ABSTRACT
The human pulmonary environment is complex, containing a matrix of cells, including fibroblasts, epithelial cells, interstitial macrophages, alveolar macrophages and neutrophils. When confronted with foreign material or invading pathogens, these cells mount a robust response. Nevertheless, many bacterial pathogens with an intracellular lifecycle stage exploit this environment for replication and survival. These include, but are not limited to, Coxiella burnetii, Legionella pneumophila, Yersinia pestis, Mycobacterium tuberculosis and Staphylococcus aureus. Currently, few human disease-relevant model systems exist for studying host–pathogen interactions during these bacterial infections in the lung. Here, we present two novel infection platforms, human alveolar macrophages (hAMs) and human precision-cut lung slices (hPCLS), along with an up-to-date synopsis of research using said models. Additionally, alternative uses for these systems in the absence of pathogen involvement are presented, such as tissue banking and further characterization of the human lung environment. Overall, hAMs and hPCLS allow novel human disease-relevant investigations that other models, such as cell lines and animal models, cannot completely provide.
Keywords: hAMs, hPCLS, pulmonary, lung, lung infection, intracellular bacteria
Utility of two human lung infection platforms, human alveolar macrophages and human precision-cut lung slices, to study multiple bacterial pathogens.
THE HUMAN ALVEOLAR ENVIRONMENT
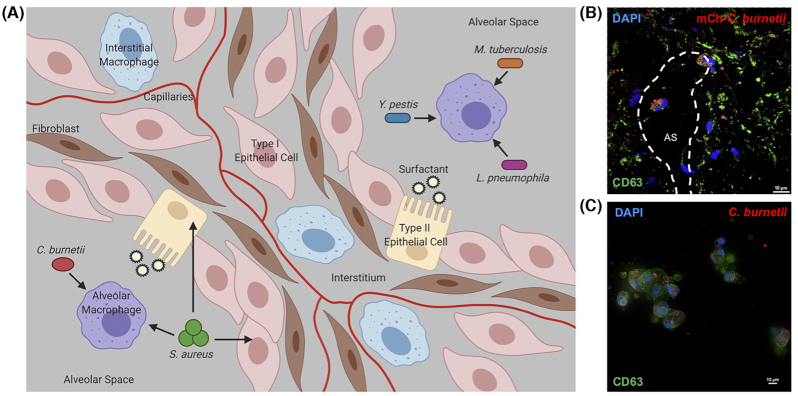
The human lung is constantly exposed to airborne material that must be efficiently recognized, processed and removed to promote a healthy respiratory system. A multitude of cell types are present in the complex human alveolar environment (Fig. 1A), each serving distinct housekeeping and defense functions. These populations include resident type I and type II epithelial cells, fibroblasts, interstitial and alveolar macrophages, and neutrophils in addition to lymphocytes recruited during immune reactions (Franks et al. 2008). Epithelial cells are one of the most abundant cell types within this environment. Type I epithelial cells are a necessary structural component that form the alveolar wall and aid gas exchange across the air–blood barrier for proper respiration (Rackley and Stripp 2012). Type II epithelial cells produce surfactant proteins that reduce respiration-related surface tension and interact with foreign pathogens and allergens. These interactions can trigger immune cell activation or increase microbial uptake by phagocytic cells, ultimately degrading the pathogen (Lawson and Reid 2000; Rackley and Stripp 2012). Fibroblasts are prominent in the interstitium, forming the extracellular matrix for pulmonary structural support. Fibroblasts also aid wound repair by remodeling the extracellular matrix and healing the alveolar epithelium (Wuyts et al. 2013; Ito et al. 2014; White 2015). In addition to structural cells, macrophages are present as the first line of defense against foreign materials in the alveolus. Interstitial macrophages are present exclusively in the interstitial region, whereas alveolar macrophages are present within alveolar spaces, often lightly adhered to the epithelial lining of the alveolus (Lambrecht 2006). Macrophages are responsible for phagocytosis of foreign material, pathogen clearance, and inflammatory cytokine and chemokine production that stimulates a robust immune response (Rubins 2003; Schyns, Bureau and Marichal 2018). Neutrophils respond to inflammatory cytokine production, aid pathogen killing and represent an essential link between innate and adaptive immunity in the lung (Liu et al. 2017). Lastly, dendritic cells recognize foreign material and pathogens, and activate the adaptive immune response (Condon et al. 2011).
Figure 1.
Preferential niche of bacteria in the lung. (A) Schematic of the human alveolar environment, cell types present and invading bacterial pathogens. Arrows = preferential niche(s) for each pathogen. Created with BioRender.com. (B) hPCLS were infected with NMII Coxiella burnetii expressing mCherry (red) for 96 h, then processed for immunofluorescence microscopy using CD63 antibody (lysosomes and PVs; green) and DAPI to stain DNA (blue). Substantially more hAMs contained C. burnetii compared with epithelial interstitial cells. AS = alveolar space. (C) Isolated hAMs were infected with NMI C. burnetii for 96 h, then processed for immunofluorescence microscopy using antibodies against CD63 (green), C. burnetii (red) and DAPI to stain DNA (blue). hAMs support PV expansion and robust C. burnetii growth.
The complex alveolar environment is well equipped to proficiently respond to foreign inhaled material, including highly infectious microbes. Even so, many respiratory pathogens exploit cells in this environment for intracellular survival and replication required to cause debilitating disease. Due to the physiological relevance deficiency in using animal models of infection to mimic the human lung environment, it is essential to use the most relevant human-derived model systems to dissect mechanisms of microbial pathogenesis. In this review, we describe two recently developed infection platforms, human precision-cut lung slices (hPCLS) and primary human alveolar macrophages (hAMs), for the study of distinct bacterial pathogens and discuss the contribution of these systems to microbial pathogenesis.
PRIMARY HUMAN LUNG PLATFORMS
Many animal models of infection have been used to investigate the pulmonary response to microbial pathogens. Although these models provide valuable mechanistic information about specific diseases, translating these findings to humans is not always possible. Establishment of an ex vivo human lung system has provided researchers with a platform to assess mechanisms of pathogenesis in distinct alveolar and interstitial cell types (Graham et al. 2016). The system developed in our laboratory uses lungs obtained post-mortem from healthy donors with no presence of asthma, pneumonia or other infection (i.e. SARS-CoV-2). Lungs are injected with low-melting-temperature agarose and incubated at 4°C until solidified, after which they are sectioned into 2.5-cm-thick slices. Using a microtome, sections are cored and cut into hPCLS (diameter = 8 mm; thickness = 750 μm). hPCLS are cultured in standard cell culture medium containing antibiotics and antimycotics [Dulbecco's modified Eagle/F-12 (DMEM/F-12) medium with 10% fetal bovine serum, penicillin (50 U/mL), streptomycin sulfate (50 μg/mL), gentamicin sulfate (60 μg/mL) and amphotericin B (0.25 μg/mL)]. Prior to infection with a microbe, antibiotic/antimycotic-containing media is replaced with non-antibiotic/antimycotic media for 2–3 days. hPCLS can then be processed for multiple readouts, including microbial growth, microscopic analysis of replication vacuole formation and cytokine production.
Although this ex vivo platform can be used for many novel applications, important caveats must be considered. Neutrophils are present in low numbers in hPCLS, and influx as a result of pathogenic infection does not occur because the tissue has been separated from the circulatory system. The absence of influx hinders studies of the entire innate immune response, but allows detailed investigation of the role of macrophages in alveolar infection. Studies are currently underway to re-populate hPCLS with exogenous neutrophils and simulate influx. A second caveat to the hPCLS system is the lack of recruited adaptive immune cells, preventing investigation of the adaptive immune response to infection. While recognizing these limitations, hPCLS provide a platform to dissect the interaction between microbes and innate immune cells in the absence of confounding host inflammation-induced effects.
In addition to using hPCLS to study the entire alveolar region, cell signaling and intracellular infection can be assessed using isolated hAMs. These cells are easily harvested from donor lungs via bronchioalveolar lavage. Prior to agarose inflation of lungs, extracted bronchioalveolar lavage fluid (BALF) is incubated with a lysis buffer to eliminate red blood cells from the mixture. BALF samples are then plated and adherent hAMs cultured as above for hPCLS (Graham et al. 2016). Following isolation, hAMs do not proliferate, indicating terminal differentiation. The quantity and viability of harvestable hAMs per lung are variable, requiring experiments to be performed using lungs from at least three donors, which also provides statistically significant and biologically relevant results. Following isolation, hAMs can be infected with a microbe and processed for numerous cell biology assays, including protein analysis, cytokine/chemokine production and fluorescence microscopy (Fig. 1B and C). It should be noted that primary hAMs respond to bacterial pathogens in a more robust manner than human macrophage cell lines, highlighting the importance of assessing infection of primary cells to detect critical events that are below the limit of detection in cell lines (Graham et al. 2016). hAMs and hPCLS have now been used to study innate interactions with diverse microbial pathogens that exploit an intracellular niche as highlighted in the following sections (Table 1).
Table 1.
Published uses of primary human lung infection systems.
| Pathogen | Model | Techniques | Reference |
|---|---|---|---|
| Coxiella burnetii | hAMs | Bacterial growth analysis, fluorescence microscopy, electron microscopy, immunoblot, multiplex cytokine assay and cell viability | Graham et al. (2013) |
| hAMs, hPCLS | Histology, confocal microscopy, immunoblot, ELISA, cell lysis and RT-PCR array | Graham et al. (2016) | |
| hAMs, hPCLS | Confocal microscopy, bacterial growth analysis and ELISA | Dragan, Kurten and Voth (2019) | |
| hAMs | Electron microscopy, confocal microscopy and immunoblot | Winchell et al. (2014) | |
| hAMs | Confocal microscopy and immunoblot | Winchell et al. (2018) | |
| hAMs | Immunoblot analysis and confocal microscopy | Colonne et al. (2016) | |
| hAMs | Fluorescence microscopy and immunoblot | MacDonald, Kurten and Voth (2012) | |
| hAMs | Fluorescence microscopy and immunoblot | MacDonald et al. (2014) | |
| Legionella pneumophila | HLTEs | Histology, immunohistochemistry, bacterial growth analysis, transcriptomics, qRT-PCR and ELISA | Jager et al. (2014) |
| HLTEs | Bacterial growth analysis | Hoppe et al. (2017) | |
| Yersinia pestis | hAMs, hPCLS | Adherence assay, epifluorescence microscopy, flow cytometry analysis, cell viability, bacterial growth analysis, confocal microscopy and cytometric bead array | Banerjee et al. (2019) |
| hAMs | Bacterial growth analysis | Crane et al. (2021) | |
| Mycobacterium tuberculosis | hAMs | Cell viability, ELISA, DNA fragmentation, electron microscopy, epifluorescence microscopy and apoptosis analysis | Keane et al. (1997) |
| hAMs | Flow cytometry, macrophage infection, cytokine bead array and RNA sequencing | Goenka et al. (2020) | |
| hAMs | Microarray, functional enrichment analysis and ingenuity pathway analysis (IPA) | Lavalett et al. (2020) | |
| hAMs | qPCR | Hackett et al. (2020) | |
| hAMs | Histology, confocal microscopy and PCR | Ufimtseva et al. (2018) | |
| Staphylococcus aureus | hAMs, hPCLS | Fluorescence microscopy, confocal microscopy, proliferation assay, ELISA, immunoblot, cyttotoxicity assay and cell viability | Brann et al. (2019) |
| Alternative | hAMs | Phase contrast microscopy, phagocytosis analysis and bacterial killing | Cohen and Cline (1971) |
| hAMs | ELISA, PGE2 assay and nitric oxide (NO) assay | Menard, Turmel and Bissonnette (2007) | |
| hPCLS | Phagocytosis analysis, flow cytometry, fluorescence microscopy, proliferation analysis and two-photon fluorescence microscopy | Bai et al. (2016) | |
| hPCLS | WST-1 assay, confocal miscroscopy, live/dead staining, bronchioconstriction analysis, ELISA and histopathology | Neuhaus et al. (2017) |
COXIELLA BURNETII
Q fever is a debilitating disease caused by the obligate intracellular bacterial pathogen C. burnetii. Acute Q fever is characterized by flu-like symptoms, whereas chronic Q fever can result in severe and potentially fatal endocarditis (Maurin and Raoult 1999; Mazokopakis, Karefilakis and Starakis 2010; Oyston and Davies 2011). Coxiella burnetii has an aerosol mode of transmission, and fewer than 10 inhaled bacteria can establish acute infection, underscoring the importance of this pathogen as a U.S. Centers for Disease Control and Prevention select agent (Oyston and Davies 2011). After human inhalation, C. burnetii is engulfed by an alveolar macrophage into a membrane-bound compartment, transitioning through the phagolysosomal maturation pathway and fusing with endosomes and acidic lysosomes that decrease the pH (pH ∼ 5.0) (Voth and Heinzen 2007; Samanta et al. 2019). During this process, C. burnetii is metabolically activated, employs a type IV secretion system (T4SS) and forms a host membrane-derived replication niche deemed the parasitophorous vacuole (PV) (Voth and Heinzen 2007; Crabill et al. 2018). Immortalized cell lines and murine models have been extensively used to study C. burnetii infection, although mice do not fully recapitulate human disease (Dragan and Voth 2020). Due to this deficiency in small animal model systems, hPCLS and hAMs provide reliable platforms for dissecting pathogenic events directly applicable to human disease.
Graham et al. (2013, 2016) established the hAM and hPCLS models of C. burnetii infection using multiple bacterial isolates. To characterize the hAM platform, cells were infected with one of three select agent isolates, Nine Mile I (NMI), a virulent strain that causes acute disease, G, a virulent strain that causes endocarditis, Dugway, a severely attenuated strain, or one avirulent isolate, Nine Mile II (NMII). All isolates form multiple PV that contain lipid droplets in hAMs and display robust growth similar to the well-established THP-1 human macrophage-like cell line. In response to C. burnetii, hAMs produce anti-apoptotic signals, including phosphorylated (activated) Akt and Erk1/2, and produce the pro-inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), and anti-inflammatory interleukin-10 (IL-10) (Graham et al. 2013). Production of interleukin-1β (IL-1β) occurs throughout NMII infection of hAMs, but not during virulent isolate infection, suggesting isolate-specific immune signaling (Graham et al. 2013). Importantly, this study was the first to show cellular infection differences between isolates, while some infection events correspond with previously established cell models, highlighting the need for further C. burnetii studies using hAMs.
In addition to robust NMII growth within hAMs (Graham et al. 2016), IL-1β is produced by both hAMs and hPCLS, suggesting a potential role for activation of the inflammasome in response to avirulent C. burnetii. This activation was confirmed by assessing caspase-1 and caspase activation and recruitment domain (ASC) co-localization with IL-1β adjacent to C. burnetii in hAMs. Moreover, upregulation of genes such as nlrp3 and nod2 further indicates that infection of hAMs specifically elicits NLRP3 inflammasome activation (Graham et al. 2016). Collectively, these results enhance understanding of the innate response to the C. burnetii.
Further hPCLS investigation by Dragan et al. showed C. burnetii replication in hAMs, while no growth occurs within interstitial macrophages, fibroblasts or epithelial cells, further showcasing the cellular specificity of infection (Dragan, Kurten and Voth 2019). However, immortalized pulmonary cell lines, including A549 alveolar epithelial cells and human lung fibroblasts, support C. burnetii replication in vitro, although growth is significantly decreased in epithelial cell lines and more substantially in macrophage-like and fibroblast cell lines. These contrasting phenotypes further highlight the utility of the hPCLS system to define infection events and cell-specific responses in a natural setting. Furthermore, addition of surfactant protein-D, typically secreted by alveolar epithelial cells to eliminate pathogens, to hAMs does not prevent PV expansion or C. burnetii growth in contrast to mouse macrophage results (Soltysiak, van Schaik and Samuel 2015). In response to C. burnetii, hAMs produce substantial levels of interleukin-8 (IL-8), a neutrophil-attracting chemokine, further demonstrating a pro-inflammatory response and correlating with mouse studies that indicate a critical role for neutrophils in Q fever (Elliott, Peng and Zhang 2013). In addition to the immune responses above, a phenotypic switch in polarization from M1 (pro-inflammatory) to M2 (anti-inflammatory) occurs in hAMs during C. burnetii infection, suggesting a more permissive environment for replication is actively promoted by the pathogen (Dragan, Kurten and Voth 2019). Significant replication of C. burnetii in M2-polarized alveolar macrophages corroborates results previously noted in murine alveolar macrophage studies (Fernandes et al. 2016). Collectively, this study uncovered novel aspects of the C. burnetii preferential growth niche in humans using hAMs and hPCLS.
The hAM system is also useful for uncovering novel cellular signaling events, as primary cells often respond to pathogen stimuli more robustly than immortalized cells. Similar to other intracellular bacteria, C. burnetii manipulates numerous host signaling cascades to promote efficient growth and progression of disease. One pathway of importance in this pathogen setting is autophagy, the removal of damaged organelles and foreign material by relocation to lysosomes for degradation. The autophagosomal protein microtubule-associated light chain 3 (LC3) associates with roughly 70% of hAM PV during infection with NMII C. burnetii in a T4SS-dependent manner, indicating the organism actively recruits autophagosomes (Winchell et al. 2014). Levels of the cargo adaptor p62/Sequestosome-1, a protein that interacts with LC3 and targets material for degradation during selective autophagy, remain constant throughout infection of hAMs with NMII, suggesting C. burnetii does not allow autophagic flux needed to revert the host cell to homeostasis (Winchell et al. 2014). p62 localizes to NMI and NMII PV, although silencing p62 expression does not significantly impact C. burnetii growth, suggesting a signaling role for the cargo adaptor. p62 contains multiple phosphorylation sites that direct protein activity and downstream signaling, with modification of S349 activating the Nuclear erythroid 2-related factor-2 (Nrf2)-Kelch-like ECH-associated protein 1 (Keap1) pathway, a major antioxidant mechanism in human cells. Nrf2 is also involved in activation of selective autophagy. Whereas total p62 levels remain constant throughout infection with NMII C. burnetii, S349-phosphorylated p62 levels increase and Nrf2 levels are stabilized, indicating activation the Nrf2-Keap1 pathway during infection (Winchell et al. 2018), which has not yet been studied using C. burnetii-infected murine models. These findings suggest important interplay between autophagy-related proteins and host signaling cascades during C. burnetii growth in hAMs.
In addition to Nrf2 signaling, C. burnetii activates the host cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) signaling pathway to form the PV and ensure host cell survival (MacDonald, Kurten and Voth 2012; MacDonald et al. 2014). The use of PKA inhibitor H-89 in hAMs infected with NMI or G C. burnetii results in defective PV formation compared with non-treated cells (MacDonald, Kurten and Voth 2012). Phosphorylated PKA levels increase from 72–96 hpi in NMI- and G-infected hAMs, suggesting PKA activity is required for optimal C. burnetii infection (MacDonald, Kurten and Voth 2012). Furthermore, a significant increase in apoptotic cell death occurs when C. burnetii-infected hAMs are treated with H-89 (MacDonald et al. 2014). Levels of phosphorylated Bad, a pro-apoptotic protein, also increase during C. burnetii infection, suggesting the pathogen uses PKA to inactivate Bad and prevent apoptosis. The use of hAMs confirmed the importance of PKA for optimal C. burnetii infection and prevention of apoptosis.
In addition to Bad, PKA activates vasodilator-stimulated phosphoprotein (VASP), which is a regulator of actin-based cytoskeleton assembly. Using hAMs, Colonne et al. showed VASP activity is essential for NMI PV formation, although C. burnetii does not use actin-based motility similar to related intracellular bacteria, but remains in the PV for replication (Colonne et al. 2016). VASP localization to the PV membrane suggests C. burnetii uses VASP to coordinate actin organization around the PV during vacuole expansion. S157- and S239-phosphorylated VASP levels also increase throughout infection, further indicating activation of the protein (Colonne et al. 2016). Together, these results suggest VASP activation is necessary for optimal C. burnetii infection, and indicate an undefined role for additional actin-related proteins during infection.
Collectively, the hPCLS and hAM systems have allowed novel investigation of the C. burnetii-host dynamic not possible using immortalized cell lines and small animal models. Understanding of the targeted cellular niche, innate pro-inflammatory response, inflammasome activation and multiple cellular signaling pathways has been enhanced using these platforms to replicate C. burnetii interactions with the human lung. These systems have been extended to additional bacteria that exploit the intracellular host cell environment as described below.
LEGIONELLA PNEUMOPHILA
Legionella pneumophila is a Gram-negative bacterium that causes Legionnaires’ disease, a type of pneumonia that presents with fever, headache, muscle pain, dyspnea and chest pain. It is difficult to distinguish this pneumonia from others, often resulting in misdiagnosis (Fields, Benson and Besser 2002; Muder and Yu 2002). Legionella pneumophila is typically found in natural fresh-water environments or can colonize man-made freshwater environments, such as cooling towers, and can replicate at a vast temperature range of 25–42°C. Here, L. pneumophila replicates within amoebae and is transmitted through infected aerosols (Castillo, Rajasekaran and Ali 2016). Once inhaled by a human, L. pneumophila is engulfed by alveolar macrophages and evades endocytic maturation that creates the degradative lysosomal environment. Instead, L. pneumophila fuses with endoplasmic reticulum-derived vesicles and deploys a T4SS to create a Legionella-containing vacuole in which to replicate (Newton et al. 2010; Castillo, Rajasekaran and Ali 2016). After replicating to high numbers, L. pneumophila causes pore formation and cell lysis to exit the host cell and infect bystander cells (Fields, Benson and Besser 2002; Newton et al. 2010). Infection systems used to model disease include cell lines, mice and guinea pigs. Primary hAMs and human lung tissue are underutilized L. pneumophila infection models, but remain the most disease-relevant systems to define pathogenic events.
Jager et al. used human lung tissue explants (HLTEs) from surgery patients to study L. pneumophila infection in the pulmonary environment (Jager et al. 2014). The HLTE model is different from the hPCLS model, as tissue samples are obtained from live patients compared with post-mortem hPCLS samples. Infection of HLTEs confirmed that alveolar macrophages are the primary target cell of L. pneumophila (Copenhaver et al. 2014). Dead macrophages and damaged tissue, with L. pneumophila close to damaged areas, are present after 48 h in HLTEs. To identify the cause of tissue damage, three conditions were used: wild type-infected, DotA−-infected (T4SS-deficient) and L. pneumophila-shed outer membrane vesicle (OMV)-stimulated samples (Jager et al. 2014). Damage increases significantly in wild type-infected and OMV-stimulated samples after 48 h, with wild-type damage more substantial than DotA−-infected samples, suggesting a functional T4SS is required for infection and resulting tissue damage. The use of multiple bacterial loads of wild type or T4SS-deficient L. pneumophila showed that bacterial inoculum size does not correlate to increased tissue damage. Bacterial colony counts confirmed HLTEs are permissible for intracellular replication of wild type but not DotA− bacteria, similar to previous studies using murine macrophage cell lines (Yan and Cirillo 2004). Transcriptional profiles of uninfected and infected HLTEs were compared and displayed a significant difference in the host response in these tissues. Expression of uteroglobulin, an airway secretory protein involved in immune cell recruitment, and the macrophage receptor with collagenous structure (MARCO), involved in bacterial uptake, decreases in wild type-infected HLTEs, though the importance of these events is unknown (Jager et al. 2014). Overall, this study opens the door to further use and characterization of disease-relevant human lung infection systems to model L. pneumophila infection.
Legionella pneumophila mutants with an insertion in Lpc2666 have decreased replication and increased association with lysosomal compartments (Shevchuk et al. 2014). Lpc2666 encodes the type IV pilus fimbrial biogenesis factor PilY1, which is necessary for proper T4SS assembly in Pseudomonas aeruginosa. Hoppe et al. expanded on this discovery by characterizing PilY1 in HLTEs (Hoppe et al. 2017). HLTEs were infected with wild type, pilY1 knockout, pilY1 complemented or T4SS-deficient L. pneumophila. A significantly decreased bacterial load is present in pilY1 knockout-infected and T4SS-deficient-infected samples over a 50 h time course compared with wild type-infected samples. Complementation of PilY1 restores replication efficiency, confirming PilY1 is needed for proper L. pneumophila infection in a human lung system (Hoppe et al. 2017). Hoppe et al. further confirmed the importance and relevance of the HLTE infection model, defining a significant role for PilY1 during infection.
Collectively, use of HLTEs has enhanced understanding of L. pneumophila infection and pathogenesis in a human disease-relevant setting. In light of large numbers of mis-diagnosed cases of Legionnaires’ disease, primary hAMs and human lung tissue represent new systems that can be used to define L. pneumophila infection events that are distinct from related pathogens that cause severe pneumonia.
YERSINIA PESTIS
Yersinia pestis is a Gram-negative facultative anaerobe that causes deadly pneumonic, bubonic and septicemic plague (Ditchburn and Hodgkins 2019). Bacterial transmission to humans occurs through an infected flea bite or contaminated aerosols, and Y. pestis is categorized as a tier 1 select agent due to the potential to cause widespread mortality. Pneumonic plague is the deadliest form of Y. pestis disease and presents with symptoms including fever, cough and shortness of breath (Bosio, Goodyear and Dow 2005). Pneumonic plague must be treated with antibiotics within 24 h after onset of symptoms to prevent lethality (Inglesby et al. 2000; Pechous et al. 2016; Ditchburn and Hodgkins 2019). After inhalation, Y. pestis can remain extracellular or infect pulmonary cells, and disease transitions from a pre-inflammatory to a pro-inflammatory phase within 2–4 days, with significant innate immune cell influx into the lungs (Bosio, Goodyear and Dow 2005). Unfortunately, early pre-inflammatory stages of Y. pestis pulmonary infection in humans are undefined. Therefore, hPCLS and hAMs represent important tools to characterize pre-inflammatory Y. pestis infection and model pneumonic plague in humans.
Recent work has established hPCLS and hAMs as reliable models of early Y. pestis infection. Plasminogen activator protease (Pla) is a virulence factor necessary for progression of pneumonic plague and is used throughout Y. pestis growth in vivo. Yersinia pestis Δpla exhibits reduced adhesion to hAMs and decreased Yersinia outer protein (Yop) effector translocation via the type III secretion system (T3SS) (Banerjee et al. 2019). In hPCLS, Yop effector translocation occurs at 2–4 hpi, demonstrating the utility of hPCLS for modeling early infection events. Yersinia pestis Δpla also displays reduced Yop translocation in hPCLS compared with wild-type bacteria, particularly in alveolar macrophages, mirroring hAM results (Banerjee et al. 2019). These data suggest Pla plays a vital role in Y. pestis effector secretion in hAMs. Pro-inflammatory cytokine expression that is more difficult to assess in murine models, due to expression being below the limit of detection in Y. pestis-infected mice (Lathem et al. 2007), is altered during infection with Y. pestis Δpla compared with wild-type bacteria. Secreted TNF-α, IL-6 and IL-8 all significantly increase during infection with Y. pestis Δpla. This result is striking, insinuating Pla is vital for early inhibition of a pro-inflammatory response, permitting Y. pestis survival and replication (Banerjee et al. 2019). Together, these results demonstrate the use of hAMs and hPCLS in assessing host–pathogen interactions during Y. pestis infection.
Regarding additional potential virulence factors, BPI-inducible protein A (BipA), a conserved GTPase, is important for virulence of many pathogens such as Escherichia coli and Pseudomonas aeruginosa; however, the role of Y. pestis BipA during infection has not been defined (Scott, Diggle and Clarke 2003). Crane et al. assessed whether BipA impacts phagocytic cells, such as primary human neutrophils, hAMs and murine alveolar macrophages (MH-S) by infecting each cell type with wild-type Y. pestis, ΔbipA bacteria or ΔbipA::bipA bacteria. No difference in Y. pestis survival in hAM or MH-S cells occurs regardless of strain. However, survival of ΔbipA Y. pestis significantly decreases in neutrophils (Crane et al. 2021). Collectively, these results show BipA is critical for bacterial survival in neutrophils, but not macrophages.
Altogether, studies using hAMs and hPCLS during Y. pestis infection have further defined parameters required for efficient infection. Though use of these models is new to this field, disease-relevant systems are necessary to derive human disease-relevant conclusions for comparison to many years of small animal model studies.
MYCOBACTERIUM TUBERCULOSIS
Mycobacterium tuberculosis is the causative agent of human tuberculosis, one of the deadliest diseases worldwide. Mycobacterium tuberculosis has an aerosol mode of transmission and results in roughly 2 million deaths annually (Ufimtseva et al. 2018). Following inhalation, alveolar macrophages engulf M. tuberculosis wherein the pathogen survives and replicates (Huang et al. 2018). Infected alveolar macrophages secrete cytokines and chemokines, attracting innate immune cells such as neutrophils, lymphocytes, dendritic cells and T cells that collectively contribute to granuloma formation (Fogel 2015). Granuloma formation typically results in an inactive or ‘latent’ phase of disease, with no symptoms or disease transmission. However, the granuloma can be disrupted, allowing M. tuberculosis to disperse throughout the pulmonary environment and infect new cells (Smith 2003; Pai et al. 2016). Due to ease of transmission and infection of human alveolar macrophages, it is necessary to expand research on new therapies and targets of M. tuberculosis using disease-relevant models such as hPCLS and primary hAMs.
Keane et al. (1997) used M. tuberculosis H37Ra and H37Rv strains that are nonpathogenic and pathogenic in mice, respectively, to study apoptotic cell death in hAMs. Both strains cause significant cell death with or without addition of TNF-α, a potentially cytotoxic cytokine, though notably higher levels of death are triggered by H37Ra. Furthermore, both isolates elicit significant production of TNF-α at 2 and 24 h, suggesting TNF-α is not the sole reason for increased cell death during H37Ra infection. Visual fragmentation and condensation of DNA is evident in M. tuberculosis H37Ra-infected samples, indicative of apoptosis. TUNEL staining also reveals a significant number of apoptotic cells in samples infected with M. tuberculosis H37Ra (Keane et al. 1997). This study established hAMs as a reliable platform to better understand the role of apoptosis during early M. tuberculosis infection.
Infants are more susceptible to tuberculosis infection, making it necessary to compare infant hAMs to adult hAMs to define mechanistic aspects of infection severity. No discernible difference in macrophage phenotype marker expression or phagocytic capacity is evident between infant and adult hAMs (Goenka et al. 2020). Even so, infant hAMs support significantly increased M. tuberculosis replication at 24 and 48 h, with increased production of neutrophil-attracting CXCL8 compared with adult hAMs. Transcriptional analysis of infant hAMs revealed significantly altered gene expression and regulation of multiple cytokines and chemokines. Specifically, lower expression of mycobactericidal genes, IFN-γ response genes, and increased expression of polymorphonuclear attractant signals occur in infant hAMs (Goenka et al. 2020). Collectively, this study revealed novel age-dependent insights into M. tuberculosis infection.
hAMs have also been used to define differential gene expression in TB patients. Lavalett et al. (2020) isolated hAMs from healthy (AMCT) and tuberculosis-positive (AMTB) donors, infected cells in vitro with one of two clinical isolates of M. tuberculosis, UT127 or UT205, and determined the differential transcriptional profiles of these samples. Differentially expressed genes are similar between control hAMs and hAMs previously exposed to M. tuberculosis after infection with UT127 or UT205. However, specific pathways, including the NF-κB pathway, TNF signaling and the overall inflammatory response, are enriched in either healthy or tuberculosis-exposed hAMs after infection with M. tuberculosis (Lavalett et al. 2020). Infection of AMCTs with M. tuberculosis UT127 or UT205 increases immune response signaling, organelle assembly and IFN-γ response gene expression. In addition, infection of AMTBs with M. tuberculosis UT127 or UT205 elicits isolate-specific differences in gene expression. For example, UT127 increases the STAT- and IL-23-related gene expression while UT205 increases IL-10 and IL-17 production. Further analysis assessed the difference in canonical pathway expression between M. tuberculosis UT127 or UT205-infected AMCTs and AMTBs. AMCTs infected with either strain results in activation of pathways such as iNOS, PI3K/AKT and apoptosis signaling. Infected AMBTs show a striking difference, with activation of interferon and IRF cytosolic pattern recognition receptor pathways (Lavalett et al. 2020). Overall, this study demonstrated important differences in transcriptional profiles between healthy and tuberculosis-infected donor hAMs when infected with distinct clinical isolates of M. tuberculosis. Ultimately, these results can be used to inform isolate-specific therapy design.
Emerging research has identified a critical role for host cell metabolism in the response to intracellular pathogens, including M. tuberculosis. Glycolysis, an important metabolic process used by all human cells, is required for protection against M. tuberculosis (Gleeson et al. 2016). To demonstrate the metabolic reprogramming ability of the pathogen, hAMs were infected with M. tuberculosis H37Rv (virulent) or γ-irradiated H37Rv (inactive). Results showed γ-irradiated H37Rv-infected samples have significantly increased production of il-1b mRNA and the glycolysis-related genes SCL2A1 and HK2 (Hackett et al. 2020). Strikingly, virulent M. tuberculosis H37Rv-infected hAMs have increased production of anti-inflammatory miRNA-21 (miR-21), suggesting a vital role for this miRNA during infection. This study then used bone marrow-derived macrophages to show miR-21 targets phosphofructokinase muscle (PFK-M) isoform to decrease glycolytic activity and program metabolism in favor of M. tuberculosis replication (Hackett et al. 2020). The use of primary hAMs strengthened these findings that show miR-21 reduces the pro-inflammatory immune response to M. tuberculosis.
In addition to defining cellular infection events, hAMs have been used to obtain information about infection severity. Ufimtseva et al. demonstrated an alternative method to obtain hAMs and other cell types from the pulmonary environment to study M. tuberculosis severity (Ufimtseva et al. 2018). Lung sections were surgically removed from tuberculosis-positive patients, processed and samples plated to define cell types within each area of the lung. Isolated cells consist of alveolar macrophages (AMs), dendritic cells, neutrophils, fibroblasts, lymphocytes, and multinucleated giant cells, although the most populous cell type is the AM (Ufimtseva et al. 2018). The majority of isolated AMs are infected with M. tuberculosis, whereas other cell types contain little to no bacteria. Some infected AMs are lipid rich, and M. tuberculosis co-localizes to these lipid body-positive regions. Strikingly, samples from patients with severe tuberculosis have increased populations of M. tuberculosis-infected AMs, suggesting a correlation between AM infection and disease severity. Infected macrophages are largely derived from cavity walls or distant sites within the pulmonary environment (Ufimtseva et al. 2018). Together, these results suggest a more direct route of AM isolation to study M. tuberculosis infection and indicate AM infection corresponds to disease severity.
Collectively, the studies above highlight the importance of using disease-relevant hAMs to fully understand M. tuberculosis pathogenesis in humans. Studies using this platform have provided apoptotic signaling discoveries, isolate-specific infection differences, age-dependent infection differences, the use of miRNA to reduce the pro-inflammatory response and an alternative route of cell isolation from tuberculosis-positive patients. The use of hAMs provides these results in a system that is immediately applicable to human disease and can be used to identify new therapeutic targets. We predict that hPCLS will also soon be used to further define early infection events in human lungs infected with M. tuberculosis.
STAPHYLOCOCCUS AUREUS
Staphylococcus aureus is a Gram-positive pathogen that causes disease in most human organs, including severe pneumonia in the lungs. Although largely considered an extracellular pathogen, S. aureus can infect and survive within epithelial cells, macrophages and neutrophils, highlighting the versatility of the pathogen and ability to resist intracellular degradation (Almeida et al. 1996; Gresham et al. 2000; Flannagan, Heit and Heinrichs 2016). Staphylococcus aureus produces numerous virulence factors that elicit distinct disease symptoms and are often isolate specific. For example, the highly pathogenic isolate LAC produces the major cytotoxins Panton–Valentine leukocidin (PVL) and α-hemolysin (Hla), whereas the less cytotoxic isolate UAMS-1 produces neither PVL or Hla (Brann et al. 2019), yet can still cause major disease. Current models of S. aureus pulmonary infection include immortalized cell lines, primary cells, and mouse, rat and rabbit models (Martínez-Olondris, Rigol and Torres 2010; Flannagan, Heit and Heinrichs 2016). However, inconsistencies when comparing results from differing disease models, and distinct human events not replicated in small animal models of pneumonia, highlight the need for improved human infection models.
To address the inconsistencies noted above, Brann et al. (2019) characterized S. aureus infection of hPCLS and hAMs. Infection of primary hAMs by LAC or UAMS-1 S. aureus indicated survival of both isolates in a late phagosome; however, replication is not efficient in this intracellular niche, suggesting hAMs do not permit S. aureus replication, a striking contrast from murine macrophage studies (Brann et al. 2019). Further assessment found S. aureus largely does not reside in hAMs in hPCLS. The pathogen is present in higher numbers within the interstitium, which contains a large population of fibroblasts, suggesting non-macrophage cells are targeted by S. aureus during lung infection (Brann et al. 2019). Further investigation of hAMs showed there is a phenotypic switch from pro-inflammatory M1 to anti-inflammatory M2 polarization during S. aureus infection, indicating a more permissive environment similar to C. burnetii (described above). hAMs produce a significant pro-inflammatory response during infection with S. aureus, and increasing multiplicities of infection of LAC results in higher cytotoxicity than UAMS-1. The cytolysin Hla is produced during LAC infection, but presence of the toxin does not correlate to increased cytotoxicity, as a Hla-deficient LAC mutant efficiently kills hAMs (Brann et al. 2019). Collectively, these results indicate S. aureus does not use hAMs as an intracellular replication niche in human lung tissue, but targets cells in the interstitium. This study demonstrates hAMs and hPCLS can now be used to further advance understanding of S. aureus pneumonia in a system that is directly applicable to the human lung setting.
ALTERNATIVE USES
In the sections above, we highlighted pathogen-specific uses for primary hAMs and hPCLS. In addition to those examples, alternative uses of these platforms are important for further characterization of these primary cellular systems and lung physiology, and to investigate the effects of drug addition to pulmonary tissue. These alternative uses further define the hAM and hPCLS platforms while expanding use of each system for disease-relevant, pathogen-free studies. We provide a sampling of these alternative uses below.
A major use for primary lung cell systems is the ability to study host–macrophage function. Cohen and Cline used lavage fluid from resected human lungs to characterize the morphology and phagocytic ability of alveolar macrophages (Cohen and Cline 1971). Three varying sizes of mononuclear cell types were noted when cells were isolated and cultured. Using Candida albicans, the ability of alveolar macrophages to phagocytose organisms was assessed in vitro. This study determined phagocytosis increases over several days post-infection and is directly dependent on macrophage size. Comparing differing lung conditions, such as healthy lungs, smokers’ lungs (cigarettes or marijuana), obstructed lungs or pneumonic lungs, researchers found smoker lung phagocytic capacity is comparable to healthy lungs whereas obstructed and pneumonic lung cells display increased phagocytosis. Specifically, hAMs display maximum ingestion ∼45 min post-infection with heat-inactivated C. albicans or Aspergillus fumigatus. Researchers also compared the bactericidal ability of polymorphonuclear cells, hAMs and monocyte-derived macrophages infected with Listeria monocytogenes. These comparative studies showed that polymorphonuclear cells have significantly increased bactericidal ability compared with hAMs and monocyte-derived macrophages (Cohen and Cline 1971). Collectively, this study revealed vital information about alveolar macrophage phagocytosis in distinct lung conditions that alters the response to foreign material.
Another nonpathogenic use for primary hAMs involves signaling molecules that regulate innate immune responses. For example, the role of serotonin in alveolar macrophage responses was first assessed in rat alveolar macrophage cell lines stimulated with lipopolysaccharide (LPS; Menard, Turmel and Bissonnette 2007). This study showed that addition of serotonin to cells decreases TNF secretion and increases IL-10 secretion. Importantly, the study was repeated and confirmed using hAMs, which display a similar decrease in TNF production with or without LPS stimulation after serotonin addition, and an increase in IL-10 with LPS stimulation after serotonin addition. Researchers also assessed the effect of indomethacin on cytokine production by hAMs, and noted indomethacin inhibits previously reported differences in TNF and IL-10 production (Menard, Turmel and Bissonnette 2007). These data suggest a major role for serotonin in regulating hAM cytokine production.
Routine use and storage of hPCLS begs the question of whether cryopreservation alters viability or functionality. Bai et al. (2016) addressed this question by evaluating cell viability and functional activities, including phagocytosis, airway contraction, intracellular calcium signaling and the effect of TAS2R agonists. These studies showed cryopreservation has no discernible effect on cell viability, lymphocyte presence or phagocytic capacity. Airway contraction in cryopreserved tissue is comparable to hPCLS that were never frozen using multiple agonists such as histamine, methacholine and β2-adrenergic receptor agonist. Additionally, and vital for smooth muscle cell research, cryopreservation has no obvious effect on intracellular calcium regulation of contractions. To further pursue the use of hPCLS as a platform for smooth muscle cell research, this study determined calcium signaling is inhibited via addition of bitter-taste receptor TAS2R (Bai et al. 2016). Overall, these results show cryopreservation has no significant effect on hPCLS and is a viable option for long-term storage.
To further expand use of hPCLS, viability and functionality of long-term cultivated tissue was assessed by Neuhaus et al. (2017). After a 15-day incubation, results showed no significant difference in metabolic activity or cell viability compared with day 1. To replicate constriction, various concentrations of methacholine were added to hPCLS over 15 days. hPCLS constrict throughout the full 15-day cultivation, though higher concentrations of methacholine are required. To further assess hPCLS functionality, the production of TNF-α with or without LPS stimulation was evaluated. TNF-α production continuously decreases over a 15-day time period in LPS-treated samples. Histology demonstrated similar maintenance of pulmonary structural components in hPCLS at day 1 and day 15 (Neuhaus et al. 2017). This study further characterized functional aspects of hPCLS as a novel platform that can be maintained for several days in vitro.
hAMs and hPCLS are useful not only to further understanding of bacterial infection but also for alternative studies, such as tissue banking, macrophage function, the immune response to hormones and culturing techniques. These studies broaden the use of these novel lung model systems and further emphasize the relevance of primary platforms.
CONCLUSIONS
Overall, the hAM and hPCLS systems have enormous potential for studying human cell activity in a physiological setting. These systems are invaluable for studying the human pulmonary innate immune response, cellular signaling and lung infection by microbial pathogens. Obtaining these tissues from human donors post-mortem provides the most human disease-relevant ex vivo lung models to date. These infection systems are gaining traction in the bacterial pathogen field and are useful for all pulmonary infection research. For example, hAMs and hPCLS have been used for influenza research, providing the groundwork for the advantages these model platforms can provide in viral research. With these systems in place, the alarming circumstances of the SARS-CoV-2 global pandemic, and the need to rapidly respond to novel microbes, hAMs and hPCLS will be reliable platforms to further understand COVID-19 disease pathogenesis and future pathogen outbreaks. We predict these platforms will enhance understanding of human pulmonary disease and physiology in several research fields over the coming years.
Contributor Information
Amanda L. Dragan, Department of Microbiology and Immunology, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Daniel E. Voth, Department of Microbiology and Immunology, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
FUNDING
This work was supported by funding to DEV from the NIH/NIAID (R21AI144508 and R21AI142056), the NIH/NIGMS (P20GM103625) and the Arkansas Biosciences Institute.
Conflict of Interest
None declared.
REFERENCES
- Almeida RA, Matthews KR, Cifrian Eet al. Staphylococcus aureus invasion of bovine mammary epithelial cells. J Dairy Sci. 1996;79:1021–6. [DOI] [PubMed] [Google Scholar]
- Bai Y, Krishnamoorthy N, Patel KRet al. Cryopreserved human precision-cut lung slices as a bioassay for live tissue banking. A viability study of bronchodilation with bitter-taste receptor agonists. Am J Respir Cell Mol Biol. 2016;54:656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee SK, Huckuntod SD, Mills SDet al. Modeling pneumonic plague in human precision-cut lung slices highlights a role for the plasminogen activator protease in facilitating type 3 secretion. Infect Immun. 2019;87:e00175–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio CM, Goodyear AW, Dow SW. Early interaction of Yersinia pestis with APCs in the lung. J Immunol. 2005;175:6750–6. [DOI] [PubMed] [Google Scholar]
- Brann KR, Fullerton MS, Onyilagha FIet al. Infection of primary human alveolar macrophages alters Staphylococcus aureus toxin production and activity. Infect Immun. 2019;87:e00167–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo NE, Rajasekaran A, Ali SK. Legionnaires' disease: a review. Infect Dis Clin Pract. 2016;24:248–53. [Google Scholar]
- Cohen AB, Cline MJ. The human alveolar macrophage: isolation, cultivation in vitro, and studies of morphologic and functional characteristics. J Clin Invest. 1971;50:1390–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonne PM, Winchell CG, Graham JGet al. Vasodilator-stimulated phosphoprotein activity is required for Coxiella burnetii growth in human macrophages. PLoS Pathog. 2016;12:e1005915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon TV, Sawyer RT, Fenton MJet al. Lung dendritic cells at the innate–adaptive immune interface. J Leukoc Biol. 2011;90:883–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver AM, Casson CN, Nguyen HTet al. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infect Immun. 2014;82:4325–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabill E, Schofield WB, Newton HJet al. Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect Immun. 2018;86:e00758–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane SD, Banerjee SK, Eichelberger KRet al. The Yersinia pestis GTPase BipA promotes pathogenesis of primary pneumonic plague. Infect Immun. 2021;89:e00673–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchburn J-L, Hodgkins R. Yersinia pestis, a problem of the past and a re-emerging threat. Biosaf Health. 2019;1:65–70. [Google Scholar]
- Dragan AL, Kurten RC, Voth DE. Characterization of early stages of human alveolar infection by the Q fever agent Coxiella burnetii. Infect Immun. 2019;87:e00028–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan AL, Voth DE. Coxiella burnetii: international pathogen of mystery. Microbes Infect. 2020;22:100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott A, Peng Y, Zhang G. Coxiella burnetii interaction with neutrophils and macrophages in vitro and in SCID mice following aerosol infection. Infect Immun. 2013;81:4604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes TD, Cunha LD, Ribeiro JMet al. Murine alveolar macrophages are highly susceptible to replication of Coxiella burnetii phase II in vitro. Infect Immun. 2016;84:2439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan RS, Heit B, Heinrichs DE. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell Microbiol. 2016;18:514–35. [DOI] [PubMed] [Google Scholar]
- Fogel N. Tuberculosis: a disease without boundaries. Tuberculosis (Edinb). 2015;95:527–31. [DOI] [PubMed] [Google Scholar]
- Franks TJ, Colby TV, Travis WDet al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc. 2008;5:763–6. [DOI] [PubMed] [Google Scholar]
- Gleeson LE, Sheedy FJ, Palsson-McDermott EMet al. Cutting edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J Immunol. 2016;196:2444–9. [DOI] [PubMed] [Google Scholar]
- Goenka A, Prise IE, Connolly Eet al. Infant alveolar macrophages are unable to effectively contain Mycobacterium tuberculosis. Front Immunol. 2020;11:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JG, MacDonald LJ, Hussain SKet al. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol. 2013;15:1012–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JG, Winchell CG, Kurten RCet al. Development of an ex vivo tissue platform to study the human lung response to Coxiella burnetii. Infect Immun. 2016;84:1438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham HD, Lowrance JH, Caver TEet al. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol. 2000;164:3713–22. [DOI] [PubMed] [Google Scholar]
- Hackett EE, Charles-Messance H, O'Leary SMet al. Mycobacterium tuberculosis limits host glycolysis and IL-1beta by restriction of PFK-M via microRNA-21. Cell Rep. 2020;30:124–136.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe J, Unal CM, Thiem Set al. PilY1 promotes Legionella pneumophila infection of human lung tissue explants and contributes to bacterial adhesion, host cell invasion, and twitching motility. Front Cell Infect Microbiol. 2017;7:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Nazarova EV, Tan Set al. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med. 2018;215:1135–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby TV, Dennis DT, Henderson DAet al. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 2000;283:2281–90. [DOI] [PubMed] [Google Scholar]
- Ito Y, Correll K, Schiel JAet al. Lung fibroblasts accelerate wound closure in human alveolar epithelial cells through hepatocyte growth factor/c-Met signaling. Am J Physiol Lung Cell Mol Physiol. 2014;307:L94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager J, Marwitz S, Tiefenau Jet al. Human lung tissue explants reveal novel interactions during Legionella pneumophila infections. Infect Immun. 2014;82:275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane J, Balcewicz-Sablinska MK, Remold HGet al. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN. Alveolar macrophage in the driver's seat. Immunity. 2006;24:366–8. [DOI] [PubMed] [Google Scholar]
- Lathem WW, Price PA, Miller VLet al. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–13. [DOI] [PubMed] [Google Scholar]
- Lavalett L, Ortega H, Barrera LF. Human alveolar and splenic macrophage populations display a distinct transcriptomic response to infection with Mycobacterium tuberculosis. Front Immunol. 2020;11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson PR, Reid KB. The roles of surfactant proteins A and D in innate immunity. Immunol Rev. 2000;173:66–78. [DOI] [PubMed] [Google Scholar]
- Liu J, Pang Z, Wang Get al. Advanced role of neutrophils in common respiratory diseases. J Immunol Res. 2017;2017:6710278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald LJ, Graham JG, Kurten RCet al. Coxiella burnetii exploits host cAMP-dependent protein kinase signalling to promote macrophage survival. Cell Microbiol. 2014;16:146–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald LJ, Kurten RC, Voth DE. Coxiella burnetii alters cyclic AMP-dependent protein kinase signaling during growth in macrophages. Infect Immun. 2012;80:1980–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Olondris P, Rigol M, Torres A. What lessons have been learnt from animal models of MRSA in the lung?. Eur Respir J. 2010;35:198–201. [DOI] [PubMed] [Google Scholar]
- Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999;12:518–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazokopakis EE, Karefilakis CM, Starakis IK. Q fever endocarditis. Infect Disord Drug Targets. 2010;10:27–31. [DOI] [PubMed] [Google Scholar]
- Menard G, Turmel V, Bissonnette EY. Serotonin modulates the cytokine network in the lung: involvement of prostaglandin E2. Clin Exp Immunol. 2007;150:340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muder RR, Yu VL. Infection due to Legionella species other than L. pneumophila. Clin Infect Dis. 2002;35:990–8. [DOI] [PubMed] [Google Scholar]
- Neuhaus V, Schaudien D, Golovina Tet al. Assessment of long-term cultivated human precision-cut lung slices as an ex vivo system for evaluation of chronic cytotoxicity and functionality. J Occup Med Toxicol. 2017;12:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, Ang DK, van Driel IRet al. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. 2010;23:274–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston PC, Davies C. Q fever: the neglected biothreat agent. J Med Microbiol. 2011;60:9–21. [DOI] [PubMed] [Google Scholar]
- Pai M, Behr MA, Dowdy Det al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. [DOI] [PubMed] [Google Scholar]
- Pechous RD, Sivaraman V, Stasulli NMet al. Pneumonic plague: the darker side of Yersinia pestis. Trends Microbiol. 2016;24:190–7. [DOI] [PubMed] [Google Scholar]
- Rackley CR, Stripp BR. Building and maintaining the epithelium of the lung. J Clin Invest. 2012;122:2724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubins JB. Alveolar macrophages: wielding the double-edged sword of inflammation. Am J Respir Crit Care Med. 2003;167:103–4. [DOI] [PubMed] [Google Scholar]
- Samanta D, Clemente TM, Schuler BEet al. Coxiella burnetii type 4B secretion system-dependent manipulation of endolysosomal maturation is required for bacterial growth. PLoS Pathog. 2019;15:e1007855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schyns J, Bureau F, Marichal T. Lung interstitial macrophages: past, present, and future. J Immunol Res. 2018;2018:5160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Diggle MA, Clarke SC. TypA is a virulence regulator and is present in many pathogenic bacteria. Br J Biomed Sci. 2003;60:168–70. [DOI] [PubMed] [Google Scholar]
- Shevchuk O, Pagelow D, Rasch Jet al. Polyketide synthase (PKS) reduces fusion of Legionella pneumophila-containing vacuoles with lysosomes and contributes to bacterial competitiveness during infection. Int J Med Microbiol. 2014;304:1169–81. [DOI] [PubMed] [Google Scholar]
- Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltysiak KA, van Schaik EJ, Samuel JE. Surfactant protein D binds to Coxiella burnetii and results in a decrease in interactions with murine alveolar macrophages. PLoS One. 2015;10:e0136699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufimtseva E, Eremeeva N, Petrunina Eet al. Ex vivo expansion of alveolar macrophages withMycobacterium tuberculosis from the resected lungs of patients with pulmonary tuberculosis. PLoS One. 2018;13:e0191918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, Heinzen RA. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9:829–40. [DOI] [PubMed] [Google Scholar]
- White ES. Lung extracellular matrix and fibroblast function. Ann Am Thorac Soc. 2015;12:S30–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchell CG, Dragan AL, Brann KRet al. Coxiella burnetii subverts p62/sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect Immun. 2018;86:e00608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchell CG, Graham JG, Kurten RCet al. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun. 2014;82:2229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts WA, Agostini C, Antoniou KMet al. The pathogenesis of pulmonary fibrosis: a moving target. Eur Respir J. 2013;41:1207–18. [DOI] [PubMed] [Google Scholar]
- Yan L, Cirillo JD. Infection of murine macrophage cell lines by Legionella pneumophila. FEMS Microbiol Lett. 2004;230:147–52. [DOI] [PubMed] [Google Scholar]