Abstract
Large-scale fluorescence calcium imaging methods have become widely adopted for studies of long-term hippocampal and cortical neuronal dynamics. Pyramidal neurons of the rodent hippocampus show spatial tuning in freely foraging or head-fixed navigation tasks. Development of efficient neural decoding methods for reconstructing the animal’s position in real or virtual environments can provide a fast readout of spatial representations in closed-loop neuroscience experiments. Here, we develop an efficient strategy to extract features from fluorescence calcium imaging traces and further decode the animal’s position. We validate our spike inference-free decoding methods in multiple in vivo calcium imaging recordings of the mouse hippocampus based on both supervised and unsupervised decoding analyses. We systematically investigate the decoding performance of our proposed methods with respect to the number of neurons, imaging frame rate, and signal-to-noise ratio. Our proposed supervised decoding analysis is ultrafast and robust, and thereby appealing for real-time position decoding applications based on calcium imaging.
1. Introduction
Pyramidal cells in the rodent hippocampus are known to encode space. Large-scale long-term calcium imaging has been used for studies of long-term memories in freely behaving or head-fixed animals (Dombeck et al., 2010; Ziv et al., 2013; Jercog et al., 2016; Mau et al., 2018). One of interesting neuroscience questions is to examine how populations of hippocampal neurons represent space at different brain states, and how these representations correlate with memory, learning and planning.
Genetically-encoded fluorescent reporters of neurobiological processes can be used to monitor biochemical events and signals in neurons and tissues of living brains. Rapid advances in neurotechnology have allowed us to simultaneously record thousands or tens of thousands of neurons using large-scale calcium imaging (e.g., Pachitariu et al., 2017; Stringer et al., 2019). However, the curse of dimensionality may arise for processing such high-throughput neural data, especially in closed-loop neuroscience experiments.
Neural decoding for position reconstruction have been well studied based on large-scale hippocampal neuronal activities or population responses. In electrophysiology, multiple sources of effort have been dedicated to position decoding based on either unsorted spikes or multi-unit activity (Chen, Kloosterman, Layton & Wilson, 2012; Kloosterman et al., 2014; Deng et al., 2015; Ciliberti et al., 2018; Hu et al., 2018), or the amplitude of ultra-high frequency band multi-site hippocampal field potentials (Cao et al., 2019). In calcium imaging, there was an approximately linear relationship between the peak amplitude of a somatic Ca2+ transient and the underlying number of spikes in the recorded brain areas (Chen et al., 2013; Jercog et al., 2016). This naturally invites the next question: can we directly estimate the position of freely behaving animals directly based on the calcium imaging data without the need of spike inference?
In the standard analysis pipeline for calcium imaging, spike inference algorithms are used to extract spiking activity from fluorescence traces (e.g., Vogelstein et al., 2010; Pnevmatikakis et al., 2016; Giovannucci, et al., 2018; Jewell & Witten, 2018; Zhou et al., 2018; Pnevmatikakis, 2019). However, spike deconvolution is a challenging and computationally demanding procedure that seeks an approximate solution to the inverse problem. However, statistical methods have also been developed to bypass spike deconvolution for subsequent analyses, such as estimating neuronal firing rates directly from calcium imaging data (Ganmor et al., 2016). In this paper, we propose a new analysis pipeline for position decoding based on in vivo calcium imaging recordings from the mouse hippocampus. We show that the preprocessed fluorescence traces (without spike inference) can be used directly in either supervised and unsupervised position decoding analysis. The proposed method not only is efficient in computational speed, but also shows comparable performance with the standard spike-inference methods. From the unsupervised learning analysis, we further recover the latent state representations that encode the animal’s position.
2. Methods
2.1. Data preprocessing and approximate spike inference
In calcium imaging, the Ca2+ dynamics are characterized by the fluorescence traces, which are typically shown in units of a percentage change of each imaged cell’s baseline fluorescence level (i.e., dF/F). At first, the standard preprocessing procedures, such as motion artifact correction, neuropil removal, and source extraction, were conducted for the raw calcium imaging data (Giovannucci, et al., 2018; Pnevmatikakis, 2019). In traditional neural data analysis, the spike inference procedure is further employed to estimate the calcium concentration and the spiking activity from the dF/F fluorescence traces. Here, we used one state-of-the-art spike deconvolution algorithm based nonnegative deconvolution (Friedrich et al., 2017; Pachitariu et al., 2018). At the end of spike inference, a time series of spiking activity (real-valued) from each fluorescence trace was obtained (Figure 1A). The fluorescence trace can be viewed as a temporal convolution between a discrete point process and low-pass filter, corrupted by an additive noise process (Vogelstein et al., 2010). Regardless the techniques or inference principle, spike deconvolution remains a computational bottleneck for processing large-scale and long-term calcium imaging recordings.
Figure 1:
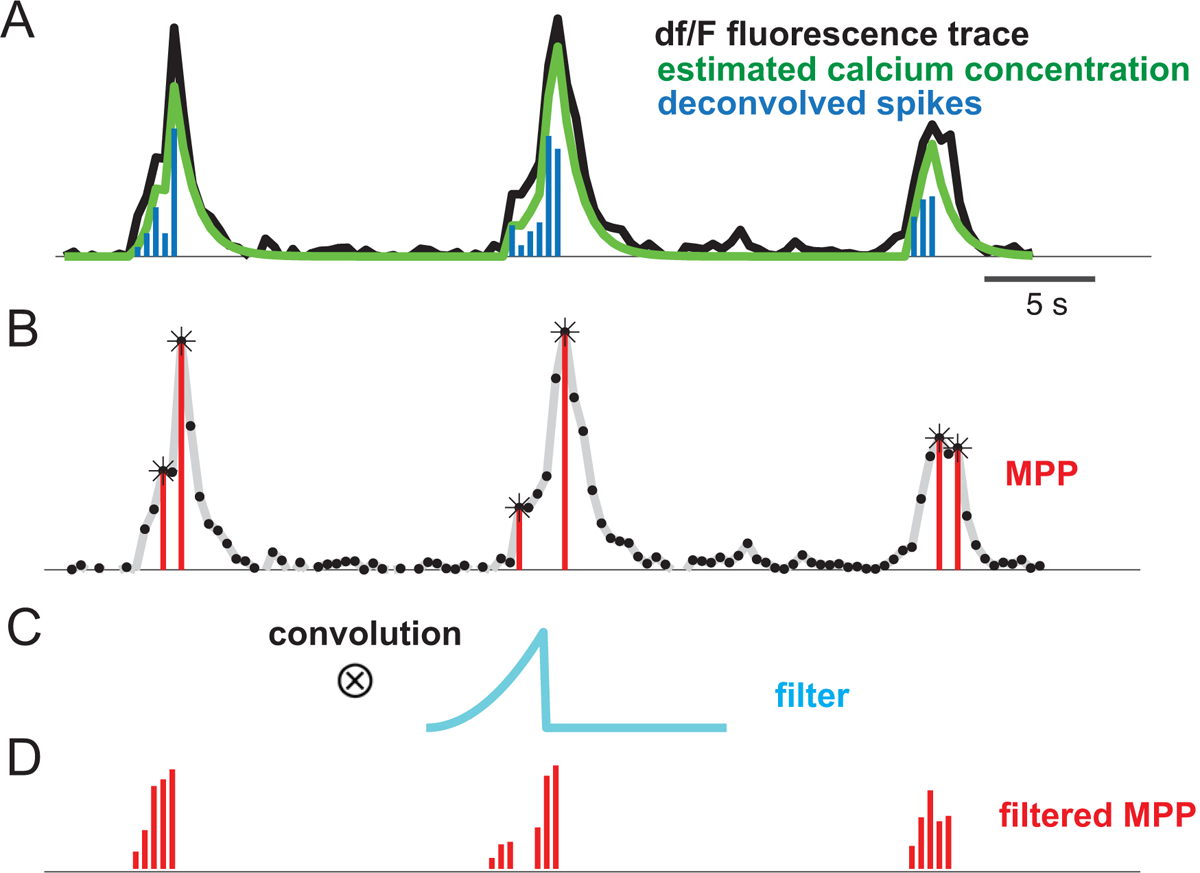
(A) Illustration of two-photon calcium imaging dF/F fluorescence trace and the inferred spiking activity. (B) Detected peaks are labeled by asterisks based on a threshold criterion, yielding a marked point process (MPP), where the mark represents the peak amplitude. (C) The asymmetric temporal filter. (D) Convolution of the MPP with the temporal filter produces a filtered MPP, which can be viewed as a proxy of the spiking activity.
To sidestep the spike deconvolution procedure, we developed a simple yet efficient strategy for inferring the “proxy” of spiking activity. The procedure consisted of three steps.
Step 1: We first estimated the positive peak amplitude from the dF/F fluorescence traces. The amplitude were identified based on a threshold-based peak detection algorithm (e.g., MATLAB function ‘findpeaks’; see Figure 1B). Depending on the noise level, the minimum peak threshold was often set as at least 10–30% peak fluorescence amplitude.
Step 2: We set the signal below the threshold to 0, and then converted the fluorescence trace into a one-dimensional marked point process (MPP; Jacobsen, 2006), where the nonzero value indicated the peak event, and the mark was defined by the peak amplitude (Figure 1B).
-
Step 3: To accommodate the spiking activity during the rise time before reaching the peak, we convolved the marked point process with an asymmetric temporal filter h (Figure 1C), resulting in a filtered MPP (Figure 1D). The custom designed filter may have the following shape
where α is the decay parameter, and A = exp(−αt0)/α is a normalization constant such that ∫ h(t)dt = 1.
The signal-to-noise ratio (SNR) of calcium imaging is relative to the fluorescent indicator dye. The measurement of fluorescence responses depends on several factors (Malik et al., 2011): (i) the nature of the stimulus and the modulation of neural activity due to the stimulus; (ii) movements due to highly structured physiological processes; (iii) spontaneous neural activity; and (iv) optical and electrical noise. To quantify the SNR of the observed fluorescence trace for each neuron, we defined an empirical measure as follows
| (2.1) |
where the denominator defines the variance of spontaneous dF/F trace (“noise”), and the numerator defines the variance of detected peak amplitudes (“signal”).
Upon feature extraction, either the deconvolved spiking activity or the filtered MPPs of selected neurons were used as the observations for the subsequent decoding analysis. Notably, the filtered MPPs can be viewed and used as the surrogate of scaled spiking activity). However, as we see below, position decoding does not require exact spike timing or scaling, implying that a proxy of spiking activity can provide a much simpler and efficient solution to decoding analysis.
2.2. Optimal linear estimation (OLE) for position decoding
For simplicity, we used the standard OLE method (Agarwal et al. 2014; Cao et al., 2019). The OLE method is based on a linear regression model. However, the principle discussed here can be adapted for other likelihood-based decoding methods, for possible incorporation of a temporal prior.
We assumed that the decoded position x can be linearly reconstructed from neural activity y = [y1, …, yC] using a set of functions {ϕc(x)}
| (2.2) |
In one-dimensional environment, we mapped the position x (with length L) via K equally spaced von Mises functions that are characterized by a circular variable θ = 2πx/L, and for k = 1, …, K. Upon representing ϕc(x) by a linear combination of circular functions, Equation 2.2 can be reformulated as
| (2.3) |
The unknown C-by-K matrix W = {wi,k} was estimated by solving the following multi-input multi-output regression problem
| (2.4) |
where yt denotes the vector containing C-dimensional observed features at time t, and denotes a K-dimensional vector that expands the position into a set of overlapping smooth basis functions (Figure 2A). The parameter K determines the number of basis functions, and κ controls the smoothness. In our data analyses, a common parameter setup was K=25–100 (step size 25), κ=25–700 (25, 50, 75, 100, 200, …, 700), and their optimal values were selected via cross-validation.
Figure 2:
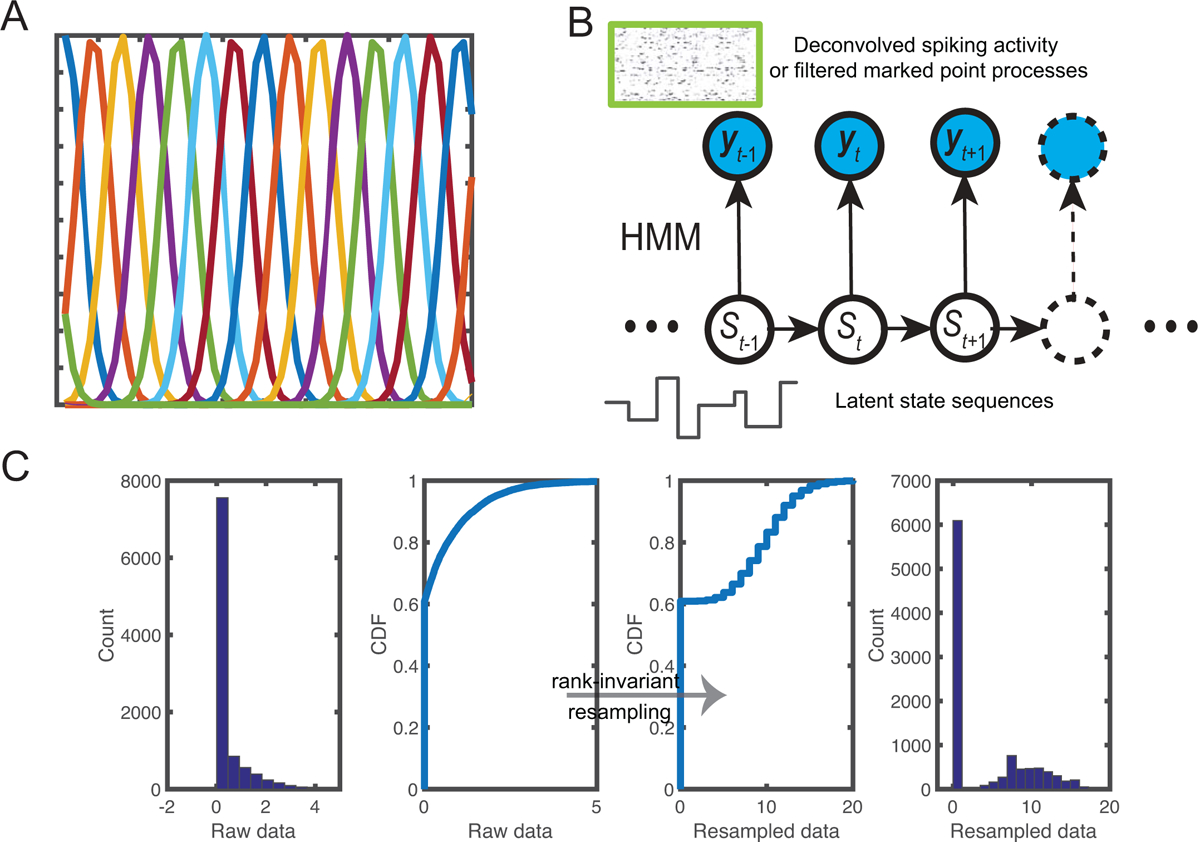
(A) A set of overlapping von Mises basis functions. (B) Schematic of a hidden Markov model (HMM) for inferring latent state sequences {St} based on either deconvolved spike activity or filtered marked point processes {yt}. (C) Illustration of rank-invariant resampling by transforming the raw data into resampled data. The rank order is preserved between two cumulative distribution functions (CDFs).
Assuming Gaussianity, the solution to Equation 2.4 was given by a least-squared (LS) estimate. In the preprocessing step, the observed features can be Z-scored across neurons. However, when the number of neurons is large (e.g., > 500), the overfitting problem will potentially arise. To resolve this issue, we imposed a sparsity constraint on the estimate and employed a variational Bayes (VB) linear regression combined with an automatic relevance determination (ARD) procedure (Bishop, 2006; Wu et al., 2016; Drugowtisch, 2017). The VB-ARD sparse regression showed improved decoding performance in the presence of high-dimensionality (see results in section 3.2).
2.3. Maximum likelihood estimation (MLE) for position decoding
In addition to OLE, we also employed MLE for position decoding (Zhang et al., 2018; Davidson et al., 2009). Let {λc(x)} denote the derived place fields (‘spatial tuning curves’ for a position variable x), of hippocampal pyramidal neurons, and let yt = {yc,t} denote the observed population vector. Assuming Poisson probability firing for each neuron, for spike count observations (where yc,t is either a positive integer or zero), the likelihood function of the observed sorted spike activity yt is given by:
| (2.5) |
where Δ denotes the temporal bin size. The maximum likelihood estimate seeks to find an optimal estimate such that
| (2.6) |
2.4. Bayesian hidden Markov model (HMM)
In our previous work, we developed a hidden Markov model (HMM) to uncover latent structures of hippocampal population activity during spatial navigation, based on observed spiking activity (Chen, Kloosterman, Brown & Wilson, 2012; Chen et al. 2014) or multichannel LFP features (Cao et al., 2019). Here we extended the idea by using the new features derived from the fluorescence calcium imaging traces.
We assumed that the latent state process follows a first-order discrete-state Markov chain {St} ∈ {1, 2, …, m} (Figure 2B). Let y1:T = {yc,t}C×T denote an observed multivariate time series of neural response vector. For spike count observations (where yc,t is zero or positive integer), we assumed that the neuronal response, conditional on the latent state, follows a Poisson distribution with an associated tuning curve function Λ = {λc} = {λc,i}. The joint probability distribution of the observed and latent variables is characterized by
| (2.7) |
where P = {Pij} denotes an m × m state transition matrix, with Pij representing the transition probability from state i to j; π = {πi} denotes a probability vector for the initial state S1. Neurons were assumed conditionally independent, so that the conditional probability distribution has a factorial form:
| (2.8) |
To accommodate automatic selection of model order m, we have developed a Bayesian nonparametric inference procedure to identify the unknown parameters {π,P,Λ} and the latent state sequences {S1:T} (Linderman et al., 2016). The Bayesian nonparametric version of the HMM—the hierarchical Dirichlet process-HMM (HDP-HMM)—generalizes the finite-state HMM with a nonparametric HDP prior. Upon completion of Bayesian inference, we estimated the posterior of latent state sequences S1:T, the tuning curve matrix Λ, and identified the correspondence between the inferred state and animal’s position (Chen et al., 2014).
2.5. Rank-invariant resampling
It is noted that the deconvolved spiking activity or filtered MPPs derived from the fluorescence traces are nonnegative real-valued, which do not satisfy the Poisson assumption in the likelihood model (Equation 2.5 or 2.8). To accommodate this change, we employed a rank-invariant resampling procedure (e.g., Honey et al., 2009) to generate the same size of random samples from a targeted Poisson distribution.
In the resampling procedure, we replaced the original ordered samples with the resampled ordered samples (by keeping the order unchanged). Specifically, we replaced the smallest raw data value with the smallest randomly sampled value, the second-smallest raw data value with the second-smallest randomly sampled value, and so on until all raw data values were replaced. A schematic illustration of the rank-invariant resampling procedure is shown in Figure 2C. Since the targeted resampled data were guaranteed to be Poisson distributed, we treated them as the pseudo spike count observations and then conducted the inference described in sections 2.3 or 2.4.
The choice of the targeted mean statistic in the targeted Poisson distribution was ad hoc, and the HMM-based decoding performance may vary according to the Poisson mean statistic. When the latent state is known, the rank-invariant resampling may be approximately scale and translationally invariant (see Appendix).
2.6. Experimental methods
Hippocampal dataset 1.
In the first dataset, male and female C57/BL6 mice with age of 6–8 weeks (Charles River Laboratory) were used in the experiments. We used AAV1-synapsin-GCaMP6s to label neurons in hippocampal dorsal CA1 area, and tracked somatic calcium activity of neurons by two-photon imaging through a chronic window as previously described (Dombeck et al., 2010). The in vivo Ca2+ imaging experiments were performed using a Bruker two-photon laser scanning system equipped with a Ti:sapphire laser (Mai Tai DeepSee; Spectra Physics) tuned to 920 nm. The average laser power on the brain sample was ~15–30 mW. All imaging was performed using a 40× objective immersed in an artificial cerebral spinal fluid (ACSF) solution and with a 1× digital zoom. Images were collected at a resolution of 473×473 pixels (Figure 3A) and a frame rate of 2 Hz. The head-restrained animal was trained to move his forelimbs to perform running forward on the treadmill (Figure 3B), while the rotation of belt provided continual changes in visual and tactile stimuli (Royer et al., 2012). Training on the 108-cm treadmill consisted of 8 trials with a continuous running for 10 laps in each trial (80 laps in total). The treadmill speed was 2.5–2.7 cm/s. The dF/F fluorescence calcium activity (Figure 3C) in trial #8 (laps 71 to 80) was analyzed. Recordings of two mice were used in the position decoding analysis (Table 1). Overall, the mean±SD cell population firing rate was 0.1440 ± 0.1121 (spike magnitude/s) for mouse 1, and 0.1438 ± 0.1187 (spike magnitude/s) for mouse 2. In addition, the mean±SD rate of fluorescence evoked events was 0.1647 ± 0.0826 Hz and 0.1642 ± 0.0901 Hz for two mice, respectively. The experimental studies were performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals to ensure minimal animal use and discomfort, and were approved by the New York University School of Medicine (NYUSOM) Institutional Animal Care and Use Committee.
Figure 3:
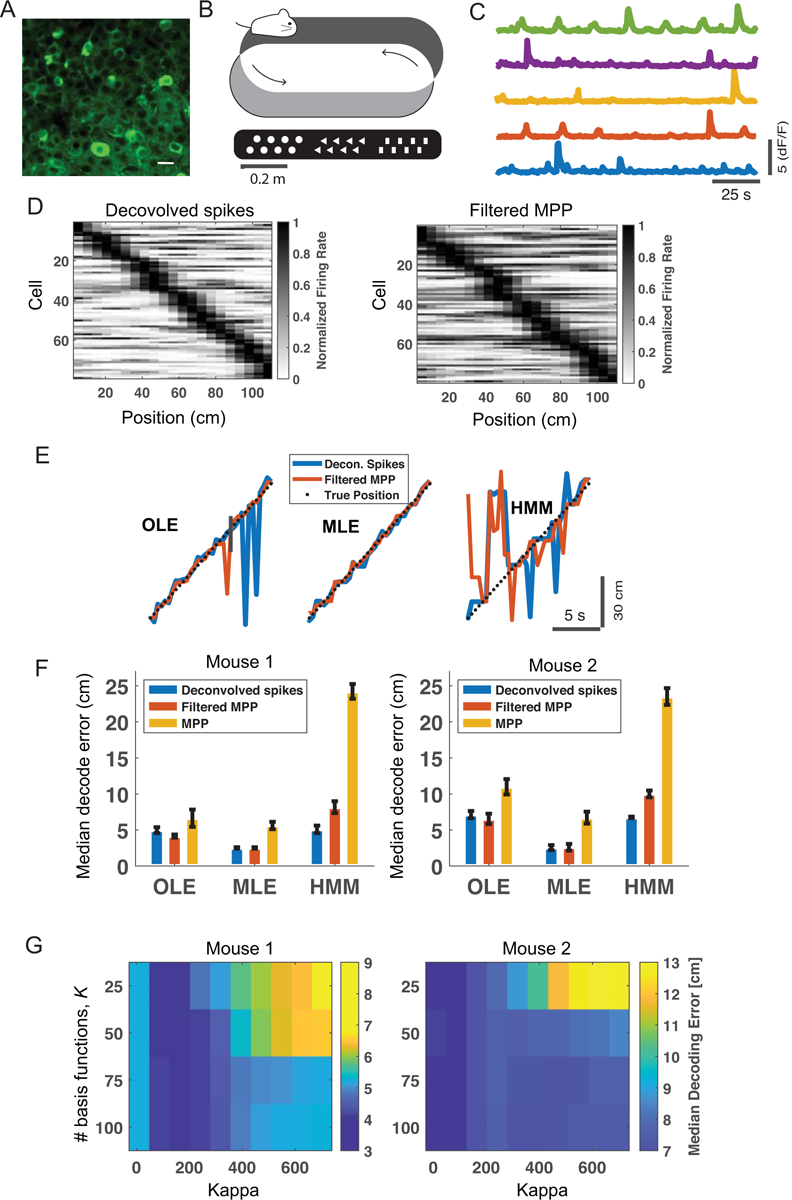
Results of hippocampal dataset 1. (A) Calcium imaging of the mouse CA1 (scale bar: 15 μm). (B) Schematic of mouse running on a sensory-cued treadmill. (C) Selective dF/F fluorescence traces from five hippocampal neurons. (D) Heat map shows a summary of peak firing rate (derived from deconvolved spikes, left or from filtered MPPs, right) location of hippocampal neurons with respect to the treadmill position (mouse 1). Neurons were sorted according to the location of the peak firing rate. Dark color shows high firing probability. All neurons were used in the decoding analyses. (E) Snapshot of supervised and unsupervised position decoding on the held-out data. Black dotted line shows the animal’s position (mouse 1). (F) Comparison of median position decoding error based on OLE, MLE, and HMM methods. Error bar shows the bootstrapped SD. (G) Cross-validation of parameters K and κ derived from the OLE-based decoding analysis using the filtered MPP features. White cross indicates the minimum value.
Table 1:
Summary of calcium imaging data from the mouse hippocampus.
| Dataset (animal) | # session | # cells | sampling rate | imaging |
|---|---|---|---|---|
| Dataset 1 (mouse 1) | 1 | 79 | 2 Hz | two-photon, GCaMP6s |
| (mouse 2) | 1 | 53 | 2 Hz | two-photon, GCaMP6s |
| Dataset 2 (mouse 3) | 4 | 1,091 | 20 Hz | one-photon, GCaMP6f |
| (mouse 4) | 4 | 835 | 20 Hz | one-photon, GCaMP6f |
Hippocampal dataset 2.
The second dataset consists of one-photon calcium imaging from the mouse hippocampus (https://data.mendeley.com/datasets/f9fmrj98n3/1). Details of experimental setup and recordings are referred to the paper (Mau et al., 2018). Briefly, mice were introduced to a 40×60 cm2 rectangular track with an embedded motorized mouse treadmill as one of its long sides. Mice were acclimated to the environment until they reliably sought 20% sucrose water solution. Then, they were trained to run in place on the treadmill for increasing time intervals (>6 s) in between laps. For the beginning sessions, running speed varied between 10–24 cm/s. Once mice would reliably run for ~30 laps per day, data were then collected for 4 days, with each session lasting approximately 30 min and consisting of ~30 laps of 10 s treadmill running and water retrieval. A miniaturized epifluorescence microscope (Inscopix) was used to collect imaging videos of hippocampal CA1 activity at a frame rate of 20 Hz. Videos were motion corrected and cropped (500×500 pixels) to exclude areas without GCaMP6f activity. The videos were then post-processed using an open-source software package (https://github.com/SharpWave/TENASPIS) to identify the regions of interest (ROIs) and further extract the fluorescence activity (dF/F; Figure 4B). Across days, imaged neurons were tracked using image registration software. We selected two mice (G45 and G48) from this public dataset for our current study. Overall, the mean±SD cell population firing rate was 0.0022 ± 0.0011 (spike magnitude/s) for mouse 3, and 0.0010 ± 0.0003 (spike magnitude/s) for mouse 4. In addition, the mean±SD rate of fluorescence evoked events was 0.0282 ± 0.0146 Hz and 0.0198 ± 0.0089 Hz for two mice, respectively.
Figure 4:
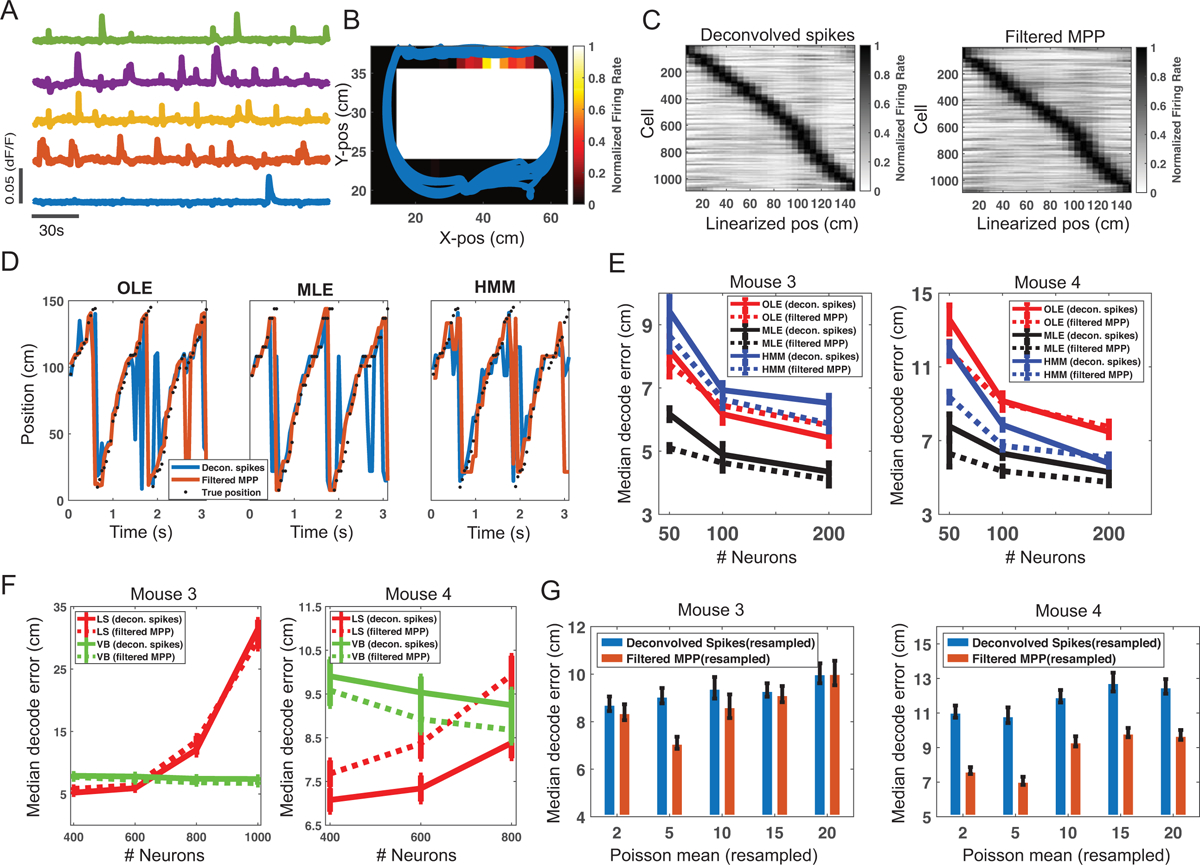
Results of hippocampal dataset 2. (A) Selected dF/F fluorescence traces from five neurons. (B) An example of hippocampal receptive field shown in the rectangular track. Warm color shows high firing probability. Animal’s trajectories were overlaid in the place receptive field. (C) Heat map shows a summary of peak firing rate location of hippocampal neurons with respect to the linearized track location (mouse 3). Left panel was derived from deconvolved spikes, and right panel was derived from filtered MPPs. (D) Snapshot of supervised and unsupervised position decoding on the held-out data. Black dotted line shows the animal’s actual position (mouse 3). Only 200 neurons were used in the decoding analysis. (E) Comparison of supervised and unsupervised decoding accuracy based on different numbers of neurons. Error bar shows the SD of median decoding error based on 20 random selections. (F) When the number of neurons was large (≥400), the least-squared (LS) solution to the OLE suffered from overfitting, whereas the variational Bayes (VB) solution did not. (G) In assessing the HMM-based decoding accuracy (randomly selected 50 neurons, resampled filtered MPP features), the optimal resampling Poisson mean varied from different features and different datasets. Error bar shows the SD.
3. Results
In the following analyses, we tested our methods on two calcium imaging datasets recorded from head-fixed or freely behaving mice. We compared the performance of supervised and unsupervised decoding analyses based on the deconvolved spiking activity, MPPs or filtered MPPs. In supervised decoding analysis, we conducted 10-fold cross-validation using OLE or MLE. In unsupervised learning analysis, we used 90% data for training the HMM, and 10% held-on data for validation. For simplicity, median position decoding errors were reported for performance assessment. However, the mean, variance and distribution statistics were also important. Custom software for all analyses was written in Python and MATLAB, and are publicly accessible (www.cn3lab.org/software.html).
3.1. Hippocampal dataset 1
Using a temporal bin size of 500 ms (i.e., 2 Hz frame rate), we compared the cross-validated median position decoding error derived from four different features: (i) deconvolved spiking activity from fluorescence traces; (ii) unfiltered MPPs derived from fluorescence traces; (iii) filtered MPPs derived from fluorescence traces; (iv) Poisson-distributed resampled data derived from feature (iii). The first three features were used in OLE, and the fourth feature was used in MLE and HMM. To obtain feature (i), we used a state-of-the-art spike nonnegative deconvolution method (Friedrich et al., 2017). From the inferred spiking activities of hippocampal cells, we constructed the normalized place fields (Figure 3D, left panel). By comparison, there was a high degree of resemblance in heat map derived from filtered MPPs (Figure 3D, right panel).
Remarkably, the MLE or OLE-based supervised decoding analysis showed that our proposed feature (ii) yielded lower median decoding error than deconvolved spiking activity in two tested animals (Figure 3E and 3F). Using the same feature, MLE produced better decoding accuracy than OLE in supervised decoding analysis. Compared with OLE, the HMM-based unsupervised decoding analysis showed comparable median decoding error based on (resampled) deconvolved spikes; however, the median decoding accuracy was worse based on the (resampled) filtered or unfiltered MPP features. In all tested methods, the filtered MPPs yielded consistently better performance than the unfiltered MPPs (especially in the HMM-based method), implying the importance of preprocessing in fluorescence traces.
It is noteworthy that the choice of the hyperparameters K and κ influenced the position decoding accuracy. The optimal values could be selected by cross-validation (Figure 3G). However, the decoding accuracy was relatively robust with a wide range of hyperparameters. For implementation simplicity, we have used a finite-length digital filter h = [h1, h2, h3, 0, 0, 0, 0] (where h1 < h2 < h3 and h1 + h2 + h3 = 1) and assumed a constant h3/h2 = α. Similarly, the shape of temporal filter, or the decay constant α, could be optimized by cross-validation.
3.2. Hippocampal dataset 2
We linearized the rectangular track and excluded the treadmill area, resulting a track length of ~150 cm (Figure 4B). We used a velocity threshold of 5 cm/s to exclude low-speed movement periods. The summary of the dataset is shown in Table 1. Similar to the first dataset, we inferred individual spiking activities and filtered MPPs, and computed the corresponding heat maps of hippocampal place fields (Figure 4C). In the remaining decoding analyses, we only focused on the results based on deconvolved spikes and filtered MPPs.
We binned the spiking activity or filtered MPPs with a bin size of 200–300 ms. We picked the bin size that yielded the better decoding performance (mouse 3: 300 ms; mouse 4: 200 ms). We repeated both supervised and unsupervised decoding analysis as before, and varied the number of neurons by randomly selecting a subset of the population. Each decoding analysis was repeated 20 times. In the OLE method, we used the same hyperparameters for all tested features (K = 75 and κ = 25 in mouse 3; K = 25 and κ = 25 in mouse 4). In the MLE method, we resampled the deconvolved spikes or filtered MPPs with Poisson count data (using the same mean statistic: 5). In the HMM method, we used the same number of latent states for all tested features (m = 50). The results are shown in Figure 4D and Figure 4E. As seen, excellent decoding performance was achieved by using only a small number of neurons, and the decoding accuracy gradually saturated with the increasing number of neurons. Comparing supervised and unsupervised decoding analyses, in the mouse-3 recording, MLE consistently yielded the overall best decoding performance, while OLE and HMM approaches had comparable decoding accuracy when the number of neurons were small; however, OLE-based decoding achieved slightly better accuracy than the HMM-based decoding with increasing number of neurons. In the mouse-4 recording, MLE still had the lowest decoding error, whereas the HMM-based strategy achieved better decoding accuracy than the OLE-based strategy. More remarkably, the simple filtered MPP (resampled) features achieved the best median decoding error in the presence of small number of neurons (C = 50 or C = 100). From Figure 4E, it seemed that the optimal number of neurons for achieving a low yet reliable decoding error was around 200, regardless of the used methods or features. Notably, when the number of neurons was more than 400, the LS estimate in the OLE suffered from overfitting, whereas the sparse VB-ARD estimate did not (Figure 4F). However, the VB-ARD estimate was biased; therefore the LS solution was slightly better when there was no overfitting. In addition, we investigated the impact of resampled Poisson mean statistic on the decoding accuracy. We found that the decoding accuracy changed according to the resampling mean statistics, and the optimal remapping value depended on the dataset (Figure 4G). The optimal resampling mean varied from different features and different datasets. The spike-based decoding accuracy was slightly more stable, possibly due to the distribution difference between the filtered MPP features and deconvolved spikes.
Next, we focused on the investigation on the OLE method. Specifically, we systematically downsampled the florescence traces (from 20 Hz to 10 Hz, to 4 Hz and to 2 Hz) and investigated the impact of sampling rate on position decoding accuracy. We considered two different downsampling conditions: (i) we took one from every N samples from the 20 Hz sampled florescence trace; (ii) we temporally averaged every N samples from the 20 Hz sampled florescence trace. Note that in each case, the decoding bin size remained the same as before resampling. Using 100 randomly selected neurons, we repeated the decoding analysis pipeline 20 times. We found that the decoding accuracy trends derived from these two downsampling operations were somewhat similar, but the accuracy was slightly better in the 2nd downsampling condition (Figure 5A). The OLE-based decoding accuracy was better with deconvolved spikes than with filtered MPPs. In addition, we observed a decreasing trend in median decoding error with a lower sampling rate. Although the result appeared counterintuitive, our downsampling operations did not apply on the raw data directly, which might deviate from the physical reality of low imaging frame rate.
Figure 5:
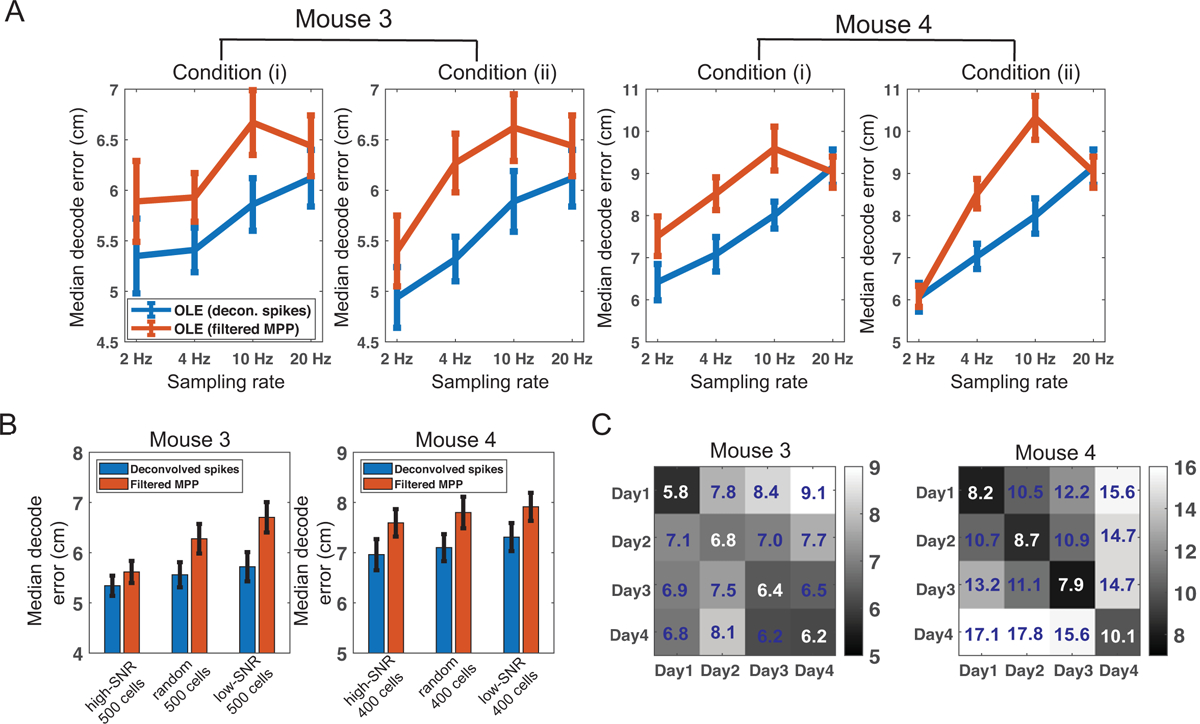
Results of hippocampal dataset 2. (A) Comparison of OLE-based decoding accuracy with four different sampling frequencies under two downsampling conditions. Error bar shows the SD based on 20 random selections of 100 neurons used for decoding analysis. (B) Comparison of OLE-based decoding accuracy using an equal number of low or high-SNR neurons. The performance derived from the same number of randomly selected neurons is shown the middle as control. Error bar denotes the bootstrapped SD. (C) Within- and between-session position decoding error matrix shows the generalization ability of the OLE-based decoding strategy in four consecutive run days (mice 3 and 4). A total of 100 randomly selected neurons were used for the decoding analysis. The number in each entry represents the averaged median decoding error.
Furthermore, we examined the relationship between the SNR and the OLE-based decoding accuracy based on deconvolved spikes and filtered MPPs. In each calcium imaging dataset, we computed and sorted the SNR (Equation 2.1), and used the top N and bottom N neurons (N = 500 for mouse 3, and N = 400 for mouse 4) in decoding, separately. We observed a decreased decoding accuracy with lower SNR, for both deconvolved spikes and filtered MPPs (Figure 5B).
To test the stability and generalization of the decoding method based on filtered MPPs, we further conducted cross-session OLE-based decoding analysis: training on one session and testing on another session, while using randomly selected (yet identical) 100 neurons from the whole population. For a fair comparison, the within-session position decoding error was computed by two-fold cross-validation. In between-session decoding, data from one complete session were trained, followed by testing on another complete session. Remarkably, we still achieved good position decoding accuracy across multiple days (Figure 5C). In general, the within-session (i.e., the diagonal) had the lowest decoding error, followed by the between-session results in neighboring days.
3.3. Computer simulations
Finally, to gain some insight into our proposed method, we conducted a computer simulation study using a setup similar to (Wei et al., 2019). Specifically, we assumed that the animal ran 20 laps on a 1-m linear track with constant velocity. We assumed 50 hippocampal place cells, with Gaussian tuning curves that uniformly cover the track. Next, assuming 20 Hz frame rate, we simulated Poisson-distributed spatially-tuned ground-truth spike trains based on the tuning curves. We then used a generative model described in (Friedrich et al., 2017; Wei et al., 2019) to generate fluorescence traces from these spike trains with varying levels of background noise (Figure 5A). Specifically, the calcium concentration traces ct are generated by a 2nd-order autoregressive (AR) model, and the fluorescence trace yt is related to calcium concentration as follows (Friedrich et al., 2017):
| (3.1) |
| (3.2) |
where si denotes the spike activity, {a, b} are two linear regression parameters, and denotes the Gaussian measurement noise with zero mean and variance σ2. Given a representative neuron’s simulated fluorescence trace, the ground-truth spike activity, deconvolved spike activity, and filtered MPPs are shown in Figure 5B. We repeated the decoding analyses based on 20 Monte Carlo runs under different noise levels. In all simulations, we used the same convolution filter h = [0.14, 0.29, 0.57, 0, 0], and the resampled Poisson mean statistic of 5. We also used the same criterion for peak detection, where the threshold was set as the 30% peak fluorescence amplitude. The decoding results are shown in Table 2. As seen in the table, our simulation decoding results were consistent with experimental data. Among three decoding methods, the MLE achieved the best decoding accuracy. Within the same decoding method, the filtered MPPs yielded the best decoding performance, followed by deconvolved spikes, and then unfiltered MPPs. In addition, the decoding accuracy reduced with decreasing SNR.
Table 2:
Summary decoding statistics of simulated data. Median±SD statistics (unit: cm) are derived from 20 Monte Carlo runs. σ denotes the measurement noise standard deviation in the simulated calcium concentration.
| Features | OLE | MLE | HMM | OLE | MLE | HMM | OLE | MLE | HMM |
|---|---|---|---|---|---|---|---|---|---|
| σ = 0.3 | σ = 0.6 | σ = 1 | |||||||
| deconvolved spikes | 7.87±0.25 | 3.64±0.07 | 4.76±0.52 | 8.30±0.27 | 3.90±0.13 | 5.13±0.47 | 9.42±0.36 | 4.71±0.09 | 6.45±0.68 |
| MPPs | 8.89±0.23 | 3.73±0.07 | 5.75±0.44 | 9.46±0.28 | 4.39±0.12 | 5.98±0.50 | 11.65±0.25 | 6.11±0.26 | 10.19±0.89 |
| filtered MPPs | 6.26±0.16 | 2.40±0.07 | 3.10±0.36 | 6.65±0.22 | 2.80±0.13 | 3.53±0.47 | 7.81±0.27 | 4.38±0.18 | 5.28±0.45 |
3.4. Computational speed
Low-latency computation and real-time scalable decoding analysis are two important factors for closed-loop neuroscience experiments. Compared to the state-of-the-art fast spike deconvolution methods, our supervised decoding strategy is simple and ultrafast. Here, we compared the CPU time between two fast spike deconvolution methods (Friedrich et al., 2017; Rahmati et al., 2018)1 and our strategy. Using custom-written MATLAB codes (MATLAB R2019, Windows 10 OS; Intel® Core™ i7–6500U CPU 2.50 GHz, 8 GM RAM), the CPU time was averaged across complete recordings and all neurons. A naive (non-optimized) implementation of our strategy already achieved significant speedup in computation. We expect an efficient C/C++ implementation will further improve the speed.
Several points are noteworthy below:
The convolution step in our proposed preprocessing procedure introduces a temporal delay and appears acausal. This issue becomes more particularly problematic when the frame rate is low (e.g., 2 Hz). However, when the frame rate is relatively high (e.g., 20 Hz), the delay and causality issue will not be the issue given the decoding bin size is 200–300 ms (i.e., 4–6 frames).
If the causation of real-time processing is a real concern, we can also discard the convolution step and only use MPPs (instead of filtered MPPs) for the subsequent decoding analysis. The decoding result will be slightly reduced (e.g., Figure 3F and Table 2), but still acceptable.
Unlike the OLE method, the MLE or HMM-based decoding strategy relies on a rank-resampling strategy, therefore limiting its real-time application.
4. Discussion
In this paper, we have proposed an efficient method to infer the proxy of spiking activity directly from dF/F florescence in calcium imaging, and used it for the subsequent position decoding analysis. Our spike inference-free procedure is simply based on a peak detection followed by a filtering operation. The temporal filter can be optimized depending on the neuron’s firing rate and the imaging frame rate. The strategy has been proven effective in both supervised and unsupervised decoding analyses for reconstruction of the animal’s position. It is demonstrated, via both simulated and experimental data, that exact spike timing is not needed for position decoding; instead, we can infer the mouse’s position directly from the proxy of spiking activity via filtered MPP.
In the literature, many methods have been developed for position decoding analysis based on calcium imaging of the hippocampus or neocortex (Ziv et al., 2013; Mao et al., 2018; Wei et al., 2019). The common idea of these methods is to establish neuronal correlates (either deconvolved spikes or their proxy) with the animal’s position. Similar rationale has also been extended for decoding time (Mau et al., 2018). Statistical modeling of deconvolved calcium signals or their preprocessed signals is critical for interpreting calcium measurements. However, some analyses, deconvolution step may turn out unnecessary, as demonstrated in our current study and other’s (e.g., Ganmor et al., 2016).
Based on the mouse hippocampal calcium imaging data, we show that a small number (~100) of hippocampal place cells is sufficient to provide a reliable spatial readout at various imaging frame rates. The unsupervised HMM-based method can also uncover spatial representations of a population of rodent hippocampal place cells from their calcium imaging activities, consistent with our prior results derived from spikes and LFP signals (Linderman et al., 2016; Cao et al., 2019). Once the temporal resolution of imaging techniques is further improved (say 50–100 Hz), the same analysis can be used to decode hippocampal memory replays (e.g., Liu et al., 2018; Cao et al., 2019; Chen et al., 2016; Chen & Wilson, 2017; Maboudi et al., 2018). Real-time calcium imaging-based decoding analyses can be readily adapted for closed-loop experiments in behaving mice.
Our calcium imaging “place” decoding analysis for the mouse hippocampus can be also extended to neocortical circuits that encode spatial information, including the neurons from the entorhinal cortex, primary visual cortex, retrosplenial cortex, parietal cortex (Hafting et al., 2005; Ji & Wilson, 2007; Whitlock et al., 2008; Haggerty & Ji, 2015; Mao et al., 2017). Extending this analysis pipeline to large-scale in vivo neocortical population calcium imaging data in other behavioral tasks will be the subject of future investigation.
Figure 6:
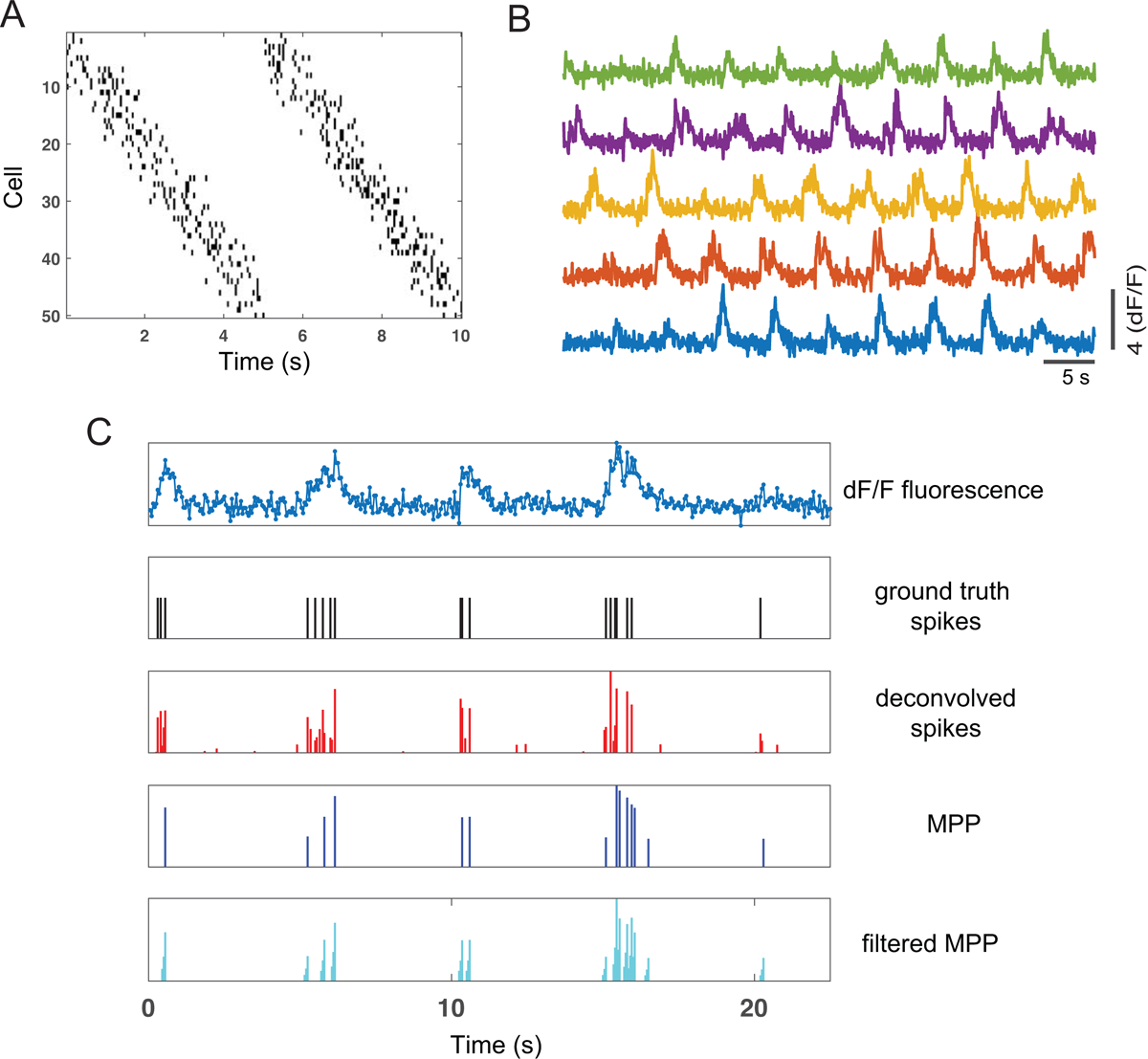
Results of computer simulations. (A) Simulated Poisson spike trains of 50 neurons in two laps. (B) Simulated fluorescence traces of selected 5 neurons in ten laps. (C) One selected simulated dF/F fluorescence trace, and its associated simulated ground truth spikes, deconvolved spike activity, inferred MPP (upon peak detection) and filtered MPP.
Table 3:
Comparison of computation speed (mean±SD) between our preprocessing method and two spike deconvolution methods. Statistics were averaged across all neurons in each dataset.
| Method | CPU time (ms per sample) | |
|---|---|---|
| Dataset 1 | Dataset 2 | |
| proposed OLE method | (1.77±0.18) × 10−4 | (1.63±0.19) × 10−4 |
| spike deconvolution (Friedrich et al., 2017) | 0.0556±0.0060 | 0.0483±0.0060 |
| spike deconvolution (Rahmati et al., 2018) | 0.0012±0.0002 | 0.0011±0.0001 |
Acknowledgments
We thank William Mau for sharing the calcium imaging data. This work was partially supported by NIH grants R01-MH118928 (Z.S.C.), R01-MH111486 (W.-B.G.) from the NIMH, R01-NS100065 (Z.S.C.), R01-NS047325 (W.-B.G.) from the NINDS, as well as an NSF grant CBET-1835000 (Z.S.C).
Appendix: Implication of rank-invariant resampling
Without the loss of generality, we assume the observed data . The goal of rank-invariant resampling is to find a linear or nonlinear transformation: ynew = ϕ(y), where ϕ(·) denotes a nonnegative monotonic increasing function, such that the rank-invariant order among the observed samples is preserved. The rank-invariant resampling procedure can be viewed as a special form of nonlinear mapping function.
For simplicity, let us first consider likelihood inference where the latent state process {St} is known. In the HMM, the model parameter related to the observed data likelihood is the rate parameter Λ = {λc,i}. The maximum likelihood estimate (m.l.e.) of the rate parameter, given the observations y1:T = {y1,1:T , …, yC,1:T}, is given by :
where denotes the mean statistic under the empirical distribution of raw data. Given the transformed observations ϕ(y1:T) in a new coordinate system, the new rate parameter is defined as
In a special case where ϕ(·) is a linear function, it is easy to verify that the parameter inference is scale and translational invariant. Let ϕ(yc) = αyc + β, the m.l.e. of new rate parameter is , where α and β are two arbitrary positive constants. When ϕ(·) is nonlinear, (the exact inequality sign depends on the convexity of ϕ). Nonetheless, the nonlinear function can be linearized, and the equality may approximately hold around the local linearized region.
The rank-invariant resampling can be approximately viewed as the implementation of a set of domain-based piecewise linear functions within the CDF interval of [0, 1]. Therefore, the local invariance property approximately holds. In the presence of latent state variables, the invariance property cannot be guaranteed.
Footnotes
The software packages are available at https://github.com/j-friedrich/OASIS and https://github.com/VahidRahmati/UFARSA.
References
- Agarwal G, Stevenson IH, Berenyi A, Mizuseki K, Buzsaki G & Sommer FT (2014). Spatially distributed local fields in the hippocampus encode rat position. Science 344: 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldler A, Zhao R, Shin ME, Yasuda R, & Gan WB (2019). Somatostatin-expressing interneurons enable and maintain learning-dependent sequential activation of pyramidal neurons. Neuron, 102: 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C (2006). Pattern Recognition and Machine Learning. Springer. [Google Scholar]
- Cao L, Varga V, & Chen Z (2019). Ultrafast readout of representations from spatially distributed rodent hippocampal field potentials. www.biorxiv.org/content/10.1101/828467v1 [Google Scholar]
- Chen TW, Wardill TJ, Pulver SR et al. (2013). Ultrasensitive fluorescence proteins for imaging neuronal activity. Nature 499: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloosterman F, Layton S & Wilson MA (2012). Transductive neural decoding for unsorted neuronal spikes of rat hippocampus. Proc. IEEE EMBC, pp. 1310–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Kloosterman F, Brown EN & Wilson MA (2012). Uncovering hidden spatial topology represented by hippocampal population neuronal codes. J. Comput. Neurosci 33: 227–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gomperts S, Yamamoto J & Wilson MA (2014). Neural representation of spatial topology in the rodent hippocampus. Neural Computation 26: 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Grosmark AD, Penagos H & Wilson MA (2016). Uncovering representations of sleep-associated hippocampal ensemble spike activity. Scientific Reports, 6: 32193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z & Wilson MA (2017). Deciphering neural codes of memory during sleep. Trends in Neurosciences, 40(5): 260–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberti D, Michon F & Kloosterman F (2018). Real-time classification of experience-related ensemble spiking patterns for closed-loop applications. eLife 7: e36275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson TJ, Kloosterman F & Wilson MA (2009). Hippocampal replay of extended experience. Neuron, 63: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Liu DF, Kay K, Frank LM & Eden UT (2015). Clusterless decoding of position from multiunit activity using a Marked point process filter. Neural Computat. 27: 1438–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, Looger LL, & Tank DW (2010). Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 13: 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drugowtisch J (2017). Variational Bayesian inference for linear and logistic regression. https://arxiv.org/pdf/1310.5438.pdf.
- Friedrich J, Zhou P & Paninski L (2017). Fast online deconvolution of calcium imaging data. PLoS Comput. Biol 13: e1005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganmor E, Krumin M, Rossi LF, Carandini M & Simoncelli EP (2019). Direct estimation of firing rates from calcium imaging data. https://arxiv.org/pdf/1601.00364.pdf
- Giovannucci A, Friedrich J, Gunn P et al. (2018). CalmAn: an open source tool for scalable calcium imaging data analysis. eLife 8: 339564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafting T, Fyhn M, Molden S, Moser M-B, & Moser E-I (2005). Microstructure of a spatial map in the entorhinal cortex. Nature 436: 801–806. [DOI] [PubMed] [Google Scholar]
- Haggerty DC & Ji D (2015). Activities of visual cortical and hippocampal neurons co-fluctuate in freely moving rats during spatial behavior. eLife 4: e08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Crmmoun L, Gigandet X, Thiran JP, Meduli R & Hagmann P (2009). Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. USA 106: 2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Ciliberti D, Grosmark AD, Michon F, Ji D, Penagos H, Buzsaki G, Wilson MA, Kloosterman F & Chen Z (2018). Real-time readout of large-scale unsorted neural ensemble place codes. Cell Reports 25: 2635–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M (2006). Point Process Theory and Applications: Marked Point and Piecewise Deterministic Processes. Birkhäuser. [Google Scholar]
- Jercog P, Rogerson T & Schnitzer MJ (2016). Large-scale fluorescence calcium-imaging methods for studies of long-term memory in behaving mammals. Cold Spring Harbor Perspectives in Biology, 8: a021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell S & Witten D (2018). Exact spike train inference via ℓ0 optimization. Ann. Appl. Statist 12: 2457–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D & Wilson MA (2007). Coordinated memory replay in the visual cortex and hippocampus during sleep. Nat. Neurosci 10: 100–107. [DOI] [PubMed] [Google Scholar]
- Kloosterman F, Layton SP, Chen Z & Wilson MA (2014). Bayesian decoding using unsorted spikes in the rat hippocampus. J. Neurophysiol 111: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderman SW, Johnson MJ, Wilson MA, & Chen Z (2016). A Bayesian nonparametric approach to uncovering rat hippocampal population codes during spatial navigation. J. Neurosci. Methods 236: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Grosmark A & Chen Z (2018). Methods for assessment of memory reactivation. Neural computation, 30: 2175–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maboudi K, Ackermann E, de Jong LW, Pfeiffer BE, Foster D, Diba K, & Kemere C (2018). Uncovering temporal structure in hippocampal output patterns. eLife 7: e34467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik WQ, Schummers J, Sur M, & Brown EN (2011). Denoising two-photon calcium imaging data. PLoS ONE 6: e20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Kandler S, McNaughton BL, & Bonin V (2017). Sparse orthogonal population representation of spatial context in the retrosplenial cortex. Nat. Commu 8: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao D, Neumann AR, Sun J, Bonin V, Mohajerani MH, & McNaughton BL (2018). Hippocampus-dependent emergence of spatial sequence coding in retrosplenial cortex. Proc. Natl. Acad. Sci. USA, 115: 8015–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau W, Sullivan DW, Kinsky NR, Hasselmo ME, Howard MW & Eichenbaum H (2018). The same hippocampal CA1 population simultaneously codes temporal information over multiple timescales. Curr. Biol, 28: 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C & Harris KD (2018). Robustness of spike deconvolution for neuronal calcium imaging. J. Neurosci 38: 7976–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Dippoppa M et al. (2017). Suite2p: beyond 10,000 neurons with standard two-photon microscopy. BioRxiv: www.biorxiv.org/content/10.1101/061507v1 [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y et al. (2016). Simultaneous denoising, deconvolution, and demixing of calcium imaging data. Neuron 89: 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA (2019). Analysis pipelines for calcium imaging data. Curr. Opin. Neurobiol 55: 15–21. [DOI] [PubMed] [Google Scholar]
- Rahmati V, Kirmse K, Holthoff K & Kiebel SJ (2018). Ultra-fast accurate reconstruction of spiking activity from calcium imaging data. J. Neurophysiol 119: 1863–1878. [DOI] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Losonczy A, Kim J, Chance F, Magee JC, & Buzsáki G. (2012). Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat. Neurosci 15: 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer S, Pachitariou M, Steinmetz N et al. (2019). Spontaneous behaviors drive multidimensional, brain-wide population activity. Science 364: eaav7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein JT, Packer AM, Machado TA, Sippy T, Babadi B, Yuste R & Paninski L (2010). Fast nonnegative deconvolution for spike train inference from population calcium imaging. J. Neurophysiol 104: 3691–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Zhou D, Grossmark A, Ajabi Z, Sparks F, Zhou P, Brandon M, Losconczy A, & Paninski L (2019). A zero-inflated gamma model for post-deconvolved calcium imaging traces. https://www.biorxiv.org/content/10.1101/637652v1.full.pdf
- Whitlock JR, Sutherland RJ, Witter MP, Moser M-B & Moser E-I (2008). Navigating from hippocampus to parietal cortex. Proc. Nat. Acad. Sci. USA 105: 14755–14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Nagarajan S & Chen Z (2016). Bayesian machine learning: EEG/MEG signal processing measurements. IEEE Signal Processing Magazine, 33: 14–36. [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, et al. (2018). Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife, 7: e28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED et al. (2013). Long-term dynamics in CA1 hippocampal place codes. Nat. Neurosci 16: 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]


