The first high-resolution structure of an adenovirus with nonmammalian host provides information on adenovirus evolution.
Abstract
Of five known adenovirus genera, high-resolution structures are available only for mammalian-infecting mastadenoviruses. We present the first high-resolution structure of an adenovirus with nonmammalian host: lizard atadenovirus LAdV-2. We find a large conformational difference in the internal vertex protein IIIa between mast- and atadenoviruses, induced by the presence of an extended polypeptide. This polypeptide, and α-helical clusters beneath the facet, likely correspond to genus-specific proteins LH2 and p32k. Another genus-specific protein, LH3, with a fold typical of bacteriophage tailspikes, contacts the capsid surface via a triskelion structure identical to that used by mastadenovirus protein IX, revealing a conserved capsid-binding motif and an ancient gene duplication event. Our data also suggest that mastadenovirus E1B-55 K was exapted from the atadenovirus-like LH3 protein. This work provides new information on the evolution of adenoviruses, emphasizing the importance of minor coat proteins for determining specific physicochemical properties of virions and most likely their tropism.
INTRODUCTION
Adenoviruses (AdVs) are nonenveloped, double-stranded DNA (dsDNA) viruses with a ~95-nm, pseudoT = 25 icosahedral capsid. Most of our current knowledge on AdVs comes from those infecting humans, in particular human AdV type 5 (HAdV-C5). Each HAdV-C5 capsid facet has 12 trimers of the major coat protein, hexon. At the vertices, five penton base subunits form the penton base, attached to trimers of the fiber protein that projects from the capsid surface to engage the primary cell receptor for entry. Minor coat proteins IIIa, VI, and VIII on the inner capsid surface and IX on the outer surface contribute to the intricate network of interactions required to assemble and stabilize an AdV capsid (1, 2). Positively charged core proteins V, VII, and μ are packed together with the 35 kilo–base pair, linear dsDNA genome within the capsid (3). Neither the core proteins, the genome, nor large parts of capsid proteins IIIa and VI follow the icosahedral symmetry. Many of these internal components are assembled in an immature form and cleaved at multiple sites by the viral protease during maturation (4). Complex composition and large size have made AdVs a challenging case for structural biology techniques (5).
Recombinant human AdVs (HAdVs) are widely used as experimental therapeutic vectors. However, their successful use in the clinic faces several obstacles, among them the preexisting immunity in the population or tropism control. A possible approach to overcome these problems is the use of nonhuman AdV (6). The International Committee for the Taxonomy of Viruses recognizes, at present, five different AdV genera: Mastadenovirus (infecting mammals), Aviadenovirus (birds), Atadenovirus (reptiles, ruminants, and birds), Siadenovirus (amphibians, birds, and reptiles), and Ichtadenovirus (a single isolate from white sturgeon) (7). A sixth genus, Testadenovirus (turtles), is under consideration (https://talk.ictvonline.org/files/proposals/animal_dna_viruses_and_retroviruses/m/animal_dna_ec_approved/10392/).
Atadenoviruses have a broad host range but are thought to have reptilian origin (8). They are very common in livestock: Approximately 50% of the known bovine AdVs are atadenoviruses, and OAdV-7 has worldwide prevalence. They have also been found in a variety of other ruminants, such as goat or deer, suggesting that a host switch from reptiles happened before the divergence of the present ruminant lineages. A common occurrence in ruminant atadenovirus genomes is duplication of genes found as single copies in the reptilian atadenoviruses (9). In cattle, some atadenoviruses have been implicated in diseases of the respiratory tract, including cases of pneumoenteritis with high mortality in calves. They have also been found to cause mild diarrhea in cattle and sheep and are often found in the animal intestines (10). An avian atadenovirus, duck AdV 1 (DAdV-1), causes the so-called egg drop syndrome, resulting in deficiencies in the egg shell and consequently decreased egg production. The presence of AdVs in reptiles was detected over 30 years ago (11). LAdV-2 was found in a Mexican beaded lizard (Heloderma horridum) dead in a zoo outbreak. It was the first lizard AdV isolated and propagated in cell culture (although several had already been detected) (12). In lizards, atadenoviruses are mostly found in the liver and intestine and cause disease in the gastrointestinal tract, liver, and neurological system. They are commonly spread through the fecal-oral route, although transmission via aerosols is also possible (13, 14).
While, at present, representatives of three different HAdV species have been solved at high resolution (1, 2, 15–17), structural analyses on nonhuman AdVs are limited to medium resolution studies on bat (BtAdV 250-A) (18), canine (CAdV-2) (19), ovine (OAdV-7) (20), and snake (SnAdV-1) AdVs (21), as well as a 4-Å resolution report on incomplete bovine AdV (BAdV-3) capsids (22). All these are mastadenoviruses, except for OAdV-7 and SnAdV-1, which are atadenoviruses. There are no high-resolution data on the complete virion structure for any AdV not belonging to the Mastadenovirus genus.
A group of genes involved in DNA replication, DNA encapsidation, and viral particle formation has been found in all members of the family Adenoviridae. These occupy the central part of the genome, while the left and right ends hold regions of higher variability with genus-specific genes (23). Among the virion components, minor coat protein IX and core protein V are unique to mastadenoviruses. In atadenoviruses, genus-specific proteins LH2, LH3, and p32K have also been found in the virion (20, 21, 24–26). LH stands for left hand, indicating genus-specific genes located near the left end of the genome (and rightward-transcribed) (27). Similarly to protein IX in mastadenoviruses, LH3 is located on the outer capsid surface, while p32k and LH2 have been tentatively assigned to positions on the inner capsid surface and may be substituting for protein V (20, 21, 24). Because of its limited sequence homology with the gene placed in a similar position in the HAdV genome, LH3 was initially referred to as E1B-55 K (26). However, protein E1B-55 K in HAdV is not part of the virion but is expressed in infected cells where it carries a large variety of functions, including promotion of genome replication and transcription, degradation of antiviral factors, or deregulation of the cell cycle (28).
Medium resolution cryo–electron microscopy (cryo-EM) combined with crystallography showed that, in SnAdV-1, LH3 has a trimeric β helix fold typical of bacteriophage host attachment proteins. This study also indicated extensive contacts between LH3 and hexons surrounding the icosahedral and local threefold axes in the capsid, corroborating a role for LH3 in the high thermostability of atadenovirus capsids (20, 21). However, it did not solve the full extent of the LH3 protein, as the form crystallized lacked ~30 residues from the N terminus. We now present the first near-atomic resolution structure of an atadenovirus, LAdV-2, showing in detail how the network of minor coat proteins differs between genera.
RESULTS
Structure determination of LAdV-2
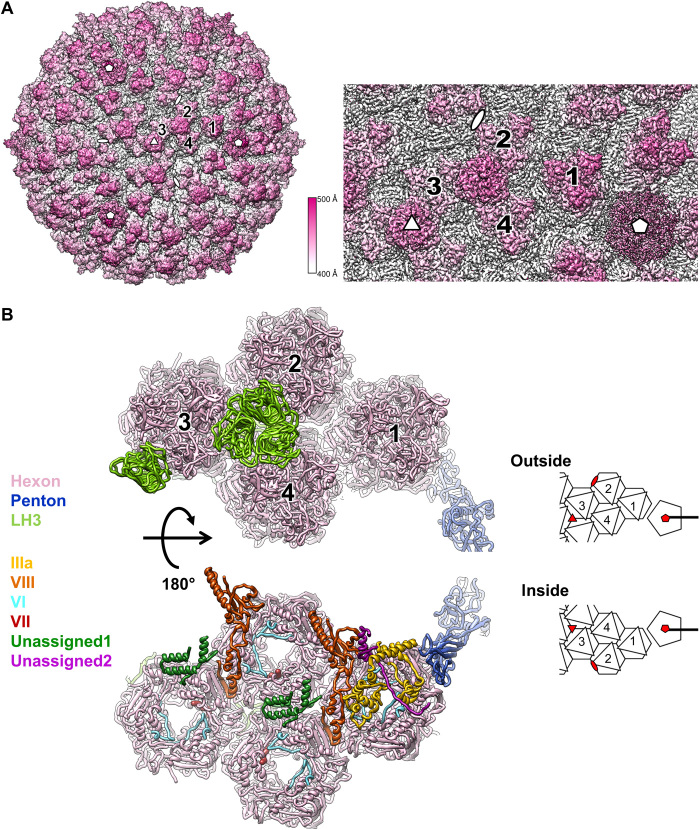
The 3.4-Å resolution cryo-EM map of LAdV-2 (Fig. 1A and fig. S1, A and B) showed the expected particle morphology, with a pseudoT = 25 icosahedral geometry, 940-Å diameter, 12 hexon trimers per facet, and outer protrusions corresponding to the genus-specific protein LH3 (21). The penton base density was weaker than the hexon shell (fig. S1C) and had lower resolution (~4 Å; fig. S1D) and higher B factors in the refined model (fig. S1E). All these observations suggested partial penton loss. Local reconstruction followed by three-dimensional (3D) classification confirmed that over 50% of the vertices were vacant (fig. S2A). Similarly to previous studies on mastadenoviruses, no indication of ordered structures or concentric shells was observed in the core (fig. S1C). We traced more than 13,400 residues in the icosahedral asymmetric unit (AU), corresponding to four independent hexon trimers, one penton base monomer, four independent copies of LH3, one copy of IIIa, and two independent copies of protein VIII. We observed density inside all hexons that we interpret as the N-terminal peptide of protein VI (nine copies per AU) and protein VII (three copies), on the basis of the fitted chain length and of the latest HAdV-C5 structure (2), although we were unable to unequivocally assign residue identities. In addition, 255 residues were modeled as poly-alanine chains in density where it was not possible to assign a sequence (Fig. 1B and table S1).
Fig. 1. LAdV-2 cryo-EM map and molecular model.
(A) Right: Surface rendering of the LAdV-2 three-dimensional (3D) map colored by radius from white to pink, as indicated by the color scale. A zoom-in on the area corresponding to the icosahedral asymmetric unit (AU) is shown at the right. The four hexon trimers in an AU are numbered 1 to 4. White symbols indicate the icosahedral fivefold (pentagon), threefold (triangle), and twofold (oval) symmetry axes. (B) Ribbon representation of the proteins traced in the AU, colored as indicated by the legend at the left. Depth cueing (fading to white) is used to give an impression of distance to the viewer. Two views are provided, as seen from outside (top) or inside (bottom) the capsid, with a cartoon at the right-hand side for guidance. In the cartoon, icosahedral symmetry axes are indicated by red symbols.
Main capsid proteins: Hexon and penton
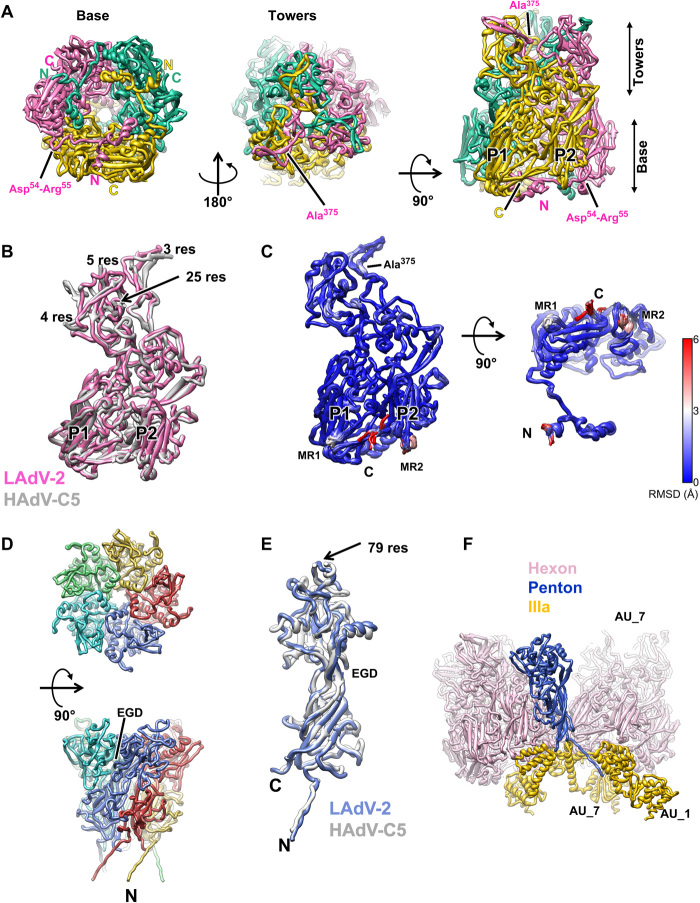
The LAdV-2 hexon protein is shorter than its HAdV-C5 counterpart (909 versus 952 residues) and could be traced almost in its entirety for all 12 monomers in the AU (table S1 and fig. S2B). As expected from previous studies (1, 15, 29), the overall structure of hexon, with a double jelly roll normal to the capsid surface, and extensive loops that imbricate to form the trimer towers, is conserved (Fig. 2A). The root mean square distance (RMSD) for all Cα atoms when comparing to HAdV-C5 hexon [Protein Data Bank (PDB) ID 6B1T] is ~4 Å. The largest differences between mast- and atadenovirus hexons occur at the towers (Fig. 2B). In HAdVs, mobile loops on the tower surface are formed by hypervariable regions (HVRs) that contribute to define the virus serotype (30). In the most recent HAdV-C5 cryo-EM structure, the hexon model contains several gaps in these loops (Ala138-Gln164, Gln253-Leu258, Thr273-Asn279, and Thr433-Asn437) where the chain could not be traced (2). The loops in the LAdV-2 hexon tower are shorter than those in HAdV-C5 and could be modeled without gaps even if the density was in some regions slightly fragmented. In LAdV-2, the hexon towers adopt very similar conformations in all hexon monomers (RMSD <2 Å for all Cα atoms) except for Ala374-Ala375, located at the valley formed by the trimer towers (Fig. 2, A and C). In HAdV-C5, residues at this valley are involved in contacts with coagulation factors (31). It is believed that the exposed HVRs have evolved in response to the evolutionary pressure of the host immune system. Simpler loops might correlate with a different immune system in reptiles (32).
Fig. 2. LAdV-2 main capsid proteins: hexon and penton.
(A) Hexon trimer as seen from inside (left) or outside (center) the capsid and in a side view (right). The capsomer base and towers are indicated, as well as the N terminus (N) and C terminus (C) of each monomer. The two β barrels forming the double jelly roll are labeled P1 and P2. (B) Superposition of the LAdV-2 and HAdV-C5 (PDB ID 6B1T) hexon monomers. The length in residues of the HAdV-C5 untraced loops is indicated. (C) Superposition of the 12 hexon monomers in the LAdV-2 AU, colored by RMSD according to the scale at the right-hand side. MR1 and MR2: mobile regions. (D) Penton base pentamer as seen from outside the capsid (top) and in a side view (bottom). The N terminus of one monomer and the location of the EGD sequence are indicated. (E) Superposition of the LAdV-2 and HAdV-C5 penton base monomers. The length in residues (res) of the HAdV-C5 penton RGD loop is indicated. (F) Interactions of one penton base monomer with other proteins. AU_1 and AU_7 indicate molecules in neighboring AUs labeled as in fig. S3.
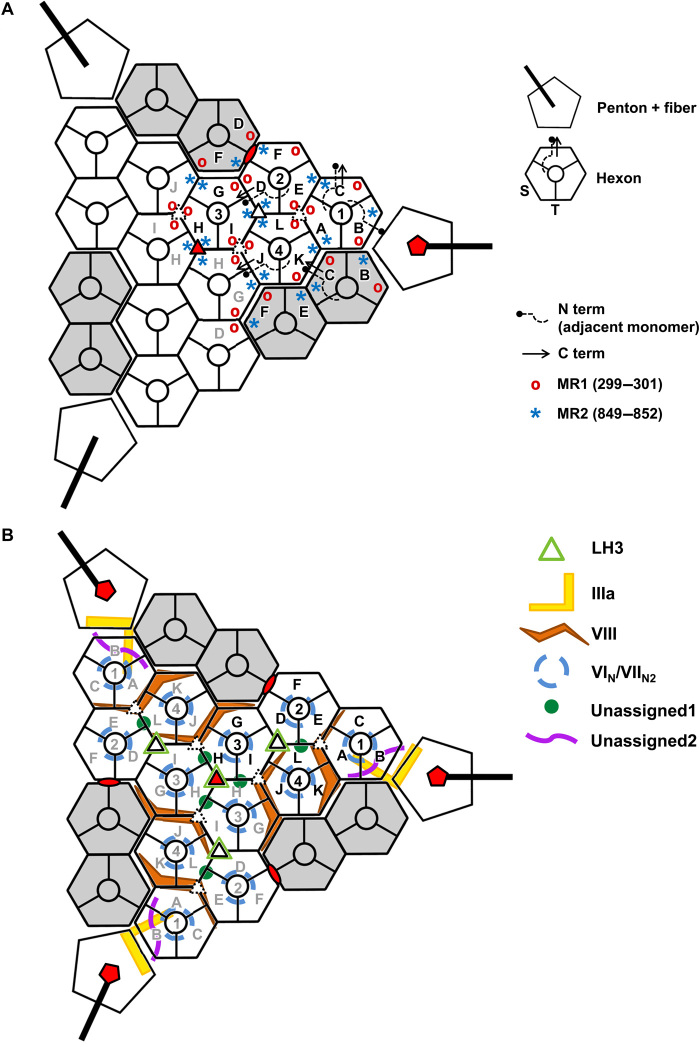
All other major differences (RMSD > 2 Å) between hexon monomers in LAdV-2 are located at the trimer base and comprise residues Met1-Glu2 (N terminus), Thr299-Gln301 [mobile region 1 (MR1)], Val849-Ala852 (MR2), and Ser904-Ala908 (C terminus) (Fig. 2C). In HAdV-C5, the N- and C-terminal regions were also observed to adopt different conformations in the different monomers, contributing to the quasi-equivalent interactions, but these regions were more extensive in the human virus (six and seven amino acids, respectively) (tables S2 to S8) (1). The other two hexon mobile regions (MR1 and MR2) are located at alternate vertices of the pseudohexagonal base, one below each β-jelly roll, and are involved in various hexon-hexon interactions and hexon interactions with penton base or minor coat proteins IIIa and VIII (mostly, MR2) (Fig. 3A and tables S2 to S8). Residues Asp54-Arg55 (Fig. 2A), also located on the inner hexon surface, can establish a pair of contiguous salt bridges at each of two local twofold symmetry axes at the facet edges: between hexon 1 (H1) in the AU and its fivefold symmetry neighbor, and between H4 in the AU and H2 in the next facet (H2_AU6 in fig. S3 and table S3, TT interactions). These residues are conserved in HAdV-C5 (Asp59-Arg60), suggesting that ionic interactions may be important for assembly of face plates onto the icosahedron. This interaction, however, is not present at the icosahedral twofold axis. The same two residues in a different monomer of H1 and H4 establish another possible pair of electrostatic interactions with the internal minor coat protein VIII (see below).
Fig. 3. Schematics illustrating the LAdV-2 protein networks.
Hexons 1 to 4 in one AU are depicted in white and labeled with black text; those in neighboring AUs in the same icosahedral facet are in white with gray labels; and those in adjacent facets are in gray. Letters A to L identify different polypeptide chains as in tables S2 to S9. Red symbols indicate the icosahedral fivefold (pentagon), threefold (triangle), and twofold (oval) symmetry axes. Closed and broken white triangles indicate two different local threefold symmetry axes. (A) Hexon conformational adaptability: location of mobile hexon regions in the capsid. Hexon mobile regions MR1 and MR2 in one AU and its immediate neighbors are indicated. A few hexon N and C termini are depicted as an example. In the hexon schematics legend, S designates the facet of the hexon pseudo-hexagonal base formed by the two β barrels in a single monomer, and T indicates the facet of the hexon pseudo-hexagonal base formed by two β barrels belonging to two adjacent hexon monomers (1). (B) Location of the minor coat proteins. Only LH3 is located on the outer capsid surface.
Despite the low occupancy, the penton base polypeptide chain could be traced in its entirety, including its N-terminal arm (Met1-Gly16) that stretches toward the viral core and corresponds to the same feature formed by residues 37 to 51 in the HAdV-C5 penton base (Fig. 2, D and E, and fig. S2B). Similarly to hexon, the penton base protein is shorter in LAdV-2 than in HAdV-C5 (451 versus 571 residues), and the main differences reside in the outer surface loops, with the rest of the structure very close to that of HAdV-C5 (2.1-Å RMSD) (1, 33). Most notably, the flexible, 79-residue-long loop containing the RGD (Arg-Gly-Asp) sequence motif in HAdV-C5 is not present in LAdV-2 (Fig. 2E). The closest sequence pattern in the LAdV-2 penton base would be EGD (Glu-Gly-Asp) at residues 133 to 135, but these are located at the interface between penton monomers and therefore not available for interactions with cell receptors (Fig. 2E). Lack of the RGD integrin binding motif suggests that the atadenovirus internalization mechanism differs from that of the extensively characterized HAdVs (34).
Penton base monomers are arranged in an oblique fashion around the fivefold symmetry axis, facilitating their bonding with multiple neighboring molecules. Each penton base monomer interacts with the two neighboring peripentonal hexon trimers (SP interactions, table S5). In addition, the N-terminal arm interacts with two different copies of protein IIIa, while the C-terminal residue is positioned within reach of interactions with a third IIIa molecule (table S6 and Fig. 2F). In HAdV-5, the first 37 residues could not be traced and were proposed to plunge into the viral core. Here we can trace the penton base chain starting from residue 1, indicating that this penton-core interaction would not be present in LAdV-2.
Weak, fragmented density was observed on the pentamer surface. This density corresponds to the fivefold averaged N-terminal peptide of the fibers (binding to the groove between penton base monomers) and the start of the shaft (fig. S2C) (35). Fiber density is poor because of the low penton occupancy in our preparation, and the symmetry mismatch between the trimeric fibers and the pentameric penton base is worsened in LAdV-2 by the presence of two different fiber proteins, one of them attached in triplets to some of the vertices (25). Consequently, fibers were not traced in this icosahedrally averaged map.
External minor coat protein: LH3
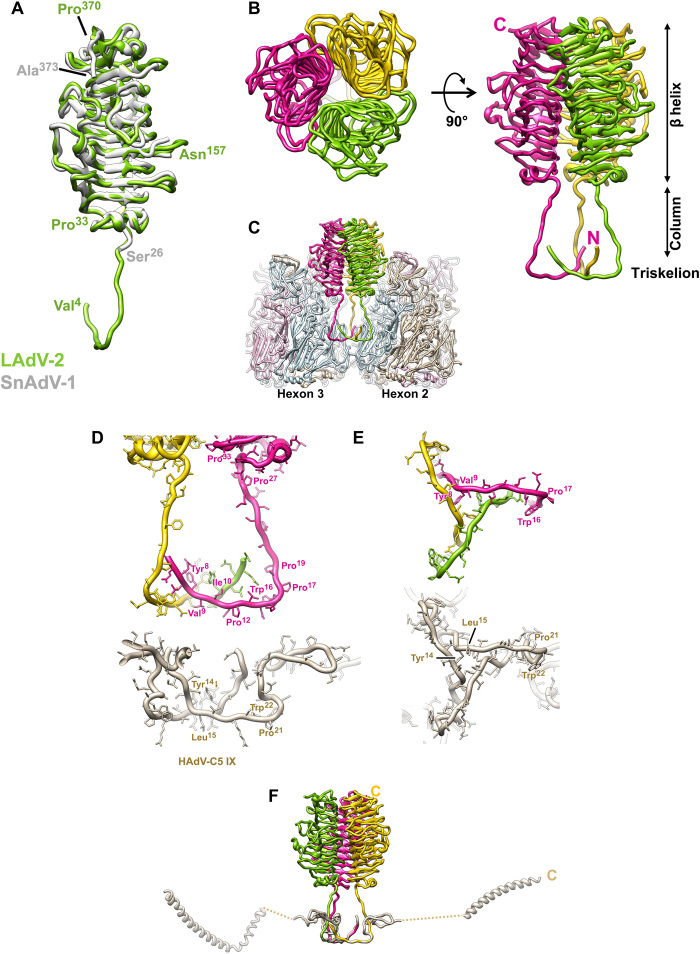
There are 12 copies of protein LH3 per icosahedral facet, organized in four trimers. One of these is located at the icosahedral threefold axis, while the other three occupy the local threefold axes between hexons 2, 3, and 4 in each AU (Figs. 1B and 3B). The structure of a stable fragment of recombinant SnAdV-1 LH3 (residues 28 to 373) was previously solved by crystallography, showing that it folds as a right-handed β helix with three strands per turn, an architecture notably similar to bacteriophage tailspikes (21). LH3 in LAdV-2 is three residues shorter (370 versus 373) and has a 62.2% sequence identity with the SnAdV-1 protein, according to the experimentally determined sequence (21). The cryo-EM map of LH3 in its native environment (the viral particle) allowed us to trace most of the chain in all four positions in the AU; only the first three residues lacked density in our LAdV-2 cryo-EM map (table S1). The overall structure of LAdV-2 LH3 is very similar to that of SnAdV-1 LH3 in the β helix domain (1.2-Å RMSD for 336 Cα atoms; Fig. 4A). A loop formed by residues Asp155-Ser162 in the outer trimer surface was absent in most of the SnAdV-1 LH3 crystal structures, indicating mobility, but could be unequivocally traced in LAdV-2, where it is fixed in place by interactions with the surrounding hexons (Fig. 4A, fig. S4A, and table S9).
Fig. 4. LAdV-2 external minor coat proteins: LH3.
(A) Superposition of the LAdV-2 and SnAdV-1 (PDB ID 5G5O) LH3 structures. The first (Val4 and Ser26) and last (Pro370 and Ala373) residues traced for each protein are indicated. Asn157 is labeled to indicate the location of the Asp155-Ser162 loop mentioned in the text. (B) Overall view of the LH3 trimer seen from outside the capsid (left) and in a side view (right). The N and C termini of the pink subunit are labeled, as well as the β helix, column and triskelion domains. (C) Hexons 2 and 3 are represented to show the LH3 trimer in the capsid context. The monomers in each neighboring hexon trimer are colored light blue, light pink, and tan. (D) Comparison between the triskelions formed by LAdV-2 LH3 and HAdV-C5 protein IX, in a side view similar to (C). (E) As in (D), but in a view from the capsid surface. In this view, the LH3 column domains would travel away from the reader. (F) LH3 trimer overlapped with the HAdV-C5 protein IX trimer, to show the large difference in the fold beyond the triskelion domain. The C terminus for one monomer of IX and one monomer of LH3 are labeled.
We can now see that each LH3 monomer has an extended N-terminal domain that lays on the capsid surface at the valley formed by three surrounding hexon trimers and, after bending almost at a square angle, goes up forming a column that reaches the β helix domain (Fig. 4, A to C). The column domain presents certain mobility, as shown by its weak density in the cryo-EM map (fig. S4B). A region rich in proline and glycine residues (27-PPGTLLPG-34) allows the formation of a sharp bend when the flexible column gives way to the β helix domain (Fig. 4, A and D).
The three N-terminal domains (4-VEELYVINPINQWPAP-19) in each LH3 trimer join to form a triskelion-shaped joint on the surface of the valley between three hexon capsomers (Fig. 4, B and C).This structure is highly similar to the one formed by the N terminus of polypeptide IX in HAdV-C5, which occupies the same positions in the capsid, stabilizing the nine central hexons in each capsid facet (Fig. 4, D and E) (1). LH3 residues Leu7, Tyr8, Val9, and Ile10 from each monomer form the triskelion hydrophobic core. A series of prolines (Pro12, Pro17, and Pro19) facilitate the sharp bend from the triskelion to the column (Fig. 4D). A hydrophobic core (Tyr14-Leu15) also underpins the polypeptide IX triskelion in HAdV-C5 (1). A conserved tryptophan (Trp16 in LH3 and Trp22 in IX) is located at the outermost vertices of the triskelion, also with a nearby proline (Pro21) in IX (Fig. 4, D and E). A core of two hydrophobic residues, as well as the tryptophan, is conserved in atadenoviruses (fig. S4C). Sequence alignment of the complete LH3 and IX proteins does not detect the similarity at the triskelion region. However, the structure that we present here reveals that such region is conserved (Fig. 4, D and E).
Previous evidence has shown that the triskelion is critical for incorporation of both protein IX and LH3 to the capsid during assembly (20, 36, 37). Conversely, a large variability is tolerated for the rest of the “triskelion containing” proteins: in HAdV-C5 IX, long, unstructured regions travel on the capsid surface all the way from the triskelions on the central plate of the facet to a C-terminal helix at the capsid edges (Fig. 4F) (1). In nonhuman mastadenovirus IX proteins, shorter connecting regions climb away from the capsid surface and end in a helix directly on top of the triskelion (18, 19, 22). Atadenovirus LH3 has also a short connecting region that moves perpendicularly to the capsid surface (the column domain) but ends in a completely different fold (the β helix) of the C-terminal domain (Fig. 4F).
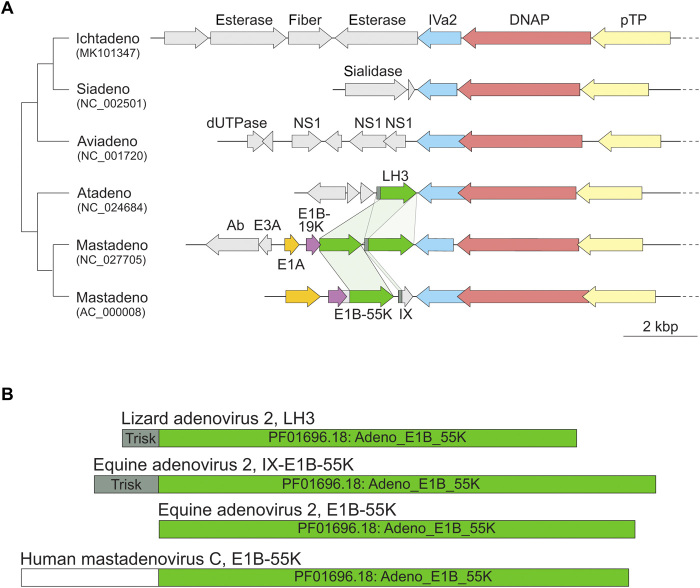
It is now interesting to consider the genomic context for proteins IX in HAdV-C5 and LH3 in LAdV-2. When atadenoviruses were first characterized, LH3 was considered a homolog for the HAdV E1B-55 K protein, because of its location at the left end of the genome and a limited similarity with mastadenovirus E1B-55 K sequences (26). However, the discovery that LH3 was present in the virion, unlike E1B-55 K that is a multifunctional protein involved in cell control and transformation, shed doubts on the homology (24, 28). Thus, we set out to reevaluate the evolutionary relationship between atadenoviral LH3 and E1B-55 K homologs conserved in mastadenoviruses. To this end, we constructed a profile hidden Markov model (HMM) of the LH3 homologs and compared this profile HMM against available HMM databases using HHsearch (38). The profile of E1B-55 K (PF01696.18; Adeno_E1B_55K) was retrieved with a highly significant probability of 99.86% and the alignment covered the near entirety of both profiles (table S10 and fig. S5). Despite relatively low pairwise sequence identity (15%), the predicted secondary structure elements showed close correspondence between the two proteins. The same result was obtained when the E1B-55 K profile was used as a query: the profile HMM of LAdV-2 LH3 was retrieved as the second best hit (following the self-hit) with the probability of 99.95%. Thus, we conclude that LH3 and E1B-55 K proteins are true homologs. However, the homologous regions do not encompass the 30 N-terminal residues of LAdV-2 LH3 (fig. S5), which correspond to the triskelion domain, consistent with the fact that E1B-55 K is not bound to the capsid. Other significant hits were to various β helix fold bacterial carbohydrate-active enzymes and phage tailspike proteins (table S10), both of which are known to form trimeric assemblies, as observed in the LH3 structure.
In the HAdV-C5 genome [National Center for Biotechnology Information (NCBI) Reference Sequence: AC_000008.1], the E1B-55 K gene is followed by the IX gene. That is, in HAdV-C5, the triskelion region is located downstream from the putative β helix fold, while in LAdV-2 LH3, the positions are reversed, with the triskelion at the N terminus of the protein. This rearrangement would suggest a swapping of gene parts or gene duplication followed by rearrangement. To investigate the evolutionary history of this locus, we compared the left genomic regions of AdVs representing all five recognized genera (Fig. 5A). The LH3/E1B-55 K homologs were identified exclusively in members of the Atadenovirus and Mastadenovirus genera, which form sister clades in phylogenetic analyses (39). Whereas all atadenoviruses share a single LH3 ortholog, we identified two mastadenoviruses [equine AdV 2 (NC_027705) and red squirrel AdV 1 (NC_035207)], which, in addition to the E1B-55 K ortholog conserved in all mastadenoviruses, encode LH3 homologs but lack gene IX (Fig. 5A). Importantly, the LH3 homolog in equine AdV 2 carries the triskelion region (40). These observations are best compatible with duplication of the ancestral LH3-like gene in the ancestor of mastadenoviruses, followed by subfunctionalization of one of the gene copies, involving loss of the triskelion region, thereby giving rise to the E1B-55 K homologs. Notably, some E1B-55 K orthologs have undergone further evolution by accreting additional domains in the N-terminal region (Fig. 5B). Addition of the large N-terminal extension, together with triskelion loss, would have abrogated binding to the capsid shell in E1B-55 K, resulting in its acquiring a whole new, nonstructural functionality. Subsequently, the β helix domain (but not the triskelion domain) in the ancestral LH3 copy has been replaced in most mastadenoviral lineages, with the notable exception of equine AdV 2, with an unrelated sequence, producing the α-helical domains in protein IX and possibly conferring new properties to the viral particle (e.g., tropism).
Fig. 5. Comparison of the E1B-55 K/LH3 locus in the family Adenoviridae.
(A) Genome alignment of AdVs representing the five genera currently recognized. The genomes are indicated with the corresponding GenBank accession numbers and the abbreviated names of the genera, Ichtadenovirus, Siadenovirus, Aviadenovirus, Atadenovirus, and Mastadenovirus. In each case, only the left extremity of the genome which in atadenoviruses and mastadenoviruses encompasses the E1B-55 K/LH3 genes is shown. The genomes are aligned according to the gene encoding the IVa2 protein. Homologous genes are colored with the same color and, in the case of E1B-55 K/LH3 homologs, are connected by light green shading. Note that mastadenovirus equine AdV 2 (NC_027705) encodes two copies of E1B-55 K/LH3 homologs. The schematic cladogram on the left represents the established phylogenetic relationships within the family Adenoviridae (39). DNAP: DNA polymerase; pTP: pre-terminal protein; dUTPase: deoxyuridine-triphosphatase; kbp: kilo-basepair. (B) Domain organization of the E1B-55 K/LH3 homologs in LAdV-2 (Atadenovirus) and two mastadenoviruses, equine AdV 2 and human mastadenovirus C. The Pfam domain Adeno_E1B_55K (accession number PF01696.18) and the triskelion domain (Trisk) are colored with different shades of green.
In our previous report on the structure of SnAdV-1, we fitted the crystal structure of recombinant LH3 (lacking the N-terminal domain) into a low-resolution cryo-EM map of the viral particle and estimated the LH3 regions interacting with hexons using a hexon homology model (21). This report indicated that each LH3 monomer could interact with three hexon monomers located in two different trimers. Now, we have the high-resolution structure of both the complete LH3 protein and the surrounding hexons, which provides a much clearer picture of the contacts between LH3 and its neighbors in their biological context. We now appreciate that the hexon-LH3 interaction network is even more complex than previously proposed (table S9 and fig. S6). On the one hand, the β helix domain of each LH3 monomer interacts with the towers of four hexon monomers from two different trimers [fig. S6A, β helix: yellow LH3 monomer (chain S) interacts with monomers tan and blue in trimer 2 and with monomers pink and tan in trimer 3]. The Asp155-Ser162 loop that was mobile in the recombinant protein is now ordered by these interactions. On the other hand, the triskelion region interacts with the base of three hexon monomers from three different trimers [fig. S6A, triskelion: yellow LH3 monomer (chain S) interacts with the blue monomer of the three surrounding hexons]. That is, each LH3 monomer bridges the surrounding capsomers by contacts with six hexon monomers located in three different trimers. These extensive interactions between LH3 and hexon contrast with those established by protein IX in HAdV-C5, located at a more basal position between the hexons except for the C-terminal helix bundle (1). Together with the strong intramolecular interactions of both hexon and LH3 trimers (21, 41), this extensive interlacing probably accounts for much of the increased thermostability of the atadenovirus capsids (fig. S6B) (21). Hexon surface charge is predominantly negative, while the LH3 surface has alternating positive and negative regions, suggesting an electrostatic component in the capsid stabilizing interaction (fig. S6C).
Internal minor coat proteins: Protein VIII
Polypeptide VIII is 50 residues longer in LAdV-2 than in HAdV-C5 (278 versus 227 residues). Assuming the same sequence specificity for the mast- and atadenovirus maturation proteases, LAdV-2 protein VIII would be cleaved after residues 121 (LHGG-A), 172 (LRGG-S), and 203 (LQGS-G). That is, the central excised region is 82 residues long, 33 residues longer than in HAdV-C5, where it stretches from residues Gly110 to Arg159 (4).
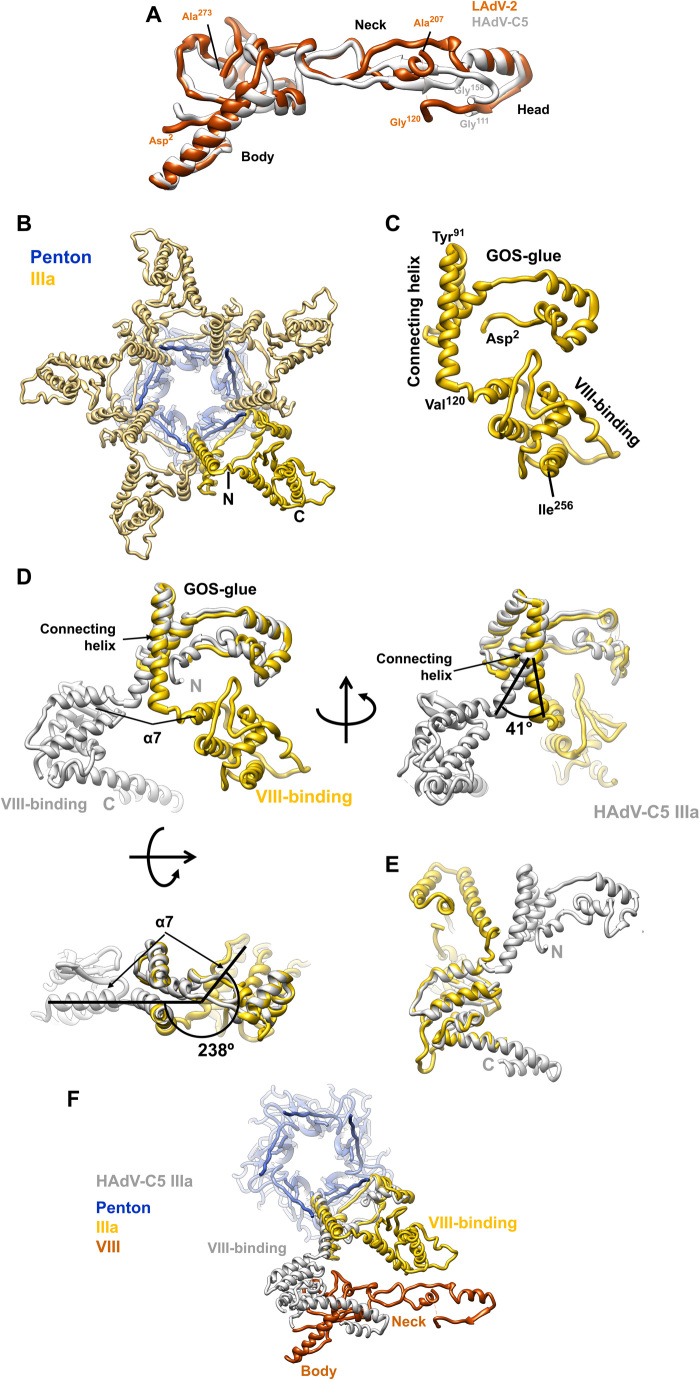
There are two copies of protein VIII per AU, located on the inner capsid surface (Fig. 1B). In both of them, we have been able to trace most of the chain for the N-terminal (residues 2 to 120) and C-terminal (207 to 273) fragments (table S1, Fig. 6A, and fig. S7A), consistently with the predicted maturation cleavages. Similarly to HAdV-C5, in LAdV-2, the protein VIII fold can be described as forming three domains: body (residues 2 to 77 and 225 to 273), neck (78 to 89 and 207 to 224), and head (90 to 120) (Fig. 6A). However, the protein folds differ more than those of hexon and penton base (RMSD, 7.75 Å for 173 Cα atoms). The largest differences are located at the neck, an extended domain which in HAdV-C5 has a small, two-stranded β sheet. In LAdV-2, however, the only secondary structure element in the neck is a three-turn α helix. The neck domain also encompasses the gap left by the maturation cleavage, unlike in HAdV-C5 where the gap is in the head domain. These differences observed in the neck domain relate to differences in the interactions between protein VIII with IIIa and to other elements of the AU that are present in LAdV-2 but not in HAdV-C5 (see below).
Fig. 6. LAdV-2 internal minor coat proteins: VIII and IIIa.
(A) Superposition of the LAdV-2 and HAdV-C5 protein VIII structures. The body, neck, and head domains are indicated, as well as the positions of the N- and C-terminal residues in the structure and the residues flanking the central gap. (B) A view from inside the capsid along a fivefold axis showing the ring of protein IIIa. One copy of IIIa is highlighted in vivid yellow. (C) Structure of the IIIa monomer. The GOS-glue and VIII-binding domains, as well as the connecting helix, the first and last residues traced, and those flanking the connecting helix are indicated. (D) Comparison between the LAdV-2 (yellow) and HAdV-C5 (gray) protein IIIa structures, presented in their original position in the capsid, in which the GOS-glue domains and part of the connecting helix overlap. Three points of view are shown, and angles between the connecting helices and between the first helix in the VIII-binding domain of each protein (α7) are indicated. (E) Superposition of the two VIII-binding domains showing the fold similarity. (F) LAdV-2 vertex proteins depicted together with HAdV-C5 protein IIIa, to show the effect of the large conformational change on its interactions with protein VIII.
One copy of protein VIII (chain P) is located beneath the central plate of the facet, around the icosahedral threefold axis, while the other one (chain O) joins the central plate to the proteins in the peripentonal region (Fig. 1B and 3B). Each copy of protein VIII interacts with four different hexon trimers, one of them located in a different facet from the other three, therefore stabilizing each facet and riveting it to the next across the icosahedron edges (tables S7 and S8 and Figs. 1B and 3B). In the head domain, protein VIII residues 108 to 113 form a β strand that interacts with one of the jelly rolls in the neighboring hexon trimer via a β sheet augmentation (tables S7 and S8, Fig. 1B, and fig. S7B). Another β sheet augmentation established between VIII and IIIa in HAdV-C5 (1) is absent in LAdV-2 (see below).
Residues Asp54-Arg55 in H1 and H4 may establish electrostatic interactions with a pair of consecutive charged amino acids (Asp100-Lys101) located at an α helix in the head domain of VIII (fig. S7, C and D, and tables S7 and S8). Only one-half of this interaction would be conserved across genera, because structure-guided sequence alignment indicates that in HAdV-C5, the same position corresponds to Glu98-Val99, lacking the basic residue in the pair (fig. S7, D and E).
Internal minor coat proteins: Protein IIIa
Five monomers of protein IIIa (one per AU) form a ring beneath each vertex, bridging the penton with the peripentonal hexons (Figs. 3B and 6B). Polypeptide IIIa is longer in LAdV-2 than in HAdV-C5 (609 versus 585 residues), but, as in HAdV-C5, only the N-terminal half appears to be ordered (table S1). Similarly to HAdV-C5, in LAdV-2 the protein IIIa fold is predominantly α-helical and can be described as forming two globular domains connected by a long α helix (residues 93 to 121) (Fig. 6C and fig. S8). By analogy with the HAdV-C5 structure (1), the N-terminal domain (residues 2 to 92) is termed GOS-glue (GOS=Group of Six, consisting of the penton base pentamer and five surrounding peripentonal hexons), and the C-terminal domain (residues 122 to 256) is designed as VIII-binding domain. When separately considered, these domains are quite similar to the corresponding ones in HAdV-C5, with RMSD values of 4.99 Å for the GOS-glue (residues 7 to 106 in HAdV-C5, 87 Ca pairs), 2.85 Å for the connecting helix (residues 107 to 134 in HAdV-C5, 28 Ca pairs), and 1.8 Å for the VIII-binding domain (residues 135 to 269 in HAdV-C5, 124 Ca pairs). However, when the complete proteins are compared, a large difference is evident (Fig. 6, D and E). The GOS-glue domain and start of the connecting helix occupy the same position in the capsid as their counterparts in HAdV-C5. However, at about half their length, the connecting helices start to become apart and end up separated by a 41° angle. In addition, the helix is half a turn shorter in LAdV-2, resulting in the VIII-binding domain swinging away from its position in the human virus by 238°. This large conformational change produces considerable differences in the network of contacts beneath the vertex.
Similarly to HAdV-C5 (1), in LAdV-2, the IIIa GOS-glue domain and connecting helix establish an extensive set of interactions that join each IIIa monomer to two different peripentonal hexons, up to three penton base monomers, and the neighboring IIIa molecule (table S6). Despite having a very similar fold, the VIII-binding domain is in a completely different location in the capsid and therefore establishes a completely different set of interactions. Instead of interacting exclusively with the body domain of the peripentonal copy of protein VIII, in LAdV-2, the VIII-binding domain of protein IIIa reaches outward to interact with hexon (table S6) and establishes no contacts with the VIII body but with the VIII neck domain (Fig. 6F). The IIIa-VIII contacts are much fewer in LAdV-2: UCSF Chimera findclash estimates 84 possible contacts in HAdV-C5 versus only 22 in LAdV-2.
Additional internal elements: Proteins VI and VII and unassigned density
The pseudo-hexagonal base of the hexon trimer encloses a central cavity that opens toward the interior of the viral particle. In HAdV-C5, weak density observed in this cavity allowed tracing of three copies per AU of the N-terminal peptide of protein VI cleaved by the protease during maturation (pVIN, residues 5 to 33), and one copy of a cleaved segment of core protein VII (pVIIN2, residues 14 to 24) (2). We observe patches of density associated with all 12 hexon monomers in the AU, in the same position as pVIN and pVIIN2 in HAdV-C5. However, the density did not have enough landmarks to unequivocally distinguish between proteins VI and VII. Following the HAdV-C5 model, and depending on the length of the peptide fragment that can be fitted in unfragmented density, we propose that three of the 12 density patches in the cavity correspond to LAdV-2 peptide pVIIN2 (residues 14 to 21), while the rest correspond to pVIN (residues 2 to 25) (table S1, Fig. 1B, and fig. S9). Weak, fragmented density indicates variable occupancy, in agreement with the recently proposed model where proteins VI and VII compete for the same hexon binding site during assembly (42).
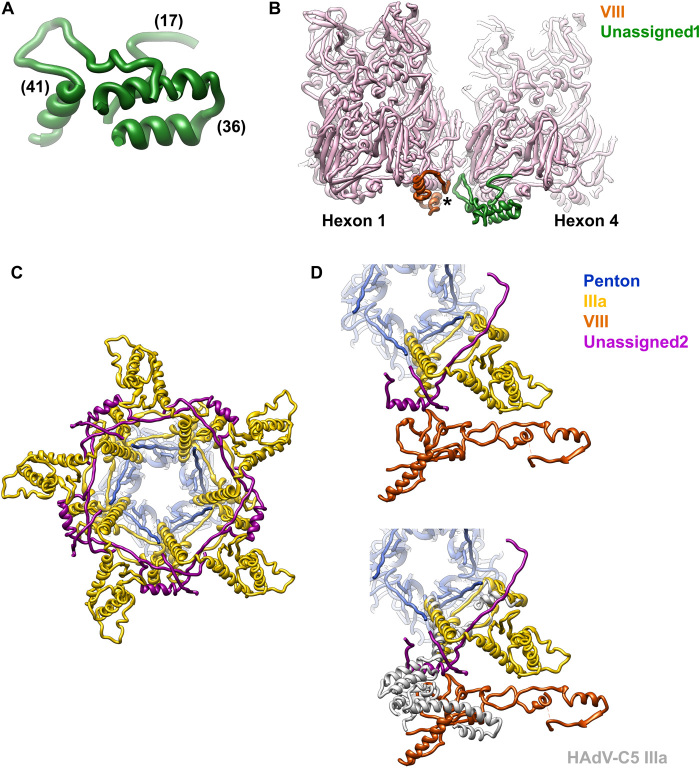
Last, we observe two additional groups of disconnected density fragments on the inner capsid surface. We tried to identify sequence regions that could fit into these fragments using the procedure described in (16). Unfortunately, no clear candidates were found. We grouped the fragments in two categories termed “unassigned1” (U1) and “unassigned2” (U2) according to their location (Fig. 1B). The U1 density can hold three peptides of lengths 17, 36, and 41 residues with predominantly α-helical structure (Fig. 7A and fig. S10A). It is present at two independent positions in the AU, near the gap left by maturation cleavages in protein VIII (Fig. 1B and 3B), and forms a wedge inserted at the local threefold axes surrounded by hexons 1, 2, and 4 in the AU and hexons 3, 4, and 3 in the neighboring AU (Fig. 7B). These are the threefold axes in the facet that do not hold an LH3 triskelion on the outer surface (Fig. 3B).
Fig. 7. Unassigned elements on the LAdV-2 internal capsid surface.
(A) Peptides modeled in the U1 density. Numbers in parentheses indicate the peptide lengths in amino acids. (B) View of the U1 peptides in the capsid context. The star indicates the location of the gap left by maturation cleavages in protein VIII. Hexon 2, which would be in front, has been removed for clarity. (C) Peptides modeled in the U2 density (magenta) form an interlaced ring with IIIa beneath the vertex. View along the fivefold symmetry axis from inside the capsid. (D) The U2 peptides are intercalated between IIIa and the body domain of VIII (top). If IIIa adopted the same conformation as in HAdV-C5, it would badly clash with U2 (bottom).
The U2 density can hold two peptides of 50 and 16 amino acids, in a mostly extended conformation except for an α helix (fig. S10B). These peptides form an interlaced ring with protein IIIa beneath the vertex (Fig. 7, C and D). A comparison with the structure of the vertex proteins in HAdV-C5 shows that the U2 peptides would clash with IIIa if this protein were not in a different conformation in the reptilian virus (Fig. 7D). U2 bridges IIIa with the body domain of VIII, compensating for the lost direct interactions. Although U2 has been modeled as a poly-alanine and therefore a proper interaction analysis cannot be carried out, it is notable that Chimera finds 24 possible contacts between U2 and proteins IIIa and VIII in the same AU, 52 with the neighboring IIIa on one side, and 11 more with the one on the other side (fig. S10C).
DISCUSSION
AdVs have been found in most types of vertebrates, but few of them have been isolated and propagated in cell culture. Therefore, little is known about the basic biology of nonhuman AdV. We present here the first high-resolution structure of an AdV infecting lower vertebrates and not belonging to the Mastadenovirus genus. The observation of the genus-specific protein LH3 in the capsid context provides detailed insights on how this protein interlocks the surrounding hexons, contributing to build a capsid more stable than those of the HAdVs. Moreover, we show that the LH3 trimeric β helix domain, folding as a bacteriophage tailspike, binds to the capsid surface using exactly the same structural motif (the triskelion) as its counterpart in mastadenoviruses, protein IX. Our sequence analyses prompted by this observation shed definitive light on the evolution of the capsid-stabilizing proteins LH3 and IX and oncoprotein E1B-55 K. Given the wide distribution of structurally similar LH3/E1B-55 K–like β helix proteins in bacteria and phages, contrasted by their restricted spread among AdVs, it is highly likely that the ancestral LH3/E1B-55 K protein has been introduced into the common ancestor of atadenoviruses and mastadenoviruses from bacteria/phages and had a structural role, akin to that of the contemporary LH3. Our data suggest that the E1B-55 K oncoprotein has evolved from a paralog of LH3 by exaptation, a common phenomenon in virus evolution (43). Subsequent evolution involved the replacement of the β helix domain of LH3 in most mastadenovirus lineages with an unrelated sequence, leading to the emergence of gene IX.
These observations indicate that the triskelion motif is critical for capsid binding, while the triskelion-containing proteins are hotspots for AdV evolution, linked to capsid stabilization, interaction with host factors (because of their accessible location on the capsid surface), and, consequently, tropism (i.e., facilitating virus propagation in a certain biological context). Protein LH3 is unique to a certain AdV genus (Atadenovirus), but even within the mastadenoviruses, there are different architectures of protein IX that set apart human from nonhuman viruses (2, 15, 18, 19, 22). Furthermore, protein IX variations in human AdV may also be related to tropism determination, as recently shown by the structure of the enteric HAdV-F41 (16, 17). HAdVs in species F have an exclusively enteric tropism, unlike the rest of HAdV species that have broader tissue targets. They have a unique conformation of minor coat protein IX and are more thermostable than the respiratory HAdV-C5. More studies are required to determine the host partners interacting with proteins IX or LH3 in the different viruses. It would also be interesting to solve structures of atadenoviruses infecting mammals or birds, as well as the equine or squirrel mastadenoviruses. The study of these mastadenoviruses, which conserve the ancestral form of LH3 with a triskelion followed by the β helix domain (Fig. 5B), would be of great interest to investigate which structural and functional features arise from the coexistence of both structural and nonstructural LH3/E1B-55 K proteins.
We show that even a protein critical for assembly and conserved throughout the AdV family, protein IIIa, can adopt very different conformations in two AdV genera. The large conformational change observed in IIIa is related to the presence of the extended, unidentified peptide U2 beneath the capsid vertex. As a result of the rotation of one of the IIIa domains, the interactions between IIIa and VIII are fewer than in the HAdVs, but U2 compensates this loss of contacts by reinforcing the interlacing of the internal vertex proteins. U2 in the vertex region, together with LH3 on the outer capsid surface, and U1 underpinning a second set of local threefold axes on the inside appear as genus-specific elements building a sturdier AdV capsid.
Possible virion components generating the U1 and U2 densities include maturation fragments or untraced regions of protein IIIa, VI, or VIII (table S1), as well as packaging proteins L1 52/55 kDa, IVa2, L4 33 kDa, or L4 22 kDa (44–48). Alternatively, U1 and U2 may correspond to the genus-specific proteins LH2 and p32k. We favor this last possibility, because the other candidates are also present in mastadenoviruses, but no equivalent densities have been detected in the structures solved so far (1, 2, 15–17). Both LH2 and p32k have positive charge (isoelectric point 11.02 for p32k and 8.59 for LH2), suggesting that they interact with the genome, and are predicted to be predominantly α-helical. Protein p32k has also large regions predicted to be unstructured, similarly to the U2 peptides, and three consensus cleavage motifs for the maturation protease, again supporting an internal location (fig. S10D) (4). Because of its proximity to the maturation gap in protein VIII, it is also possible that the excised peptides of VIII, which are 1.7 times longer in LAdV-2 than in HAdV-C5, form part of the U1 density (Fig. 7B and fig. S10D). The two genus-specific proteins and the longer central part of protein VIII would all be contributing to enhance the interactions stabilizing the atadenovirus capsid.
In summary, our work provides new information on the structure and evolution of AdVs, emphasizing the importance of minor coat proteins for determining specific physicochemical properties of the virions and most likely their host specificity. Knowing the structure of uncommon, nonhuman AdVs will facilitate their development as vectors. Ovine atadenoviruses have already been considered as alternative vectors, free of preexisting immunity issues that hinder the use of HAdV-C5 in the clinic. Three dispensable, nonessential regions of the viral genome, where foreign genes can be inserted, have been delineated in the OAdV-7 genome and used to insert transgenes that proved to be expressed and generate immunity in mice. Gene transfer was not abrogated by previous exposure of the animals to HAdV-C5 (49). Lack of an integrin-binding RGD motif in penton base, the presence of two different fibers in LAdV-2, and the prominent LH3 knobs on the capsid surface suggest that interactions with the host for entry and trafficking are different in atadenoviruses and HAdV. The increased capsid stability may also allow transduction in harsh environments, such as intestinal cells. The conservation of the triskelion motif and the existence of mastadenoviruses with bona fide LH3 proteins open the possibility to construct hybrid vectors incorporating the atadenovirus protein on the HAdV chassis. Last, the surface-exposed C terminus of LH3 would constitute an interesting locale for display of peptides with biomedical interest (50).
MATERIALS AND METHODS
Virus production
LAdV-2 (25) was propagated in iguana heart epithelial cells (IgH-2, American Type Culture Collection: CCL-108) (51) at 37°C by amplification from one to 184 culture plates (10-cm diameter). The cells and supernatant from previous infections were used for the next infection round. Infected cells were frozen and thawed four times to release the viral particles before each infection step. Cell growth and virus propagation were carried out at 37°C. At 2 days after infection in the last round, when the cells showed cytopathic effect, they were collected and viral particles purified by centrifugation in two consecutive CsCl gradients, as previously described (44).
Thermostability assays were carried out as described (21). Briefly, purified LAdV-2, SnAdV-1, or HAdV-C5 were heated at 37°, 45°, 50°, or 60°C for 5 min and inoculated onto replicate cell cultures (human embryonic kidney–293 for HAdV-C5, and IgH-2 for SnAdV-1 and LAdV-2). The end point dilution assay was used to measure infectious titer and compared to the titer of control virus (kept in ice before infection). The data for HAdV-C5 and SnAdV-1 have been reported previously (21). The data for LAdV-2 were collected in the same time period, as part of a study comparing the thermostability of reptilian atadenoviruses to that of HAdV-C5.
Cryo–electron microscopy
Purified LAdV-2 was dialyzed for 1 hour at 4°C against phosphate-buffered saline [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 (pH 7.4)] and concentrated by spinning in a Microcon YM-100 device for 6 min at 4°C, for a final estimated concentration of 7 × 1012 viral particles/ml. Samples were deposited in glow discharged Quantifoil R2/4 300 mesh Cu/Rh grids and vitrified in liquid ethane after manual blotting in a Leica CPC device. Cryo-EM images (table S11) were recorded using a 300-kV Titan Krios microscope equipped with a Falcon II detector (MRC-LMB, Cambridge, UK), with a total dose of 54 e−/Å2 distributed over 34 frames, at nominal pixel size 1.34 Å and defocus range between −1 and −3 μm.
Image processing
All image processing and 3D reconstruction tasks were performed within the Scipion framework (table S11) (52). Frames 2 to 24 of each movie were aligned using whole-image motion correction implemented in Xmipp, followed by correction of local movements using Optical Flow (53). The contrast transfer function (CTF) was estimated using CTFFIND4 (54). Particles were semiautomatically picked from micrographs corrected for the phase oscillations of the CTF (phase-flipped), extracted into 780 × 780 pixel boxes, normalized, and downsampled by a factor of 2, using Xmipp (55). All 2D and 3D classifications and refinements were performed using RELION (56). Two-dimensional classification was used to discard low-quality particles and was run for 25 iterations, with 50 classes, angular sampling 5, and regularization parameter T = 2. Classification in 3D was run for 40 iterations, with three classes, starting with an angular sampling of 3.7° and sequentially decreasing to 0.5, and regularization parameter T = 4. Icosahedral symmetry was imposed throughout the refinement process. The initial reference for 3D classification was the SnAdV-1 cryo-EM map (21), low-pass–filtered to 60-Å resolution. The class yielding the best resolution was individually refined using the original 780-pixel boxed particles and the map obtained during the 3D classification as a reference, producing a final map at 3.4-Å resolution, as estimated according to the gold-standard Fourier Shell Correlation (FSC) = 0.143 criterion implemented in RELION autorefine and postprocess routines (57). A global B factor was estimated after dividing the map Fourier transform by the modulation transfer function of the Falcon II detector. The actual sampling for the map was estimated by comparison with the SnAdV-1 crystallographic and homology models (21) in UCSF Chimera (58), yielding a value of 1.35 Å per pixel. Local resolution was calculated with ResMap (59). Localized reconstruction of the penton region followed by 3D classification in three classes, without orientation change or symmetry enforcement, was carried out as described (17), using RELION as implemented in Scipion (35).
Model building and analysis
The initial model for each polypeptide chain was predicted with MODELLER (60), using as input template the structure of the respective homolog chain in HAdV-C5 (hexon, PDB ID 1P30; penton base, PDB ID 1X9P; and IIIa and VIII, PDB ID 3IYN) or SnAdV-1 (LH3, PDB ID 5G5O) (1, 21, 33, 61). UCSF Chimera (58) was used to perform a rigid fitting of each chain initial model into the sharpened map. Next, the fitted model of each chain was refined using Coot (62), REFMAC (63), and Phenix real space refine (64). Validation metrics to assess the quality of the atomic structure were computed with the Phenix comprehensive validation (cryo-EM) algorithm (table S11). Once we generated the whole structure of the AU, the nearest neighboring molecules were generated with Chimera (sym #0 group i,222r contact 3). Possible contacts between each molecule in the AU and all its neighbors were identified with Chimera findclash, listed and grouped using a protocol integrated in the Scipion molecular modeling workflow (tables S2 to S9) (65).
Sequence alignments were carried out with Clustal O 1.2.4 (66) and displayed as text or with JalView (67). The ClustalX color scheme (68) was used in fig. S7E. Surface coloring by electrostatic potential was carried out with APBS and Chimera (58, 69). Chimera matchmaker was used for RMSD calculation and structure guided alignment (followed by match-align). Secondary structure and disorder predictions were carried out with PsiPred and Disopred (70, 71).
To test possible sequence identities of density fragments U1 and U2, we used a procedure similar to that described in (16). This procedure uses the model to map correlation coefficient provided by Phenix real space refinement to evaluate the fit of different regions in the candidate protein sequences (in both direct and reverse orientation) to the different density fragments. We tested the fit of proteins p32k, LH2, and the excised central peptide of VIII to the U1 and U2 densities. This required testing over 7400 possibilities, even without considering other possible candidates such as the packaging proteins (L1 52/55 kDa, IVa2, and L4 22k/33k), or untraced regions of capsid (IIIa and VI) and core (VII and μ) proteins. To facilitate the execution, we have incorporated this procedure as a new protocol (SearchFit) in the Phenix plugin of Scipion (65).
Sequence analysis
Profile HMM of LAdV-2 LH3 was constructed and compared against the Pfam, PDB, CDD, and SCOPe databases of profile HMMs using HH-suite v3 (38). LH3 and EB1-55 K homologs were searched for using BLASTP against the nonredundant protein database at NCBI.
Acknowledgments
We thank T. Papp (Institute for Veterinary Medical Research, Budapest, Hungary) and R. E. Marschang (Laboklin GmbH & Co. Bad Kissingen, Germany) for the initial LAdV-2 virus and IgH-2 cell seeds; S. Chen, S. Scheres, C. Savva, G. McMullan (MRC-LMB, Cambridge, UK), and F. de Haas (Thermo Fisher Scientific, formerly FEI, Eindhoven, NL) for cryo-EM data collection; and the INSTRUCT Image Processing Center (I2PC) for advice on data processing (INSTRUCT Access Project PID: 1395). Funding: This work is supported by the Spanish Agencia Estatal de Investigación and European Regional Development Fund (BFU2016-74868-P and PID2019-104098GB-I00/AEI/10.13039/501100011033), the Ministerio de Economía y Competitividad of Spain (BFU2013-41249-P and BIO2015-68990-REDT), and the Agencia Estatal CSIC (2019AEP045). The CNB-CSIC is further supported by a Severo Ochoa Excellence grant (SEV 2017-0712). M.K. was supported by the Emergence(s) program from City of Paris (grant MEMREMA). Author contributions: G.N.C. and R.M.-C. prepared virus samples. G.N.C., R.M., and C.S.M. collected, processed, and analyzed cryoEM data. R.M., G.N.C., J.G.-B., and C.S.M. modeled, refined, and analyzed the structure. R.M.-C. carried out thermostability assays. M.K. performed sequence analyses of the E1B-55 K/LH3 proteins. C.S.M. designed the study and wrote the paper with contributions from the rest of the authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The LAdV-2 cryoEM map and model are deposited at the Electron Microscopy Data Bank (EMDB; www.ebi.ac.uk/pdbe/emdb) and the Protein Data Bank (PDB, www.ebi.ac.uk/pdbe) with accession numbers EMD-4551 and 6QI5, respectively.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabe6008/DC1
REFERENCES AND NOTES
- 1.Liu H., Jin L., Koh S. B., Atanasov I., Schein S., Wu L., Zhou Z. H., Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329, 1038–1043 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai X., Wu L., Sun R., Zhou Z. H., Atomic structures of minor proteins VI and VII in human adenovirus. J. Virol. 91, e00850-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez-Berná A. J., Marion S., Chichón F. J., Fernández J. J., Winkler D. C., Carrascosa J. L., Steven A. C., Šiber A., San Martín C., Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 43, 4274–4283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangel W. F., San Martín C., Structure, function and dynamics in adenovirus maturation. Viruses 6, 4536–4570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San Martín C., Latest insights on adenovirus structure and assembly. Viruses 4, 847–877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopez-Gordo E., Podgorski I. I., Downes N., Alemany R., Circumventing antivector immunity: Potential use of nonhuman adenoviral vectors. Hum. Gene Ther. 25, 285–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.B. Harrach, M. Benkő, G. Both, M. Brown, A. Davison, M. Echavarría, M. Hess, M. Jones, A. Kajon, H. Lehmkuhl, V. Mautner, S. Mittal, G. Wadell, Family Adenoviridae, in Virus Taxonomy: Classification and Nomenclature of Viruses. Ninth Report of the International Committee on Taxonomy of Viruses, A. King, M. Adams, E. Carstens, E. Lefkowitz, Eds. (Elsevier, 2011), pp. 95–111. [Google Scholar]
- 8.Harrach B., Reptile adenoviruses in cattle? Acta Vet. Hung. 48, 485–490 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Harrach B., Tarjan Z. L., Benko M., Adenoviruses across the animal kingdom: a walk in the zoo. FEBS Lett. 593, 3660–3673 (2019). [DOI] [PubMed] [Google Scholar]
- 10.A. Gaba, L. E. Ayalew, S. K. Tikoo, Animal Adenoviruses, in Recent Advances in Animal Virology, Y. Malik, R. Singh, M. Yadav, Eds. (Springer, 2019), pp. 3–20. [Google Scholar]
- 11.Jacobson E. R., Gaskin J. M., Gardiner C. H., Adenovirus-like infection in a boa constrictor. J. Am. Vet. Med. Assoc. 187, 1226–1227 (1985). [PubMed] [Google Scholar]
- 12.Papp T., Fledelius B., Schmidt V., Kaján G. L., Marschang R. E., PCR-sequence characterization of new adenoviruses found in reptiles and the first successful isolation of a lizard adenovirus. Vet. Microbiol. 134, 233–240 (2009). [DOI] [PubMed] [Google Scholar]
- 13.D. P. O’Rourke, K. Lertpiriyapong, in Laboratory Animal Medicine (Third Edition), J. G. Fox, L. C. Anderson, G. M. Otto, K. R. Pritchett-Corning, M. T. Whary, Eds. (Academic Press, 2015), pp. 967–1013. [Google Scholar]
- 14.T. Hyndman, R. E. Marschang, in Reptile Medicine and Surgery in Clinical Practice, B. Doneley, D. Monks, R. Johnson, B. Carmel, Eds. (John Wiley & Sons Incorporated, 2018), pp. 197–216. [Google Scholar]
- 15.Yu X., Veesler D., Campbell M. G., Barry M. E., Asturias F. J., Barry M. A., Reddy V. S., Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses. Sci. Adv. 3, e1602670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafie K., Lenman A., Fuchs J., Rajan A., Arnberg N., Carlson L. A., The structure of enteric human adenovirus 41—A leading cause of diarrhea in children. Sci. Adv. 7, eabe0974 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Illana M., Martínez M., Condezo G. N., Hernando-Pérez M., Mangroo C., Brown M., Marabini R., San Martín C., Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses. Sci. Adv. 7, eabd9421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackenbrack N., Rogers M. B., Ashley R. E., Keel M. K., Kubiski S. V., Bryan J. A., Ghedin E., Holmes E. C., Hafenstein S. L., Allison A. B., Evolution and cryo-EM capsid structure of a North American bat adenovirus and its relationship to other mastadenoviruses. J. Virol. 91, e01504-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoehn G., El Bakkouri M., Fabry C. M., Billet O., Estrozi L. F., Le L., Curiel D. T., Kajava A. V., Ruigrok R. W., Kremer E. J., Three-dimensional structure of canine adenovirus serotype 2 capsid. J. Virol. 82, 3192–3203 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantelic R. S., Lockett L. J., Rothnagel R., Hankamer B., Both G. W., Cryoelectron microscopy map of Atadenovirus reveals cross-genus structural differences from human adenovirus. J. Virol. 82, 7346–7356 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menéndez-Conejero R., Nguyen T. H., Singh A. K., Condezo G. N., Marschang R. E., van Raaij M. J., San Martín C., Structure of a reptilian adenovirus reveals a phage tailspike fold stabilizing a vertebrate virus capsid. Structure 25, 1562–1573.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Cheng L., Huang X., Li X., Xiong W., Sun W., Yang C., Zhang K., Wang Y., Liu H., Huang X., Ji G., Sun F., Zheng C., Zhu P., Cryo-EM structures of two bovine adenovirus type 3 intermediates. Virology 450-451, 174–181 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Davison A. J., Benkő M., Harrach B., Genetic content and evolution of adenoviruses. J. Gen. Virol. 84, 2895–2908 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Gorman J. J., Wallis T. P., Whelan D. A., Shaw J., Both G. W., LH3, a "homologue" of the mastadenoviral E1B 55-kDa protein is a structural protein of atadenoviruses. Virology 342, 159–166 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Pénzes J. J., Menéndez-Conejero R., Condezo G. N., Ball I., Papp T., Doszpoly A., Paradela A., Pérez-Berná A. J., López-Sanz M., Nguyen T. H., van Raaij M. J., Marschang R. E., Harrach B., Benkö M., San Martín C., Molecular characterization of a lizard adenovirus reveals the first atadenovirus with two fiber genes and the first adenovirus with either one short or three long fibers per penton. J. Virol. 88, 11304–11314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vrati S., Brookes D. E., Strike P., Khatri A., Boyle D. B., Both G. W., Unique genome arrangement of an ovine adenovirus: identification of new proteins and proteinase cleavage sites. Virology 220, 186–199 (1996). [DOI] [PubMed] [Google Scholar]
- 27.G. W. Both, in The Springer Index of Viruses, C. A. Tidona, G. Darai, C. Büchen-Osmond, Eds. (Springer Berlin Heidelberg, 2001), pp. 2–8. [Google Scholar]
- 28.Hidalgo P., Ip W. H., Dobner T., Gonzalez R. A., The biology of the adenovirus E1B 55K protein. FEBS Lett. 593, 3504–3517 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Xu L., Benson S. D., Burnett R. M., Nanoporous crystals of chicken embryo lethal orphan (CELO) adenovirus major coat protein, hexon. J. Struct. Biol. 157, 424–431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford-Miksza L., Schnurr D. P., Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70, 1836–1844 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alba R., Bradshaw A. C., Parker A. L., Bhella D., Waddington S. N., Nicklin S. A., van Rooijen N., Custers J., Goudsmit J., Barouch D. H., McVey J. H., Baker A. H., Identification of coagulation factor (F)X binding sites on the adenovirus serotype 5 hexon: Effect of mutagenesis on FX interactions and gene transfer. Blood 114, 965–971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. M. Zimmerman, in Encyclopedia of Immunobiology, M. J. H. Ratcliffe, Ed. (Academic Press, 2016), pp. 493–497. [Google Scholar]
- 33.Zubieta C., Schoehn G., Chroboczek J., Cusack S., The structure of the human adenovirus 2 penton. Mol. Cell 17, 121–135 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Wickham T. J., Mathias P., Cheresh D. A., Nemerow G. R., Integrins αvβ3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell 73, 309–319 (1993). [DOI] [PubMed] [Google Scholar]
- 35.Abrishami V., Ilca S. L., Gómez-Blanco J., Rissanen I., de la Rosa-Trevin J. M., Reddy V. S., Carazo J. M., Huiskonen J. T., Localized reconstruction in Scipion expedites the analysis of symmetry mismatches in cryo-EM data. Prog. Biophys. Mol. Biol. , (2020). [DOI] [PubMed] [Google Scholar]
- 36.Vellinga J., van den Wollenberg D. J., van der Heijdt S., Rabelink M. J., Hoeben R. C., The coiled-coil domain of the adenovirus type 5 protein IX is dispensable for capsid incorporation and thermostability. J. Virol. 79, 3206–3210 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosa-Calatrava M., Grave L., Puvion-Dutilleul F., Chatton B., Kedinger C., Functional analysis of adenovirus protein IX identifies domains involved in capsid stability, transcriptional activity, and nuclear reorganization. J. Virol. 75, 7131–7141 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinegger M., Meier M., Mirdita M., Vohringer H., Haunsberger S. J., Soding J., HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20, 473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doszpoly A., Harrach B., LaPatra S., Benko M., Unconventional gene arrangement and content revealed by full genome analysis of the white sturgeon adenovirus, the single member of the genus Ichtadenovirus. Infect. Genet. Evol. 75, 103976 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Giles C., Vanniasinkam T., Barton M., Mahony T. J., Characterisation of the Equine adenovirus 2 genome. Vet. Microbiol. 179, 184–189 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Rux J. J., Burnett R. M., Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1, 18–30 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Hernando-Pérez M., Martín-González N., Pérez-Illana M., Suomalainen M., Condezo G. N., Ostapchuk P., Gallardo J., Menéndez M., Greber U. F., Hearing P., de Pablo P. J., San Martín C., Dynamic competition for hexon binding between core protein VII and lytic protein VI promotes adenovirus maturation and entry. Proc. Natl. Acad. Sci. U.S.A. 117, 13699–13707 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koonin E. V., Krupovic M., The depths of virus exaptation. Curr. Opin. Virol. 31, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Condezo G. N., Marabini R., Ayora S., Carazo J. M., Alba R., Chillón M., San Martín C., Structures of adenovirus incomplete particles clarify capsid architecture and show maturation changes of packaging protein L1 52/55k. J. Virol. 89, 9653–9664 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostapchuk P., Almond M., Hearing P., Characterization of Empty adenovirus particles assembled in the absence of a functional adenovirus IVa2 protein. J. Virol. 85, 5524–5531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu K., Guimet D., Hearing P., The adenovirus L4-33K protein regulates both late gene expression patterns and viral DNA packaging. J. Virol. 87, 6739–6747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guimet D., Hearing P., The adenovirus L4-22K Protein has distinct functions in the posttranscriptional regulation of gene expression and encapsidation of the viral genome. J. Virol. 87, 7688–7699 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustin K. E., Imperiale M. J., Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72, 7860–7870 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Both G. W., Ovine atadenovirus: A review of its biology, biosafety profile and application as a gene delivery vector. Immunol. Cell Biol. 82, 189–195 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Matteson N. L., Barry M. A., Reddy V. S., Structure-based assessment of protein-protein interactions and accessibility of protein IX in adenoviruses with implications for antigen display. Virology 516, 102–107 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Clark H. F., Cohen M. M., Karzon D. T., Characterization of reptilian cell lines established at incubation temperatures of 23 to 36°. Proc. Soc. Exp. Biol. Med. 133, 1039–1047 (1970). [DOI] [PubMed] [Google Scholar]
- 52.de la Rosa-Trevin J. M., Quintana A., Del Cano L., Zaldivar A., Foche I., Gutierrez J., Gomez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V., Vargas J., Oton J., Sharov G., Vilas J. L., Navas J., Conesa P., Kazemi M., Marabini R., Sorzano C. O., Carazo J. M., Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Abrishami V., Vargas J., Li X., Cheng Y., Marabini R., Sorzano C. O., Carazo J. M., Alignment of direct detection device micrographs using a robust Optical Flow approach. J. Struct. Biol. 189, 163–176 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de la Rosa-Trevin J. M., Oton J., Marabini R., Zaldivar A., Vargas J., Carazo J. M., Sorzano C. O., Xmipp 3.0: An improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen S., McMullan G., Faruqi A. R., Murshudov G. N., Short J. M., Scheres S. H., Henderson R., High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rux J. J., Kuser P. R., Burnett R. M., Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 77, 9553–9566 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallograph. Sec. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown A., Long F., Nicholls R. A., Toots J., Emsley P., Murshudov G., Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Afonine P. V., Poon B. K., Read R. J., Sobolev O. V., Terwilliger T. C., Urzhumtsev A., Adams P. D., Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallograph. Sec. D 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martínez M., Jiménez-Moreno A., Maluenda D., Ramirez-Aportela E., Melero R., Cuervo A., Conesa P., Del Cano L., Fonseca Y. C., Sanchez-García R., Strelak D., Conesa J. J., Fernández-Giménez E., de Isidro F., Sorzano C. O. S., Carazo J. M., Marabini R., Integration of Cryo-EM model building software in Scipion. J. Chem. Inf. Model. 60, 2533–2540 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Soding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G., Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L. E., Brookes D. H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D. W., Dolinsky T., Konecny R., Koes D. R., Nielsen J. E., Head-Gordon T., Geng W., Krasny R., Wei G. W., Holst M. J., McCammon J. A., Baker N. A., Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones D. T., Cozzetto D., DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857–863 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buchan D. W. A., Jones D. T., The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/14/eabe6008/DC1