alanis-Lobato G., Andrade-Navarro M. A., Schaefer M. H., HIPPIE v2.0: Enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 45, D408–D414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosevitsky M. I., Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 245, 245–325 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Meyer H., Bug M., Bremer S., Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Guo X., Sun X., Hu D., Wang Y. J., Fujioka H., Vyas R., Chakrapani S., Joshi A. U., Luo Y., Mochly-Rosen D., Qi X., VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 7, 12646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ducker C. E., Stettler E. M., French K. J., Upson J. J., Smith C. D., Huntingtin interacting protein 14 is an oncogenic human protein: Palmitoyl acyltransferase. Oncogene 23, 9230–9237 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R. R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C. A., Hayden M. R., El-Husseini A., Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron 44, 977–986 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R., Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9, 824–831 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adibekian A., Martin B. R., Chang J. W., Hsu K. L., Tsuboi K., Bachovchin D. A., Speers A. E., Brown S. J., Spicer T., Fernandez-Vega V., Ferguson J., Hodder P. S., Rosen H., Cravatt B. F., Confirming target engagement for reversible inhibitors in vivo by kinetically tuned activity-based probes. J. Am. Chem. Soc. 134, 10345–10348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin D. T., Conibear E., ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4, e11306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vartak N., Papke B., Grecco H. E., Rossmannek L., Waldmann H., Hedberg C., Bastiaens P. I., The autodepalmitoylating activity of APT maintains the spatial organization of palmitoylated membrane proteins. Biophys. J. 106, 93–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menalled L. B., Sison J. D., Dragatsis I., Zeitlin S., Chesselet M.-F., Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 465, 11–26 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., Khudayberdiev S., Leuschner P. F., Busch C. J., Kane C., Hubel K., Dekker F., Hedberg C., Rengarajan B., Drepper C., Waldmann H., Kauppinen S., Greenberg M. E., Draguhn A., Rehmsmeier M., Martinez J., Schratt G. M., A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moutaux E., Christaller W., Scaramuzzino C., Genoux A., Charlot B., Cazorla M., Saudou F., Neuronal network maturation differently affects secretory vesicles and mitochondria transport in axons. Sci. Rep. 8, 13429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liot G., Zala D., Pla P., Mottet G., Piel M., Saudou F., Mutant Huntingtin alters retrograde transport of TrkB receptors in striatal dendrites. J. Neurosci. 33, 6298–6309 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brito V., Giralt A., Enriquez-Barreto L., Puigdellivol M., Suelves N., Zamora-Moratalla A., Ballesteros J. J., Martin E. D., Dominguez-Iturza N., Morales M., Alberch J., Gines S., Neurotrophin receptor p75(NTR) mediates Huntington’s disease-associated synaptic and memory dysfunction. J. Clin. Invest. 124, 4411–4428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutekunst C. A., Li S. H., Yi H., Mulroy J. S., Kuemmerle S., Jones R., Rye D., Ferrante R. J., Hersch S. M., Li X. J., Nuclear and neuropil aggregates in Huntington’s disease: Relationship to neuropathology. J. Neurosci. 19, 2522–2534 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinckelmann M. V., Zala D., Saudou F., Releasing the brake: Restoring fast axonal transport in neurodegenerative disorders. Trends Cell Biol. 23, 634–643 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Sleigh J. N., Rossor A. M., Fellows A. D., Tosolini A. P., Schiavo G., Axonal transport and neurological disease. Nat. Rev. Neurol. 15, 691–703 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Ho G. P. H., Ramalingam N., Imberdis T., Wilkie E. C., Dettmer U., Selkoe D. J., Upregulation of cellular palmitoylation mitigates α-synuclein accumulation and neurotoxicity. Mov. Disord. 36, 348–359 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao J. W., Wang J., Guo H., Zhao Y. Y., Sun H. H., Li Y. F., Lai X. Y., Zhao N., Wang X., Xie C., Hong L., Huang X., Wang H. R., Li C. B., Liang B., Chen S., Zhao T. J., CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 11, 4765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei X., Adak S., Zayed M., Yin L., Feng C., Speck S. L., Kathayat R. S., Zhang Q., Dickinson B. C., Semenkovich C. F., Endothelial palmitoylation cycling coordinates vessel remodeling in peripheral artery disease. Circ. Res. 127, 249–265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gok C., Plain F., Robertson A. D., Howie J., Baillie G. S., Fraser N. J., Fuller W., Dynamic palmitoylation of the sodium-calcium exchanger modulates its structure, affinity for lipid-ordered domains, and inhibition by XIP. Cell Rep. 31, 107697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H., Yan P., Ren J., Wu C., Yuan W., Rao M., Zhang Z., Kong E., Identifying the potential substrates of the depalmitoylation enzyme Acyl-protein thioesterase 1. Curr. Mol. Med. 19, 364–375 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Arteaga-Bracho E. E., Gulinello M., Winchester M. L., Pichamoorthy N., Petronglo J. R., Zambrano A. D., Inocencio J., De Jesus C. D., Louie J. O., Gokhan S., Mehler M. F., Molero A. E., Postnatal and adult consequences of loss of huntingtin during development: Implications for Huntington’s disease. Neurobiol. Dis. 96, 144–155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molero A. E., Arteaga-Bracho E. E., Chen C. H., Gulinello M., Winchester M. L., Pichamoorthy N., Gokhan S., Khodakhah K., Mehler M. F., Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc. Natl. Acad. Sci. U.S.A. 113, 5736–5741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behan A. T., Byrne C., Dunn M. J., Cagney G., Cotter D. R., Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol. Psychiatry 14, 601–613 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Novak M. J., Tabrizi S. J., Huntington’s disease: Clinical presentation and treatment. Int. Rev. Neurobiol. 98, 297–323 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., Cuervo A. M., Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 13, 567–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochaba J., Lukacsovich T., Csikos G., Zheng S., Margulis J., Salazar L., Mao K., Lau A. L., Yeung S. Y., Humbert S., Saudou F., Klionsky D. J., Finkbeiner S., Zeitlin S. O., Marsh J. L., Housman D. E., Thompson L. M., Steffan J. S., Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc. Natl. Acad. Sci. U.S.A. 111, 16889–16894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong Y. C., Holzbaur E. L., The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J. Neurosci. 34, 1293–1305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rui Y. N., Xu Z., Patel B., Chen Z., Chen D., Tito A., David G., Sun Y., Stimming E. F., Bellen H. J., Cuervo A. M., Zhang S., Huntingtin functions as a scaffold for selective macroautophagy. Nat. Cell Biol. 17, 262–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang W. K., Xia D., Mutations in the human AAA+ Chaperone p97 and related diseases. Front. Mol. Biosci. 3, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wheeler V. C., Gutekunst C.-A., Vrbanac V., Lebel L.-A., Schilling G., Hersch S., Friedlander R. M., Gusella J. F., Vonsattel J. P., Borchelt D. R., MacDonald M. E., Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum. Mol. Genet. 11, 633–640 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Zala D., Hinckelmann M. V., Yu H., Lyra da Cunha M. M., Liot G., Cordelieres F. P., Marco S., Saudou F., Vesicular glycolysis provides on-board energy for fast axonal transport. Cell 152, 479–491 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Metz G. A., Whishaw I. Q., Cortical and subcortical lesions impair skilled walking in the ladder rung walking test: A new task to evaluate fore- and hindlimb stepping, placing, and co-ordination. J. Neurosci. Methods 115, 169–179 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Pla P., Orvoen S., Benstaali C., Dodier S., Gardier A. M., David D. J., Humbert S., Saudou F., Huntingtin acts non cell-autonomously on hippocampal neurogenesis and controls anxiety-related behaviors in adult mouse. PLOS ONE 8, e73902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gribaudo S., Tixador P., Bousset L., Fenyi A., Lino P., Melki R., Peyrin J. M., Perrier A. L., Propagation of α-synuclein strains within human reconstructed neuronal network. Stem Cell Rep. 12, 230–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicoleau C., Varela C., Bonnefond C., Maury Y., Bugi A., Aubry L., Viegas P., Bourgois-Rocha F., Peschanski M., Perrier A. L., Embryonic stem cells neural differentiation qualifies the role of Wnt/β-Catenin signals in human telencephalic specification and regionalization. Stem Cells 31, 1763–1774 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabb0799/DC1