Abstract
Course-based undergraduate research experiences (CUREs) provide the same benefits as individual, mentored faculty research while expanding the availability of research opportunities. One important aspect of CUREs is students’ engagement in collaboration. The shift to online learning during the COVID-19 pandemic created an immediate need for meaningful, collaborative experiences in CUREs. We developed a partnership with the Caenorhabditis elegans (C. elegans) database, WormBase, in which students submitted annotations of published manuscripts to the website. Due to the stress on students during this time of crisis, qualitative data were collected in lieu of quantitative pre- and postanalyses. Most students reported on cognitive processes that represent mid-level Bloom’s categories. By partnering with WormBase, students gained insight into the scientific community and contributed as community members. We describe possible modifications for future courses, potential expansion of the WormBase collaboration, and future directions for quantitative analysis.
INTRODUCTION
Course-based undergraduate research experiences (CUREs) are effective ways to include students in primary research as part of their coursework. Students who participate in research have strengthened science identities and are more likely to persist in science (1–3). CUREs provide an equitable way for students to participate in research, regardless of whether they are able to join a laboratory (4). CUREs include five essential elements of scientific research as part of their curriculum: use of scientific practices, a focus on broadly relevant or important work, discovery, collaboration, and iteration (5). Thus, students who participate in CUREs are introduced to a wide variety of activities that professional researchers do regularly.
Collaboration is a central aspect of scientific research and often occurs between students in CUREs (5, 6). However, students can collaborate with the wider scientific community by contributing to intramural research projects and adding their data to existing databases (7, 8). We developed a partnership that allows students in a CURE to collaborate directly with researchers by submitting annotations for previously published data to the WormBase data curation project. This collaboration was designed as part of a shift from in-person laboratory work to online learning due to the COVID-19 pandemic. It represents a chance for students to participate in a collaborative research activity without requiring the in-person lab facilities that remain closed due to the pandemic.
WormBase (www.wormbase.org) is an open access, publicly funded genomic database for the nematode (roundworm) Caenorhabditis elegans and related nematodes (9, 10). WormBase curators extract knowledge about C. elegans genes and their biological functions from the research literature and create annotations about connections between genes and functional information, e.g., gene expression patterns, phenotypic variations in response to genetic mutation, and genetic contributions to biological processes. WormBase staff rely on the C. elegans research community to get full coverage of information published each year in relevant articles. Curated annotations can be accessed through WormBase by researchers and the public. Anyone with sufficient knowledge of the basic cell and molecular biology of C. elegans may contribute annotations to WormBase via community curation.
The cessation of in-person teaching due to the COVID-19 pandemic in March 2020 meant that our entire Advanced Cell Biology CURE ran as an online course. The course focuses on the cell biology and neuroscience of the model organism C. elegans and usually runs as a one-quarter, 10-week intensive laboratory (10 contact hours per week). Students’ usual activities include a scientific collaboration with another C. elegans laboratory on campus and presenting at a local C. elegans conference, neither of which were available. However, an open invitation to the C. elegans community to annotate data for WormBase initiated the collaborative partnership that we describe below. Students contributed to the C. elegans community by collaborating with WormBase as annotators for published C. elegans papers.
Intended audience, learning time, and prerequisite student knowledge
This collaborative activity was used for an advanced (senior) level course, but any molecular genetics course could use it as a scaffolded exercise. In this collaboration, students participated in an introductory workshop or lecture about annotations in WormBase (90 minutes) and completed at total of nine annotations as three homework assignments over the course of a week (60 to 120 minutes). Students were familiar with C. elegans as a model system and with the nomenclature of C. elegans genetics (11, 12).
Learning objectives
The overarching learning goals for this activity were for students to:
Appreciate the role of community members in maintaining a scientific database
Participate meaningfully in the collaborative process of community curation
At the end of this activity students will be able to:
Navigate published C. elegans literature to find manuscripts in need of annotation
Use an online portal for submitting annotations
Identify and compare controls with experimental conditions
Identify and differentiate between genetic and phenotypic concepts in the context of an experiment (especially alleles, genetic mutations, transgenic perturbations)
Identify key phrases in manuscripts that discuss individual pieces of data
Data presented in this manuscript were collected in accordance with Western Washington University’s IRB exemption EX16-094.
PROCEDURE
Materials
Students will need a networked computer to access the Wormbase homepage and publication databases, preferably PubMed. WormBase administrators curate different data types from the published literature using at least three elements: publication or source, entity1 (e.g., genetic perturbation, such as an allele, transgene, RNA interference [RNAi], etc.), and entity2 (e.g., phenotype or change in gene expression exhibited upon genetic perturbation), where entity1 and entity2 are associated by an inferred or explicit relationship. Annotations are created by filling in text boxes and/or choosing descriptions from drop-down menus as described in Appendix 1. WormBase curators tend to add supplementary metadata and context in addition to the three elements listed above. For phenotype curation, curators or students submit a publication, an allele or transgene, and an observed (or unobserved) phenotype.
Detailed instructions and definitions for annotations are included in Appendix 1.
Student instructions
Use PubMed or Google Scholar to find a paper or author whose work is interesting to you.
Determine whether your paper needs curation: Navigate to the Submit Data page on WormBase, then “Fill out online form” for Phenotype data and enter an author’s name.
Click on “Click here to review your publications and see which are in need of phenotype curation” and make sure that the paper you are interested in reads “Needs curation” or “Curation in progress.” For the purposes of this exercise, do not worry about the RNAi phenotype status. If your paper has been curated, check with your instructor to make sure that it is okay for you to submit annotations for this paper.
Use the annotation guide (Appendix 1) to help you fill out the annotation for a single piece of data from your paper. Note: this could mean comparing two panels of a figure, or two bars from a 10-bar graph!
Be sure that you change the “your name” field back to your instructor’s name.
Leave a comment in the Notes section for WormBase with your own name so that the administrators can keep track of who in the class sends in what annotations.
When you are done with your annotation, select preview and download the image for grading or marking by your instructor; this can be turned in in-person or online.
Submit your annotation by clicking Submit.
Repeat steps 4 to 8 two more times, so that you have annotated three pieces of data. If you want to annotate data from a different paper, you will also need to repeat steps 1 to 3 for each additional paper.
Students did three iterations of the assignment (Appendix 1). For annotation assignment #1, they chose one of two papers that were used as examples during the annotation workshop with a WormBase administrator (13, 14). For annotation assignment #2, they chose a paper that related to their own research project for the course. For annotation assignment #3, students had the option of choosing a third paper or revising a previous set of annotations.
Students read the entire manuscript and chose three pieces of data to annotate using the online webform (Fig. 1). The steps for annotation and definitions for genetic and phenotypic identifiers are provided in Appendix 1. Students turned in the preview of their work to the instructor for grading.
FIGURE 1.
Image of the annotation webform for submitting annotations. Annotators fill out the input fields to generate a phenotype annotation. Most fields can bring up a pull-down menu for confirmation of the term, ID, number, etc. Annotators fill in their instructor’s name in the “Your Name” box and their own name in the “Comment” box to ensure that the contribution can be tracked by WormBase administrators.
Faculty instructions
Instructors should reach out to WormBase before having undergraduates submit annotations as part of a class exercise (help@wormbase.org).
This activity assumes that instructors are aware of the importance of community involvement and collaboration in the scientific process. The C. elegans community has active fora through WormBase as well as on Slack, Facebook, and Twitter. Instructors should be comfortable with C. elegans nomenclature and genetics. “A Transparent Window into Biology: A Primer on Caenorhabditis elegans” provides a succinct overview of the worm as a model system (12). Working with Worms: Caenorhabditis elegans as a Model Organism includes a step-by-step worksheet activity for becoming familiar with the bioinformatics tools available in WormBase (11). There are also written introductions to WormBase (9, 10, 15). A WormBase administrator provided an introduction workshop to annotation that used papers that students had already read (13, 14). This ensured that students did not have to learn new nomenclature and new gene names at the same time. Providing tutorials or orientations for many large classes in parallel may be beyond the current capacity of WormBase curators, so instructors should contact WormBase (help@wormbase.org). Increased support may be considered in the future, should demand warrant it.
Students choose papers to annotate, and in some cases they may choose papers that have already been annotated or partially annotated. Generally, redundant annotations are not a problem, and they may provide helpful confirmations. Instructors or students may want to check with WormBase (help@wormbase.org) if they have questions.
Instructors will receive the preview image of the annotation to review for grading or marking. These can be done directly on the electronic copy using a tablet and stylus or by printing out and writing directly on the document, which can then be photographed or scanned for upload for student review. The annotation will include the PubMed ID of the paper, so finding the data that is being annotated is not difficult; students should include direct quotes to support their annotations (Appendix 1), which allows instructors to search directly for the figure number, if necessary. Instructors will also receive a confirmation of each annotation via email, assuming that students use the instructor’s WormPerson ID for the submission.
This activity is scalable, especially if students are working on the same paper or in groups. By using a single paper, many different annotations could be produced without requiring instructors to read many different papers. If students work in groups, group members could each individually produce a practice annotation for the same data, which could be peer-reviewed and refined into a single annotation for each group. This would reduce the burden on WormBase staff by minimizing redundant submissions. Students could also be graded on practice annotations for early (or formative) assessment, so that final and/or revised submissions would be less likely to include errors or misunderstandings for WormBase staff to correct.
Suggestions for determining student learning
Assessment of this assignment during the course was low-stakes, with all three annotations totaling less than 10% of the course grade. Each annotation was essentially unique, but point breakdown had approximately 50% of the point total going to understanding the structure of the annotation, for example, filling out the relevant fields with the correct type of information; not filling out fields that were not represented; using the correct e-mail address and including a note about who was curating (Appendix 2).
The remaining 50% of the point total was awarded based on the correctness of individual fields. For example, choosing the correct background to represent the control state, identifying the correct allele or transgene, determining whether the data showed an “observed phenotype” or a “not observed phenotype” (negative result). One way to assess student learning is to follow students’ revisions of earlier annotations. Students generally revised their annotations to be correct and wrote cogent explanations for why they changed their answers in the way(s) that they did (Appendix 2).
After students completed their first annotation, they were given an online quiz that included a novel (to them) piece of data and questions about how it could be annotated in WormBase (Appendix 3). At the time of the quiz, students were generally able to use their understanding of molecular genetics to answer questions about complementation and rescue (14 of 16 correct answers for questions 1 and 2). Students were less able to correctly parse the choices for the annotation example (8 of 16 full credit for question 3), though all students were able to correctly identify the allele name, and most (15 of 16) correctly identified the phenotype. Due to ongoing crises in spring 2020, a planned end-of-the-quarter quiz with similar questions was excluded from the course.
Sample data
Students uploaded their submissions via the webform (Fig. 1). An example of a student’s “preview” and instructor comments is provided in Appendix 2. For the final iteration, students were invited to revise an original submission. An example of an original submission with instructor comments and a revision is provided here (Appendix 2).
Safety issues
There are no physical safety issues to address for this activity because the activity and training were completed entirely online. While some students did choose to apply for WormBase IDs (identifier codes in the form: WBPersonXXXX), which requires the input of an e-mail address and affiliation, they were not required to do this. Most students chose to submit annotations using the instructor’s WormBase ID (Appendices 2 and 4). This also allowed the WormBase administrators to more easily group all submissions from the course together (Appendix 5).
WormBase curation of annotations
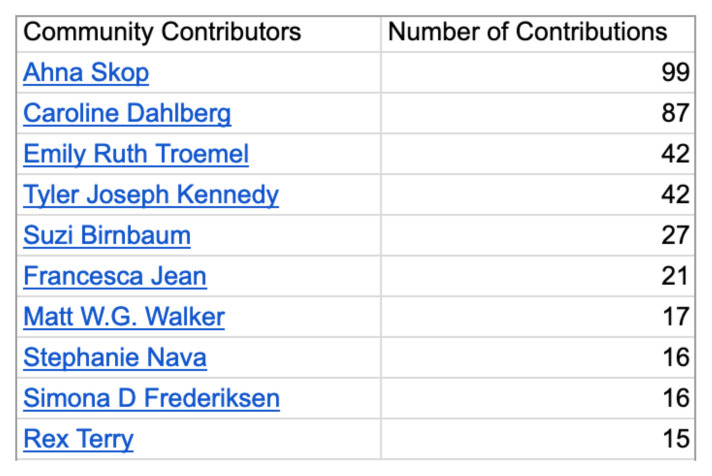
Students’ phenotype annotation submissions were checked for accuracy, including that the submitted phenotype was actually from the publication indicated, that mutations and alleles were correct, and that phenotype identifiers were reasonable. They were also tracked in a spreadsheet for redundancy with other students’ submissions. Distinct, verified annotations were marked as good submissions (Appendix 5) and have been subsequently entered into the official WormBase release (WS278, released in October 2020). The output for the community includes URLs of vetted annotations by undergraduates, as well as a standing or listing of undergraduate annotators (Fig. 2, Appendix 4).
FIGURE 2.
Screen shot from the WormBase homepage, September 2020. The WormBase website tracks annotation contributions by WBPersonID. The CURE had the second highest number of annotations. One student used their own WBPersonID and was also listed in the top ten annotators for the same time period.
DISCUSSION
Field testing
Sixteen senior-level students in a research-based advanced cell biology laboratory course annotated manuscripts in WormBase using the online webform (Fig. 1). Students were introduced to members of the C. elegans community through synchronous online visits that included seminars and discussions with scientists at a variety of different research and outreach jobs. Prior to annotating, students were introduced to C. elegans as a model system (11) and to the WormBase platform (www.wormbase.org). They received a video tutorial from a WormBase administrator along with a sheet of guidelines (Appendix 1). For each assignment, students submitted three separate annotations to WormBase and uploaded a screenshot of their submission to the instructor for feedback and grading. Students completed this assignment three times, for a total of nine separate annotations. Based on this single iteration, annotations by the class and individual students resulted in two of the top 10 annotators’ places on the WormBase homepage (Fig. 2).
Evidence of student learning
In lieu of a final quiz (see above, Suggestions for determining student learning), we used an anonymous survey to assess student perceptions and learning through this assignment. We asked students to describe their learning in four categories by comparing the annotation activity with other biology classes they have taken. The four categories were: distinguishing between genotype and phenotype, distinguishing between control and experimental conditions, identifying different alleles, and distinguishing between transgenic animals and genetic mutants. We also asked students to describe other ways in which annotating helped or hindered their learning (Appendix 6). The responses to these questions were coded by three independent coders for student descriptions of the activity (Appendix 7, Table 1) (16). Coders also reviewed answers for student perceptions of the assignment’s utility and the sophistication of student answers.
TABLE 1.
WormBase survey code book.
| Code Label | Code Name | Code Definition | Percentage of Students whose Responses Included this Code |
|---|---|---|---|
| A. Student perceptions of learning | |||
| Theme definition: Responses that include descriptions of how a person’s learning was impacted. | |||
| A.1 | Making connections* | When a respondent describes that the activity helped them “tie [ideas] together” or connect two processes (for example: experimental conditions and change in phenotype), or when the response shows the ability to do so. | 42% |
| A.2 | Deeper or better understanding* | When a respondent writes “I have a better understanding,” shows an ability to name, [describes their learning], places detail, describes a process. | 92% |
| A.3 | Proficiency* | When a respondent describes how the activity intersected, or didn’t, with “prior knowledge” [which made the activity not useful]. | 57% |
| A.4 | Skills and competency* | When a respondent identifies particular skills that they got better at: for example, reading scientific literature, “paying attention” to details; can also include referring to practicing. | 64% |
| B. Cognition and metacognition | |||
| Theme definition: Responses that describe a person’s awareness of their thinking or changes in their thinking. | |||
| B.1 | Different perspective | When a respondent describes gaining a “different perspective” or recognizing a gap in knowledge. | 43% |
| B.2 | New use of resources* | When a respondent describes how they used resources that they already knew about, for example to “look and find … in papers.” | 36% |
| B.3 | Emotion | When a respondent explains why they felt good or bad about the assignment, for example: overcoming confusion (if there’s a negative: not enjoying or not overcoming confusion), feeling helped or not helped, describing feelings of independence. | 50% |
| C. Collaborative science | |||
| Theme definition: Responses that describe a meaningful connection to science outside of the virtual classroom | |||
| C.1 | Community* | When a respondent writes about connecting to the scientific community, understanding how the scientific community works. | 21% |
| C.2 | Real-world application* | When a respondent relates the activity to the “real-world.” | 29% |
*Indicates a code that directly addresses Learning Goals or Objectives for this activity.
Qualitative responses regarding learning goals and objectives
Student responses to the survey questions (Appendices 6 and 7) included descriptions of their interactions with collaborative science, their perceptions of impact on learning, and their cognition and metacognition (Table 1). Many of these corresponded our learning objectives and learning goals. While these self-reported descriptions are indirect measures of learning, they show that students’ engagement with an online, collaborative activity allowed them to address a variety of competencies and content (17–19). In the future, pre- and posttesting can more definitively determine the extent of student learning.
Collaboration and real world scientific interaction were learning goals for this activity. Students described, without direct prompting, how they connected to the scientific community, gained an understanding of how a scientific community functions (Table 1, code C.1) or how “real world” science works (Table 1, code C.2).
Annotating helped me learn more about the scientific community and how things are actually annotated and that it’s not just done by a computer system or by the researchers themselves… It gives a deeper connection with the scientific community because you can be annotating a gene of interest from a paper, then when you see that author or gene of interest again you will be able to remember what the paper is talking about.
I enjoyed being able to contribute, at least in some small way, to the curation of Worm data. It is cool! And accessible which is nice.
The fact that students reported on these without being prompted suggests that working as part of a scientific community on a real life project was indeed a particularly important aspect of this experience.
Students also described whether and how their learning was impacted, in both positive and negative terms. For example, a student describing proficiency (Table 1, code A.3) could discuss building on prior knowledge, or that they were “already aware of how to pick out…conditions [that] qualify as a control.” Students also reported specifically on activities that directly addressed the learning objectives by making connections (Table 1, code A.1) or building skills and competency (Table 1, code A.4):
I felt like I already had a pretty solid understanding of the differences between genotype and phenotype, but I guess WormBase really solidified that idea. Lots of the data was hard for me to dissect and filter out from the paper itself.… WormBase did help me realize that there are such things as Not Observed Phenotypes, that was a new concept for me.
This student recalled the process of using the annotation submission portal and connected it with the differentiation of genotype and phenotype, suggesting that a quantitative analysis of student learning could be used during future implementations of this activity. Importantly, students reported gaining a deeper understanding (Table 1, code A.2) of genotype vs. phenotype (64% of respondents), control vs. experimental (29% of respondents), how to identify alleles (50% of respondents), and mutations vs. transgenic (71% of respondents), compared with their experiences in other courses.
Finally, students reported about awareness of and changes in their own thinking, including recognizing gaps in knowledge, using resources in a new way, explaining why they felt good or bad about the assignment, or overcoming confusion (Table 1, codes B.1, B.2, and B.3, respectively).
Yes, many times I was confused about whether a strain was a control/background, or a mutant inducing a specific phenotype. I enjoyed annotating in the sense that I had to pay particular attention to these details.
WormBase helped me realize what I did and did not know about the paper I had just read. When I realized I didn’t know what over-expressed gene caused the phenotype, I would have to go back to the paper and re-read it until I understood.
In both of these statements, students recognized their own gaps in knowledge and reported on their emotional responses to filling the gaps or using available resources to finish the assignment. Although addressing metacognition was not an explicit goal of this activity, these students were able to describe how it encouraged them to assess their own learning and redirect to find solutions.
Student reactions to the WormBase annotation exercise
We classified the responses of students for the first four questions (Appendix 6) using five categories (very negative, slightly negative, neutral, slightly positive, and very positive) that described their experiences with the WormBase annotation activity. The majority of students rated the activity positively when they compared it with previous biology classes (Fig. 3A). Students who did not have a positive experience often reported feeling either unsupported in their learning, which may have been due in part to the online format, or as though the activity was unnecessary. Nonetheless, negative responses sometimes included evidence of student learning. For example, one student wrote:
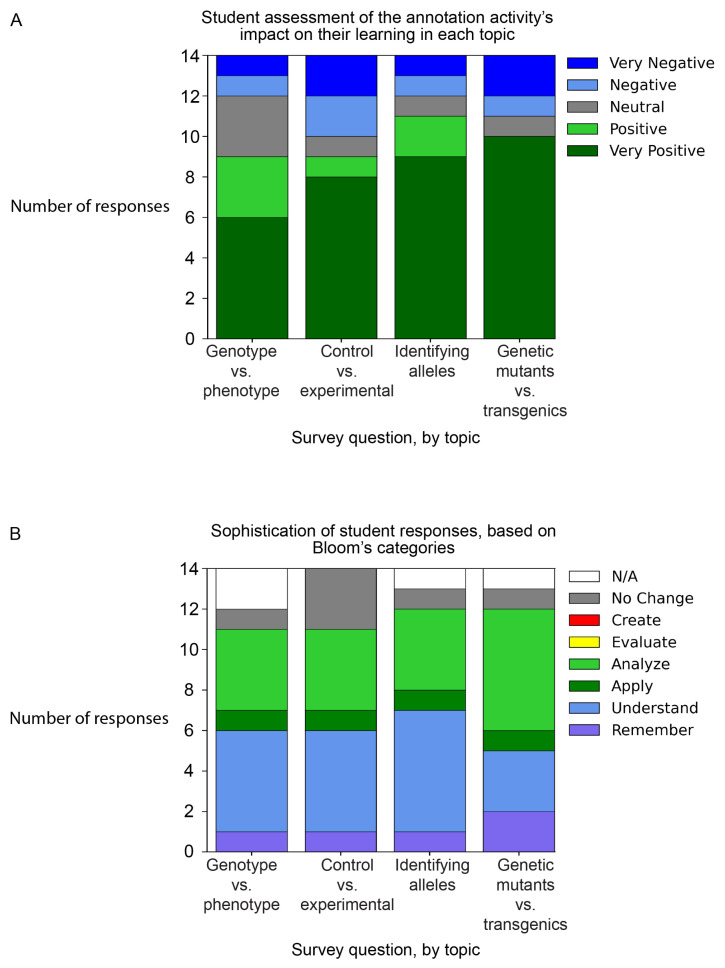
FIGURE 3.
Self-reported impacts of the WormBase annotation activity on student learning. Students were asked whether the activity helped them in four different categories, compared with other biology courses they had taken. Questions asked about: distinguishing between genotype and phenotype, distinguishing between control and experimental conditions, identifying different alleles, and distinguishing between transgenic organisms and genetic mutants. The questions, in full, are found in Appendix 6. A. student responses were categorized as Very Negative, Negative, Neutral, Positive, or Very Positive. B. Student responses to the questions described in (A) were scored using Bloom’s taxonomy categories. Responses that indicated no change in self-reported knowledge are shown as No Change (gray). Responses with single word affirmatives or negatives were categorized as n/a (white). No students described their learning in terms of the two highest Bloom’s categories, Evaluate and Create.
WormBase confused me more than it helped… it would make sense if the control condition were always N2 worms, it was difficult for me to realize that transgenic animals like nuIs24 could also be considered control conditions.
Even in describing the experience with the activity in a negative light, the student shows improved understanding of C. elegans nomenclature and genetic terminology. This lack of self-reflective metacognition during productive problem solving is not unique (20), and even students who understand the value of metacognition may not practice it (21, 22). It is therefore likely to be important for instructors to remind students of their gains during this challenging activity (23, 24).
Bloom’s categorization of student responses
We also coded student responses by revised Bloom’s categories (25, 26) (Fig. 3B). The majority of student responses describe mid-level Bloom’s thinking: understanding, applying, analyzing. The high-level Bloom’s categories (“create” and “evaluate”) were absent. For responses that used more than one category, the higher one was adopted for the purposes of quantification. Answers that did not address students’ thinking were categorized as n/a (no answer, or one-word answers, such as “no”) or no change, which indicates that a student only responded that they already knew the content. Other assignments in this course, including an analysis and write-up of primary data as a microPublication (15, 27), suggest that both of these higher-level Bloom’s categories were attained (not shown) in part through practice with the annotation activity. WormBase annotation may be a foundation on which students build up to higher-level activities, in the same way that more elementary in-person research tasks (for example, setting up reactions) may provoke deeper conversation or exploration in experimentation.
Not all students can participate in extracurricular research. CUREs provide all students access to the critical elements of research experiences that immerse them in inquiry and experimentation regardless of their background, socioeconomic status, or previous interest in research science (1, 2, 28, 29). One of the important elements of experimentation and the process of science is collaboration, in part because students receive peer input and practice communication skills (30). This activity connected students directly to the larger scientific community at precisely the same time as opportunities for in-person collaborations were lost. The learning goals focus on collaboration and were identified by students following the activity. Even under the potentially isolating circumstances of an online CURE, students appreciated that collaboration does not have to occur in person. Indeed, many scientific collaborations are intermural and international, so this activity presents a realistic and authentic opportunity for student participation in the scientific process.
Students reported that their repeated work on this activity helped them make novel connections and apply their knowledge in new and meaningful ways. Their responses show that they were thoughtfully engaged with mid- and lower-level Bloom’s activities, and this was apparent in their correct use of terminology in other aspects of the course, such as final reports and presentations (not shown). Activities that provide repeated opportunities for students to work on a real world scientific task can solidify their content knowledge and their science process skills (6, 24, 30). In this CURE, the annotations were completed alongside writing and primary (unpublished) data analysis, which may have made the annotations seem less “important” to students, even though they used the annotation skills in other aspects of the course.
Possible modifications
In the future, a more rigorous pre- and postanalysis will be useful in determining precisely what tasks and content are learned through the annotation activity. However, given the crisis conditions that intensified during spring 2020 (pandemic, all-online format for a laboratory course, and civil rights protests in or very near students’ hometowns in Seattle and Portland), nearly all quantitative assessments were removed from the course as a whole. In the future, questions such as the quiz example (Appendix 3) could be used in a pre- and postformat. In addition, other implementations of this activity could include a class wide focus on one particular cellular pathway or phenomenon (apoptosis, tissue specification, developmental progression, etc.) or a biotechnology technique (RNAi or CRISPR).
Although a tutorial was presented to the class before the annotation exercise, it became clear after the first round of submissions that, while the students had a good sense as to how to proceed, certain elements of the curation should be clarified so as to avoid confusion in the submission and vetting process. WormBase administrators found that many misunderstandings were due to the particular manner in which WormBase curates annotations and did not reflect a misunderstanding of the biology reported in the article. Some other common problems included differentiation between “true” wild-type (N2) controls and transgenic (protein expression) controls for protein abundance, annotations submitted for one publication but originating in a different publication, alleles submitted as transgenes, incorrect controls, reporter gene expression induced by environmental conditions submitted as a mutant phenotype, phenotypes attributed to background mutations, and trouble differentiating between a phenotype-causing transgene and a reporter transgene. Time permitting, a second tutorial later in the academic session could provide students with feedback from their collaborators on what works well and what does not. It could also enhance the metacognitive aspects of the process for students, who could directly address what they do and do not understand with their collaborators. This would be particularly helpful for students who may not appreciate the gains that they have made in their learning, as we described (Student reactions to the WormBase annotation exercise).
This was the first attempt by WormBase to have undergraduate students submit phenotype annotations. Our experience suggests that while undergraduates have limited experience performing and reporting experiments, they can quickly be trained to use proper annotation via an initial tutorial. Misconceptions that arise can be addressed in future tutorials. Once an undergraduate student has undergone multiple rounds of annotation and assessment, they could be identified as a trusted curator for future submissions which would not require detailed verification by WormBase staff.
A major concern of graduating seniors during this period is their ability to be hired in the absence of hands-on laboratory experience. Demonstrating a clear understanding of molecular biology and genetics literature via WormBase annotation submissions should be a noteworthy accomplishment that could be recognized on one’s resume/CV or through digital merit badges, like those generated by the Badgr resource (https://info.badgr.com). If such digital badges could be appropriately recognized as indicators of significant skill sets, they could be shared via social media and job search sites, reported on CV’s and potentially used to advance early-stage careers.
Another potential avenue to explore is to specifically engage early-stage graduate students who are beginning the literature research process so they can provide annotations to WormBase as they discover what is not already available in the database. This could also allow for a more engaging annotation submission experience by particularly focusing on publications and annotations of interest for that particular graduate research project. Graduate student submissions could be summarized and provided alongside one’s dissertation as a record of service to the C. elegans research community.
Collaboration between students and the scientific community was a major goal of this activity and the C. elegans research community greatly benefits from the curated annotations made to WormBase curators and contributors. Validated annotations are made visible on the WormBase website, on the FTP site of data files, and via several data mining tools. They also save researchers valuable time piecing together the collective information from the literature themselves. Annotations will also be made available at the Alliance of Genome Resources (www.alliancegenome.org) (31). There are other sites dedicated to facilitating undergraduate and community contributions. For example, SUPRdb (run by the Tetrahymena thermophila community) has a contribution portal and webform, and entries to SUPRdb are linked to the Tetrahymena Genome Consortium, though they are not curated after submission (8, SUPRdb | Welcome [http://suprdb.org/]). Broader curation projects, such as BioGRID and CACAO also solicit input from the community in a variety of formats (32; Contribute to the BioGRID | BioGRID [https://wiki.thebiogrid.org/doku.php/contribute], Category:CACAO Spring 2019 – GONUTS [https://gowiki.tamu.edu/wiki/index.php/Category:CACAO_Spring_2019]). Databases for other model organisms (for example, TAIR for Arabidopsis thaliana, FlyBase for Drosophila melanogaster, and SGD for Saccharomyces cerevisiae) also provide annotated genomic and genetic information and could be the basis for similar annotation activities. However, to our knowledge, the same kind of webform and menu-based input is not available through these sites so the direct community collaboration aspect of our activity might be harder to achieve.
Even in the best of times, results of student efforts are often isolated from the scientific community. Our students’ submissions of annotations to WormBase represent real and productive collaborations that can have an immediate and lasting impact on C. elegans research, model organism research, and research on human health and disease while furthering students’ education.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
We thank Ben Wiggins and Kelly Hennessey for their critical reading of this manuscript. This teaching and research took place in the homelands of the Lummi Nation, the Nooksack Tribe, the Kizh Nation, and their ancestors. This material is based upon work supported by the National Science Foundation under grant no. 1612252. The authors have no conflicts of interest to declare.
Footnotes
Supplemental materials available at http://asmscience.org/jmbe
REFERENCES
- 1.Hurtado S, Cabrera NL, Lin MH, Arellano L, Espinosa LL. Diversifying science: underrepresented student experiences in structured research programs. Res High Educ. 2008;50:189–214. doi: 10.1007/s11162-008-9114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopatto D. Undergraduate research experiences support science career decisions and active learning. CBE Life Sci Educ. 2007;6:297–306. doi: 10.1187/cbe.07-06-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dolan E. Course-based undergraduate research experiences: current knowledge and future directions. National Research Council; Washington, DC: 2016. [Google Scholar]
- 4.Bangera G, Brownell SE. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ. 2014;13:602–606. doi: 10.1187/cbe.14-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ. 2014;13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corwin LA, Runyon CR, Ghanem E, Sandy M, Clark G, Palmer GC, Reichler S, Rodenbusch SE, Dolan EL. Effects of discovery, iteration, and collaboration in laboratory courses on undergraduates’ research career intentions fully mediated by student ownership. CBE Life Sci Educ. 2018;17:ar20. doi: 10.1187/cbe.17-07-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahlberg L, Groat Carmona AM. CRISPR-Cas technology in and out of the classroom. CRISPR J. 2018;1:107–114. doi: 10.1089/crispr.2018.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley EA, Stover NA. Immediate dissemination of student discoveries to a model organism database enhances classroom-based research experiences. CBE Life Sci Educ. 2014;13:131–138. doi: 10.1187/cbe.13-07-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grove C, Cain S, Chen WJ, Davis P, Harris T, Howe KL, Kishore R, Lee R, Paulini M, Raciti D, Tuli MA, Van Auken K, Williams G WormBase Consortium. Using WormBase: a genome biology resource for Caenorhabditis elegans and related nematodes. Methods Mol Biol. 2018;1757:399–470. doi: 10.1007/978-1-4939-7737-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, Lee RYN, Muller H-M, Nakamura C, Nuin P, Paulini M, Raciti D, Rodgers FH, Russell M, Schindelman G, Auken KV, Wang Q, Williams G, Wright AJ, Yook K, Howe KL, Schedl T, Stein L, Sternberg PW. WormBase: a modern model organism information resource. Nucleic Acids Res. 2020;48:D762–D767. doi: 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meneely PM, Dahlberg CL, Rose JK. Working with worms: Caenorhabditis elegans as a model organism. Cur Prot Essent Lab Tech. 2019;19:e35. [Google Scholar]
- 12.Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. 2015;200:387–407. doi: 10.1534/genetics.115.176099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlberg CL, Juo P. The WD40-repeat proteins WDR-20 and WDR-48 bind and activate the deubiquitinating enzyme USP-46 to promote the abundance of the glutamate receptor GLR-1 in the ventral nerve cord of Caenorhabditis elegans. J Biol Chem. 2014;289:3444–3456. doi: 10.1074/jbc.M113.507541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldherr SM, Strovas TJ, Vadset TA, Liachko NF, Kraemer BC. Constitutive XBP-1s-mediated activation of the endoplasmic reticulum unfolded protein response protects against pathological tau. 1. Nat Comm. 2019;10:4443. doi: 10.1038/s41467-019-12070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee RYN, Howe KL, Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Davis P, Gao S, Grove C, Kishore R, Muller H-M, Nakamura C, Nuin P, Paulini M, Raciti D, Rodgers F, Russell M, Schindelman G, Tuli MA, Van Auken K, Wang Q, Williams G, Wright A, Yook K, Berriman M, Kersey P, Schedl T, Stein L, Sternberg PW. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018;46:D869–D874. doi: 10.1093/nar/gkx998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percy WH, Kostere K, Kostere S, Percy WH, Kostere K, Kostere R. Generic qualitative research in psychology. Qual Rep. 2015;20(2):76–85. [Google Scholar]
- 17.American Association for the Advancement of Science. Vision and change in undergraduate biology education: a call to action: a summary of recommendations made at a national conference organized by the American Association for the Advancement of Science; July 15–17, 2009; Washington, DC. 2011. [Google Scholar]
- 18.Clemmons A, Timbrook J, Herron J, Crowe A. BioSkills Guide. 2019. [DOI] [PMC free article] [PubMed]
- 19.Brownell SE, Freeman S, Wenderoth MP, Crowe AJ. BioCore guide: a tool for interpreting the core concepts of vision and change for biology majors. CBE Life Sci Educ. 2014;13:200–211. doi: 10.1187/cbe.13-12-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlberg C, Lee S, Leaf D, Lily L, Wiggins B, Jordt H, Johnson T. A short, course-based research module provides metacognitive benefits in the form of more sophisticated problem solving. J Coll Sci Teach. 2019;48(4):22–30. doi: 10.2505/4/jcst19_048_04_22. [DOI] [Google Scholar]
- 21.Dye KM, Stanton JD, Tomanek D. Metacognition in upper-division biology students: awareness does not always lead to control. LSE 16:ar31. 2017 doi: 10.1187/cbe.16-09-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanton JD, Neider XN, Gallegos IJ, Clark NC, Tomanek D. Differences in metacognitive regulation in introductory biology students: when prompts are not enough. CBE Life Sci Educ. 2015;14:ar15. doi: 10.1187/cbe.14-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry MA, Shorter S, Charkoudian L, Heemstra JM, Corwin LA. FAIL is not a four-letter word: a theoretical framework for exploring undergraduate students’ approaches to academic challenge and responses to failure in STEM learning environments. CBE Life Sci Educ. 2019;18:ar11. doi: 10.1187/cbe.18-06-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanner KD. Promoting student metacognition. CBE Life Sci Educ. 2012;11:113–120. doi: 10.1187/cbe.12-03-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen L, Krathwohl D, editors. A taxonomy for learning, teaching, and assessing: a revision of Bloom’s taxonomy of educational objectives Complete ed. Longman; New York: 2001. [Google Scholar]
- 26.Armstrong P. Bloom’s taxonomy. Vanderbilt University; 2010. [Google Scholar]
- 27.Raciti D, Yook K, Harris TW, Schedl T, Sternberg PW. Micropublication: incentivizing community curation and placing unpublished data into the public domain. Database (Oxford) 20182018 doi: 10.1093/database/bay013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Academies of Sciences, Engineering, and Medicine. Undergraduate research experiences for STEM students: successes, challenges, and opportunities. 2017. [Google Scholar]
- 29.Estrada M, Burnett M, Campbell AG, Campbell PB, Denetclaw WF, Gutiérrez CG, Hurtado S, John GH, Matsui J, McGee R, Okpodu CM, Robinson TJ, Summers MF, Werner-Washburne M, Zavala M. Improving underrepresented minority student persistence in STEM. CBE Life Sci Educ. 2016;15:es5. doi: 10.1187/cbe.16-01-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brownell SE, Kloser MJ. Toward a conceptual framework for measuring the effectiveness of course-based undergraduate research experiences in undergraduate biology. Stud Higher Educ. 2015;40:525–544. doi: 10.1080/03075079.2015.1004234. [DOI] [Google Scholar]
- 31.Alliance of Genome Resources Consortium. Alliance of genome resources portal: unified model organism research platform. Nucleic Acids Res. 2020;48:D650–D658. doi: 10.1093/nar/gkz813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oughtred R, Rust J, Chang C, Breitkreutz B, Stark C, Willems A, Boucher L, Leung G, Kolas N, Zhang F, Dolma S, Coulombe-Huntington J, Chatr-aryamontri A, Dolinski K, Tyers M. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic and chemical interactions. Protein Science pro. 2020:3978. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.