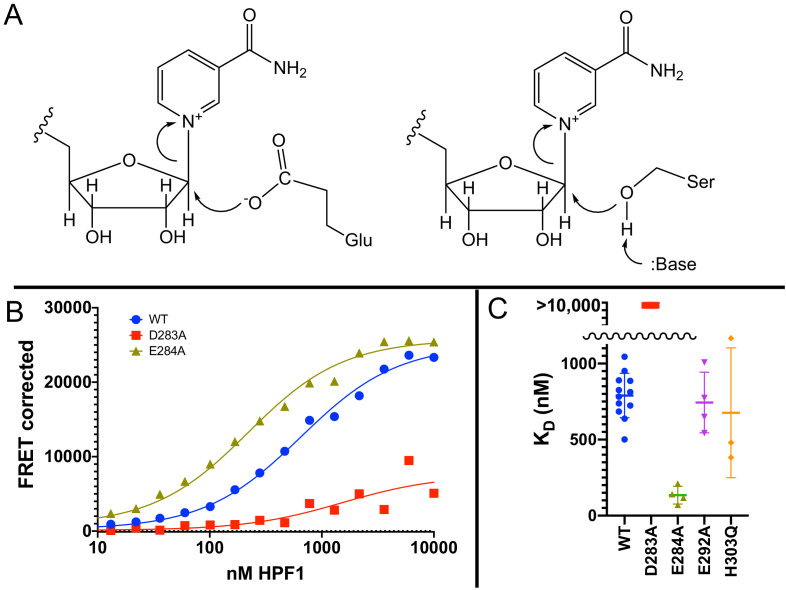
Figure 4. Glu284 of Histone PARylation Factor 1 (HPF1) is the catalytic base required for transPARylation.
(A) Chemical mechanism of PARylation of glutamate (Glu, on left) does not require a catalytic base, whereas PARylation of serine (Ser, on right) requires deprotonation of serine by a catalytic base. (B) Representative curves demonstrating the binding of HPF1 (WT, D283A, and E284A) to the poly(ADP-ribose) polymerase 1 (PARP1)–Nuc165 complex using FRET between labeled HPF1 and labeled PARP1. (C) Bar graph for binding of HPF1 to PARP1–Nuc165 complex as determined by FRET assay demonstrating that the E284A mutant of HPF1 binds more tightly than WT, and that the D283 mutant does not bind with measurable affinity. The E292A and H303Q mutants of HPF1 are shown to bind with similar affinity as WT HPF1. Data for these findings with standard deviations and number of replicates can be found in Table 2.