Abstract
The Kidney Precision Medicine Project (KPMP) and other national efforts are collecting and integrating large disparate clinical, biotechnology, and imaging datasets to better understand and stratify kidney disease. Enabling these efforts, ontologies are powerful tools for organizing and making sense of different data elements and their relationships. Ontologies are critical for supporting the types of big data analysis necessary to conduct kidney precision medicine, where heterogeneous clinical, imaging, and biopsy data from diverse sources must be combined to define a patient’s phenotype. In this article, we demonstrate how reference ontologies and two KPMP-developed ontologies, the Kidney Tissue Atlas Ontology (KTAO) and the Ontology of Precision Medicine and Investigation (OPMI), will be used to support the creation of the Kidney Tissue Atlas. The KPMP ontologies can improve the concepts available for annotating kidney data, and revise existing definitions of kidney disease in support of precision medicine. We also provide a roadmap for how various ontologies, including KTAO and OPMI, can be used to support kidney disease modeling by the broader nephrology community.
1. Introduction
Precision medicine is broadly defined as the delivery of tailored interventions or treatments to individual patients, or “the right drug for the right patient at the right time.”1 The practice of precision medicine depends acutely on emergent high-throughput technologies capable of generating detailed molecular phenotypes in clinically obtained human biosamples. Molecular phenotypes provide an opportunity for more nuanced descriptions of disease, and methods for systematically incorporating this information for clinical care and discovery are needed.
Currently, kidney disease is frequently classified as acute kidney injury (AKI)2 or chronic kidney disease (CKD).3 These terms provide information about the duration of decreased kidney function or kidney damage but do not provide diagnostic specificity. Instead, these terms are better characterized as syndromes with many underlying causes. Existing classification criteria for AKI and CKD are designed to provide a standardized way to stage the severity of disease and cover a broad range of cases based on changes in serum creatinine, proteinuria, and urine output, but these criteria do not help clinicians identify causal factors that can be targeted for precision treatments. For example, a clinician could conclude that a patient has CKD Stage 3B based on their serum creatinine and urinary protein excretion, but the cause of the patient’s CKD could vary widely from diabetic kidney disease to medication side effects or myeloma-related kidney disease.
To reassess definitions of kidney disease and enable precision medicine, molecular phenotypes derived from high-throughput omics technology and detailed histopathological assessments must be combined with traditional clinical measurements. Harmonization and integration of these data require the development of common languages or ontologies. Ontologies adopted by the biomedical sciences provide computer-readable representations of entities of interest, such as anatomical structures, cells, molecules, genes, phenotypes, and diseases. These representations can be leveraged by scientists and engineers to build computational models and systems for knowledge integration and discovery. In other words, ontologies help to bridge the language barrier between humans and computers by encoding knowledge in a form that is accessible to both. By providing a controlled vocabulary, standardized definitions, and explicit relationships between terms, ontologies enable validation of new relationships and leverage structured terms and relationships in the development of predictive models. Ontology definitions in the form of both natural language and logical expressions are created and agreed upon by members of the community, and represent the state of shared knowledge within a field.
The Kidney Precision Medicine Project (KPMP) (https://kpmp.org/) aims to accelerate our understanding of the most common forms of kidney disease. The KPMP was initiated in 2017 with funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) to generate molecular and 3D imaging maps of reference kidneys and kidneys with AKI and CKD. Participants with AKI or CKD consent to generously provide biopsy tissue solely for research, along with detailed demographic, clinical, pathology, social history, and follow-up data.
A goal of the consortium is to create the Kidney Tissue Atlas, a comprehensive molecular, cellular, and anatomic map of the kidney, by combining state-of-the-art molecular and cellular analyses of kidney tissue with demographic, clinical, and histopathological data elements collected from participants. The Kidney Tissue Atlas will complement concurrent atlas projects, such as the Allen Brain Atlas,4 The Cancer Genome Atlas,5 Human BioMolecular Atlas Program (HuBMAP),6 and Human Cell Atlas.7 The integration of clinical and histopathological characteristics with deep molecular data in both reference and diseased tissue collected from participants will be used to define novel kidney disease categories and discover mechanistic drivers of these diseases. Ultimately, the Atlas is expected to provide the foundational knowledge necessary to develop new diagnostic tools and targeted therapies for the most common forms of kidney disease and injury.
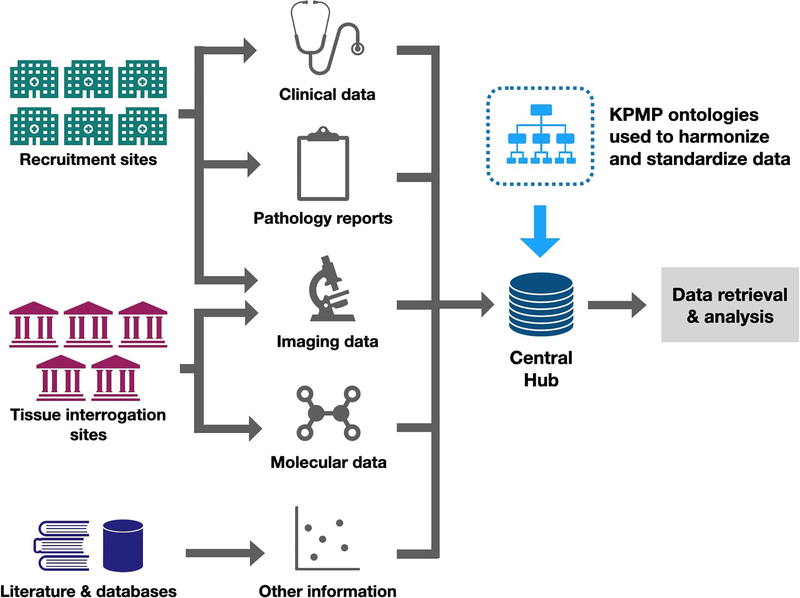
Ontologies are at the core of data integration and accessibility for the KPMP. To enable novel discoveries about kidney disease, the KPMP manages and integrates various types of data collected at its recruitment and tissue interrogation sites. FIG. 1 shows different data types collected by KPMP sites, and where these data are integrated and standardized using the KPMP suite of ontologies at the central hub (details in Sections 5 and 6). The data will be made publicly available at the Kidney Tissue Atlas data portal (https://atlas.kpmp.org/) following standardization. Ontologies will facilitate user access and analysis of the data, and allow data collected by KPMP to be searchable comprehensively and flexibly.
FIG. 1: Overview of KPMP centers and the flow of KPMP data from different provenances.
Clinical data and pathology reports from recruitment centers, and molecular data and imaging data from tissue interrogation sites are integrated with data from the scientific literature and omics databases at the KPMP central hub using KPMP ontologies. Six recruitment sites enroll participants with common forms of acute and chronic kidney disease, and collect biosamples including a research kidney biopsy. Five tissue interrogation sites process and perform molecular analysis of participant biosamples. The central hub manages and aggregates data from all sites for systematic analysis, develops visualization tools and Application Programming Interfaces (APIs) to facilitate community access to data. Data harmonization and standardization with KPMP ontologies facilitate flexible data retrieval and analysis.
Due to the novel nature of the collected data and proposed analyses, new ontology terms and relationships must be introduced to model them. To bridge the gap in existing ontologies for annotating kidney-specific data, the KPMP is developing the Kidney Tissue Atlas Ontology (KTAO)8 for describing kidney anatomy, phenotypes, diseases, molecular features, and other kidney-related concepts. KTAO imports and seamlessly aligns terms from many pre-existing open biomedical ontologies and deepens the granularity of kidney-specific terms to facilitate biomedical research in kidney diseases. The KPMP also leads the effort in developing the community-based Ontology of Precision Medicine and Investigation (OPMI) to support data operations in the domain of precision medicine.
This article offers an introduction to ontologies for nephrology clinicians and researchers as well as a broad overview of ontological resources in the nephrology domain, which have not been used extensively by the nephrology community thus far. We then describe the development of the KTAO and OPMI ontologies to support kidney disease modeling. In this article, we:
Define ontologies and their role in the biomedical sciences, and how interoperable ontologies can support data integration in large biomedical projects (Sections 2 and 3),
Discuss existing ontology resources in the kidney domain and domains relevant to modeling kidney disease (Section 4),
Describe how KPMP ontologies and extensions to other ontologies are created to fill the gaps in kidney-specific data representation (Section 5), and
Provide an example of how these ontologies will be used to harmonize data and revise existing definitions of kidney disease (Section 6).
As ontologies are shared resources, we also discuss how the broader nephrology community can contribute to their development and use. We encourage others to adopt these open biomedical ontologies to annotate their data, making this data more interoperable (i.e., interconnected in a computer-understandable format) with other community resources, with the goal of increasing shared knowledge and producing rapid advancements in the diagnosis and treatment of common forms of kidney disease.
2. Ontologies and their roles in the biomedical sciences
Ontology is the study of the nature of entities and their relations in the real world.9 With the advent of “big data”, computer scientists and informaticists have adopted ontologies as a means to create computationally tractable models of entities and relationships within a domain. Ontology is a formal, structured, domain-specific, human- and computer-interpretable representation of these entities and relationships.10 Ontology is a foundation of knowledge representation and reasoning (KR2, KR&R), a major field of artificial intelligence (AI). Ontologies can be used to:
Represent established knowledge within a domain,
Maintain standardized vocabulary within a specific field of study, across multiple locations and datasets, as well as across different consortial efforts,
Allow automated computation and decision support over structured data, and
Facilitate the integration of data from distinct knowledge domains.
Ontologies share similarities with, but differ from, controlled vocabularies (which provide a set of terms used for document indexing) and taxonomies (which are controlled vocabularies with a hierarchical structure indicating subclass relationships between entities). Ontologies not only include a controlled vocabulary and a hierarchy, but also incorporate other semantic relationships, such as part_of or located_in, that provide additional information about the nature of a relationship between entities. Each term (or entity) in an ontology is described by its name, synonyms, attributes, and relationships to other concepts.
Though most nephrology clinicians and researchers do not currently interface with ontologies, they are incorporated seamlessly into biomedical research. For example, the Gene Ontology (GO)11 systematically classifies about 45,000 entities under biological processes, cellular components, and molecular functions of gene products for various organisms. GO was originally developed at the end of the 1990s by a consortium of researchers studying the genomes of three model organisms: fruit fly, mouse, and yeast. It was later also used to annotate genes from other organisms, including humans, plants, animals, and microbes. Since its publication in 2000, the first GO paper has been cited over 26,000 times. Without GO, it is impossible to generate consistent representation and annotation of the gene products from different organisms.
In addition to annotations, GO has been used in a variety of different applications, including the integration of annotated genomic data curated from the literature, the development of novel genomic analytic approaches such as gene expression functional enrichment analysis12 or Gene Set Enrichment Analysis (GSEA),13 and literature mining.14 Enrichment analyses similar to GSEA allow for better interpretation of otherwise uninterpretable or difficult to interpret big data. For example, high-throughput experiments for gene expression generate hundreds or sometimes thousands of differentially expressed genes (DEGs), and their associated biological functions can be summarized through enrichment analysis over GO terms.12 Gene-level annotations defined in GO can also be further elaborated into a network of biological pathway annotations via the recently developed GO Causal Activity Modeling (GO-CAM) models to represent the integrative effect of DEGs on the level of biological pathways. GO demonstrates the value of ontologies in establishing consistent annotation schemes for a class of biomedical entities. These annotations are able to interoperate and can be used to derive benefit in downstream analyses. The successes of GO have spurred the development of many hundreds of ontologies15 in other domains such as anatomy,16,17 proteins,18 or disease.19,20
3. Development and usage of interoperable Open Biomedical Ontologies
Given that biomedical ontologies have proliferated, issues of redundancy and diminished interoperability frequently occur.21 Although ontology matching algorithms22 have been developed with some success to map terms between different ontologies, this matching is usually based on features such as entity name or description rather than relational semantics. As a solution for this issue, the Open Biomedical Ontologies (OBO) Foundry consortium23 was established to achieve better ontology interoperability and resolve problems of overlapping representations across different biomedical ontologies. OBO ontologies are created and formatted following a set of shared principles maintained by the OBO consortium. These principles ensure that OBO ontologies remain open, orthogonal, interoperable, and logically well-formed with a well-specified syntax.
OBO currently includes over 160 biomedical ontologies in domains such as phenotype,24 disease,19,20 anatomy,16 genetics,11 and proteomics,18 among others. The OBO ontologies have been successfully applied to research questions in the biomedical domain.25 For example, the Human Phenotype Ontology (HPO) supports a deep phenotyping approach to define human diseases.24 The comparison of a patient’s phenotypic profile to the phenotypic elements of HPO supports the computational prediction of the likelihood that an individual has a particular disease.
Ontologies are commonplace in big biomedical projects. For example, The Library of Integrated Network-Based Cellular Signatures (LINCS) Program aims to create a network-based understanding of biology by profiling changes in gene expression and various cellular processes that occur after cells are exposed to perturbing agents.26 To systematically study these perturbed cell responses, LINCS heavily relies on ontologies to support standard representation and analysis.27,28 Another example is the Encyclopedia of DNA elements (ENCODE) project,29 which aims to identify and annotate functional elements in the human genome. It was a huge challenge to organize and standardize all ENCODE experimental data, metadata, and associated computational analyses. Ontologies played a significant role in organizing this data, and are used to support ontology-driven searches at the ENCODE portal.29,30
Interoperable, reliable, and community-driven OBO ontologies are also critical to support the seamless assembly and integration of kidney data from heterogeneous resources and domains. Because of its status as a sizable multi-center project, the KPMP faces challenges coordinating data across multiple sites and groups of personnel. Having a shared vocabulary for annotating patient information and tissue specimens at recruitment, and for summarizing molecular features and analytic results, is vital for maintaining the quality, interpretability, and reusability of KPMP data. The KPMP focuses on adapting and extending OBO ontologies to achieve this goal. Linking KPMP ontologies to other ontology resources (through observing OBO consortium principles and reusing terms in existing ontologies) will allow the work of the KPMP to benefit not only the kidney precision medicine study, but also the broader biomedical community.
4. Ontologies for modeling kidney disease
We provide a summary of ontological resources relevant to the kidney domain and how KPMP ontologies relate to these resources. We also describe gaps in existing ontologies and justify the creation and extension of resources to achieve the goals of the KPMP. In this section, we describe:
Existing kidney ontologies and classification/categorization systems,
Reference ontologies (community resources designed to be reused by multiple groups and stakeholders) relevant to modeling kidney knowledge, and
Gaps in existing ontological resources.
4.1. Prior work in kidney ontologies and classification/categorization systems
A number of kidney ontologies and classification systems have been introduced to support kidney research. We describe a few of these below, specifically the ontology of the Genitourinary Development Molecular Anatomy Project (GUDMAP),31 the Chronic Kidney Disease Ontology (CKDO),32 and the Renal Pathology Society (RPS)33–35. These projects have developed standardized terms in highly specific subareas of kidney physiology and disease modeling, like cell types (GUDMAP), or clinical diagnostic criteria of CKD (CKDO); however, none of them currently provide the framework to integrate the data types needed for precision medicine diagnostics and treatment.
The Genitourinary Development Molecular Anatomy Project (GUDMAP) consortium was formed in 2004 to create a molecular anatomical atlas of the developing mouse kidney and urogenital tract.36 One component of this project was the creation of an ontology of genitourinary developmental cell types anchored to mouse anatomy.31 The initial ontology was released in 200737 and has been created as an expansion of the Edinburgh Mouse Atlas Project (EMAP) ontology.38 The GUDMAP ontology was developed primarily to facilitate the annotation of murine cell types and has also evolved to include human fetal kidney and urinary tract data. Molecular cell types described in KPMP ontologies can be mapped to GUDMAP classes to facilitate the bridging of data collected across the lifespans of human and murine specimens. This mapping is part of future work to be done in collaboration with curators of the GUDMAP ontology.
The Chronic Kidney Disease Ontology (CKDO) is a clinically-oriented ontology designed to assist in the characterization and staging of CKD.32 The ontology primarily describes clinical features associated with CKD. For example, the editors define CKD based on associated clinical diagnostic codes, as well as based on abnormal laboratory/observational values such as decreased/elevated estimated glomerular filtration rate (eGFR) and proteinuria. The CKDO is a useful ontology for discovering and classifying patients in a clinical setting using defined stages of CKD. However, it lacks the ability to connect clinical descriptions to molecular phenotypes or anatomy.
While ontologies are not conventionally used by renal pathologists in clinical practice, the Renal Pathology Society (RPS) has undertaken several initiatives to standardize language and reporting, organize, categorize and stage kidney biopsies.33–35 These initiatives, although not formal ontologies, they may provide a helpful roadmap for the use of ontologized pathology features to drive novel classifications while maintaining an objective comparison to existing disease schemes. For example, a RPS working group has recently conducted an international consensus process to harmonize language, definition and metric (when relevant) for histologic and ultrastructural parameters across all currently used classification and scoring systems (manuscript under review). The RPS working group has worked closely with the KPMP pathology working group, and RPS harmonized terminology and definitions were then used, and enriched when needed, by the KPMP pathology working group, to provide the framework for anthologizing histologic and ultrastructural features.
4.2. Reference ontologies for the kidney precision medicine study
In TAB. 1, we provide a list of reference ontologies relevant to kidney anatomy, function, and disease. Reference ontologies are community resources designed to be reused by multiple groups and stakeholders. Each of these reference ontologies focuses upon a group of entities relevant to a particular subdomain. For example, human phenotypes are ontologized in the HPO24; Uber-anatomy ontology (UBERON)16 focuses on anatomical structure; and biological processes are represented by the GO11 (which connects molecular entities to cellular and tissue processes) and the Molecular Biology of the Cell Ontology (MBCO)39 (which describes interactions between molecular entities and subcellular processes). The content of clinical terminologies (SNOMED, ICD-9, etc.) is also mapped to KTAO through reference ontologies such as the Mondo Disease Ontology (MONDO),19 which includes mappings to these terminologies. When data are annotated with reference ontology terms, they can be easily integrated into the ecosystem of other datasets annotated with terms from the same ontologies.
TAB. 1:
Reference ontologies helpful for modeling the kidney.
| Domain | Ontology | Application to KPMP | # of entities | # of relationships |
|---|---|---|---|---|
| Phenotype | HPO (Human Phenotype Ontology)24 | Describe patient clinical and pathological phenotypes | 26,578 | 61,665 |
| Disease | MONDO (Mondo Disease Ontology)19 | Describe relations between patient observations and disease terminology | 111,478 | 136,833 |
| Anatomy | UBERON (Uber-anatomy ontology)16 | Describe aspects of renal anatomy | 15,183 | 43,082 |
| Cells and cell types | CL (Cell Ontology)40 | Describe cell types and cellular components relevant to modeling | 10,630 | 35,916 |
| Proteins | PRO (Protein Ontology)18 | Describe protein-related entities and the relations between these entities | 317,974 | 919,192 |
| Biological processes | GO (Gene Ontology)11 | Describe the association of molecular features and biological processes | 50,255 | 106,149 |
| Subcellular processes | MBCO (Molecular Biology of the Cell Ontology)39 | Describe subcellular processes (pathways) and their interactions leading to cell level functions | 6,136 | 19,932 |
| Lab measurements | OBI (Ontology for Biomedical Investigations)41 | Describe laboratory values related to patient diagnostics | 3,584 | 7,228 |
| Clinical measurements | CMO (Clinical measurement ontology)42 | Describe clinical measurements related to patient diagnostics | 3,054 | 3,718 |
| Adverse events | OAE (Ontology of Adverse Events)43 | Describe clinical features and comorbidities associated with patients | 5,700 | 11,572 |
| Chemical compounds | ChEBI (Chemical Entities of Biological Interest)44 | Describe metabolites and other chemical entities | 137,894 | 266,753 |
| Drugs | DrON (Drug Ontology)45 | Describe patient medications | 554,934 | 1,112,074 |
| Kidney (KPMP) | Kidney Tissue Atlas Ontology (KTAO) * | Facilitate data collection, integration, and analysis for a comprehensive kidney precision medicine studies | 5,593 | 10,838 |
| Precision medicine (KPMP) | Ontology of Precision Medicine Investigation (OPMI)* | Describe general precision medicine projects | 2,896 | 4,413 |
KPMP-initiated ontologies
Note: entity and relation counts are reported as of May 8, 2020.
Each of these ontologies contains representations that are relevant to the nephrology community. For example, UBERON contains references to kidney anatomy and HPO has terms representing abnormalities in urine microscopy and electrolyte abnormalities. While these reference ontologies are extensive, the definitions and terms relevant to the nephrology field have not necessarily been reviewed by nephrologists or researchers in the nephrology community. To improve the value of these ontologies for the nephrology community, KPMP has identified teams of subject matter experts who have reviewed these terms and carefully curated their definitions. In circumstances where the terms are agreed upon by the appointed subject matter experts to be inaccurate or incomplete, the KPMP collaborates with curators of the existing ontology to either modify or add terms as appropriate (see Section 4.3).
4.3. Gaps in existing ontologies
Although an abundance of ontology resources are available for reuse, some necessary entities are not represented and must be either defined in a KPMP-specific ontology or added to an existing reference ontology. Kidney specific terms may be inaccurately represented in existing reference ontologies, synonyms may be missing, or taxonomic classification may need to be reorganized. For example, existing reference ontologies lack the detailed catalog of descriptive cell type and pathological terms that will be used in the KPMP. A number of phenotypes related to the renal disease are described in HPO, such as HP:0000097, “Focal segmental glomerulosclerosis.” However, insufficient details are available for annotating the full breadth of kidney pathological observations, in which glomerulosclerosis can also present globally (there is not an HPO term for “Global glomerulosclerosis”). Similarly, while the Cell Ontology (CL)40 classifies some kidney-specific cell types, it lacks the terminology to describe molecular kidney cell phenotypes, which are key for the goals of the KPMP. Furthermore, clinical data representation in kidney precision medicine also has big gaps, which the KPMP hopes to address through the integration and expansion of terms in ontologies such as MONDO or OAE.
Based on these gaps, the KPMP aims to create ontological resources for annotating the kidney pathological and molecular features currently undescribed or under-described by existing ontologies, and collaborate with curators of existing ontologies to improve ontology representation for the nephrology community. Suggested changes to reference ontologies are documented and shared with the curators of each reference ontology for review and incorporation into that ontology. Similarly, if terms are missing, they are created by working with the reference ontologies to develop a definition, synonyms, and hierarchical classification. We anticipate that this will be an ongoing process as new technologies are developed and novel data becomes available.
Additionally, entities from different ontologies may not yet be semantically linked, and one of the tasks of KPMP ontology development is to provide links between existing terms where appropriate. For example, a gene marker in a specific kidney cell type may not be semantically linked to its related phenotypes in another ontology. When the KPMP discovers such missing or novel associations -- for example, a novel gene variant is found to be associated with CKD progression -- the relationship is added to the KPMP ontologies. The KTAO (described in greater detail in the next Section) provides an integrative ontology framework in which to import and link these terms.
5. KPMP ontologies: a semantic framework to integrate KPMP data
In this section, we describe the two KPMP-initiated ontologies, KTAO and OPMI, as the shared ontological resources for kidney disease during the preliminary stages of the project. The KTAO is an application ontology designed to integrate the data collected by the KPMP (example provided in Section 6). Application ontologies are usually derived from reference ontologies, with the addition of highly-specific terms and relationships applicable to a single project or end use. The purpose of KTAO is to facilitate comprehensive KPMP studies and support the needs of participating institutions within the KPMP consortium. The Ontology of Precision Medicine Investigation (OPMI) is a reference ontology of concepts used to describe data for precision medicine, and is designed to support better data harmonization and integration for precision medicine projects beyond KPMP.
These two ontologies are used to annotate and standardize KPMP data at various stages of data management (FIG. 2), including collection, analysis, and long-term storage and retrieval as part of the Kidney Tissue Atlas. For example, KPMP ontologies are used to standardize clinical report forms (CRFs) and the data elements collected using these forms, and unify clinical data with kidney disease biomarkers, cell types, and anatomic entities. These ontologies are also integrated with OBO ontologies and shared with the community to promote broad adoption and reuse of standardized structured knowledge. Integrated data can then be queried to answer questions over the data of the KPMP Kidney Tissue Atlas. For example, a researcher may want to determine the unique genes expressed in the proximal tubule of the kidney from patients with diabetic nephropathy, in an effort to identify novel gene markers or targets for treatment. This question can only be answered by combining clinical features with pathological images, transcriptomic, proteomic, and metabolomic studies. Data from these studies must be annotated using a shared ontological framework in order to be combined and analyzed. It is anticipated that the shared Kidney Tissue Atlas data platform, supported by the KPMP ontologies, will facilitate future nephrology research for the community at large.
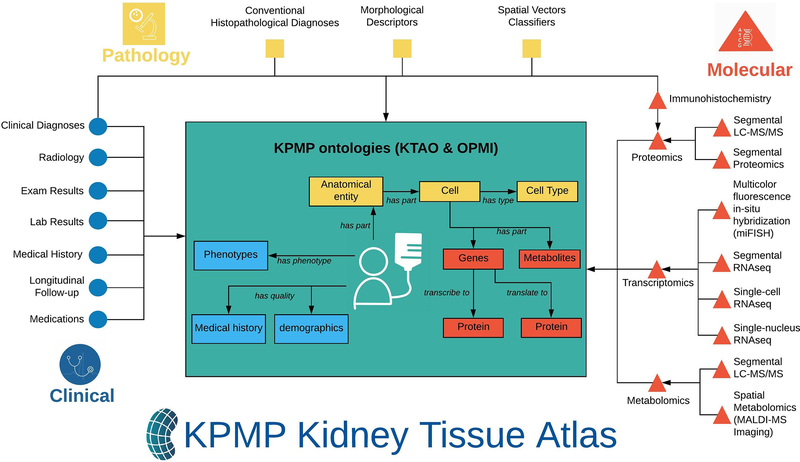
FIG. 2. The KPMP ontology framework for supporting data representation, integration, and analysis.
Clinical, pathology, and molecular data collected from KPMP recruitment sites and tissue interrogation sites will be deposited in the KPMP Kidney Tissue Atlas. Arrows outside of the box indicate different types of data flowing into the KPMP ontology environment. Two KPMP ontologies, KTAO and OPMI, provide the semantic framework for modeling relations among the heterogeneous data in the atlas. Arrows inside the box represent ontological relations among entity types represented in the KPMP ontologies.
5.1. KTAO – Kidney Tissue Atlas Ontology
The KTAO was developed in 2018 as an application ontology to support the KPMP, and was designed to logically represent the relations among gene markers, phenotypes, diseases, cell types, and anatomic entities to support modeling of common forms of kidney disease.8 The KTAO was developed using both a top-down and bottom-up approach. The top-down approach allows the ontologists to define the basic structure of the ontology and populate it with initial terms and relationships. The bottom-up approach allows for the incorporation of term recommendations and edit suggestions from the end-users of the ontology. To avoid reinventing the wheel, KTAO applied the top-down approach by reusing appropriate terms from existing OBO ontologies, including the GO,11 HPO,24 MONDO,19 Ontology for Biomedical Investigations (OBI),46 UBERON,16 and CL,40 and other reference ontologies like OPMI. The KTAO is strongly linked to the open biomedical ontology ecosystem, following the OBO principles of reuse and repurposing. KPMP has also been collaborating with the pre-existing ontologies to help deepen the granularity of terms describing kidney structure, function, and disease, which are often incompletely represented in existing ontologies.
As KPMP is undertaking an unparalleled assessment of reference and diseased kidney tissue, new knowledge will be added and linked within the KTAO to ensure the availability of a set of well-defined kidney disease-related entities or phenomena. This will enable the integration of distinct data types and support user-defined searches or clustering of participants based upon a panel of clinically-relevant features. Developing capability for user-defined searches and user-directed clustering is an important component of the KPMP mission and is anticipated to be an important driver of new knowledge discovery. It is expected that new entities and relationships will be identified and added to the KTAO, and existing entities and relationships will also be modified through the course of the study. Examples include the molecular definition of kidney cell types or cell states, the refinement of existing anatomical entities, or the creation of new kidney disease classifications. New entities and relationships developed during the course of KPMP studies will initially be added to the KTAO and, when suitable, will be submitted to the corresponding reference ontologies to benefit the broader scientific community.
5.2. OPMI – Ontology of Precision Medicine Investigation
KPMP faces the challenges of big data standardization and integration, which requires the synthesis of high-throughput multi-scale (clinical, pathological, and molecular) data into knowledge. The OPMI is designed as a community-based open source biomedical ontology to address this challenge. The proper formal representation and integration of basic research results can be impacted by various factors, such as technical factors, including the instrument used to generate the data or the methods used to collect the biosamples, as well as clinical and pathological factors unique to individual participants. Precision medicine data must be captured and modeled accurately to facilitate robust analysis, and the OPMI has been developed to address these challenges. For example, as data are collected in the KPMP, the data elements and their relationships in the OPMI can be used to validate values during data entry, and errors can be flagged before they are stored, thereby improving the quality and reliability of the data collected by the KPMP.
OPMI is developed as a community-based open source biomedical reference ontology following OBO Foundry principles such as openness and collaboration. OPMI has been accepted as an OBO Foundry ontology (http://obofoundry.org/ontology/opmi.html). The ontology is designed as a data integration platform for general precision medicine projects including the KPMP. OPMI reuses many terms and relations from existing ontologies, including the Ontology of General Medical Science (OGMS) (https://github.com/OGMS/ogms), Ontology for Biomedical Investigations (OBI), HPO, UBERON multi-species anatomy ontology, OAE,47 and Informed Consent Ontology (ICO).48 Meanwhile, OPMI represents many precision medicine-specific terms that can be imported to KTAO and other clinical ontologies. The KPMP developed approximately 30 Clinical Report Forms (CRFs) that include over 2,500 clinical questions. We used OPMI to standardize the major metadata types and clinical factors derived from these CRFs, which significantly improves ontology-based data integration across different institutes.49 In addition to supporting KPMP, OPMI has also been used by other biomedical projects. For example, OPMI has been used as an ontology platform to model the metadata shown in clinicaltrials.gov and other clinical trial repositories.50
6. The vision: using interoperable ontologies to support kidney disease research
Below, we define the use cases that illustrate the central role of ontologies in enhancing our current understanding of kidney disease. This includes using deep phenotyping to identify new classifications and subclassifications of common kidney diseases, as well as previously unrecognized relationships between clinical, anatomic, pathological, and molecular phenotypes. The following examples illustrate 1) why ontologies are an essential element for kidney precision medicine, 2) how they will enable the data analytic goals of the KPMP, and 3) the rationale behind our ontology design choices. We also aim to provide inspiration for how these ontologies can be used to solve practical challenges in nephrology.
The current clinical approach to diagnose kidney disease is based on patient demographics, medical history of past and present illness, physical exam, and lab tests. One of the first goals of clinical evaluation is to establish a cause for kidney disease. While nephrologists often use their clinical judgement to infer the cause of kidney disease from the patient’s past medical history, laboratories and other clinical features, a kidney biopsy is sometimes necessary to establish the underlying cause of kidney disease. The biopsy is routinely evaluated with standard histopathologic approaches, including light microscopy with specialized staining, immunofluorescence microscopy, and electron microscopy. The incorporation of molecular features captured by high-throughput evaluation of renal biopsies is not currently standard of care. Combining these molecular features with the standard clinical, laboratory, and pathology data may allow us to distinguish previously unrecognized subtypes of kidney diseases. Transcriptomic, proteomic, and metabolomic data can also be integrated to redefine the classification/categorization of kidney disease and identify driver cell types and potential therapeutic targets.
Cell type-specific gene, protein, and metabolite expression profiles translate into cell type-specific physiology that generates tissue, organ, and finally, whole organism function. While cell ontologies allow the characterization of pathways that underlie cellular physiology from molecular profiles, the integration of cell physiology with kidney physiology and pathophysiology, as well as whole body function, depends on an integrated ontology that spans multiple levels. For example, the ZMPSTE24 gene51 in podocytes is linked to the basement membrane organization pathway, which plays a role in barrier formation and glomerular filtration related to the pathophysiology of the minimal change glomerulopathy. Ontology links genes in specific cell types (obtained from the single nucleus and single cell transcriptomic data) to cellular pathways and to cell physiological function with whole body physiology, allowing for connectivity between pathway activity changes and cellular dysfunction caused by disease.
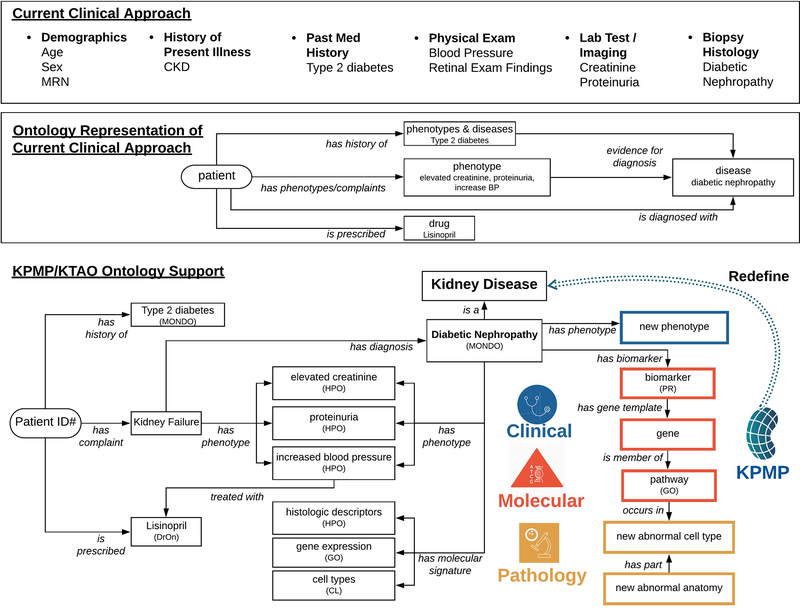
Ontologies already support clinical and translational examination of kidney diseases, illustrated in FIG 3 using diabetic nephropathy as an example. The top layer of FIG. 3 illustrates the variety of data types collected by clinicians in standard clinical practice, such as demographic data, clinical history, physical exam findings, and diagnostic testing. The clinician then uses all of this information to arrive at a diagnosis and treatment plan for a patient. For example, a clinician evaluates a 63-year-old man with a 40-year history of poorly controlled type 2 diabetes, a slowly increasing serum creatinine over the course of several years, and 2 grams of proteinuria per day; a comprehensive serologic evaluation for non-diabetic kidney pathologies was normal. Based on these observations, the clinician determines that the patient most likely has diabetic nephropathy and most clinicians would opt not to biopsy the patient. If this patient chose to participate in KPMP, their pathologic and molecular data would be collected and available to facilitate deeper understanding of their disease.
FIG. 3. The KPMP KTAO ontology supports molecular and histopathologic extensions to current clinical approaches to kidney disease diagnosis.
The top layer lists different data types and example data fields collected during current clinical practice. The middle layer provides a representation of how these data elements can be used to model or support disease diagnosis and treatment with current ontologies. The bottom layer provides an integrative ontology-based point of view that also incorporates molecular and pathologic data in addition to clinical measures, and demonstrates the data harmonization goals of the KPMP. The diagnosis of diabetic nephropathy is used as an example. The KPMP Kidney Atlas, supported by ontologies (e.g., KTAO and OPMI), will be used to redefine kidney diseases (bottom layer) based on molecular mechanisms combined with traditional clinical and histopathologic features to identify critical cells, pathways, and targets for the development of novel diagnosis and therapies.
As shown in the middle layer of FIG. 3, the KPMP ontological framework can capture these clinical data, linking them with molecular and imaging data, as well as other sources of background knowledge, to allow for more nuanced assessment of the individual’s disease presentation in the context of other reference and disease tissues. The ontology framework can also be easily adapted to enable computational phenotyping of patients and the development of decision support systems to assist clinicians in diagnosis and treatment.
The current clinical model does not integrate molecular and pathologic data, which are key components for precision medicine. As defined in the lower layer of FIG. 3, a central goal of the KPMP is to develop an integrative KTAO ontology framework to standardize and harmonize data obtained in standard clinical practice with the novel molecular and histopathologic data that will be generated by the KPMP. In this example, diabetic nephropathy may be associated with specific biomarkers, encoded by genes and linked to particular biological pathways and functions. Single-cell and single-nucleus sequencing technologies may enable us to identify kidney cell types that drive disease and identify new kidney disease subtypes based on the results of molecular and cellular phenotyping. The hierarchical structure and semantic relations provided by KPMP ontologies are used to link diverse data types and make such discoveries possible. Integrated representation of clinical and molecular features will enable the redefinition of our understanding of kidney disease, provide clinicians with novel diagnostic and treatment options for their patients, and facilitate novel discoveries in the field of nephrology research.
7. Summary & future work
Successful applications of precision medicine require that large numbers of phenotypic traits (molecular, genomic, clinical, and otherwise) be documented about each individual. The relationships between various traits and treatment outcomes allow us to predict the needs of each individual and select the best treatment plan in each specific case. To observe these patterns, data must be collected from a large, diverse group of individuals and standardized to allow for proper comparison. Ontologies provide both a means for the terminological standardization of data, enhancing our ability to analyze and learn from the data, as well as support for the incorporation of structured terms and relationships into predictive models for clinical deployment.
By creating a resource like the Kidney Tissue Atlas, the KPMP aims to create a repository of clinical and bio-specimen data that can be used to support kidney precision medicine. A goal of the KPMP is to use these data, particularly molecular data, to define novel subtypes of current (and currently insufficient) disease classifications. With these new disease subtypes, clinicians and researchers can discover more targeted and effective therapies. The molecular phenotypes needed for these novel disease classifications are especially challenging to describe, as the molecular features used to define these phenotypes are continuous, whereas traditional phenotypes are discrete. How best to define novel molecular and cellular phenotypes is an open question that the KPMP hopes to contribute to answering as we acquire more data and insight into this issue. This challenge mirrors the global challenge faced by precision medicine: the disconnect between the recognition of each individual as a unique case deserving of specialized treatment, and the need to classify individuals into groups in order to assess the statistical efficacy of those treatments.
Ontologies play a critical infrastructural role for the above tasks. Ontologies provide a mechanism for harmonizing and integrating data collected from disparate centers and organizations across different categories and domains. When analyzing large combined datasets, computational methods are necessary for discovering correlations and relationships between input features. Manual harmonization of large datasets is impractical and expensive; built-in annotation to shared ontology terms is critical and makes these sorts of analyses feasible.
Due to the novelty of data generated by the KPMP, additions must be made to existing open biomedical ontologies to support data annotation. Members of the KPMP are working with other ontology groups and developers to incorporate kidney-specific terminology and relationships into reference ontologies like the HPO. A standard operating procedure has been established to support the collaboration between the HPO (and other ontologies) and KTAO development teams. The introduction of new terms and ontologies allows for rich annotation of kidney data, which benefits both KPMP and other research groups. As the KPMP ramps up tissue collection and analysis, ontology terms will be used to annotate patient data, specimens, and analysis results. Development of KPMP ontologies and suggested additions and changes to references ontologies are ongoing as annotation needs are continuously re-evaluated.
In addition to assisting in data annotation and analysis, the KPMP’s integrative KTAO ontology framework will also become a living representation of our understanding and knowledge of kidney diseases. KPMP studies are expected to generate new kidney disease subtypes, biomarkers, and disease-specific pathways, which will be integrated into the KTAO and other reference ontologies. These updated ontologies can be used to further improve kidney-specific data annotation and analysis. We also expect that the KPMP ontology framework can be used to support more productive tool development. For example, the Kidney Tissue Atlas visualization tool can use the KTAO entity hierarchy to provide better browsing and query of tissue samples. Molecular data and pathways annotated using KTAO terms can also be used for advanced biomarker and pathway analysis.
In this article, we described how ontologies are essential for kidney precision medicine and some practical benefits of ontologies for data organization and knowledge discovery. We provided a summary of two ontology resources (KTAO and OPMI) built specifically by the KPMP to aid the collection and analysis of KPMP data. We also described how we are working with the ontology community to add or enhance kidney disease representations in various open biomedical ontologies.
The strength of a shared data resource depends on the contributions and efforts of its surrounding research community. Ontological improvements made by the KPMP are meant to help enable standardized data sharing for the nephrology clinical research community. By making data more interoperable through annotation with the same shared ontologies, the pool of data that can be harnessed for research grows significantly. It is our hope that you, as members of this community, will support this shared ecosystem, and take on these ontologies as the fundamental organizational layer in your data. Only through consistent investment in data interoperability can we derive greater gains from the resources we so laboriously build and share.
Key points.
Ontologies are powerful tools for organizing, integrating, and linking heterogeneous data types, especially in the biomedical sciences.
Significant additions to biomedical ontologies are necessary to enable the definition of kidney molecular and histopathological phenotypes, which are critical for kidney precision medicine.
The Kidney Precision Medicine Project (KPMP) is creating a community-based Kidney Tissue Atlas to integrate molecular, cellular, and anatomic knowledge about the kidney.
The KPMP is developing the Kidney Tissue Atlas Ontology (KTAO) and Ontology of Precision Medicine and Investigation (OPMI) for data collection, harmonization, and analysis in support of kidney precision medicine.
Community-based reference ontologies have been extensively adopted, reused, and extended to support community kidney data annotations.
Acknowledgment
This KPMP project is supported by the NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) U2C Project #: 1U2CDK114886. We appreciate the discussion, editing, and support from Dr. Deborah Hoshizaki and the KPMP consortium.
Footnotes
Competing interests
The authors declare no conflicts of interest.
Bibliography
- 1.Abrahams E Right drug-right patient-right time: personalized medicine coalition. Clin. Transl. Sci 1, 11–12 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khwaja A KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin. Pract 120, c179–84 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Stevens PE, Levin A & Kidney Disease: Improving Global Outcomes Chronic Kidney Disease Guideline Development Work Group Members. Evaluation and management of chronic kidney disease: synopsis of the kidney disease: improving global outcomes 2012 clinical practice guideline. Ann. Intern. Med 158, 825–830 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Hawrylycz M et al. The Allen Brain Atlas. Springer Handbook of Bio-/Neuroinformatics 1111–1126 (2014) doi: 10.1007/978-3-642-30574-0_62. [DOI] [Google Scholar]
- 5.Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regev A et al. The Human Cell Atlas. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y et al. KTAO: A Kidney Tissue Atlas Ontology to Support Community-Based Kidney Knowledge Base Development and Data Integration. in ICBO (2018). [Google Scholar]
- 9.Cimiano P Ontologies. in Ontology Learning and Population from Text: Algorithms, Evaluation and Applications (ed. Cimiano P) 9–17 (Springer; US, 2006). doi: 10.1007/978-0-387-39252-3_2. [DOI] [Google Scholar]
- 10.Gruber TR Toward principles for the design of ontologies used for knowledge sharing? International Journal of Human-Computer Studies vol. 43 907–928 (1995). [Google Scholar]
- 11.Ashburner M et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DW, Sherman BT & Lempicki RA Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doms A & Schroeder M GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 33, W783–6 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whetzel PL et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 39, W541–5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mungall CJ, Torniai C, Gkoutos GV, Lewis SE & Haendel MA Uberon, an integrative multi-species anatomy ontology. Genome Biol. 13, R5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosse C & Mejino JLV Jr. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform 36, 478–500 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Natale DA et al. Protein Ontology (PRO): enhancing and scaling up the representation of protein entities. Nucleic Acids Res. 45, D339–D346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kibbe WA et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 43, D1071–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shefchek KA et al. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Research (2019) doi: 10.1093/nar/gkz997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamdar MR, Tudorache T & Musen MA A Systematic Analysis of Term Reuse and Term Overlap across Biomedical Ontologies. Semant Web 8, 853–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Euzenat J & Shvaiko P Ontology Matching. (2013) doi: 10.1007/978-3-642-38721-0. [DOI] [Google Scholar]
- 23.Smith B et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 25, 1251–1255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson PN & Mundlos S The Human Phenotype Ontology. Clinical Genetics vol. 77 525–534 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Haendel MA, Chute CG & Robinson PN Classification, Ontology, and Precision Medicine. N. Engl. J. Med 379, 1452–1462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keenan AB et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst 6, 13–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vempati UD et al. Metadata Standard and Data Exchange Specifications to Describe, Model, and Integrate Complex and Diverse High-Throughput Screening Data from the Library of Integrated Network-based Cellular Signatures (LINCS). J. Biomol. Screen 19, 803–816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong E et al. Ontological representation, integration, and analysis of LINCS cell line cells and their cellular responses. BMC Bioinformatics 18, 556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malladi VS et al. Ontology application and use at the ENCODE DCC. Database 2015, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMahon AP et al. GUDMAP: the genitourinary developmental molecular anatomy project. J. Am. Soc. Nephrol 19, 667–671 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Cole NI et al. An ontological approach to identifying cases of chronic kidney disease from routine primary care data: a cross-sectional study. BMC Nephrol. 19, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bajema IM et al. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int. 93, 789–796 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Leung N et al. The evaluation of monoclonal gammopathy of renal significance: a consensus report of the International Kidney and Monoclonal Gammopathy Research Group. Nat. Rev. Nephrol 15, 45–59 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethi S et al. Mayo Clinic/Renal Pathology Society Consensus Report on Pathologic Classification, Diagnosis, and Reporting of GN. J. Am. Soc. Nephrol 27, 1278–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harding SD et al. The GUDMAP database--an online resource for genitourinary research. Development 138, 2845–2853 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little MH et al. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns 7, 680–699 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayamizu TF et al. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J. Biomed. Semantics 4, 15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen J, Meretzky D, Woldesenbet S, Stolovitzky G & Iyengar R A flexible ontology for inference of emergent whole cell function from relationships between subcellular processes. Sci. Rep 7, 17689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diehl AD et al. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics 7, 44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandrowski A et al. The Ontology for Biomedical Investigations. PLoS One 11, e0154556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith JR et al. The clinical measurement, measurement method and experimental condition ontologies: expansion, improvements and new applications. J. Biomed. Semantics 4, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Y et al. OAE: The Ontology of Adverse Events. J. Biomed. Semantics 5, 29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hastings J et al. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 44, D1214–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna J, Joseph E, Brochhausen M & Hogan WR Building a drug ontology based on RxNorm and other sources. J. Biomed. Semantics 4, 44 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters B, Peters B & The OBI Consortium. Ontology for Biomedical Investigations. Nature Precedings (2009) doi: 10.1038/npre.2009.3623. [DOI] [Google Scholar]
- 47.Kang Y, Fink JC, Doerfler R & Zhou L Disease Specific Ontology of Adverse Events: Ontology extension and adaptation for Chronic Kidney Disease. Comput. Biol. Med 101, 210–217 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Lin Y, Harris MR, Manion FJ, Eisenhauer E, Zhao B, Shi W, et al. Development of a BFO-based Informed Consent Ontology (ICO). in Proceedings of the 5th International Conference on Biomedical Ontologies (ICBO). [Google Scholar]
- 49.He Y et al. OPMI: the Ontology of Precision Medicine and Investigation and its support for clinical data and metadata representation and analysis. in ICBO (2019). [Google Scholar]
- 50.Hripcsak G et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud. Health Technol. Inform 216, 574–578 (2015). [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy HJ et al. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin. J. Am. Soc. Nephrol 8, 637–648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]