Reconstruction of deep-sea cephalopod communities in cetacean foraging zones indicates differentiation on prey size, not species.
Abstract
Fundamental insight on predator-prey dynamics in the deep sea is hampered by a lack of combined data on hunting behavior and prey spectra. Deep-sea niche segregation may evolve when predators target specific prey communities, but this hypothesis remains untested. We combined environmental DNA (eDNA) metabarcoding with biologging to assess cephalopod community composition in the deep-sea foraging habitat of two top predator cetaceans. Risso’s dolphin and Cuvier’s beaked whale selectively targeted distinct epi/meso- and bathypelagic foraging zones, holding eDNA of 39 cephalopod taxa, including 22 known prey. Contrary to expectation, extensive taxonomic overlap in prey spectra between foraging zones indicated that predator niche segregation was not driven by prey community composition alone. Instead, intraspecific prey spectrum differences may drive differentiation for hunting fewer, more calorific, mature cephalopods in deeper waters. The novel combination of methods presented here holds great promise to disclose elusive deep-sea predator-prey systems, aiding in their protection.
INTRODUCTION
The pelagic deep sea is the largest and least explored habitat on the planet, harboring a large and unexplored biodiversity (1, 2). Cumulative impacts of climate change and industrial exploitation impose increasing pressure on deep-sea systems, challenging marine ecosystem health and services at a global scale (1, 3). While key to our understanding of food web dynamics, interactions between elusive and sometimes giant deep-sea predators and prey occur outside of the range of human observations and remain virtually unknown, limiting effective conservation (3). Advanced on-animal recorders have revealed extensive use of meso- and bathypelagic waters as foraging grounds by multiple species of cetaceans with diverse, often cephalopod-dominated diets (4–7). As top predators, cetaceans are essential in maintaining marine diversity and ecosystem functioning (8). Their exploitation of the deep sea may have extensive effects on deep-sea prey populations and food webs and constitutes direct ecological linkage between deep and shallow ocean systems.
Despite its apparent homogeneity, the pelagic deep sea hosts a myriad of foraging niches (9). As air-breathing marine predators, cetaceans have evolved a suite of specialized morphological and physiological traits, enabling extreme diving and localization and capture of deep-sea prey (10, 11). Optimal foraging theory predicts that these traits and associated behavioral strategies evolve toward maximization of foraging performance (i.e., net energetic gain) (12). In contrast to their terrestrial counterparts, cetacean predators face a trade-off between selective forces arising from the dependency on air at the surface and prey at depth. Their hunting strategy on remote deep-sea prey puts stringent pressure on the need for efficient foraging, balancing oxygen use (i.e., from modulation of dive depth, duration, and movement energetics) with energetic return [calorific intake per dive (13)]. Hence, in line with their specialized adaptations, deep-diving cetaceans may optimize foraging performance by selective targeting of distinct foraging zones that hold specific prey communities.
Despite the high global biomass and pivotal role of cephalopods in oceanic food webs, knowledge on deep-sea cephalopod community composition is still very limited (14). Many cephalopod species have never been observed alive in their habitat or captured as adults [e.g., (14, 15)]. Cetacean cephalopod prey spectra can be assessed using nets, stomach content analysis, or optical methods (16). Two issues associated with physical and optical sampling of cephalopod diversity are avoidance behavior and patchiness, resulting in sampling bias toward less mobile and more abundant specimens (17). Alternative methods are needed for an efficient and complete assessment of regional cephalopod biodiversity. Environmental DNA (eDNA) metabarcoding enables the detection of species on the basis of genetic material (e.g., mucus and feces) that they release in their environment (18). eDNA has been successfully used to reconstruct the horizontal distribution, diversity and migration of open-ocean nekton (19, 20). Yet, to the best of our knowledge, it has not been used to investigate cephalopod biodiversity in the deep sea.
We investigated the foraging zones and prey spectra in the habitats of two co-occurring, deep-diving cetaceans, Risso’s dolphin (Grampus griseus) and Cuvier’s beaked whale (Ziphius cavirostris), representing two distinct deep-sea foraging strategists, targeting epi/meso- and bathypelagic waters, respectively (4, 6). Stomach content analyses of both species, based on relatively few specimens, show diverse, partially overlapping diets dominated by oceanic deep-sea squids (7). Risso’s dolphins belong to a group of deep-diving cetaceans foraging at depths between the surface and around 800 m (6). Individuals can actively switch between mesopelagic and near-surface foraging, targeting often dynamic prey patches or scattering layers (6, 21).
Beaked whales (Ziphiidae), along with members of the sperm whale family, are the deepest diving cetaceans, routinely foraging at depths beyond 1000 m. Cuvier’s beaked whale holds the current world record for extreme diving, at depths to 2992 m during dives that may exceed 2 hours (4, 22). Prey search is not initiated until several hundreds of meters deep (4) and may continue to the bathyal seafloor (23), but zones of prey capture within this foraging habitat spanning several kilometers have rarely been reported [only in (4)]. Cetacean foraging depth is often inferred from maximum diving depth. Characterization of target prey layers, however, requires assessment of the target foraging zone, which may vary between dives as a function of dynamic variation in the presence of prey [e.g., (21)].
Whereas we expect prey community composition to be a main driver of observed niche segregation in deep-sea predators, methodological challenges associated with sampling cetacean foraging zones in the extreme deep-sea environment have thus far prevented rigorous testing of this hypothesis in the field. Here, we pioneer a combination of methods that enabled matching of prey community composition with deep-sea predator foraging behavior. We combined cephalopod eDNA analysis with biologging of cetacean diving and acoustic behavior and hypothesize that, to efficiently capitalize on nonuniformly distributed deep-sea prey, (i) cetacean predators target discrete foraging zones that (ii) hold specific prey spectra.
RESULTS
Deep-sea foraging niche segregation
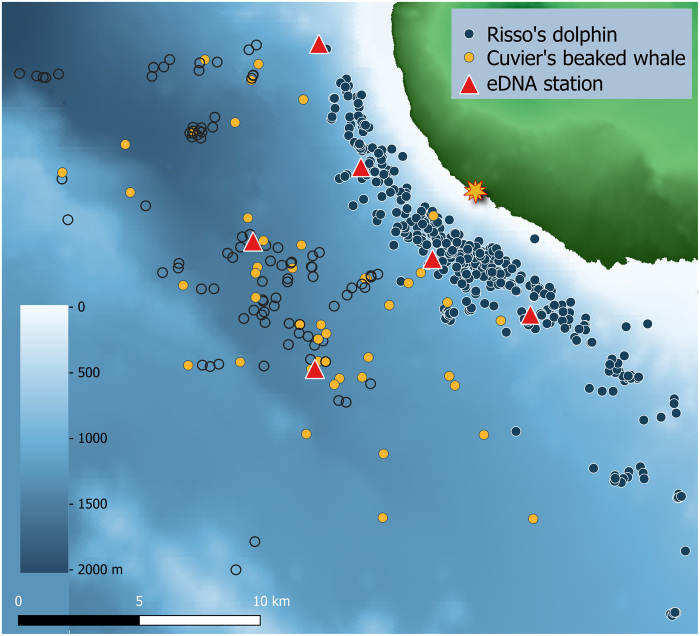
Risso’s dolphins and Cuvier’s beaked whales targeted distinct foraging zones that were spatially segregated in both horizontal and vertical space (Figs. 1 to 4). Risso’s dolphin foraging habitat off Terceira Island (Azores) was situated along a narrow zone over the island slope, at a mean (SD) distance of 3.1 (1.6) km from shore (range, 0.7 to 13.0 km; n = 134 groups, 455 foraging dive observations) and mean bottom depth of 811 (203) m (range, 81 to 1261 m). Cuvier’s beaked whale foraging habitat was located offshore [mean (SD) = 8.8 (3.3) km; range, 4.1 to 21.9 km; n = 47 groups, 148 observations], over deep waters of the bathyal seafloor [bottom depth, 1411 (186) m; Fig. 2 and fig. S1].
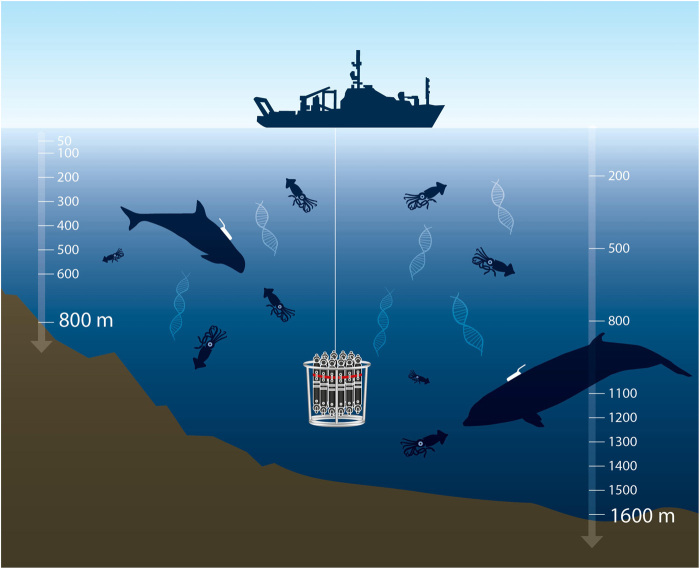
Fig. 1. Pelagic sampling of cephalopod eDNA in cetacean foraging zones.
Targeted sampling of cephalopod eDNA across the foraging zones of two cetacean deep-sea predators, Risso’s dolphin (G. griseus; left) and Cuvier’s beaked whale (Z. cavirostris; right), as determined from biologging of their diving and biosonar foraging behavior using noninvasive sound and movement recording tags (24).
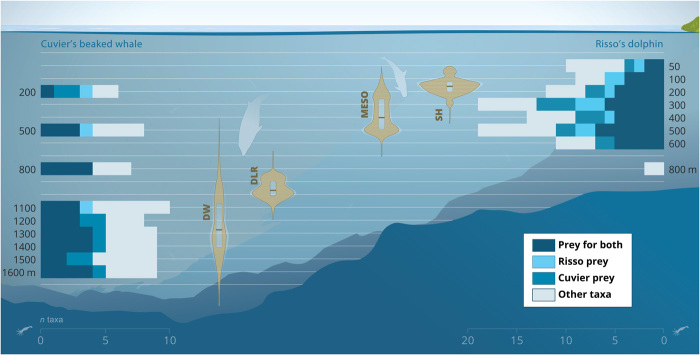
Fig. 4. Deep-sea foraging niche differentiation in cetacean top predators.
Risso’s dolphin (G. griseus) and Cuvier’s beaked whale (Z. cavirostris) depth distributions of prey capture attempts (buzzes; violin plots) show discrete foraging zones for the two cetacean predators. Both predators perform two foraging dive types targeting different zones: mesopelagic (MESO) and shallow (SH) for Risso’s dolphin and deep-wide (DW) and deep layer-restricted (DLR) into the lower meso- and bathypelagic for Cuvier’s beaked whale. Foraging zones of both predators match with the presence of diverse cephalopod prey communities (bar plots; color indicates the number of taxa that are prey for both or either predator).
Fig. 2. Risso’s dolphin and Cuvier’s beaked whale foraging habitat and associated eDNA sampling stations.
eDNA surface to deep-sea CTD sampling locations (red triangles) placed centrally in the foraging habitats of Risso’s dolphin (G. griseus; blue circles, foraging dive locations) and Cuvier’s beaked whale (Z. cavirostris; orange circles, group sighting location; open circles, sequential observations of sighted group), off Terceira Island, Azores. Bathymetry derived from (50).
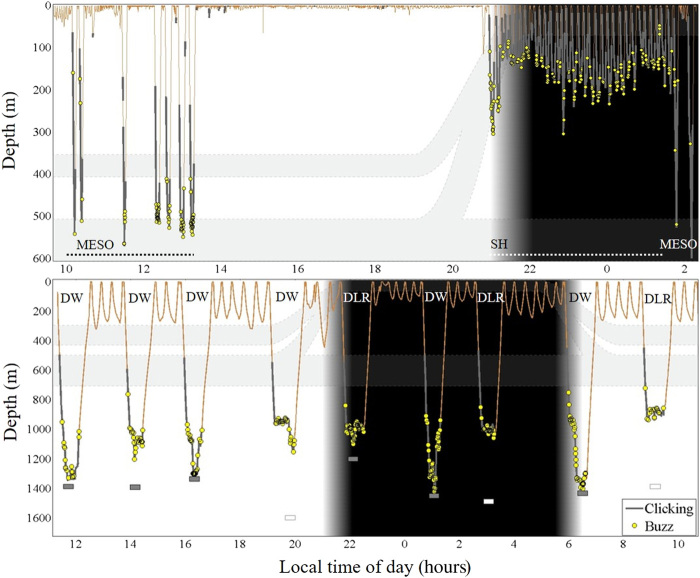
Risso’s dolphin mesopelagic (MESO) and shallow (SH) foraging dive types [mean maximum (SD) depth, 508 (52) and 204 (81) m, respectively], as recorded from eight noninvasive, dive and acoustic recording tags [Dtags ((24)], characterized two foraging zones (Figs. 3 and 4 and tables S1 and S2). Foraging effort during dives was determined from echolocation vocalizations (buzzes; n = 1188 in 145 dives). Depth distribution of buzzes indicated that individuals performed prey capture attempts between depths of 12 and 623 m. Mesopelagic dives targeted prey across a wide zone (66 to 623 m; within-dive foraging depth SD = 126 m) with main effort in the mesopelagic, between 450 and 570 m. Shallow dives targeted a relatively narrow near-surface zone with main effort between 130 and 250 m (within-dive foraging depth SD = 60 m; Figs. 3 and 4).
Fig. 3. Risso’s dolphin and Cuvier’s beaked whale foraging behavior off Terceira Island, Azores.
Example 17-hour and 24-hour dive profile (orange line) and associated foraging vocalizations of Risso’s dolphin (G. griseus; gg15_229a; top) and Cuvier’s beaked whale (Z. cavirostris; zc18_185a; bottom). Echolocation signals (clicking and buzzes) emitted by the tagged individual define foraging effort. Clicking (gray thicker outline) indicates prey search. Buzzes (yellow circles) are emitted at close approach of a target, indicating a prey capture attempt. Both species perform different foraging dive types: Risso’s dolphin, mesopelagic (MESO) and shallow (SH); Cuvier’s beaked whale, deep-wide (DW) and deep layer–restricted (DLR). Gray rectangles, bottom depth at nearest distance from foraging whale; white rectangle, bottom not detected, minimum depth of nearest bottom; gray bands, schematic representation of depth of the deep scattering layers at the Azores (25).
Cuvier’s beaked whale foraging zone was deeper, between 911 and 1782 m [n = 8 tag records and n = 60 dives, 2068 buzzes; mean diving depth (SD) = 1420 (255) m; Figs. 3 and 4 and table S1]. Individuals performed three foraging dive types, based on maximum dive depth and width of the within-dive foraging zone (table S2). Dive type I, deep layer–restricted (DLR; n = 12), were relatively shallow dives [mean diving depth (SD) = 1042 (66) m], with layer-restricted foraging [within-dive foraging depth SD (SD) = 37 (11) m]. Foraging occurred in a narrow-depth zone around the meso-bathypelagic boundary (850 to 1050 m; Figs. 3 and 4). Deep layer-restricted dives were typically pelagic, with 70% of dives remaining at least 450 m above the seafloor. In contrast, dive types II and III showed foraging across a wide depth zone [within-dive foraging depth SD (SD): type II, 113 (42) and type III, 203 (37) m], into the bathypelagic [mean diving depth (SD): type II, 1299 (148) and type III, 1609 (109) m]. Type II represented an intermediate strategy, comparable to type III but over shallower bottom depths (bottom depth restricts both maximum dive depth and potential width of the foraging zone). These types were therefore merged into one dive category: deep-wide (DW; n = 48). Deep-wide dives reached the seafloor (38%), near-bottom waters within 200 m above the seafloor [31%; mean (SD) = 132 (47) m] or remained pelagic [31%; mean (SD) = 317 (72) m]. Deep-wide foraging occurred in pelagic, near-bottom, and bottom habitats, with main effort between 800 and 1700 m (Figs. 3 and 4). In all dives to the bottom, individuals performed foraging buzzes and thus prey capture attempts, up to and after reaching the seafloor.
Cephalopod diversity and zonation identified from eDNA
To sample prey spectra of the two deep-sea predators, we performed CTD (conductivity, temperature, depth) casts at the locations of known foraging habitat of Risso’s dolphin (n = 4) and Cuvier’s beaked whale (n = 2; Figs. 1 and 2). eDNA sampling was conducted at 50- to 300-m intervals between the surface and the mesopelagic or bathyal seafloor at six stations (45 sampling records; 50 to 1600 m; table S3). This enabled the reconstruction of cephalopod community composition spanning the water column (Figs. 4 and 5). Analysis of the resulting nuclear 18S ribosomal RNA (rRNA) and mitochondrial 16S rRNA gene sequences revealed 39 cephalopod taxa, representing 17 families. Most taxa (79%) could be identified to genus (n = 8) or species level (n = 23; Fig. 5 and table S4). The most widely detected taxa were Enoploteuthis leptura (60% of sampling records), Liocranchia reinhardti (38%), Pterygioteuthis sp. (36%), Abralia redfieldi (33%), and Histioteuthis reversa (33%). All remaining taxa were detected in less than 25% of sampling records. Additional cephalopod taxa that could not be identified to family level (Teuthida and Cephalopoda) were present in 64 and 4% of sampling records, respectively (Fig. 5).
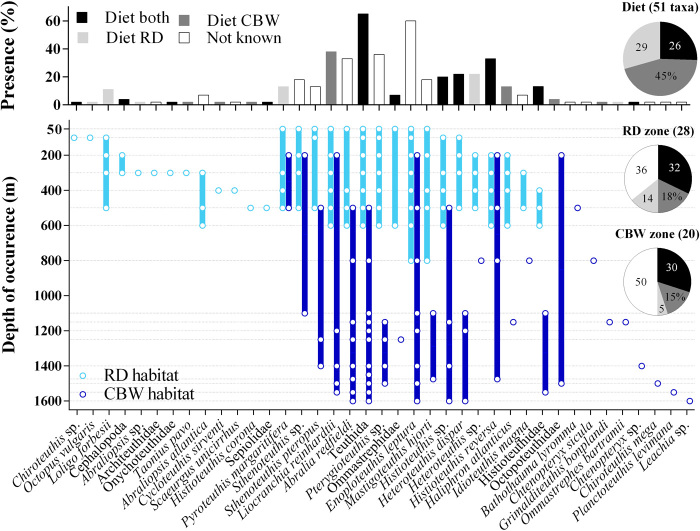
Fig. 5. Cephalopod species community and depth distribution from eDNA reveal cetacean deep-sea prey spectra.
(Top) Frequency of presence (percentage of sampling records) of the 39 cephalopod taxa identified from eDNA. The color indicates occurrence in the diet of both predators (black), one of the predators (dark/light gray), or not reported in the diet (white; table S5). (Bottom) Cephalopod taxon detection depth and range of occurrence (open circles, filled bar) in Risso’s dolphin (G. griseus, RD; light blue) and Cuvier’s beaked whale (Z. cavirostris, CBW; dark blue) habitat, with respective bottom depths of ~800 and ~1600 m. Dotted lines represent sampling depths (Risso’s dolphin habitat, 50 to 800 m; Cuvier’s beaked whale habitat, 200, 500, 800, and 1100 to 1600 m). Pie graphs show the percentage of cephalopod species reported in the diet of both predators, that are prey of both or either predator (top) and the percentage of taxa recorded in the foraging zones that are prey for both or either predator. Numbers indicate percentage of total taxa.
Cephalopod diversity in the Risso’s dolphin zone increased with depth from the surface to 300 to 500 m (9 to 19 taxa; Figs. 4 and 5). This pattern matches the depth of the deep scattering layers (DSLs) at the Azores, which occur at depths between ~300 and 700 m during the day, partially migrating to surface waters at dusk (25). Cuvier’s beaked whale habitat also contained a diverse cephalopod community in the mesopelagic (6 to 8 taxa), which diversified further in the bathypelagic and through to the bottom (8 to 10 taxa). Highest diversity was recorded around 1100 m (Figs. 4 and 5). Taxa showed strong variation in their spatial distribution, ranging from a single depth horizon, to a confined depth range spanning several hundreds of meters, to most of the water column (Fig. 5). Thirteen taxa (37%, at family or lower taxonomic level) occurred in both foraging zones. Fifteen (43%) and seven taxa (20%) were restricted to Risso’s dolphin and Cuvier’s beaked whale foraging zone, respectively (Fig. 5 and fig. S2). Sampling records from the two foraging zones were all classified as part of the same community, i.e., this spatial variation did not translate to significant differences in cephalopod species composition between foraging zones. Moreover, taxonomic overlap between the two habitats was not restricted to the same depth zone, with overlapping community composition between epi/mesopelagic (50 to 600 m) and lower meso/bathypelagic (800 to 1600 m) waters in Risso’s dolphin and Cuvier’s beaked whale habitat, respectively (Fig. 5).
Sampling effort was larger in Risso’s dolphin habitat than in Cuvier’s beaked whale habitat (four versus two casts), and it is possible that more taxa would have been detected with additional casts in the latter. However, additional sampling would result only in few additional detections (mean of two new detections of species with lower eDNA density presence per cast after second cast; fig. S3 and table S4). Total cephalopod diversity in the two foraging habitats was in the same range (14 to 21 versus 18 to 20 taxa per cast, 30 and 26 taxa in total, for Risso’s dolphin and Cuvier’s beaked whale habitat, respectively).
Deep-sea predator-prey dynamics
Whereas the predators displayed strict niche segregation, prey spectra in both foraging zones could provide ample foraging opportunity for either predator (Figs. 4 and 5). Known prey species of both predators were present from near-surface to the deep sea, with overlapping prey spectra recorded across the foraging zones. Presence of suitable prey in the foraging zones was confirmed by consistent, ample recordings of prey capture attempts during foraging dives. On average, Cuvier’s beaked whale performed 30 and 34 prey capture attempts per hour of foraging effort in deep layer-restricted and deep-wide dives. Risso’s dolphin rate of prey capture attempt was higher, with a mean of 41 and 51 buzzes per hour in shallow and mesopelagic dives.
The literature review showed that both cetacean species have diverse diets (31 and 36 cephalopod taxa reported from the North Atlantic and Mediterranean Sea for Risso’s dolphin and Cuvier’s beaked whale, respectively; tables S5 and S6) that vary between geographic locations, in terms of detected prey species and their importance in the diet. However, most cephalopod families are consistently preyed upon, and diet shows a considerable degree of taxonomic overlap between areas (table S5). Both habitats in our study area held a diverse prey community for the cetacean predators, harboring 83% (10 of 12) and 47% (8 of 17) of prey families recorded in Risso’s dolphin and Cuvier’s beaked whale diet, respectively (Fig. 5 and table S4). In total, 46% of taxa recorded by eDNA in the cetacean foraging zones represented known prey [13 of 28 for Risso’s dolphin (46%) and 9 of 20 for Cuvier’s beaked whale (45%)]. The most commonly detected families—Enoploteuthidae, Histioteuthidae, Pyroteuthidae, and Cranchiidae—include main dietary components of Risso’s dolphin (Histioteuthidae) and Cuvier’s beaked whale (Histioteuthidae and Cranchiidae; tables S5 and S6). Accordingly, eDNA of main prey species was detected at high frequency [e.g., H. reversa (33% of sampling records) and Heteroteuthis sp./Heteroteuthis dispar (22%/22%) for Risso’s dolphin and Cranchiidae (two species; 40%) and Histioteuthis sp./H. reversa (20%/33%) for Cuvier’s beaked whale; Fig. 5 and table S4). Thus, combined overlap in diet (Risso’s dolphin, 13 of 31; Cuvier’s beaked whale, 13 of 36 shared known prey taxa; Fig. 5) and prey spectra (Risso’s dolphin prey in Cuvier’s beaked whale habitat, 35% of taxa; Cuvier’s beaked whale prey in Risso’s dolphin habitat, 50% of taxa) resulted in the presence of potentially suitable prey for either predator across the discrete foraging zones.
DISCUSSION
Whereas terrestrial foraging niches are often delineated by structural components, such as the different parts of a tree, habitat structuring in the open ocean is governed by gradients in environmental and oceanographic conditions, including light, pressure, and nutrient and oxygen availability (26). We demonstrate that the resulting zonation and prey distribution can offer specialized foraging niches for mammalian deep-sea predators. We present the first reconstruction of cephalopod communities in the three dimensions of the pelagic environment, using eDNA analysis. Matched with high-resolution data on cetacean foraging behavior, this enabled examination of the relation between the fine-scale distribution of cephalopod prey and top predator foraging zones. Confirming our first hypothesis, the two co-occurring cetaceans exploited entirely discrete deep-sea foraging niches. Their target zones held diverse, overlapping cephalopod species communities, largely composed of known preferred prey. Contrary to expectation, cephalopod community composition alone did not fully explain the strict niche segregation observed between the two deep-sea predators. Instead, cephalopod life history patterns, and the observation of lower prey capture rates by the predator foraging at the largest depths, support an alternative hypothesis. Through the process of ontogenetic migration, performed by several of the most frequently detected cephalopods in the foraging zones, deeper waters may contain larger, more calorific individuals of the same prey (27). Hence cetacean top predators may forage on similar species, but differentiate by targeting individuals of different size and maturity.
Cetacean deep-sea foraging niche segregation
For every hunt, mammalian deep divers need to access a remote foraging zone from the surface. Optimal foraging theory predicts that their foraging strategy should balance the cost of travel in such a way that net energetic return from prey patches is maximized (12). Hence, deep-diving cetaceans have evolved specialized energy- and oxygen-conserving locomotion strategies, modulating speed, fluking patterns, dive duration, and depth as a function of target prey (11).
Risso’s dolphins foraged in the top 600 m and relatively close to shore, targeting layers and patches that can show dynamic patterns in time and space, such as the DSL (6, 21). Although their foraging zone also held suitable prey species for Cuvier’s beaked whale (50% of recorded taxa), they did not capitalize on the dense DSL resources. Instead, Cuvier’s beaked whale targeted prey at greater depths, between 800 m deep in the pelagic and the bathyal seafloor. Dives either targeted relatively narrow, pelagic prey layers (layer-restricted foraging) or covered a wide foraging zone, across the pelagic, benthic boundary layer (up to 200 m from the seafloor) and bottom habitat, with individuals foraging directly at the seafloor.
The extreme diving strategy of Cuvier’s beaked whale may help avoid competition with other mammalian predators. The highly social Risso’s dolphin can benefit from long-term stable associations and large numbers for competition and social defense (28, 29). In contrast, Cuvier’s beaked whale is a cryptic flight strategist that occurs in small, likely ephemeral groups with limited capability for interspecific competition or defense (30). Whereas the shallower foraging zone of Risso’s dolphin is also accessible to many potential cetacean competitors and other marine predators, only few large predators are capable of targeting larger prey at beaked whale foraging depths (30). Extreme breath-hold dives, however, require a high calorific intake to render deep foraging effort energetically rewarding (12).
Deep-sea predator-prey dynamics
Energetic return from prey can be modulated from the number, volume, catchability, and calorific content of individuals targeted. In our study, Cuvier’s beaked whale, on average, targeted 7 to 21 fewer prey per hour of foraging effort than Risso’s dolphin. The combination of a larger body size and higher energetic requirements through more extreme diving predicts that Cuvier’s beaked whale targets larger or more calorific prey (31). Comparative data on calorific content of deep-sea cephalopods of different size and maturity are limited. However, five of seven shared prey families plus another four families in the diet of Cuvier’s beaked whale are known to migrate ontogenetically, including important prey such as H. reversa and cranchiid squids (Fig. 6) (32, 33). Many squids spend the paralarval and juvenile phase in surface waters to profit from increased primary productivity (14, 34). As part of ontogenetic migration, larger and more mature individuals descend to deeper layers for reproduction and better protection against predators (35). For example, mature females of H. reversa have not been captured above 800 m (36), indicating that sexual maturation of this species takes place beyond Risso’s dolphin but inside Cuvier’s beaked whale hunting zone.
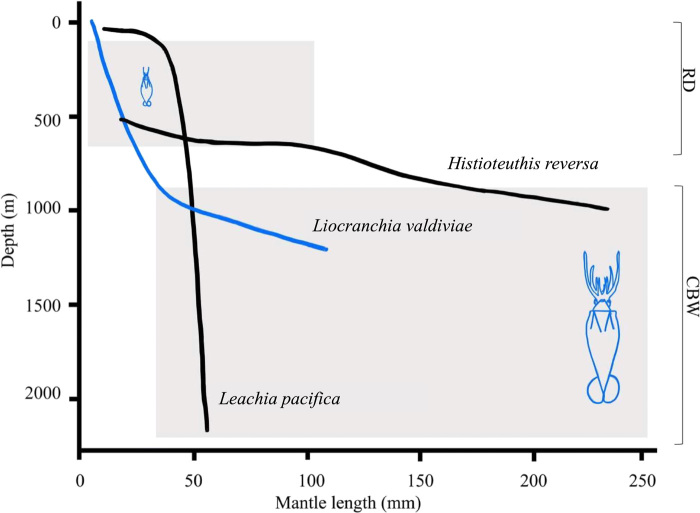
Fig. 6. Deeper waters offer larger, more mature cephalopod prey through vertical ontogenetic migration.
Literature-derived data of ontogenetic migration from epi- and mesopelagic waters to lower meso- and bathypelagic waters for three cephalopod genera predated by both Risso’s dolphin (G. griseus, RD) and Cuvier’s beaked whale (Z. cavirostris, CBW). All three genera were detected in the predator foraging zones off Terceira Island, Azores. Data derived from the Mediterranean Sea (H. reversa) and Pacific Ocean (Hawaii; Leachia pacifica and Liocranchia valdiviae). Juveniles occur shallow, or in the upper 1000 m, and migrate deeper when maturing (36, 63). Inset: Size comparison of juvenile (26 mm) versus adult (240 mm) cranchiid L. valdiviae, occurring in the epi-and mesopelagic versus meso-and bathypelagic zone, respectively [data from the Pacific (64)]. The sister species L. reinhardti was detected off the Azores and is a known prey for Cuvier’s beaked whales. Drawings of L. reinhardti adapted from (64). Gray boxes indicate range of prey sizes present in the respective foraging zones.
Segregation of foraging niches by targeting different ontogenetic stages of deep-sea cephalopods would allow Risso’s dolphin and Cuvier’s beaked whale to feed on different life stages and sizes of the same abundant species in the region, while reducing interspecific competition (Fig. 6). Cephalopods have only one reproductive episode before death (semelparity), leading to a relatively high gonadal investment (14, 16) and mature individuals of enhanced energetic value and volume (i.e., carrying ripe eggs). Moreover, mating and brooding squids can be compromised in their escape responses (35, 37). Whereas Risso’s dolphin foraging was predominantly pelagic, Cuvier’s beaked whale also targeted benthic habitat, suggesting beneficial and possibly enhanced foraging opportunity at and near the bathyal seafloor. Hence, given extensive overlap in diet (and prey spectra in the two predator’s foraging zones), this indicates that enhanced energetic demands from extreme deep dives may be balanced by a prey community offering high-calorific prey, that may be easier to catch.
eDNA elucidates deep-sea cephalopod community
eDNA proved to be an efficient and potent technique to establish diversity and distribution of cephalopods in the deep sea, in particular using a vertically stratified approach. We detected 21 of the 83 cephalopod species that have been reported in waters around the Azores to date, plus an additional two new species for the region (Chiroteuthis mega and Cycloteuthis sirventi) (38), as well as giant squid (Architeuthis; second eDNA detection worldwide (39)). Moreover, cephalopod distribution patterns were biologically meaningful. Risso’s dolphin prey veined squid (Loligo forbesii), for example, was only detected over the island slope at relatively shallow depths (100 to 500 m), matching its known habitat, as well as depth of catches from local fisheries (33, 40). As a confirmed prey species of the Azorean population of Risso’s dolphins (41), its absence offshore may help explain the species’ preference for mesopelagic foraging over slope versus bathyal waters. Strictly deep-sea species such as Planctoteuthis levimana and Chtenopteryx sp. were only recorded at large depth (1600 m) (33). These data show that cephalopod eDNA is not a homogeneous mixture as a result of currents, upwelling, or biological vectors such as whale defecation (42).
Trophic coupling in changing oceans
The predator-prey systems revealed here play a key ecological role in deep-sea food webs. Yet, they have thus far remained largely undocumented because of the challenging deep-sea environment and the elusive nature of both foraging whales and cephalopods. As top predators, cetaceans capture many and large cephalopods and shape the population size and structure of their prey (8). The coexisting predators have differentiated into entirely discrete foraging niches. Their foraging zones, however, are linked by common occurrence of shared cephalopod prey species, creating interdependent food web dynamics between the two systems through the processes of carbon and nutrient transport and emergent facilitation (43). The cephalopods in our study area likely perform considerable migration between depth zones and transport biomass into deeper waters through ontogenetic migration. In combination with the deposition of carcasses after terminal reproduction (44), this implies considerable fluxes of nutrients between depth zones driven by cephalopod movements (45) and predator consumption and defecation (7, 42). Combined data on deep-sea prey spectra available in cetacean target foraging zones also represent critical knowledge aiding in the understanding of marine top predator foraging performance and how this may change under disturbance settings. Absence of knowledge on prey communities has been identified as a limiting factor in the understanding of population-level effects of predator behavioral responses causing impeded foraging, particularly for beaked whales, which are highly sensitive to disturbance from anthropogenic noise (46, 47).
Unravelling the specifics and magnitude of predation coupled with prey distributions and population composition is pivotal for an integrative understanding of the food webs and carbon budgets of deep-sea waters, which cannot directly benefit from nutrient input through primary production. The combination of methodologies pioneered here can be transferred to other predator-prey systems, thus creating major opportunity for the advancement of our knowledge of open-ocean and deep-sea food web processes.
MATERIALS AND METHODS
Foraging habitat
Annual field effort was conducted off Terceira Island (Azores, Portugal), between May and August 2013 to 2019 (Fig. 2). Shore- and vessel-based observations were conducted to record the locations of Cuvier’s beaked whale groups and Risso’s dolphin foraging dives. Risso’s dolphin daytime foraging dive starts are energetic and can be identified from visual observation at the surface. Cuvier’s beaked whales forage throughout the 24-hour day whereby foraging dives are alternated with short series of non-foraging dives (Fig. 3) with limited movement between consecutive surfacing locations (i.e., typically <2 km off Terceira Island; Fig. 2).
Observations were conducted from shore-based lookouts elevated at 65 m or higher above sea level or from the research vessel and comprised (i) standardized surveys, recording all groups present and their location, and (ii) focal follow observations, tracking one group to record location and behavior following a standardized protocol (48). We recorded all foraging dive starts (Risso’s dolphin) and surfacing or dive locations (Cuvier’s beaked whale). Location was recorded using a theodolite linked to a computer running visual tracking software, VADAR (shore based) (49), or using a hand-held GPS (vessel based). Area bathymetry and bottom depth was derived from EMODnet bathymetry data (50) and matched to sighting and dive locations using QGIS (51).
Foraging zones and diet
To identify foraging zone depths, dive and acoustic data were collected from individuals instrumented with noninvasive, high-resolution digital acoustic recording tags [Dtag version 3; 240-kHz sound, 200-Hz accelerometer, magnetometer, and pressure sensor (24)]. Dtags were attached to the dorsal area of individuals using suction cups, with a 6- to 8-m-long hand-held pole. Risso’s dolphin and Cuvier’s beaked whale forage using sound. They detect and track prey by emission of echolocation click series (biosonar) (10). Following the dive start, individuals initiate the search phase, emitting broadband click series at regular intervals [interclick interval (ICI); e.g., ICI mean = 143 ms for Risso’s dolphin (52)]. Upon selection and close approach of a suitable prey item, the click train transitions into a discrete, rapid click series at lower amplitude termed “buzz” (mean ICI = 3.6 ms for Risso’s dolphin), indicating a prey capture attempt (10, 52). Buzzes therefore form accurate indicators of foraging effort and presence of prey. In combination with the dive profile, buzzes were used to define (i) foraging dives (all dives deeper than 20 m with one or more buzz) and (ii) foraging zones (range of buzz depths). The timing of the start and end of echolocation click series and foraging buzzes of the tagged animals were obtained manually through customized auditing scripts from the DTAG toolbox (soundtags.st-andrews.ac.uk) using MATLAB 2014b (MathWorks, MA, USA). Tagged whale clicks can be readily distinguished from clicks produced by nearby conspecifics by their fairly consistent angle of arrival on the two tag hydrophones and the existence of artificial low-frequency energy (<15 kHz), which is absent in clicks produced by conspecifics (53).
The presence of differential dive types in both species was assessed using HMM [package momentuHMM using R (54, 55)]. HMM models classifying foraging dives were run for one to four states, with the two covariates maximum dive depth and SD of within-dive buzz depth (proxy for width of the foraging zone). Parameters improving model fit were retained in the final fitted models. The model with the lowest value of the Akaike information criterion was selected as the best model.
For Cuvier’s beaked whale, we analyzed the closest distance to the bottom for each foraging dive from the echoes of clicks emitted by the tagged whale, which were recorded on the tag (because of physical properties of Risso’s dolphin, returning click-echoes are likely blocked by the melon and rarely recorded on the tag). We used customized scripts from the DTAG toolbox to visualize the echogram, composed of a window of 50 consecutive clicks with all returning echoes up to 0.6 s (mean click train ICI; using MATLAB 2014b). Echoes originating from the seafloor are reverberant and stronger than from objects passing by in the water column (56). The ICI to the next click defines the maximum time window for a returning echo detection and thus closest distance at which the seafloor can be detected, in this case 450 m. The moment of closest proximity to the seafloor was recorded for every dive with clear echo patterns from visual inspection of the echogram. The distance to the seafloor was calculated using the time delay between the produced click and its returning echo [two-way travel time (TWT)] as follows: sound speed in water × TWT/2. This represents maximum closest distance to the seafloor as the angle at which the seafloor is hit by the echolocation beam (and thus distance of travel) will depend on the orientation of the individual with respect to the seafloor.
To test whether Risso’s dolphin and Cuvier’s beaked whale foraging zones held differential prey spectra, we performed a random forest (RF) classification (57). The RF model was set up to aim to discriminate between sampling records originating from the Risso’s dolphin (n = 28; 50 to 600 m) or Cuvier’s beaked whale (n = 10; 900 to 1600 m) foraging zone. The RF model was run using 1000 trees, with random selection of 20 predictor variables (taxa) at each node and using a subsample of two-thirds of the dataset. Model selection was performed by running the full model without the variable with the lowest variable importance. Diet data were derived from the literature, extracting all taxa occurring in the diet of Risso’s dolphin and Cuvier’s beaked whale in comparable regions (North Atlantic Ocean and Mediterranean Sea).
eDNA sample collection, filtration, and extraction
eDNA was sampled centrally in the foraging habitats of Risso’s dolphin (n = 4) and Cuvier’s beaked whale (n = 2; Fig. 1 and table S3) at maximum bottom depths of 922 and 1600 m, respectively. Sampling was conducted from the RV Pelagia in July 2018, overlapping the annual period of tag data collection. At each station, water was collected using Niskin bottles mounted on a CTD rosette (24 × 12-liter bottles) at seven or eight specific depths, in biological triplicates, resulting in a total of 144 discrete water collections. Water was immediately transferred from the Niskin bottles to 2-liter sterile single-packed urine bags and stored at 4°C. Water was then filtered through 0.22-μm Sterivex-GP filters (Merck Millipore) using sterile 60-ml syringes. The Sterivex filters were stored in −80°C until further processing. DNA was extracted from the filters using the QIAGEN DNeasy Blood and Tissue Kit (modified protocol). DNA extracts were quantified using a Qubit fluorometer and the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific) and stored at −20°C. To identify potential contaminations, filtration negative controls (Milli-Q), DNA extraction negative controls (using elution buffer from the extraction kit) and polymerase chain reaction (PCR) negative controls (PCR water instead of DNA extract) were included. Detailed description of all our procedures, including reduction of contamination risks, is provided in Supplementary Methods.
Library preparation and sequencing
Two universal cephalopod primer sets were applied. The first primer set targeted the nuclear 18S rRNA gene yielding an amplicon of 140 to 190 base pairs (bp) (Ceph18S_forward CGCGGCGCTACATATTAGAC and Ceph18S_reverse, GCACTTAACCGACCGTCGAC) (58). The second primer set targets the mitochondrial 16S rRNA gene yielding an amplicon of 212 to 244 bp [CephMLS_forward, TGCGGTATTWTAACTGTACT and CephMLS_reverse, TTATTCCTTRATCACCC (59)]. For PCR amplification, a two-step PCR protocol was used. The first PCR amplified the cephalopod DNA sequences present in the sample with the universal primer sets mentioned above including a sequencing tail, and the second PCR attached a unique indexing primer combination to every PCR product of each sample to be able to pool the samples. PCRs were carried out in duplicate for every biological replicate (resulting in six replicates per site and depth). On every 96-well plate, one negative control and three positive controls were added in duplicate. The first PCR had a total volume of 20 μl and included 7 μl of PCR-grade water, 4 μl of 5× KAPA HiFi Buffer (Roche), 0.6 μl of 10 mM deoxynucleotide triphosphates (dNTPs; Roche), 1 μl of dimethyl sulfoxide (DMSO), 1 μl each of the 10 μM forward and reverse primers, 0.4 μl of KAPA HiFi polymerase (5 U/μl; Roche), and 5 μl of the DNA extract. The PCR program started with an initial denaturing step at 95°C for 5 min, 35 cycles of 98°C for 20 s, annealing temperature of primer for 15 s (62°C for 18S and 55°C for 16S), 72°C for 1 min, and a final extension step of 72°C for 10 min. Fragment sizes were verified on a 2% agarose gel stained with GelRed (Biotium). The PCR product of the first PCR was diluted 1:100 and used as a template for the second PCR. During the second PCR, a unique indexing primer combination was used for every sample. The second PCR was performed in 10-μl volumes of 1 μl of PCR-grade water, 2 μl of 5× KAPA HiFi Buffer (Roche), 0.3 μl of 10 mM dNTPs (Roche), 0.5 μl of DMSO, 0.2 μl of KAPA HiFi polymerase (5 U/μl; Roche), 0.5 μl of a reverse and forward indexing primer, and 5 μl of template.
The PCR products were pooled to equimolar concentrations according to the DNA concentrations measured using a Qubit fluorometer. This resulted in two libraries, one for each primer set. The fragment size of the libraries was validated on a 2% agarose gel and stained with GelRed; the correct bands were cut out and purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research) following the manufacturer’s protocol. The library pool was quantified with the Qubit dsDNA HS Assay Kit (Molecular Probes, Life Technologies). Insert size distribution was determined with the 4200 Tapestation D5000 ScreenTape (Agilent). The working solution was diluted to 2 nM, and loading solution was prepared according to protocol. The library pools were loaded with 8 pM and 20% PhiX spike-in to increase diversity. Sequencing was done on the Illumina MiSeq with the MiSeq Reagent Kit v3 (600 cycles).
Cephalopod reference database
To complement public databases for the targeted mitochondrial 16S rRNA and nuclear 18S rRNA gene, additional cephalopod reference tissue samples, collected during WH383 on RV Walther Herwig III, were barcoded. For the nuclear 18S rRNA gene, the sequences barcoded in (58) were used. For Sanger sequencing of the mitochondrial 16S rRNA gene, the same DNA extracts were used as for the 18S rRNA gene, resulting in 33 successful cephalopod voucher sequences, including 32 different species and 15 different families. Tissue samples had been stored in 70% ethanol at −20°C. DNA was extracted using QIAGEN DNeasy Blood and Tissue following the manufacturer’s protocol. PCRs were performed with the CephMLS primer sets used for metabarcoding. Forward and reverse strand sequencing of the PCR products was performed using the Sanger Sequencing Kit (Applied Biosystems). Primers and low-quality ends were trimmed from the sequences, checked manually, edited, and assembled using CodonCode Aligner (version 3.7.1).
These sequences were added to the public sequence databases used for training IDTAXA that were based on the MIDORI (16S) and SILVA (18S) databases (60), which were updated with sequences retrieved from the National Center for Biotechnology Information (NCBI) GenBank database in June 2020. Briefly, the taxonomic information of sequences contained in the MIDORI (version 20180221 unique) and SILVA (version SILVA_138_SSURef_NR99) databases was updated to reflect the taxonomy assigned by NCBI as of June 2020. Cephalopod sequences were extracted from the databases and used as input for the “eukref_gbretrieve.py” program from the EukRef project (https://unieuk.org/) to recursively query GenBank until no new sequences were retrieved. This resulted in a total of 2116 and 169 sequences representing 144 and 81 genera for 16S and 18S, respectively. These cephalopod sequences were then merged with the full MIDORI or SILVA databases by removing duplicates, and these databases used to generate two IDTAXA training sets for 16S and 18S.
Bioinformatic analysis
After sequencing, the sequences were demultiplexed and sorted by sample without indexing primer. The primer sequences were removed using cutadapt (version 1.18). Untrimmed sequences were discarded, and the maximum accepted error rate was set to 0.1. Allowed errors are mismatches, insertions, or deletions. The pipeline used for data analysis is summarized in Supplementary Methods. The sequencing analysis was conducted with the Divisive Amplicon Denoising Algorithm with an implemented quality-aware model of Illumina amplicon errors (DADA2, version 1.15.0), an R package that corrects for amplicon errors without constructing operational taxonomic units, in RStudio version 1.1.463 (54, 61). Forward and reverse reads were truncated after a quality score ≤2 and merged with at least 80-bp overlap. Only merged sequences ranging from 150 to 300 bp for CephMLS and 80 to 215 bp for Ceph18S were retained (table S7). The taxonomic assignment of the environmental samples and all controls against the training set was performed by IDTAXA with the R package DECIPHER version 2.6.0. IDTAXA was used because it combines features of phylogenetic, distance-based, and machine learning classification methods, which is especially suitable for incomplete training sets and has been shown to have higher accuracy than popular classifiers (62). The confidence threshold for accepting a classification was set to 60%, providing a conservative classification with relatively low mis- and overclassifications. In addition, the BLAST classifier was applied, which assigns a sequence based on its nearest neighbor in a training set.
Ethical statement
Fieldwork was conducted under scientific permits issued by the Direção Regional dos Assuntos do Mar, Secretaria Regional do Mar, Ciência e Tecnologia (Regional Directorate for Science and Technology). Access and Benefit Sharing (ABS) Regulation: Portugal is party to the Nagoya Protocol but does not regulate access to genetic resources. The ABS regulations of the Autonomous Region of the Azores were followed by obtaining the required declaration of conformity, establishing informed consent for the collection and export of biological material from the Direção Regional da Ciência e Tecnologia. Cabo Verde (where some reference tissue samples were collected) has not ratified the Nagoya protocol. To fulfill the national ABS regulations of Cabo Verde, we obtained the required permit for the publication of results based on samples collected in Cabo Verde waters from the Direcção Nacional do Ambiente (National Directorate for the Environment of Cabo Verde).
Acknowledgments
We thank all field team members, particularly E. Falcone, G. Schorr, A. Kok, O. Keller, L. Barcelos, E. Speelman, the OceanEmotion team, T. Morato, F. Reis, H. Slabbekoorn, P. Kraal, S. Gollner, and all scientists and crew of RV Pelagia. P. Tyack and M. Johnson provided tagging equipment and analytical support. Scientists and crew of Walther Herwig (WH383), particularly H. Fock and S. Czudaj, performed cephalopod sampling. The Institute of Clinical Molecular Biology in Kiel provided Sanger sequencing, as supported, in part, by the DFG Clusters of Excellence “Precision Medicine in Chronic Inflammation” and “ROOTS.” T. Naujoks, D. Langfeldt, B. Löscher, and H. de Haas provided technical support. We thank E. B. de Azevedo (ITTAA), R. Gabriel, P. Borges, and J. P. Barreiros of GBA (CE3C) of the University of the Azores for research support and collaboration. Visual data were collected using Logger 2000, developed by the International Fund for Animal Welfare (IFAW) to promote benign and noninvasive research. Graphic design was by C. Kersten (Fig. 1) and S. Oudejans (Fig. 4). Funding: This project was funded by the Office of Naval Research Marine Mammal Biology Program, USA (ONR; grant numbers N00014-15-1-2341 and N00014-17-1-2715; program manager, M. Weise), the Dutch Research Council (NWO; Veni grant 016.Veni.181.086), the German Research Foundation [DFG; Emmy Noether Independent Junior Research Group grant of H.J.T. Hoving (HO 5569/2-1)], and GEOMAR’s POF III OCEANS program. The Netherlands Initiative Changing Oceans (NICO) expedition on RV Pelagia, was funded by NWO and the Royal Netherlands Institute for Sea Research (NIOZ). Author contributions: F.V. and H.J.T.H. conceived the study. F.V., H.J.T.H., and V.J.M. designed the study. F.V., H.J.T.H., V.J.M., D.S.W.d.J., T.B., and M.G.O. conducted the investigation process, performed formal analysis and processing, and drafted the manuscript. T.B., T.B.H.R., and O.P. provided critical support during eDNA analysis and data collection from samples. J.F. performed the sequencing. All authors critically revised the manuscript. All authors gave final approval for publication and agreed to be held accountable for the work performed therein. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials or are available from the PANGAEA repository at https://doi.pangaea.de/10.1594/PANGAEA.926840. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf5908/DC1
REFERENCES AND NOTES
- 1.Danovaro R., Gambi C., Dell’Anno A., Corinaldesi C., Fraschetti S., Vanreusel A., Vincx M., Gooday A. J., Exponential decline of deep-sea ecosystem functioning linked to benthic biodiversity loss. Curr. Biol. 18, 1–8 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Ramirez-Llodra E., Brandt A., Danovaro R., De Mol B., Escobar E., German C. R., Levin L. A., Martinez Arbizu P., Menot L., Buhl-Mortensen P., Narayanaswamy B. E., Smith C. R., Tittensor D. P., Tyler P. A., Vanreusel A., Vecchione M., Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences. 7, 2851–2899 (2010). [Google Scholar]
- 3.Danovaro R., Fanelli E., Aguzzi J., Billett D., Carugati L., Corinaldesi C., Dell’Anno A., Gjerde K., Jamieson A. J., Kark S., Clain C. M., Levin L., Levin N., Ramirez-Llodra E., Ruhl H., Smith C. R., Snelgrove P. V. R., Thomsen L., Van Dover C. L., Yasuhara M., Ecological variables for developing a global deep-ocean monitoring and conservation strategy. Nat. Ecol. Evol. 4, 181–192 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Tyack P. L., Johnson M., Soto N. A., Sturlese A., Madsen P. T., Extreme diving of beaked whales. J. Exp. Biol. 209, 4238–4253 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Watwood S. L., Miller P. J. O., Johnson M., Madsen P. T., Tyack P. L., Deep-diving foraging behaviour of sperm whales (Physeter macrocephalus). J. Anim. Ecol. 75, 814–825 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Arranz P., Benoit-Bird K. J., Southall B. L., Calambokidis J., Friedlaender A. S., Tyack P. L., Risso’s dolphins plan foraging dives. J. Exp. Biol. 221, jeb165209 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Clarke M. R., Cephalopods as prey. III. Cetaceans. Philos. Trans. Biol. Sci. 351, 1053–1065 (1996). [Google Scholar]
- 8.Heithaus M. R., Frid A., Wirsing A. J., Worm B., Predicting ecological consequences of marine top predator declines. Trends Ecol. Evol. 23, 202–210 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Benoit-Bird K. J., Southall B. L., Moline M. A., Predator-guided sampling reveals biotic structure in the bathypelagic. Proc. R. Soc. B Biol. Sci. 283, 20152457 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson M., Madsen P. T., Zimmer W. M. X., Aguilar de Soto N., Tyack P. L., Beaked whales echolocate on prey. Proc. R. Soc. Lond. B Biol. Sci. 271, S383–S386 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.G. L. Kooyman, Diving physiology, in Encyclopedia of Marine Mammals, W.F. Perrin, B. Würsig, J.G.M. Thewissen, Eds. (Elsevier, 2009), pp. 327–332. [Google Scholar]
- 12.Thompson D., Fedak M. A., How long should a dive last? A simple model of foraging decisions by breath-hold divers in a patchy environment. Anim. Behav. 61, 287–296 (2001). [Google Scholar]
- 13.Hazen E. L., Friedlaender A. S., Goldbogen J. A., Blue whales (Balaenoptera musculus) optimize foraging efficiency by balancing oxygen use and energy gain as a function of prey density. Sci. Adv. 1, e1500469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.P. R. Boyle, P. Rodhouse, Oceanic and deep-sea species, in Cephalopods: Ecology and Fisheries (Blackwell Science, 2005), pp. 176–204. [Google Scholar]
- 15.Laptikhovsky V. V., Fock H., Piatkowski U., Schwarz R., Hoving H. J. T., Reproductive strategies of deep-sea squid (Mastigoteuthidae, Chiroteuthidae, Batoteuthidae and Cranchiidae). Mar. Biol. 166, 85 (2019). [Google Scholar]
- 16.H. J. T. Hoving, J. A. A. Perez, K. S. R. Bolstad, H. E. Braid, A. B. Evans, D. Fuchs, H. Judkins, J. T. Kelly, J. E. A. R. Marian, R. Nakajima, U. Piatkowski, A. Reid, M. Vecchione, J. C. C. Xavier, The Study of Deep-Sea Cephalopods (Advances in Marine Biology, 2014), vol. 67; https://linkinghub.elsevier.com/retrieve/pii/B9780128002872000032. [DOI] [PubMed]
- 17.Wormuth J. H., Roper C. F. E., Quantitative sampling of oceanic cephalopods by nets: Problems and recommendations. Biol. Oceanogr. 2, 357–377 (1983). [Google Scholar]
- 18.Ruppert K. M., Kline R. J., Rahman M. S., Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 17, e00547 (2019). [Google Scholar]
- 19.Thomsen P. F., Møller P. R., Sigsgaard E. E., Knudsen S. W., Jørgensen O. A., Willerslev E., Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLOS ONE 11, e0165252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigsgaard E. E., Nielsen I. B., Bach S. S., Lorenzen E. D., Robinson D. P., Knudsen S. W., Pedersen M. W., Jaidah M. A., Orlando L., Willerslev E., Møller P. R., Thomsen P. F., Population characteristics of a large whale shark aggregation inferred from seawater environmental DNA. Nat. Ecol. Evol. 1, 0004 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Benoit-Bird K. J., Southall B. L., Moline M. A., Dynamic foraging in Risso’s dolphins revealed in 4-dimensions. Mar. Ecol. Prog. Ser. 632, 10.3354/meps13157 (2019). [Google Scholar]
- 22.Schorr G. S., Falcone E. A., Moretti D. J., Andrews R. D., First long-term behavioral records from Cuvier’s beaked whales (Ziphius cavirostris) reveal record-breaking dives. PLOS ONE 9, e92633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barlow J., Schorr G. S., Falcone E. A., Moretti D. J., Variation in dive behavior of Cuvier’s beaked whales with seafloor depth, time-of-day, and lunar illumination. Mar. Ecol. Prog. Ser. 644, 199–214 (2020). [Google Scholar]
- 24.Johnson M. P., Tyack P. L., A digital acoustic recording tag for measuring the response of wild marine mammals to sound. IEEE J. Ocean. Eng. 28, 3–12 (2003). [Google Scholar]
- 25.Cascão I., Domokos R., Lammers M. O., Santos R. S., Silva M. A., Seamount effects on the diel vertical migration and spatial structure of micronekton. Prog. Oceanogr. 175, 1–13 (2019). [Google Scholar]
- 26.D. C. B. Miller, P. A. Wheeler, Biological Oceanography (Wiley-Backwell, ed. 2, 2012). [Google Scholar]
- 27.Arkhipkin A. I., Bjørke H., Ontogenetic changes in morphometric and reproductive indices of the squid Gonatus fabricii (Oegopsida, Gonatidae) in the Norwegian Sea. Polar Biol. 22, 357–365 (1999). [Google Scholar]
- 28.Shane S. H., Relationship between pilot whales and Risso’s dolphins at Santa Catalina Island, California, USA. Mar. Ecol. Prog. Ser. 123, 5–11 (1995). [Google Scholar]
- 29.Hartman K. L., Visser F., Hendriks A. J. E., Social structure of Risso’s dolphins (Grampus griseus) at the Azores: A stratified community based on highly associated social units. Can. J. Zool. 86, 294–306 (2008). [Google Scholar]
- 30.Aguilar de Soto N., Visser F., Tyack P. L., Alcazar J., Ruxton G., Arranz P., Madsen P. T., Johnson M., Fear of killer whales drives extreme synchrony in deep diving beaked whales. Sci. Rep. 10, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carbone C., Houston A. I., The optimal allocation of time over the dive cycle: An approach based on aerobic and anaerobic respiration. Anim. Behav. 51, 1247–1255 (1996). [Google Scholar]
- 32.Clarke M. R., Oceanic cephalopod distribution and species diversity in the eastern North Atlantic. Arquipél. Life Mar. Sci. 23A, 27–46 (2006). [Google Scholar]
- 33.Jereb P., Roper C. F. E., Cephalopods of the world. An annotated and illustrated catalogue of cephalopod species known to date. FAO Species Cat. Fish. Purp. 2, 605 (2010). [Google Scholar]
- 34.C. F. E. Roper, R. E. Young, Vertical Distribution of Pelagic Cephalopods (Smithsonian Contributions to Zoology, Smithsonian Institution Press, 1975); 10.5479/si.00810282.209. [DOI]
- 35.Seibel B. A., Robison B. H., Haddock S. H. D., Post-spawning egg care by a squid. Nature 438, 929 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Quetglas A., de Mesa A., Ordines F., Grau A., Life history of the deep-sea cephalopod family Histioteuthidae in the western Mediterranean. Deep Sea Res. Part Oceanogr. Res. Pap. 57, 999–1008 (2010). [Google Scholar]
- 37.Hoving H. J. T., Vecchione M., Mating behavior of a deep-sea squid revealed by in situ videography and the study of archived specimens. Biol. Bull. 223, 263–267 (2012). [DOI] [PubMed] [Google Scholar]
- 38.K. E. Carpenter, N. De angelis, Eds. The Living Marine Resources of the Eastern Central Atlantic. FAO Species Identification Guide for Fishery Purposes, vol. 1 (FAO, Rome, 2014), pp. 1–663. [Google Scholar]
- 39.Wada T., Doi H., Togaki D., Kaida R., Nagano M., Katano I., Suzuki M., Ohtani T., Mitsuhashi H., Exploring a legendary giant squid: An environmental DNA approach. Mar. Biol. 167, 160 (2020). [Google Scholar]
- 40.Porteiro F. M., The present status of the squid fishery (Loligo forbesi) in the Azores archipelago. Fish. Res. 21, 243–253 (1994). [Google Scholar]
- 41.Cruz M. J., Jordao V. L., Pereira J. G., Santos R. S., Silva M. A., Risso’s dolphin depredation in the Azorean hand-jig squid fishery: Assessing the impacts and evaluating effectiveness of acoustic deterrents. ICES J. Mar. Sci. 71, 2608–2620 (2014). [Google Scholar]
- 42.Doughty C. E., Roman J., Faurby S., Wolf A., Haque A., Bakker E. S., Malhi Y., Dunning J. B., Svenning J.-C., Global nutrient transport in a world of giants. Proc. Natl. Acad. Sci. 113, 868–873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. M. de Roos, L. Persson, Population and Community Ecology of Ontogenetic Development (Monographs in Population Biology, Princeton Univ. Press, 2013). [Google Scholar]
- 44.Hoving H. J. T., Bush S. L., Haddock S. H. D., Robison B. H., Bathyal feasting: Post-spawning squid as a source of carbon for deep-sea benthic communities. Proc. R. Soc. B Biol. Sci. 284, 20172096 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arkhipkin A. I., Squid as nutrient vectors linking Southwest Atlantic marine ecosystems. Deep Sea Res. Part II Top. Stud. Oceanogr. 95, 7–20 (2013). [Google Scholar]
- 46.Southall B. L., Benoit-Bird K. J., Moline M. A., Moretti D., Quantifying deep-sea predator–prey dynamics: Implications of biological heterogeneity for beaked whale conservation. J. Appl. Ecol. 56, 1040–1049 (2019). [Google Scholar]
- 47.Benoit-Bird K. J., Southall B. L., Moline M. A., Claridge D. E., Dunn C. A., Dolan K. A., Moretti D. J., Critical threshold identified in the functional relationship between beaked whales and their prey. Mar. Ecol. Prog. Ser. 654, 1–16 (2020). [Google Scholar]
- 48.Visser F., Miller P. J. O., Antunes R. N., Oudejans M. G., Mackenzie M. L., Aoki K., Lam F. P. A., Kvadsheim P. H., Huisman J., Tyack P. L., The social context of individual foraging behaviour in long-finned pilot whales (Globicephala melas). Behaviour 151, 1453–1477 (2014). [Google Scholar]
- 49.E. Kniest, Visual Detection and Ranging (VADAR), version 1.45.06 (Univ. Newctle. Callaghan Aust., 2012). [Google Scholar]
- 50.EMODnet Bathymetry Consortium, EMODnet Digital Bathymetry (EMODnet Bathymetry Consort., 2016); 10.12770/c7b53704-999d-4721-b1a3-04ec60c87238. [DOI]
- 51.QGIS.org, 2020. QGIS Geographic Information System. QGIS Association. http://www.qgis.org.
- 52.Arranz P., DeRuiter S. L., Stimpert A. K., Neves S., Friedlaender A. S., Goldbogen J. A., Visser F., Calambokidis J., Southall B. L., Tyack P. L., Discrimination of fast click-series produced by tagged Risso’s dolphins (Grampus griseus) for echolocation or communication. J. Exp. Biol. 219, 2898–2907 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Zimmer W. M. X., Johnson M. P., Madsen P. T., Tyack P. L., Echolocation clicks of free-ranging Cuvier’s beaked whales (Ziphius cavirostris). J. Acoust. Soc. Am. 117, 3919–3927 (2005). [DOI] [PubMed] [Google Scholar]
- 54.McClintock B. T., Michelot T., momentuHMM: R package for generalized hidden Markov models of animal movement. Methods Ecol. Evol. 9, 1518–1530 (2018). [Google Scholar]
- 55.R Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018); www.R-project.org/.
- 56.Arranz P., de Soto N. A., Madsen P. T., Brito A., Bordes F., Johnson M., Following a foraging fish-finder: Diel habitat use of Blainville’s beaked whales revealed by echolocation. PLOS ONE 6, e28353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breiman L., Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 58.de Jonge D. S. W., Merten V., Bayer T., Puebla O., Reusch T. B. H., Hoving H.-J. T., A novel metabarcoding primer pair for environmental DNA analysis of Cephalopoda (Mollusca) targeting the nuclear 18S rRNA region. R. Soc. Open Sci. 8, 201388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jarman S. N., Redd K. S., Gales N. J., Group-specific primers for amplifying DNA sequences that identify Amphipoda, Cephalopoda, Echinodermata, Gastropoda, Isopoda, Ostracoda and Thoracica. Mol. Ecol. Notes. 6, 268–271 (2006). [Google Scholar]
- 60.Leray M., Ho S.-L., Lin I.-J., Machida R. J., MIDORI server: A webserver for taxonomic assignment of unknown metazoan mitochondrial-encoded sequences using a curated database. Bioinformatics 34, 3753–3754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murali A., Bhargava A., Wright E. S., IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 6, 140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young R. E., Vertical distribution and photosensitive vesicles of pelagic cephalopods from Hawaiian Waters. Fish. Bull. 76, 583–615 (1978). [Google Scholar]
- 64.A. Evans, “A systematic review of the squid family Cranchiidae (Cephalopoda: Oegopsida) in the Pacific Ocean,” thesis, Auckland University of Technology, Auckland, New Zealand (2018). [Google Scholar]
- 65.Blanco C., Raduán M. Á., Raga J. A., Diet of Risso’s dolphin (Grampus griseus) in the western Mediterranean Sea. Sci. Mar. 70, 407–411 (2006). [Google Scholar]
- 66.Bloch D., Life history of Risso’s dolphin (Grampus griseus) (G. Cuvier, 1812) in the Faroe Islands. Aquat. Mamm. 38, 250–266 (2012). [Google Scholar]
- 67.Clarke M. R., Pascoe P. L., The stomach contents of a Risso’s dolphin (Grampus griseus) stranded at Thurlestone, South Devon. J. Mar. Biol. Assoc. U. K. 65, 663–665 (1985). [Google Scholar]
- 68.Milani C. B., Vella A., Vidoris P., Christidis A., Koutrakis E., Frantzis A., Miliou A., Kallianiotis A., Cetacean stranding and diet analyses in the North Aegean Sea (Greece). J. Mar. Biol. Assoc. U. K. 98, 1011–1028 (2018). [Google Scholar]
- 69.Oztürk B., Salman A., Öztürk A. A., Tonay A., Cephalopod remains in the diet of stripped dolphins (Stenella coeruleoalba) and Risso’s dolphins (Grampus griseus) in the eastern Mediteranean. Vie Milieu - Life Environ. 57, 53–59 (2007). [Google Scholar]
- 70.Würtz M., Poggi R., Clarke M. R., Cephalopods from the stomachs of a Risso’s dolphin (Grampus griseus) from the Mediterranean. J. Mar. Biol. Assoc. UK 72, 861–867 (1992). [Google Scholar]
- 71.Spitz J., Cherel Y., Bertin S., Kiszka J., Dewez A., Ridoux V., Prey preferences among the community of deep-diving odontocetes from the Bay of Biscay, Northeast Atlantic. Deep Sea Res. Part Oceanogr. Res. Pap. 58, 273–282 (2011). [Google Scholar]
- 72.Blanco C., Raga J. A., Cephalopod prey of two Ziphius cavirostris (Cetacea) stranded on the western Mediterranean coast. J. Mar. Biol. Assoc. UK 80, 381–382 (2000). [Google Scholar]
- 73.Kovačić I., Đuras M., Gomerčić H., Lucić H., Gomerčić T., Stomach contents of two Cuvier’s beaked whales (Ziphius cavirostris) stranded in the Adriatic Sea. Mar. Biodivers. Rec. 3, E19 (2011). [Google Scholar]
- 74.Santos M. B., Pierce G. J., Herman J., López A., Guerra A., Mente E., Clarke M. R., Feeding ecology of Cuvier’s beaked whale (Ziphius cavirostris): A review with new information on the diet of this species. J. Mar. Biol. Assoc. UK 81, 687–694 (2001). [Google Scholar]
- 75.Santos M. B., Martin V., Arbelo M., Fernández A., Pierce G. J., Insights into the diet of beaked whales from the atypical mass stranding in the Canary Islands in September 2002. J. Mar. Biol. Assoc. U. K. 87, 243–251 (2007). [Google Scholar]
- 76.Andruszkiewicz E. A., Starks H. A., Chavez F. P., Sassoubre L. M., Block B. A., Boehm A. B., Biomonitoring of marine vertebrates in Monterey Bay using eDNA metabarcoding. PLOS ONE 12, e0176343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pedersen M. W., Overballe-Petersen S., Ermini L., Der Sarkissian C., Haile J., Hellstrom M., Spens J., Thomsen P. F., Bohmann K., Cappellini E., Schnell I. B., Wales N. A., Carøe C., Campos P. F., Schmidt A. M. Z., Gilbert M. T. P., Hansen A. J., Orlando L., Willerslev E., Ancient and modern environmental DNA. Philos. Trans. R. Soc. B Biol. Sci. 370, 20130383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.M. Vecchione, R. E. Young, Ancistrocheiridae Pfeffer 1912, Ancistrocheirus lesueurii (Orbigny 1842). Ancistrocheirus Gray 1849 (2016); http://tolweb.org/Ancistrocheirus_lesueurii/19632/2016.11.16, in The Tree of Life Web Project; http://tolweb.org/.
- 79.M. Vecchione, T. Kubodera, R. E. Young, Taningia Joubin 1931, Taningia danae Joubin 1931 (2010); http://tolweb.org/Taningia_danae/19840/2010.08.22, in The Tree of Life Web Project, http://tolweb.org/.
- 80.Pinfield R., Dillane E., Runge A. K. W., Evans A., Mirimin L., Niemann J., Reed T. E., Reid D. G., Rogan E., Samarra F. I. P., Sigsgaard E. E., Foote A. D., False-negative detections from environmental DNA collected in the presence of large numbers of killer whales (Orcinus orca). Environ. DNA. 1, 316–328 (2019). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabf5908/DC1