ABSTRACT
Biofilm-forming bacteria have the potential to contribute to the health, physiology, behavior and ecology of the host and serve as its first line of defense against adverse conditions in the environment. While metabarcoding and metagenomic information furthers our understanding of microbiome composition, fewer studies use cultured samples to study the diverse interactions among the host and its microbiome, as cultured representatives are often lacking. This study examines the surface microbiomes cultured from three shallow-water coral species and two whale species. These unique marine animals place strong selective pressures on their microbial symbionts and contain members under similar environmental and anthropogenic stress. We developed an intense cultivation procedure, utilizing a suite of culture conditions targeting a rich assortment of biofilm-forming microorganisms. We identified 592 microbial isolates contained within 15 bacterial orders representing 50 bacterial genera, and two fungal species. Culturable bacteria from coral and whale samples paralleled taxonomic groups identified in culture-independent surveys, including 29% of all bacterial genera identified in the Megaptera novaeangliae skin microbiome through culture-independent methods. This microbial repository provides raw material and biological input for more nuanced studies which can explore how members of the microbiome both shape their micro-niche and impact host fitness.
Keywords: bacteria, SSU rRNA, coral, whale, microbiome, skin
This study captures a diverse range of cultured microorganisms from the surface microbiomes of marine animals, which fills necessary gaps in the availability of cultured isolates from this important environment.
INTRODUCTION
The epidermis is an animal's first line of defense against the external environment, yet not impervious to environmental influences. The dermis of an organism can be thought of as its own ecosystem, hosting a diverse microbial milieu, where the composition of communities is driven by both endogenous host factors and the exogenous environment. In marine ecosystems, surface-associated microbes must contend with unpredictable external variables including temperature, pH, salinity fluctuations and skin/mucus shedding by the host. The animal surface microbiome plays a significant role in the health of the host by protecting the body against transient pathogenic microorganisms, selecting for antibiotic-producing commensal strains, playing critical roles in host nutrition and significantly impacting immune system development (Nelson et al. 2015; Bourne, Morrow and Webster 2016; Apprill 2017; Ross, Rodrigues Hoffmann and Neufeld 2019). Often marine animal surfaces are referred to as ‘hot spots’ of microbial diversity. These environments are dominated by unique assemblages of host-specific bacteria capable of generating specific microenvironments that promote higher-level microbial community organization, including the production of shielding biofilm matrices, antiprotozoal factors and chemical compounds that assist in protection from predators, viruses and environmental stressors (Bik et al. 2016; Dang and Lovell 2016). The uniqueness of surface-associated microbiota in both their genetic composition and functional roles in comparison to their free-living counterparts (Burke et al. 2011) indicates either an active role of the hosts in recruitment of epibiotic bacteria or a passive mechanism of colonization based on the eukaryotic surface and exudates (Wahl et al. 2012).
The surface microbiomes of both cetaceans (Nelson et al. 2015) and coral (Rosenberg et al. 2007; Zaneveld et al. 2016) have been studied to elucidate the role of the microbial community on the health, persistence and resilience of these keystone and habitat-forming species. In corals, it is well known that their resident microalgal symbionts mediate host physiology. However, it has been suggested that bacteria, archaea and fungi participate in cycling of nutrients and organic matter in coral as well (Apprill 2017). While coral microbial communities are often host specific, spatially restrictive and can be highly stable across geographic and environmental conditions, oversimplification of host–microbe association via broad profiling studies can miss low abundance taxa that contribute to physiologically significant interactions with their hosts (Bourne, Morrow and Webster 2016). Similarly, marine mammals, particularly cetaceans, are considered sentinel species in marine ecosystems (Bossart 2011). There are few microbiome studies in cetaceans and still fewer that target members of the skin microbiome (Chiarello et al. 2017; Hooper et al. 2019; Apprill et al. 2020). Recent investigations into the skin microbiome of cetaceans have suggested the existence of species-specific assemblages that can change seasonally (Bierlich et al. 2017). These assemblages can also be linked to the presence of epiphytic diatoms (Hooper et al. 2019) and are shown to be distinct from surrounding planktonic samples (Chiarello et al. 2017), suggesting ecological and evolutionary forces including the unique features of an animal's epidermis have shaped cetacean skin microbiomes.
The availability of sequence data is rapidly readjusting our view of how bacterial communities associated with marine organisms vary in composition depending on host type and environment (Vega Thurber et al. 2009; Sunagawa, Woodley and Medina 2010; Barott et al. 2011; Hentschel et al. 2012; Sison-Mangus et al. 2014; Cooper and Smith 2015; Rouco, Haley and Dyhrman 2016). However, there is a gap in microbial diversity obtained between sequencing-based techniques and culture-dependent isolation of strains. This study therefore sought to identify key features of animal surfaces and marine environments in order to recapitulate environmental conditions with the goal of isolating as diverse a complement of surface microbiome representatives possible from the dermis of two species of whale (Delphinapterus leucas and Megaptera novaeangliae) and the mucus and tissue from three species of coral (Porites astreoides, Acropora palmata and Millepora alcicornis). Six culture conditions were used in this study and chosen based on either previous success in culturing marine microbes or were developed here using knowledge of features associated with the dermis surface chemistry and microbial signaling processes. A total of three media variations used marine agar (MA), combining an agar and a commercially available marine broth base, and is a widely used media type demonstrated to yield biologically diverse microbial flora because of its nutrient richness (Mincer et al. 2002). One marine agar variation contained the antimicrobial enzyme, lysozyme (LYS), which is produced by cetaceans within the upper lamellae of the stratum corneum of the dermis and may select for microbes resistant to this non-specific defense mechanism (Seegers and Meyer 2004; Mouton and Botha 2012). Another marine agar variation contained both the secondary messenger cyclic adenosine monophosphate, shown to regulate bacterial metabolism, virulence gene expression, flagella mobility, surface attachment and biofilm formation (Smith, Wolfgang and Lory 2004; Fuchs et al. 2010; Ono et al. 2014; Dang and Lovell 2016), and N-(oxo-hexanoyl)-homoserine lactone, a homoserine lactone used by the majority of Gram-negative bacteria with quorum sensing ability (HSL-AMP; Bruns, Cypionka and Overmann 2002). Acylhomoserine lactone signals are known to mediate interspecies communication, biofilm formation and community structure (Wang et al. 2020). Lower nutrient media types included Actinobacteria-selecting media (AC; Okami and Hotta 1988) and a low nutrient media (R2A) containing chitin as both an energy source and substrata, that facilitates slower-growing, oligotrophic microbes (Reasoner and Geldreich 1985). The final media type selected for marine fungi (KJ; Kjer et al. 2010).
Here we demonstrate the cultivation of 592 isolates contained within 15 bacterial orders representing 50 bacterial genera and two fungal species isolated from 25 cetacean and coral samples, indicating targeted cultivation by use of media additives and intensive microbial cultivation efforts yields high microbial diversity in comparison to other published efforts. Multivariate analyses demonstrate that microbial diversity is associated with sampled species rather than media variation, although different media variations and host surfaces were successful in culturing potentially phylogenetically distinct strains based on SSU rRNA variation. With this new microbial repository from diverse animal hosts, we can now begin to explore how interactions among microbial community members can impact host fitness.
MATERIALS AND METHODS
Ethics statement
Collection of coral tissue and mucus samples were conducted under permits issued by the Government of the Virgin Islands (permit #DFWCZM17003J). Humpback whale skin samples were collected under permits issued by the U.S. National Marine Fisheries Service (#16325 and #18786).
Sample collection
Skin samples were obtained from North Atlantic humpback whales (Megaptera novaeangliae) on the Gulf of Maine feeding ground in May and June, 2016 as part of a long-term population research program led by the Center for Coastal Studies (CCS). Samples were collected from twelve free-ranging individuals as either sloughed skin or biopsy sampling techniques. Biopsy samples were collected from the dorsal flank using a crossbow equipped with a hollow biopsy dart capable of obtaining a 1 cm3 skin sample (Palsbøll, Larsen and Sigurd Hansen 1991) and subsampled for analysis. All samples were placed in 10 mL of 0.2 µm filter-sterilized natural seawater, transported on ice back to the laboratory for culturing within 5 h of collection, and processed immediately. In July 2016, a 14-year-old beluga whale, Delphinapterus leucas, under professional care at the Mystic Aquarium in Mystic, Connecticut was sampled at five sites (right-side front, right-side back, ventral chest, left-side back and left-side peak) with sterile cotton swabs during a routine blood draw procedure. Freshly collected skin swabs were placed in 10 mL of 0.2 µm filter-sterilized natural seawater, transported on ice back to the laboratory within 6 h of collection and processed immediately. Coral tissue + mucus, collected via surface swabs, and mucus samples, collected via syringe, were sampled underwater using SCUBA at 2–3 m from three species of coral, Porites astreoides, Acropora palmata and Millepora alcicornis (two individuals, one individual and one individual, respectively) in St. John, U.S.V.I. (18.31°N, −64.76°W). Mucus was collected by gentle syringe pulls (which may have dislodged some coral tissue), and mucus + tissue samples were obtained by swabbing the coral surface using a sterile cotton swab. For all colonies, approximately the same surface area of coral was sampled. Upon surfacing, the syringe samples were held upside down to gravity-concentrate the mucus. Then, the mucus was released from the syringe into the cryovial. The swabs were directly placed in 4% glycerol and frozen in liquid nitrogen vapors and then held at −80°C until use in culturing experiments. Although it is difficult to distinguish coral-associated bacteria from potentially water-borne bacteria using this sampling method, water immediately surrounding tropical coral contains more coral-associated bacteria, as well as surface-associated genes, than water more distantly related to the reefs (1 m off the reef), suggesting a coral association even for potentially water-borne microbes (Weber et al. 2019). Sample details are provided in Table S1 (Supporting Information).
Microbial enumeration and characterization
Aliquots of environmental samples were plated on five selective media types and variations including Marine Agar (MA; agar, Remel, Lenexa, KS Cat. No. R451012; marine broth base, Difco, Franklin Lakes, NJ Cat. No. 2216); Marine Agar + cAMP/AHL (HSL-AMP) containing both cyclic adenosine monophosphate (cAMP, Sigma-Aldrich, A9501, St Louis, MO) and N-(oxo-hexanoyl)-homoserine lactone (OHHL, Sigma, K3255) at 10 µM final concentration; Marine Agar + lysozyme (LYS) containing lysozyme (Fisher BioReagents, BP535, Waltham, MA) at a final concentration of 1 µg/mL; Actinobacteria-selecting (AC) media containing 10 g starch, 0.3 g casein, 2 g KNO3 in 1 L of natural seawater and nalidixic acid (Sigma, N8878) at a final concentration of 50 µg/mL; R2A broth + chitin (R2A) containing 2 g of chitin (Sigma, C7170) in 1 L of (3:1) natural seawater: MilliQ water. Inclusion of chitin in culture media has been previously successful in isolating bacteria of the genus, Tenacibaculum, a prominent skin microbiome member of M. novaeangliae (Sheu et al. 2007; Bierlich et al. 2017). All bacterial agar plates also contained cycloheximide at a final concentration of 100 µg/mL. Marine fungi (KJ) were isolated as described in (Kjer et al. 2010) on Medium A plates containing 15 g malt extract (Sigma 70167) in 1 L of natural seawater and chloramphenicol at a final concentration of 50 µg/mL. All natural seawater was filtered using a 0.2 µm filter.
Humpback whale sloughed skin and biopsy samples were vortexed in 10 mL of 0.2 µm filter sterilized natural seawater and 100 µL aliquots were plated at multiple dilutions (varying from undiluted to 10−6) on six types of selection plates in order to obtain single, morphologically distinguishable colonies. Aliquots plated on AC media plates were heat shocked at 70°C for 10 min prior to spreading. Plates were incubated at temperatures reflecting natural conditions (20°C for whale samples and 23°C for coral samples) until growth of distinguishable colonies was detected. Plates were incubated and monitored for a maximum of 15 days after plating. For AC media, four colonies were picked with an average of 11.25 days of growth; for HSL-AMP media, 165 colonies were picked with an average of 5.9 days of growth; for KJ media, five colonies were picked with an average of 11.4 days of growth; for LYS media, 228 colonies were picked with an average of 5.7 days of growth, for MA media, 257 colonies were picked with an average of 5.1 days of growth; for R2A media, 150 colonies were picked with an average of 8.8 days of growth. For each media type, the number of distinguishable colonies and colony morphology were recorded (Table S2, Supporting Information). Colonies were differentiated based on colony morphology (e.g. size, color, edge/margin, surface and elevation), pigmentation and growth characteristics (e.g. time of colony appearance and media type used for isolation) and were observed using a dissecting light microscope. For each media variation, the number of distinguishable colonies and colony morphology were recorded (Table S2, Supporting Information). Distinct colonies were streaked onto new selection plates corresponding to the original isolation media type to confirm purity, and colonies from multiple dilutions from the same sample and media type were streaked to promote isolation of different strains with similar morphologies. However, due to time constraints, the microbial biomass that had grown on selection plates containing D. leucas aliquots was cryopreserved in 15% glycerol and Marine broth (Difco 2216), and frozen at −80°C for 3 months. These cryopreserved samples were then spread again on selection plates, and distinct microbial colonies were picked and streaked onto subculture plates. In sum, of the 809 colonies streaked onto subculture plates, only 30 colonies failed to grow. The microbial culture collection is held at both Woods Hole Oceanographic Institution by Dr Amy Apprill and at Haverford College by Dr Kristen Whalen, and inquiries to obtain isolates can be directed to either investigator.
Once purity on subculture plates was confirmed, a single colony was picked and used to inoculate 7 mL of Marine broth (37.4 g Marine broth base (Difco 2216) to 1 L MilliQ water) and allowed to grow at room temperature at 100 rpm for 3 days. An aliquot of the inoculum was preserved in 15% glycerol and stored at −80°C, while an additional aliquot was centrifuged at 13 000 × g for 5 min to obtain a microbial pellet for genomic DNA isolation. Genomic DNA from bacterial and fungal samples were isolated using the Qiagen's DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) following the manufacturer protocol with enzymatic lysis buffer containing 50 mM Tris-Cl, 10 mM Sodium-EDTA, 1.2% Triton X-100 and lysozyme at a final concentration of 20 mg/mL. Bacterial SSU rRNA genes were amplified through PCR using oligonucleotide primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′), with an expected amplicon of ∼1400 bp. Fungal ITS1 region was amplified via nested PCR, first using oligonucleotide primers ITS5F (5′-GGAACAATGCTGAAAATGAAGG-3′) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′) followed by ITS4R and ITS1 (5′-GGCGTCCAAGTGGATGCCT-3′) primer pair, with an expected amplicon of ∼500 bp. Each 50 µL PCR reaction contained 1.25 U of GoTaq® G2 Flexi DNA polymerase (Promega, Fitchburg, WI), 1 X GoTaq Flexi Buffer, 1.25 mM MgCl2, 200 µM of each dNTPs, 200 nM of each primer and 250 ng of genomic template and was performed in a MyCycler thermal cycler (Bio-Rad Laboratories, Hercules, CA). Amplification of PCR products was carried out according to the GoTaq® Flexi Kit and cycling parameters were as follows: 95°C for 2 min; 40 cycles of 95°C for 20 s, 54°C for 30 s, 72°C for 1.5 min; 1 cycle of 72°C for 5 min. Amplification products were subjected to gel electrophoresis in 1% agarose gels and purified using the QIAquick PCR purification kit (Promega, Madison, WI). Products were sequenced in a single direction using the Sanger method by Eurofin MWG Operon Biotech. Sequences were manually inspected using Sequencher 4.8 (Gene Codes, Ann Arbor, MI), and ambiguous nucleotides on the ends of the sequence were excluded from further analysis.
Phylogenetic analysis of bacterial SSU rRNA gene sequences
Taxonomic identifications of each bacterial sequence to the genus level were conducted in ARB (Ludwig 2004). First, the sequences were aligned using SINA web aligner v.1.2.11. This alignment was then imported into ARB software, and sequences were aligned to the SILVA (Pruesse et al. 2007) non-redundant database using default parameters to identify taxonomy to the genus level and in some cases to the species level. The taxonomic identity of the fungal sequences was determined through the ISHAM ITS database (Irinyi et al. 2015). Of the 592 sequenced isolates (i.e. 587 bacterial and five fungal), another 46 sequences with poor quality (i.e. lengths shorter than 600 nucleotides) were removed from the dataset before downstream phylogenetic analysis and tree generation. Of these 46 sequences that were removed, ARB analysis indicated that none represented genera that were not already present among the remaining 546 sequences used to construct the final phylogenetic tree. The remaining sequences >600 bp in length were aligned using MUSCLE alignment in Mega7 using -400 gap open and UPGMB clustering parameters (Edgar 2004). This alignment was used to construct a maximum likelihood phylogenetic tree with 402 informative nucleotide positions in the final dataset. Confidence intervals of the phylogenetic relationships were determined using 1000 bootstrap samples. To minimize sequence redundancy and to enable comparison of taxonomic identity vs. sampled surface and taxonomic identity vs. media type, a second tree was generated by first selecting a single representative sequence from a node of sequences sharing ≥ 99% similarity, and then realigning these 109 representative sequences in Mega7 as described above. The resulting alignment was used to construct a maximum likelihood tree with 417 informative nucleotide positions in the final data set (Fig. 1). Confidence intervals of the phylogenetic relationships were determined using 1000 bootstrap samples. Both trees were edited in FigTree version 1.4.3 and Adobe Illustrator CS6.
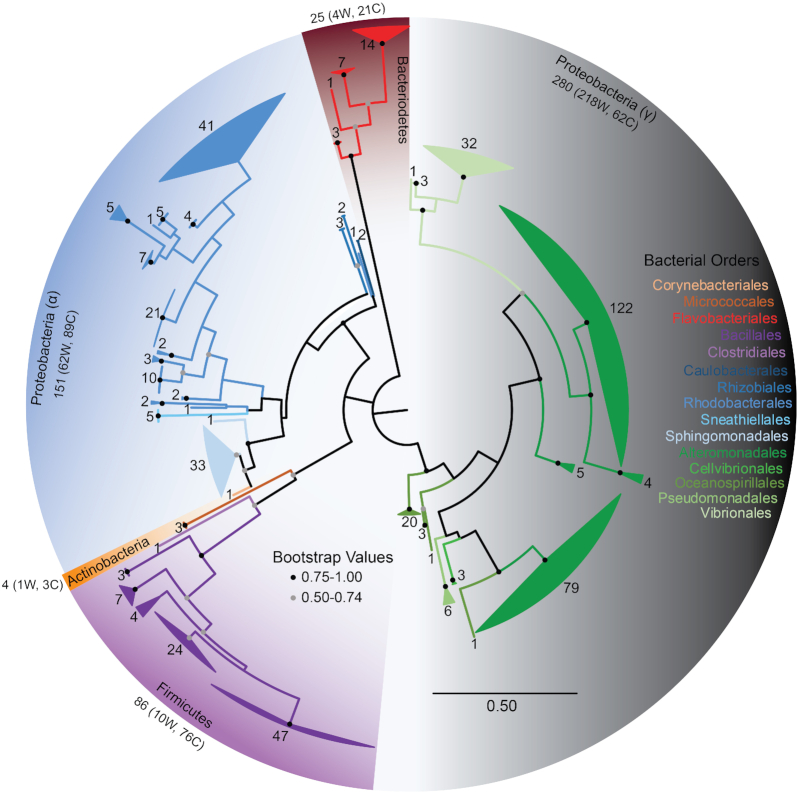
Figure 1.
Phylogenetic relationship of marine surface-associated bacteria. Maximum likelihood phylogenetic tree of bacterial SSU rRNA gene sequences from all twenty-five environmental samples constructed in MEGA7 and modified through FigTree v1.4.3. The phylogenetic tree includes 546 bacterial nucleotide sequences used to infer evolutionary history of the microorganisms isolated in this study. Fungal isolates are not included. The analysis involved 402 nucleotide positions. Branches are collapsed by Genera and colored by Order. The number of sequences at each node is noted. Orders within the same class are colored in a similar hue.
GreenGenes OTU Assignment in QIIME
OTUs were assigned for bacterial isolates. A total of 587 sequences passed a preliminary check for quality for the downstream analyses. OTU assignment was performed using Quantitative Insights Into Microbial Ecology (QIIME, version 1) software running on an Ubuntu virtual machine with default parameters using two algorithms (UCLUST or BLAST) to assign our sequences to fully sequenced microbial genomes sharing 99% similarity to the 16S rRNA gene (GreenGenes 13.5 reference database; Caporaso et al. 2010). The operational taxonomic unit formation was first performed using the QIIME closed reference picking command with default parameters, which uses UCLUST taxon assignment method (Edgar 2004), version 1.2.22q to perform the assignment. The UCLUST algorithm was not successful in assigning 12.9% (75 of 587 sequences) of the sequences; therefore, the OTU assignment of the remaining sequences was performed in QIIME using the BLAST assignment method at 99% similarity to the GreenGenes (13.5 reference database). The OTUs assigned using BLAST were manually added to the OTU table constructed through closed reference picking. Both algorithms together were successful in assigning all sequences to OTUs in the GreenGenes database (Table S4, Supporting Information). Sample diversity was demonstrated through OTU richness by sample type (Smith and Wilson 1996; Table 1).
Table 1.
Bacterial richness by sample. Observed unique OTUs (grouped by 99% similarity to the 13.5 GreenGenes 13.5 reference database through closed-reference OTU picking via QIIME software package) from 583 bacterial 16S sequences for the sample categories listed. Only the media types yielding more than five isolates are included. For each media type or surface type, the total number of isolates, the number of unique OTUs identified through GreenGenes closed-reference picking, and the total number of genera identified through BLAST alignment are included.
| Sample | Total isolates | Number of unique GGs OTUs | Total number of genera represented | |
|---|---|---|---|---|
| Media type | MA | 177 | 84 | 37 |
| HSL-AMP | 132 | 62 | 28 | |
| LYS | 164 | 72 | 28 | |
| R2A | 110 | 54 | 25 | |
| M. novaeangliae | 220 | 80 | 29 | |
| D. leucas | 99 | 26 | 6 | |
| A. palmata (all) | 63 | 28 | 15 | |
| A. palmata (syringe) | 41 | 25 | 13 | |
| A. palmata (swab) | 25 | 8 | 9 | |
| P. astreoides (all) | 139 | 69 | 26 | |
| P. astreoides (syringe) | 81 | 49 | 24 | |
| Host organism | P. astreoides (swab) | 58 | 33 | 16 |
| M. alcicornis (all) | 66 | 35 | 25 | |
| M. alcicornis (syringe) | 32 | 20 | 14 | |
| M. alcicornis (swab) | 34 | 17 | 16 | |
| Coral mucus (all species) | 153 | 75 | 34 | |
| Coral surface (all species) | 115 | 48 | 26 |
Multivariate analyses of cultured microbial community composition
Multivariate statistical approaches were used to examine cultured microbial community variation among sample types and media variations. All data manipulation and analysis were conducted in R version 3.6.3 using the packages ‘vegan’ (Oksanen 2018) and ‘Biostats’ (McGarigal 2009). The multivariate analysis compared 102 objects, representing the microbial community composition from every media variation and surface sample (17 whale skin swab samples, eight coral syringe samples, eight coral swab samples) combination. Microorganisms cultured using KJ (fungi-selecting) and AC (Actinobacteria-selecting) media were removed from the analysis because their low microbial culture yield and low OTU richness would skew comparisons of group differences, therefore reducing the analysis to 97 objects. Microbial community composition was analyzed using the presence and absence of 21 microbial families to avoid the disproportionate influence of microbes exhibiting rapid growth on culture media. Similarity between objects was computed using a Jaccard coefficient, an asymmetrical binary coefficient that treats double-zeros in a differently than the other data (Baroni-Urbani and Buser 1976). This asymmetry is important in a culture-dependent experimental context, since a microbe's observed absence does not necessarily denote a true absence in the sample.
A non-metric multidimensional scaling (NMDS) ordination approach was used to visualize microbial community variation among samples and media variations. NMDS attempts to locate objects in a low-dimensional ordination space such that the inter-object distances have the same rank order as the inter-object dissimilarities in the original dissimilarity matrix (Digby and Kempton 1987). The NMDS ordination approach was chosen because of its flexibility in choice of similarity coefficient and because it avoids the assumption of linear relationships among variables by using ranked distances. NMDS was used to ordinate three sets of objects: (1) all sampled species to compare microbial variation between species and media variation (97 objects), (2) sampled species yielding high OTU richness (excluding D. leucas) to compare microbial variation between species and media variation (79 objects), and (3) coral species to compare microbial variation between syringe and swab samples (32 objects). Due to low OTU richness in the D. leucas samples, the ordination of the first set of objects was performed only on microbial families present in at least 5% of samples, reducing the dataset to 11 microbial families.
To determine the optimum number of dimensions, a scree plot was generated comparing the first 10 NMDS dimensions and their associated stress. To validate the ordination, a stress plot was generated to compute the correlation between the original object dissimilarities and the ranked Euclidean distances in the ordination. To calculate and depict microbial family loadings on each derived axis from the NMDS ordination, the envfit() function was used to perform a simple linear regression between each of the original descriptors (microbial families) and the scores from each NMDS axis. A permutation test was used to assess the statistical significance of each microbial family to the ordination. All ordinations and variable loadings were visualized using the ‘ggplot2’ package (Wickham et al. 2020), and figures were modified in Adobe Illustrator (v24.3).
A permutational multivariate analysis of variance (perMANOVA) was used to determine if microbial community composition in multivariate space was significantly different between groups (species, media variation, coral sample type). perMANOVA partitions the within- and among-group sums of squares of the Jaccard similarity matrix and is permuted 1000 times to test for significance (Anderson 2001). A series of pair-wise perMANOVA tests were also conducted to compare each of the groups directly against each other, and a test of multivariate homogeneity of group dispersions was conducted both globally and pair-wise to determine if the dispersion of one or more groups were significantly different (Anderson, Ellingsen and McArdle 2006).
Nucleotide sequence accession number
The SSU rRNA and ITS1 sequences have been deposited in NCBI and Genbank Accession numbers are listed in Table S2 (Supporting Information).
RESULTS
Characterization of the cultured bacterial communities
The intense cultivation effort used in this study yielded a phylogenetically diverse array of cultured microbial isolates. A total of 779 pure microbial strains were isolated from the surface microbiomes from 25 animal surface samples representing three coral and two cetacean species (Table S1, Supporting Information). The SSU rRNA gene or ITS1 region was successfully sequenced for 587 bacterial isolates and 5 fungal isolates, respectively, allowing for taxonomic identification of 592 microbial isolates in total. Table S2 (Supporting Information) reports the bacterial and fungal taxonomic identification, Genbank Accession number, and culturing condition that led to their isolation. The remaining unsequenced 187 isolates endure in the culture collection for further genetic and functional exploration. The sequenced culture collection yielded five microbial phyla: Proteobacteria (78.5% of all isolates), Firmicutes (15.9%), Bacteroidetes (3.62%), Actinobacteria (0.987%) and Ascomycota (0.987%; Fig. 1 and Table S3, Supporting Information). Alignment of these sequences within the SILVA 106 database in ARB (Ludwig 2004; Quast et al. 2012) revealed 50 bacterial genera contained within 15 bacterial orders, and alignment with the ISHAM ITS database (Irinyi et al. 2015) revealed two fungal genera found within two additional orders represented in the culturable surface microbiomes of two whale species and three coral species.
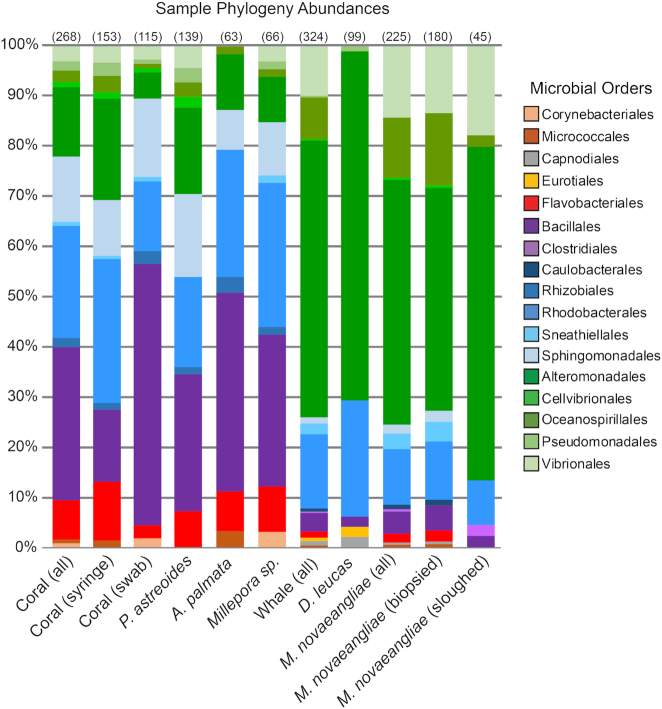
Most prevalent in coral samples were the presence of members of the Flavobacteriales, Bacillales, Rhodobacterales and Sphingomonadales microbial orders (Fig. 2). In the whale samples, Alteromonadales were the most prominent (Fig. 2). Of the 50 bacterial genera identified in this study, 36% are unique to a single animal surface. However, commonalities exist between sampled species, including genera Sulfitobacter, Alteromonas, Marinobacter and Neptuniibacter, which were found to occur in between 60 and 80% of all samples examined (Fig. 3). Microorganisms associated with the skin of D. leucas were observed to record the lowest OTU richness (Fig. 2).
Figure 2.
Percent abundance of microbial orders for each sample type. Abundances of operational taxonomic units at the level of microbial order are shown. Orders of the same class are clustered by color. Only OTUs with abundance values above 0.1% are shown. The number in parentheses above each column indicates the number of SSU rRNA sequences used in the analysis for each sample type. The number in parentheses next to each sample type is the number of samples. See Table S1 (Supporting Information) for more sample information.
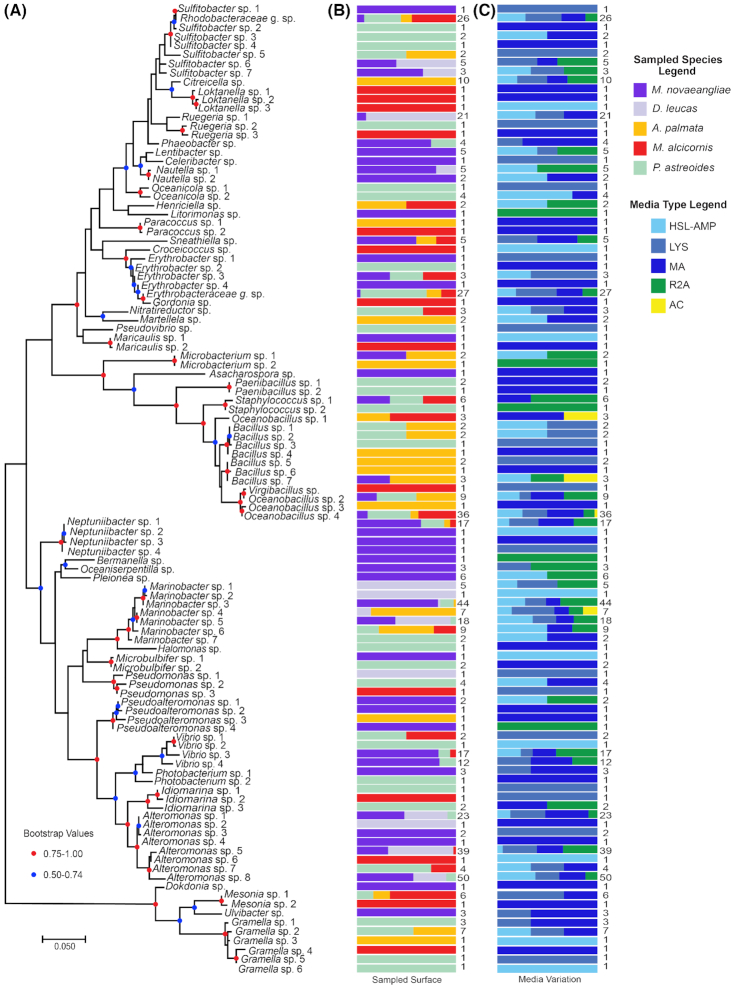
Figure 3.
Collapsed bacterial phylogenetic tree comparing sample type and selection media. Maximum likelihood phylogenetic tree with condensed external nodes, representing the 587 bacterial SSU rRNA sequences in the dataset. The analysis involved 109 sequences and 417 informative nucleotide positions. (A) Each node is designated by a bacterial genus or family that represents the taxonomic composition of the node. (B) Colored bars indicate the % contribution of each sampled surface type attributed to the isolates represented by the external node to the left. The number to the right of each bar indicates the number of bacterial isolates represented by each node in part A. (C) Colored bars indicate the % contribution of each bacterial selection media attributed to the isolates represented in part A. The number to the right of each bar is the number of bacterial isolates represented by each node.
Cultured isolates associated with the coral syringe sampling method were more diverse and had higher OTU richness than cultured isolates associated with coral surface swab isolates. This diversity and richness are found both within and across the three coral species (Table 1). Firmicutes dominated the syringe samples while surface swabs hosted more bacteria in the Bacteroidetes and Proteobacteria phyla.
Of the 109 OTUs isolated in this study, 54.1% were isolated from the MA media variation only (Fig. 3C). Additionally, the culture media selected for potentially phylogenetically distinct microorganisms, even within the same genus. For example, Gramella sp. 5 and Gramella sp. 6 were both isolated from P. astreoides, but the use of ‘LYS’ and ‘HSL-AMP’ media variations selected for strains with distinct SSU rRNA sequences (Fig. 3C). Bacillus sp. 4 and Bacillus sp. 5 were both isolated from A. palmata, but the use of ‘MA’ and ‘LYS’ media selected for strains with distinct SSU rRNA sequences (Fig. 3C), suggesting closely related isolates could be responding to unique attributes of the culture media.
MA media type consistently supported the growth of diverse isolates (Fig. 3). For example, many isolates within genera Paracoccus and Paenibacillus were isolated from MA alone. Among the four media types producing the greatest abundance and diversity of bacterial isolates, MA media produced the highest OTU richness and R2A produced the lowest OTU richness (Table 1).
Phylogenetic analyses of host-microorganism associations
For numerous bacterial genera where multiple OTUs are present, OTUs with divergent SSU rRNA sequences were isolated from distinct animal hosts (Fig. 3B). For example, 14 isolates can be binned into one of six unique OTUs within the genus Gramella. A total of two OTUs within this genus were isolated exclusively from either A. palmata or M. alcicornis from HSL-AMP and MA media types, respectively. A total of 12 genera were cultivated distinctly from a single surface/sample type. For example, the genus Citreicella was isolated only from A. palmata. The media variation used, however, did not select for significantly different strains of Citreicella, as isolates from this genus were cultured from four of the five bacterial media types (Fig. 3). Additionally, isolates within genera Neptuniibacter, Bermanella, Oceaniserpentilla, Pleionea, Litorimonas, Asacharospora, Lentibacter, Celebribacter and Ulvibacter were isolated from only humpback whales, highlighting the potential uniqueness of this surface type (Fig. 3).
Multivariate analyses of cultured microbial community composition
For the first NMDS ordination of objects from all sampled species, the ordination was performed on a reduced dataset containing only 11 microbial families, since the low OTU richness in the D. leucas samples inhibited model convergence with all 21 microbial families (Figure S1, Supporting Information). Due to the loss of data from the resulting reduced dataset and potential skewed multivariate tests of group differences, downstream analyses focused on species yielding high OTU richness, therefore excluding the D. leucas samples. The second NMDS analysis ordinated 79 objects from four sampled species (A. palmata, M. alcicornis, M. novaeangliae andP. astreoides) cultured with four media conditions (HSL-AMP, LYS, MA and R2A; Fig. 4). The scree plot indicated three NMDS dimensions as the optimum number of dimensions, and the ordination containing three dimensions produced a stress value of 0.106. NMDS validation showed a close relationship between ordination distance and observed dissimilarity with a non-metric R2 of 0.989 and a linear R2 of 0.927.
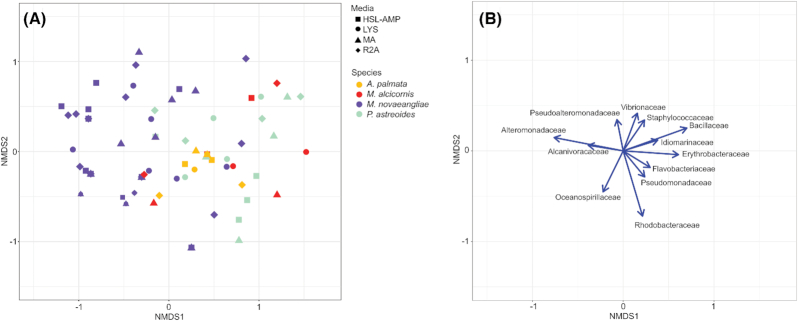
Figure 4.
Non-metric multidimensional scaling (NMDS) visualization of cultured microbial community composition across sampled species. A. Ordination of the 79 species/media combination objects in multivariate space of sampled species yielding high OTU richness. Each object color indicates the sampled species, and the object shape indicates the associated culture condition. B. Bacterial family variable loadings on NMDS axes 1 and 2. The length of the vectors indicates the strength of the associated variable for describing the NMDS axes. The direction of the vectors indicates the direction of the associated variable gradients in ordination space. Only variable loadings (microbial families) contributing significantly (P < 0.01) to the ordination are included.
Alteromonadaceae, Bacillaceae, Erythrobacteraceae, Idiomarinaceae, Oceanospirillaceae, Rhodobacteraceae, Staphylococcaceae and Vibrionaceae families contributed most significantly to the NMDS ordination of all species samples (P < 0.001; Fig. 4B). Microbial communities cultured from M. novaeangliae samples more often contained microbes from Alteromonadaceae and Alcanivoracaceae families, while microbial communities cultured from the three corals species more often contained microbes from Erythrobacteraceae and Bacillaceae families (Fig. 4B).
The global perMANOVA test comparing cultured microbial communities across the four species (A. palmata, M. alcicornis, M. novaeangliae andP. astreoides) showed statistically significant differences in microbial community composition between species (0.001 P-value and 0.185 R2). Additionally, pair-wise perMANOVA tests comparing each species directly found statistically significant differences (P-value < 0.05) in microbial community composition between M. novaeangliae and each coral species, as well as between P. astreoides and A. palmata (Table 2). The multivariate test of dispersion for the global comparison between the four species indicated a significant difference in dispersion between groups (0.004 P-value and 4.93 F-statistic). Pair-wise multivariate tests of dispersion directly comparing each species indicated significant differences in dispersion between A. palmata and all three other species (Table 2).
Table 2.Multivariate tests for comparison of group differences and multivariate dispersion. Results of pair-wise perMANOVA tests and pair-wise multivariate dispersion tests between four sampled species and four media variations. Bottom diagonals show the results of pair-wise perMANOVA tests between each group. Each cell contains the R2 and the associated P-value in parenthesis. Upper diagonals show the results of pair-wise multivariate tests of dispersion between each group. Each cell contains the t-statistic and the associated P-value in parenthesis. Significant and marginally significant test statistics are bolded (P-value < 0.05*; P-value < 0.001**) A. Multivariate tests comparing the four sampled species. B. Multivariate tests comparing the four media variations producing the highest OTU richness.
| A | A. palmata | M. alcicornis | M. novaeangliae | P. astreoides | |
|---|---|---|---|---|---|
| A. palmata | n/a | −2.97 (0.010)* | −3.40 (0.001)** | −3.14 (0.005)* | |
| M. alcicornis | 0.084 (0.192) | n/a | 0.71 (0.482) | 0.79 (0.436) | |
| M. novaeangliae | 0.098 (0.001)** | 0.086 (0.001)** | n/a | 0.05 (0.959) | |
| P. astreoides | 0.085 (0.015)* | 0.019 (0.974) | 0.145 (0.001)** | n/a | |
| B | HSL-AMP | LYS | MA | R2A | |
| HSL-AMP | n/a | 0.75 (0.458) | 0.12 (0.907) | −1.22 (0.230) | |
| LYS | 0.030 (0.328) | n/a | −0.62 (0.536) | −2.27 (0.029)* | |
| MA | 0.015 (0.800) | 0.017 (0.673) | n/a | −1.36 (0.181) | |
| R2A | 0.017 (0.731) | 0.041 (0.105) | 0.023 (0.498) | n/a |
The global perMANOVA test comparing cultured microbial communities across the four media variations (HSL-AMP, LYS, MA and R2A) showed no significant difference in microbial community compositions between media variation (0.555P-value, 0.036 R2). Additionally, pair-wise perMANOVA tests comparing each media variation directly found no significant differences between media variations (Table 2). The global multivariate test of dispersion found no significant difference in dispersion between media variations (0.277 P-value, 1.41 F-statistic).
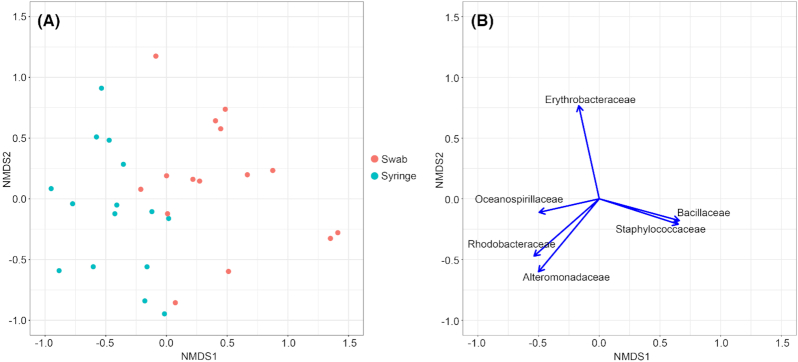
The third NMDS analysis ordinated 32 objects from the three sampled coral species (A. palmata, M. alcicornis and P. astreoides) to compare differences in microbial composition based on coral sampling method (Fig. 5). The scree plot indicated three NMDS dimensions as the optimum number of dimensions, and the ordination containing three dimensions produced a stress value of 0.120. NMDS validation showed a close relationship between ordination distance and observed dissimilarity with a non-metric R2 of 0.987 and a linear R2 of 0.905. Alteromonadaceae, Bacillaceae, Erythrobacteraceae, Rhodobacteraceae and Staphylococcaceae families contributed most significantly to the NMDS ordination of coral samples (P < 0.001; Fig. 5B). Microbial communities cultured from coral swab samples more often contained microbes from Oceanospirillaceae, Rhodobacteraceae and Alteromonadaceae families, while microbial communities cultured from coral syringe samples more often contained microbes from Bacillaceae and Staphylococcaceae families (Fig. 5B).
Figure 5.
Non-metric multidimensional scaling (NMDS) visualization of cultured microbial community composition across coral sampling methods. A. Ordination of the 32 species/media combination objects in multivariate space of coral species. Each object color indicates the coral surface sampling method. B. Bacterial family variable loadings on NMDS axes 1 and 2. The length of the vectors indicates the strength of the associated variable for describing the NMDS axes. The direction of the vectors indicates the direction of the associated variable gradients in ordination space. Only variable loadings (microbial families) contributing significantly (P < 0.01) to the ordination are included.
The perMANOVA test comparing cultured microbial communities between the two coral sampling methods (syringe and swab) showed statistically significant differences in microbial community composition between sample methods (0.001 P-value, 0.151 R2). The multivariate test of dispersion found no significant difference in dispersion between sampling methods (0.397 P-value, 0.74 F-statistic).
DISCUSSION
This study uses an intensive culturing effort to isolate 592 microbes comprising 50 bacterial genera within 15 orders, from the surface microbiomes of two species of whales and three species of corals, demonstrating the effectiveness of using our methodology and suite of culture conditions in capturing microbial diversity from diverse marine animals (Table S2, Supporting Information). A wealth of knowledge can be gained by using bacterial strains in manipulative experiments to understand disease origin; community dynamics relating to resistance, resilience and persistence; functional roles of microbiome members; and impacts on host health. However, investigations have been severely limited by the availability of phylogenetically diverse microbes in culture as efforts to recover and isolate marine microbes from environmental samples struggle to overcome microbial culturing challenges (Dance 2020). This study is also the first to develop a comprehensive collection of cultured bacteria associated with the skin of healthy cetaceans, supplementing the growing body of literature examining the culturable bacteria of cetacean respiratory and digestive systems (Buck et al. 2006; Venn-Watson, Smith and Jensen 2008; Morris et al. 2011; Godoy-Vitorino et al. 2017). Additionally, efforts outlined here yielded a cultured microbial collection, from coral tissue and mucus, greater in phylogenetic diversity and total number of strains recovered than catalogued previously (Lampert et al. 2006; Chimetto et al. 2008; Shnit-Orland and Kushmaro 2009; Galkiewicz et al. 2011). Moreover, 36% of the isolates were unique to a single animal surface, indicating our culturing technique is capable of capturing highly diverse bacterial isolates from diverse host organisms.
Strain specificity is a common feature in many symbiotic relationships, and strain-level genetic differences can exist in symbionts, particularly for bacteria (Bongrand and Ruby 2019; Apprill 2020). Culture-independent analyses of coral-associated bacteria find sub-genus phylogenetic clusters represented exclusively in one clade of Scleractinian coral, including strains within the genus Ruegeria, isolated in this study (Huggett and Apprill 2018). The potential for strain specificity to host origin is found in many instances in this study, since OTUs grouped by 99% similarity within the same bacterial genus were isolated solely from separate hosts (Fig. 3). These potential host-specific strains include bacteria within the genera Ruegeria, Paracoccus, Maricaulis, Pseudomonas, Pseudoalteromonas, Idiomarina and Gramella. A total of four of these genera—Ruegeria, Paracoccus, Idiomarina and Gramella—may contain distinct bacterial strains isolated exclusively from different coral species in the same environment. Further examination is needed to indicate if these associations are host-driven rather than environment-driven, and understanding the factors driving host-specific strain association requires interrogation of the host microscale environmental and bacterial species-specific metabolic characteristics and exchange to more fully resolve stable host-bacterial relationships (Ziemert et al. 2014; Mönnich et al. 2020). The microbial library produced in this study is poised to investigate the potential for metabolically selective, strain-specific symbioses. Future work should include a FISH-microscopy based approach to elucidate the micro-scale interactions these cells have with their animal surfaces.
Acquisition of bacteria detected in previous culture-independent efforts
Cetacean skin microbes isolated here resemble the main phyla identified through culture-independent methods from Tursiops sp. blow (Bik et al. 2016; Nelson et al. 2019), skin microbiota from Tursiops truncatus and Orcinus orca (Chiarello et al. 2017) and the oral microbiome of Delphinus delphis, Stenella coeruleoalba and Phocoena phocoena (Soares-Castro et al. 2019) which are also dominated by Proteobacteria, Firmicutes and Bacteroidetes. Additionally, at the family level, our study was successful in isolating representatives from the Roseobacter clade, one of the most dominant groups of surface colonizers previously identified from the skin of killer whales (Chiarello et al. 2017). However, isolates that we obtained from humpback whale skin through cultivation are phylogenetically distinct from core members of the skin of this same species as identified through previous cultivation-independent surveys. Although phylogenetic analyses of humpback whale microbiomes reveal the ubiquitous and abundant presence of Tenacibaculum sp. and Psychrobacter sp. across humpback whale populations, other less-abundant microbial genera exhibit temporal shifts in presence and absence, throughout the foraging season and between whales inhabiting different geographic regions (Apprill et al. 2014; Bierlich et al. 2017). Of these less abundant genera, we did have success in cultivating 14 of the 43 (approx. 32.5%) differentially present bacterial genera found to be distinct from the core microbial members (14 of 49 (29%) of all bacterial genera identified); (Bierlich et al. 2017). Understanding these temporally and geographically variable bacterial–host associations can provide insight into the ecological roles of the microorganisms and the biochemical and environmental drivers of microbial community shifts.
We were also able to culture 268 bacterial isolates from coral surface swabs and syringe samples (115 and 153 isolates, respectively). Of these isolates, the most frequently recovered species from coral swab and syringe samples were members in the orders Flavobacteriales, Bacillales, Rhodobacterales, Alteromonadales and Sphingomonadales (Fig. 2). Each of these bacterial orders have been previously identified in surveys of 16S rRNA genes from the microbiomes of Caribbean corals (Sunagawa, Woodley and Medina 2010; Morrow et al. 2012; Kimes et al. 2013; Ainsworth et al. 2015; Apprill, Weber and Santoro 2016). Furthermore, representatives of the family Rhodobacteraceae and the genera Bacillus, Erythrobacter, Gramella, Marinobacter, Neptuniibacter and Oceanobacillus were recovered from all three coral species studied, and each of these genera have been documented on P. astreoides and A. palmata (Fig. 3 and Table S2, Supporting Information; Sharp, Distel and Paul 2012; McDevitt-Irwin et al. 2017). Roseobacter clade-associated bacteria, members of the family Rhodobacteraceae, and the genus Marinobacter are known to associate with P. astreoides throughout the coral life cycle, and these bacteria may be passed down from coral parent to larvae (Sharp, Distel and Paul 2012), which could be a mechanism to retain select metabolically distinct bacterial strains. Marine Roseobacter strains are known to facilitate coral settlement (Sharp et al. 2015). Additionally, we were able to isolate representatives from Halomonas, Hyphomonas, Microbulbifer, Paenibacillus, Photobacterium, PseudovibrioandOceanicola (Table S2, Supporting Information) frequently found on both healthy and diseased P. astreoides colonies (McKew et al. 2012; Meyer, Paul and Teplitski 2014; Staley et al. 2017).
Multivariate analyses highlight variation in cultured microbial communities
The non-metric multidimensional (NMDS) ordination and perMANOVA tests of group differences indicated that cultured microbial community composition differs across sampled species rather than across culture condition (Fig. 4A). However, the global group difference across sampled species is largely driven by the difference between M. novaeangliae and the three coral species. Significant differences between coral species are likely driven by differences in multivariate dispersion, since cultured microbial communities from A. palmata samples were relatively similar and less disperse between samples. This result suggests that variation in cultured microbial community composition is associated with differences in environment and biological characteristics of the species surface, rather than culture media composition, like nutrient availability or the presence of quorum sensing molecules, for example. Despite their use for targeted cultivation, media variations did not select for particular bacterial clades (Fig. 4A). Additionally, the AC media, designed to select for microorganisms within the Actinobacteria phylum, did not select for any bacteria within this phylum and instead allowed for the growth of three OTUs within the order Bacillales. The Bacillales order contains many spore-producing organisms. It is possible that the heat shock may have selected for heat-resistant spores that structurally, metabolically and functionally differ from vegetative cells (Gopal et al. 2015).
Due to time constraints, microbial biomass from D. leucas samples were cryopreserved before colony picking, likely selecting for microbes with rapid growth. The bias induced by the microbial isolation method for the D. leucas samples produced low microbial OTU richness and resulted in a less robust dataset that inhibited model convergence in multivariate analyses. This bias underscores the importance of immediate microbial isolation upon surface sampling.
The NMDS ordination and perMANOVA test of group differences indicated that cultured microbial community composition differs between coral sampling method, with swab samples enriched with microbes in the Oceanospirillaceae, Rhodobaccteraceae and Alteromonadaceae families and syringe samples enriched with microbes from Bacillaceae and Staphylococcaceae families (Fig. 5). The syringe sampling method more likely recovers microorganisms contained in the coral mucus, while the swabbing method more likely recovers microorganisms attached to the coral surface. Previous work suggests the existence of specific coral mucus-associated bacteria that regulate mucus layer bacterial populations through antimicrobial activities (Ritchie and Smith 1997; Shnit-Orland and Kushmaro 2009), and future inquiries should investigate the mechanisms that mediate divergent microbial communities across coral surface micro-environments. Additionally, coral syringe samples maintain greater phylogenetic diversity than coral swabs, potentially because the mucosal layer is frequently colonized by bacteria carried to the animal via sediments (Apprill, Weber and Santoro 2016; Glasl, Herndl and Frade 2016). This greater phylogenetic diversity in the syringe samples (e.g. mucosal layer only) is reflected in the orders of bacteria most frequently recovered from each set of samples (Fig. 2). Greater microbial diversity in the coral mucus-only samples may also be due to the syringe method of collection. Coral micro-habitats sampled using this method may dislodge Symbiodinaceae from the coral surface, and therefore bacteria associated with these cells may also be represented in the data.
Culture-dependent design introduces bias in recovered microorganisms
Many of the culturable bacteria from both whale and coral species included Alteromonas sp. (Fig. 2), which are globally distributed marine copiotrophic bacteria that dominate in heterotrophic blooms and outcompete other bacteria due to their rapid growth when organic nutrients are readily available (Math et al. 2012). The ubiquity of this genus in our whale samples could have resulted due to the nutrient-rich media base used during cultivation, which has previously led to an overestimation of their abundance in the environment due to the ease at which the cells form colonies on agar plates and the voracity with which they take advantage of sporadic inputs of organic matter (Behringer et al. 2018). However, our data indicate that a fraction of the 127 Alteromonas strains isolated here were only found on coral or on cetaceans, indicating potential host specificity for particular strains. Additionally, isolates that are phylogenetically similar can exhibit different biochemical profiles and functional roles (Lampert et al. 2006). Culture independent studies by themselves would likely fail to link varied contributions of similar strains to larger implications, such as their effects on host fitness. Future studies should integrate more diverse media types, particularly low nutrient conditions, to target more slower growing microorganisms that might have been overlooked in this study.
CONCLUSION
The dermis represents both a first line of defense for marine animals, and an environment uniquely colonized by phylogenetically and functionally diverse bacterial representatives. The challenge for marine microbial ecology remains in linking the phylogenetic diversity of host-associated microbes to their functional roles within the community. Progress will depend on merging data from sequencing-based studies with manipulative experiments using cultured isolates, thereby providing insight on nutrient cycling capacity, interspecies communication, defense against pathogens and parasites, host immune response, wound repair, microbial community succession and benefits to host development, health and survival (Krediet et al. 2013; Meyer, Paul and Teplitski 2014; Medina et al. 2017; Bell, Garland and Alford 2018; Longford et al. 2019; van Oppen and Blackall 2019). Moreover, culture-based methods have already proven successful in conservation efforts in terrestrial ecosystems (Daskin et al. 2014; Loudon et al. 2014) and could prove equally as valuable in marine systems (van Oppen and Blackall 2019). Our culturable library holds great promise for future manipulative experiments that can examine the metabolic factors and surface properties influencing complementary associations between animal host and microbe, as our study was able to capture a rich repository of marine microbes and inspires new questions that will undoubtedly fuel future research into the varied and elusive roles of marine microbes.
Supplementary Material
ACKNOWLEDGEMENTS
Funding was provided by the National Science Foundation (Biological Oceanography) award #1657808 and National Institutes of Health grants 1R21-AI119311–01 to K. E. Whalen, as well as funding from the Koshland Integrated Natural Science Center and Green Fund at Haverford College. This constitutes scientific manuscript #298 from the Sea Research Foundation. The authors would also like to thank Jenn Tackaberry, Scott Landry and the CCS Marine Animal Entanglement Response Program for their assistance with sample collection.
Contributor Information
Abigail G Keller, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Amy Apprill, Marine Chemistry & Geochemistry Department, Woods Hole Oceanographic Institution, 266 Woods Hole Road, Woods Hole, MA, 02543, USA.
Philippe Lebaron, Laboratoire de Biodiversité et Biotechnologies Microbiennes, USR 3579 Sorbonne Université (UPMC) Paris 6 et CNRS Observatoire Océanologique, Banyuls-sur-Mer, France.
Jooke Robbins, Center for Coastal Studies, 5 Holway Ave., Provincetown, MA, 02657, USA.
Tracy A Romano, Mystic Aquarium, a division of Sea Research Foundation Inc., 55 Coogan Blvd., Mystic, CT, 06355, USA.
Ellysia Overton, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Yuying Rong, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Ruiyi Yuan, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Scott Pollara, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Kristen E Whalen, Department of Biology, Haverford College, 370 Lancaster Ave., Haverford, PA, 19041-1392, USA.
Conflicts of interest
None declared.
REFERENCES
- Ainsworth TD, Krause L, Bridge Tet al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol Lett. 2006;9:683–93. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for nonparametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Apprill A, Miller CA, Van Cise AMet al. Marine mammal skin microbiotas are influenced by host phylogeny. R Soc Open Sci. 2020;7:192046. 10.1098/rsos.192046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill A, Robbins J, Eren AMet al. Humpback whale populations share a core skin bacterial community: towards a health index for marine mammals?. PLoS One. 2014;9:e90785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill A, Weber LG, Santoro AE. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. MSystems. 2016;1:e00143–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apprill A. Marine animal microbiomes: toward understanding host–microbiome interactions in a changing ocean. Front Mar Sci. 2017;4:222. [Google Scholar]
- Apprill A. The role of symbioses in the adaptation and stress responses of marine organisms. Ann Rev Mar Sci. 2020;12:291–314. [DOI] [PubMed] [Google Scholar]
- Baroni-Urbani C, Buser MW. Similarity of binary data. Syst Zool. 1976;25:251–9. [Google Scholar]
- Barott KL, Rodriguez-Brito B, Janouškovec Jet al. Microbial diversity associated with four functional groups of benthic reef algae and the reef-building coral Montastraea annularis: microbial diversity on benthic algae and corals. Environ Microbiol. 2011;13:1192–204. [DOI] [PubMed] [Google Scholar]
- Behringer G, Ochsenkühn MA, Fei Cet al. Bacterial communities of diatoms display strong conservation across strains and tme. Front Microbiol. 2018;9:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SC, Garland S, Alford RA. Increased numbers of culturable inhibitory bacterial taxa may mitigate the effects of Batrachochytrium dendrobatidis in Australian wet tropics frogs. Front Microbiol. 2018;9:1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierlich KC, Miller C, DeForce Eet al. Temporal and regional variability in the skin microbiome of humpback whales along the Western Antarctic Peninsula. Appl Environ Microbiol. 2017;84:e02574–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bik EM, Costello EK, Switzer ADet al. Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat Commun. 2016;7:10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongrand C, Ruby EG. Achieving a multi-strain symbiosis: strain behavior and infection dynamics. ISME J. 2019;13:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart GD. Marine mammals as sentinel species for oceans and human health. Wildl Mar Anim. 2011;48:676–90. [DOI] [PubMed] [Google Scholar]
- Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol. 2016;70:317–40. [DOI] [PubMed] [Google Scholar]
- Bruns A, Cypionka H, Overmann J. Cyclic AMP and Acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the Central Baltic Sea. Appl Environ Microbiol. 2002;68:3978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck JD, Wells RS, Rhinehart HLet al. Aerobic microorganisms associated with free-ranging bottlenose dolphins in coastal gulf of Mexico and Atlantic ocean waters. J Wildl Dis. 2006;42:536–44. [DOI] [PubMed] [Google Scholar]
- Burke C, Thomas T, Lewis Met al. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011;5:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh Jet al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarello M, Villéger S, Bouvier Cet al. Captive bottlenose dolphins and killer whales harbor a species-specific skin microbiota that varies among individuals. Sci Rep. 2017;7:15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimetto LA, Brocchi M, Thompson CCet al. Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst Appl Microbiol. 2008;31:312–9. [DOI] [PubMed] [Google Scholar]
- Cooper MB, Smith AG. Exploring mutualistic interactions between microalgae and bacteria in the omics age. Curr Opin Plant Biol. 2015;26:147–53. [DOI] [PubMed] [Google Scholar]
- Dance A. The search for microbial dark matter. Nature. 2020;582:301–3. [DOI] [PubMed] [Google Scholar]
- Dang H, Lovell CR. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev. 2016;80:91–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskin JH, Bell SC, Schwarzkopf Let al. Cool temperatures reduce antifungal activity of symbiotic bacteria of threatened amphibians – implications for disease management and patterns of decline. PLoS One. 2014;9:e100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digby PGN, Kempton RA. Multivariate Analysis of Ecological Communities. Chapman and Hall, 1987. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs EL, Brutinel ED, Klem ERet al. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J Bacteriol. 2010;192:2779–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkiewicz JP, Pratte ZA, Gray MAet al. Characterization of culturable bacteria isolated from the cold-water coral Lophelia pertusa: lophelia cultured bacteria. FEMS Microbiol Ecol. 2011;77:333–46. [DOI] [PubMed] [Google Scholar]
- Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Vitorino F, Rodriguez-Hilario A, Alves ALet al. The microbiome of a striped dolphin (Stenella coeruleoalba) stranded in Portugal. Res Microbiol. 2017;168:85–93. [DOI] [PubMed] [Google Scholar]
- Gopal N, Hill C, Ross PRet al. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front Microbiol. 2015;6. DOI: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SMet al. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10:641–54. [DOI] [PubMed] [Google Scholar]
- Hooper R, Brealey JC, Valk Tet al. Host-derived population genomics data provides insights into bacterial and diatom composition of the killer whale skin. Mol Ecol. 2019;28:484–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett MJ, Apprill A. Coral microbiome database: Integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ Microbiol Rep. 2018;11:372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irinyi L, Serena C, Garcia-Hermoso Det al. International society of human and animal mycology (ISHAM)-ITS reference DNA barcoding database—The quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol. 2015;53:313–37. [DOI] [PubMed] [Google Scholar]
- Kimes NE, Johnson WR, Torralba Met al. The Montastraea faveolata microbiome: ecological and temporal influences on a Caribbean reef-building coral in decline: the Montastraea faveolata microbiome. Environ Microbiol. 2013;15:2082–94. [DOI] [PubMed] [Google Scholar]
- Kjer J, Debbab A, Aly AHet al. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat Protoc. 2010;5:479–90. [DOI] [PubMed] [Google Scholar]
- Krediet CJ, Ritchie KB, Paul VJet al. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc R Soc B Biol Sci. 2013;280. 10.1098/rspb.2012.2328. DOI: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert Y, Kelman D, Dubinsky Zet al. Diversity of culturable bacteria in the mucus of the Red Sea coral Fungia scutaria. FEMS Microb Ecol. 2006;58:99–108. [DOI] [PubMed] [Google Scholar]
- Longford SR, Campbell AH, Nielsen Set al. Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci Rep. 2019;9:1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon AH, Woodhams DC, Parfrey LWet al. Microbial community dynamics and effect of environmental microbial reservoirs on red-backed salamanders (Plethodon cinereus). ISME J. 2014;8:830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Math RK, Jin HM, Kim JMet al. Comparative genomics reveals adaptation by alteromonas sp. SN2 to marine tidal-flat conditions: cold tolerance and aromatic hydrocarbon metabolism. PLoS One. 2012;7:e35784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt-Irwin JM, Baum JK, Garren Met al. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci. 2017;4:262. [Google Scholar]
- McGarigal K. “Biostats” R package. 2009. https://www.umass.edu/landeco/teaching/ecodata/labs/biostats.pdf [Google Scholar]
- McKew BA, Dumbrell AJ, Daud SDet al. Characterization of geographically distinct bacterial communities associated with coral mucus produced by Acropora spp. and Porites spp. Appl Environ Microbiol. 2012;78:5229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D, Walke JB, Gajewski Zet al. Culture media and individual hosts affect the recovery of culturable bacterial diversity from amphibian skin. Front Microbiol. 2017;8:1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One. 2014;9:e100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Jensen PR, Kauffman CAet al. Widespread and persistent populations of a major new marine Actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris PJ, Johnson WR, Pisani Jet al. Isolation of culturable microorganisms from free-ranging bottlenose dolphins (Tursiops truncatus) from the southeastern United States. Vet Microbiol. 2011;148:440–7. [DOI] [PubMed] [Google Scholar]
- Morrow KM, Moss AG, Chadwick NEet al. Bacterial associates of two caribbean coral species reveal species-specific distribution and geographic variability. Appl Environ Microbiol. 2012;78:6438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton M, Botha A. Cutaneous lesions in cetaceans: an indicator of ecosystem status?In: Romero A, Keith EO (eds), New Approaches to the Study of Marine Mammals, 2012, 123–51. [Google Scholar]
- Mönnich J, Tebben J, Bergemann Jet al. Niche-based assembly of bacterial consortia on the diatom Thalassiosira rotula is stable and reproducible. ISME J. 2020;14:1614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson T, Wallen M, Bunce Met al. Detecting respiratory bacterial communities of wild dolphins: implications for animal health. Mar Ecol Prog Ser. 2019;622:203–17. [Google Scholar]
- Nelson TM, Apprill A, Mann Jet al. The marine mammal microbiome: current knowledge and future directions. Microbiol Aust. 2015;36:8. [Google Scholar]
- Okami Y, Hotta K. Search and discovery of new antibiotics. In: Goodfellow M, Williams ST, Mordarski M (eds), Actinomycetes in Biotechnology, 1988, 33–67. [Google Scholar]
- Oksanen J. Vegan: Ecological Diversity. R Package Version 2.5-2. 2018, https://cran.rproject.org/web/packages/vegan/index.html. [Google Scholar]
- Ono K, Oka R, Toyofuku Met al. CAMP signaling affects irreversible attachment during biofilm formation by Pseudomonas aeruginosa PAO1. Microb Environ. 2014;29:104–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsbøll PJ, Larsen F, Sigurd Hansen E. Sampling of skin biopsies from free-ranging large cetaceans in West Greenland: development of new biopsy tips and bolt designs. Rep Int Whal Commn (Special Issue). 1991;13:71–9. [Google Scholar]
- Pruesse E, Quast C, Knittel Ket al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz Pet al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie KB, Smith GW, Physiological comparison of bacterial communities from various species of scleractinian corals. In: Proceedings of the Eight International Coral Reef Symposium, 1. Panama City, PA: Smithsonian Tropical Research Institute, 1997, 521–6. [Google Scholar]
- Rosenberg E, Koren O, Reshef Let al. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol. 2007;5:355–62. [DOI] [PubMed] [Google Scholar]
- Ross AA, Rodrigues Hoffmann A, Neufeld JD. The skin microbiome of vertebrates. Microbiome. 2019;7:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouco M, Haley ST, Dyhrman ST. Microbial diversity within the Trichodesmium holobiont: trichodesmium holobiont diversity. Environ Microbiol. 2016;18:5151–60. [DOI] [PubMed] [Google Scholar]
- Seegers U, Meyer W. A preliminary approach to epidermal antimicrobial defense in the Delphinidae. Mar Biol. 2004;144:841–4. [Google Scholar]
- Sharp KH, Distel D, Paul VJ. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 2012;6:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Sneed JM, Ritchie KBet al. Induction of larval settlement in the reef coral Porites astreoides by a cultivated marine Roseobacter Strain. Biol Bull. 2015;228:98–107. [DOI] [PubMed] [Google Scholar]
- Sheu S-Y, Lin K-Y, Chou J-Het al. Tenacibaculum litopenaei sp. Nov., isolated from a shrimp mariculture pond. Int J Syst Evol Microbiol. 2007;57:1148–53. [DOI] [PubMed] [Google Scholar]
- Shnit-Orland M, Kushmaro A. Coral mucus-associated bacteria: a possible first line of defense: coral mucus-associated bacteria. FEMS Microbiol Ecol. 2009;67:371–80. [DOI] [PubMed] [Google Scholar]
- Sison-Mangus MP, Jiang S, Tran KNet al. Host-specific adaptation governs the interaction of the marine diatom, pseudo-nitzschia and their microbiota. ISME J. 2014;8:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Wilson JB. A consumer's guide to evenness indices. Oikos. 1996;76:70. [Google Scholar]
- Smith RS, Wolfgang MC, Lory S. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect Immun. 2004;72:1677–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Castro P, Araújo-Rodrigues H, Godoy-Vitorino Fet al. Microbiota fingerprints within the oral cavity of cetaceans as indicators for population biomonitoring. Sci Rep. 2019;9: 13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C, Kaiser T, Gidley MLet al. Differential impacts of land-based sources of pollution on the microbiota of southeast Florida coral reefs. Appl Environ Microbiol. 2017;83:e03378–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5:e9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen MJH, Blackall LL. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol. 2019;17:557–67. [DOI] [PubMed] [Google Scholar]
- Vega Thurber R, Willner-Hall D, Rodriguez-Mueller Bet al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11:2148–63. [DOI] [PubMed] [Google Scholar]
- Venn-Watson S, Smith C, Jensen E. Primary bacterial pathogens in bottlenose dolphins Tursiops truncatus: needles in haystacks of commensal and environmental microbes. Dis Aquat Organ. 2008;79:87–93. [DOI] [PubMed] [Google Scholar]
- Wahl M, Goecke F, Labes Aet al. The second skin: ecological role of epibiotic biofilms on marine organisms. Front Microbiol. 2012;3. DOI: 10.3389/fmicb.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ding W, Long Let al. Exploring the influence of signal molecules on marine biofilms development. Front Microbiol. 2020;11. DOI: 10.3389/fmicb.2020.571400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L, Gonzalez-Díaz P, Armenteros Met al. The coral ecosphere: a unique coral reef microbial habitat that fosters coral-microbial interactions. Limnol Oceanogr. 2019;64:2373–88. [Google Scholar]
- Wickham H, Chang W, Henry Let al. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. R Package Version 3.3.3. 2020, https://cran.r-project.org/web/packages/ggplot2/index.html [Google Scholar]
- Zaneveld JR, Burkepile DE, Shantz AAet al. Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nat Commun. 2016;7:11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz Met al. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci. 2014;111:E1130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.