Abstract
To generate physiologically relevant experimental models, the study of enteric diarrheal diseases is turning increasingly to advanced in vitro models that combine ex vivo, stem cell-derived “organoid” cell lines with bioengineered culture environments that expose them to mechanical stimuli, such as fluid flow. However, such approaches require considerable technical expertise with both microfabrication and organoid culture, and are, therefore, inaccessible to many researchers. For this reason, we have developed a perfusion system that is simple to fabricate, operate, and maintain. Its dimensions approximate the volume and cell culture area of traditional 96-well plates and allow the incorporation of fastidious primary, stem cell-derived cell lines with only minimal adaptation of their established culture techniques. We show that infections with enteroaggregative E. coli and norovirus, common causes of infectious diarrhea, in the system display important differences from static models, and in some ways better recreate the pathophysiology of in vivo infections. Furthermore, commensal strains of bacteria can be added alongside the pathogens to simulate the effects of a host microbiome on the infectious process. For these reasons, we believe that this perfusion system is a powerful, yet easily accessible tool for studying host-pathogen interactions in the human intestine.
Keywords: Escherichia coli, bacteria, norovirus, enteroid, organoid, microfluidic, shear, dynamic, microbiome
Introduction
Diarrheal diseases and other infections of the intestine are a primary cause of death worldwide3, especially in children, in whom repeated diarrhea-causing infections lead to significant malnutrition, growth stunting, and cognitive impairment.15 Investigations of these diseases, however, are complicated by the lack of reliable and relevant models for many of their pathogens. Small animal models poorly mimic human physiology and disease pathophysiology due to inter-species differences in physiologically-important proteins, host polymorphisms that underlie differences in susceptibility to infection, and the strict tropisms of many clinically important mucosal pathogens for humans. While large animal models may demonstrate pathologies that are more similar to those found in humans, their use is severely limited by high costs and low scalability.16 Finally, traditional in vitro models lack functional similarity with the human intestine, often lacking the full diversity of epithelial cell types and surface proteins.1 As a result, many enteric pathogens are simply unable to be studied in vitro.6
The most promising alternatives are ex vivo models of the human intestinal epithelium derived from intestinal epithelial stem cells.25 These cultures, known as human intestinal enteroids (HIEs), contain the major cell types and recapitulate many important physiological features of the native intestinal epithelium.24 HIEs are typically grown as three-dimensional aggregates embedded in Matrigel, but can be converted to two-dimensional monolayers to facilitate access to their apical surface.30
HIE models have contributed significantly to our understanding of diarrheal diseases and other intestinal infections, but fail to capture important aspects of host-pathogen interactions. For example, physical forces such as fluid shear affect the behavior of intestinal cells,8 as well as how these cells interact with luminal contents,17 and hence are important elements of intestinal host-pathogen interactions. Although most infectious disease research is conducted in mechanically static environments, such as Transwell inserts and multiwell plates, certain newer models utilize mechanically dynamic environments mimicking the human intestine.13,28 However, these technologies may require sophisticated equipment and/or extensive bioengineering and microfluidic expertise to build and operate,11 and thus adoption of these powerful experimental tools has been limited.
To address this barrier, we have developed a millifluidic perfusion cassette (mPC). To fabricate the device, we eschewed traditional microfabrication approaches, and instead used relatively inexpensive, consumer-level, three-dimensional printing for construction. The wells of these devices are easily accessible by micropipette, require only minimal adaptation of traditional cell culture technique, and accommodate diverse bioanalytical techniques. Here, we demonstrate the utility of these devices by modeling infections with both human norovirus (HuNoV) and Enteroaggregative Escherichia coli (EAEC) and show that flow significantly affects the infectious process. For these reasons, we believe that the mPC is a powerful, yet easily accessible tool for studying enteric infections.
Materials and Methods
Fabrication of millifluidic perfusion chambers (mPCs)
The mPC master mold was designed with CAD software (OpenSCAD), and 3D printed with poly(lactic acid) filament (Ultimachine) on an Ultimaker 2+ (Ultimaker). They were affixed to a 3 in by 4 in glass slide using GO2 Glue (Loctite) and treated with Sigmacote (Sigma Aldrich) to reduce adhesion to poly(dimethyl siloxane), PDMS. PDMS precursor (Sylgard 184, Dow Corning) was poured over the mold, degassed under vacuum, and partially cured overnight at room temperature. The mPCs were then removed, cured at 90°C for 2 hours, and punched with a 1.5 mm biopsy punch to create inlet and outlet ports. The mPCs were bonded to a 35 mm by 50 mm #2 glass coverslip (Fisher Scientific) with a corona generator (BD20AC, Electrotechnic Products, and then treated with 2% volume 3-aminopropyl(triethoxy)silane solution (Sigma Aldrich) in 95% ethanol for 15 minutes at room temperature. This silane treatment enhanced the hydrophobicity of the glass coverslip to promote subsequent protein binding. The wells of the mPCs were then washed twice with 95% ethanol, baked at 90°C for 1 hour, and sterilized under UV light for 1 hour. Finally, each well was filled with 50 μL of 0.125 mg mL−1 solution of human collagen IV (Sigma Aldrich) in PBS, and incubated overnight at 4°C.
Simulation of flow and shear stress in the mPCs
Flow through one mPC chamber was simulated using computational fluid dynamics software (ANSYS Fluent). The density and viscosity of cell culture media were assumed to be those of water. Flow in the chamber at both 200 μL hr−1 and 400 μL hr−1 was laminar, with Reynolds numbers calculated to be less than 1. No-slip boundary conditions were set at the chamber walls (dimensions shown in Figure 1).
Figure 1:
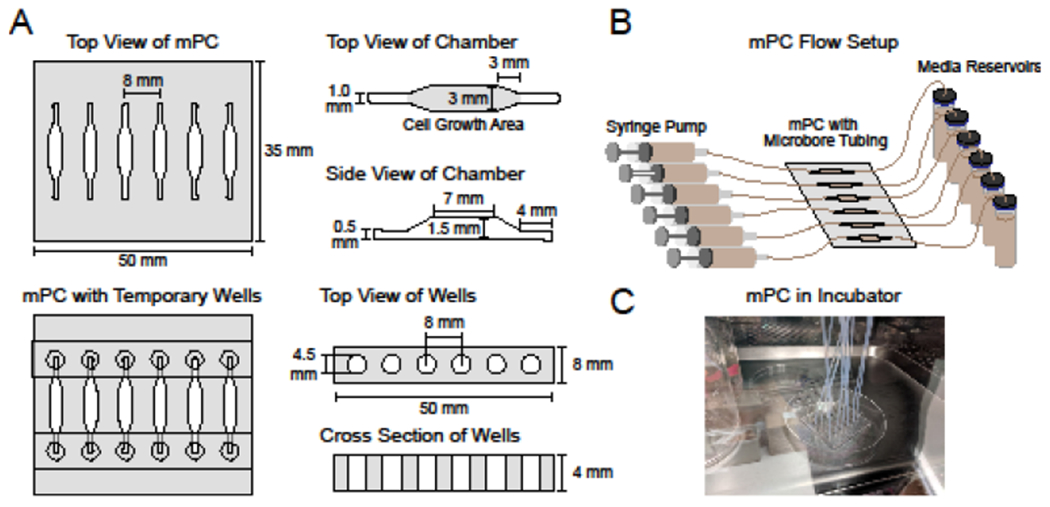
(A) Dimensions of the millifluidic perfusion cassette (mPC) and temporary PDMS wells. (B) Schematic of mPC assembled with syringe pump, tubing and reservoirs. (C) Image of the mPC in cell culture incubator.
Isolation and culture conditions of human intestinal enteroids
As established previously,26 HIE cultures were produced from small intestinal biopsies from adults undergoing routine endoscopy or from surgical specimens according to a protocol approved by the Institutional Review Board at Baylor College of Medicine; established cultures were grown as multi-lobular forms termed 3D HIEs in growth factor-reduced, phenol-free Matrigel (Corning). The cultures used in this work were derived from jejunal and duodenal intestinal stem cells, and included the lines J25 and D10922 that we have previously reported.
Three different formulations of media were used to establish, maintain, and differentiate the HIEs. These were referred to as complete medium without growth factors (CMGF−), complete medium with growth factors (CMGF+), and differentiation medium. CMGF−, used to establish HIEs, consisted of Advanced DMEM/F12 (Invitrogen) supplemented with 100 U mL−1 of penicillin-streptomycin solution (Invitrogen), 10 mM HEPES buffer, and 1x GlutaMAX (Invitrogen). CMGF+, used to maintain cultures, was prepared in 2 steps. In the first step, CMGF-medium was supplemented with 50 ng mL−1 epidermal growth factor (Invitrogen), 10 mM nicotinamide (Sigma-Aldrich), 10 nM gastrin I (Sigma-Aldrich), 500 nM A-83-01 (Tocris Bioscience), 10 μM SB202190 (Sigma-Aldrich, 1 mM N-acetylcysteine (Sigma Aldrich), 1X B27 supplement (Invitrogen), and 1X N2 supplement (Invitrogen). In the second step, the supplemented CMGF+ medium contained medium conditioned by culture of cells expressing growth factors. Fifty percent of the CMGF+ was conditioned with Wnt3A-producing ATTC CRL-2847 cells (ATCC), 20% was conditioned with R-spondin-producing cells (gift of Calvin Kuo, Palo Alto, CA), and 10% was conditioned with Noggin-producing cells (gift of G. R. van der Brink, Amsterdam, The Netherlands). Differentiation medium was prepared in the same 2-step method as the CMGF+ medium, but it was not supplemented with SB202190 and nicotinamide and was not conditioned by the Wnt3A-producing cells. Additionally, the concentrations of Noggin and R-spondin conditioned media were halved for the differentiation medium.
Establishment of HIE monolayers in mPCs and application of flow
Established cultures of HIEs were expanded as multi-lobular forms, termed 3D HIEs, in growth factor-reduced, phenol-free Matrigel with CMGF+ medium. All experiments were performed with HIEs below P20. To create a HIE monolayer in the mPCs, 3D HIEs were digested to single cells and seeded into the mPC wells in an adaptation of a previous method.30 Briefly, the HIEs were released from Matrigel by washing with 500 μL of cold PBS with 0.5 mM EDTA pH 7.4, followed by centrifugation at 175 rcf for 5 min at 4°C. The cells were treated with 0.05% w/v trypsin-0.48 mM EDTA (Gibco) for 4 min at 37°C, and the trypsin was deactivated with basal media containing 10% serum. The cells were strained (40 μm) and the flow-through was centrifuged at 250 rcf for 5 min at room temperature. Finally, the cell pellet was resuspended in CMGF+ with 10 μM Y-27632 (EMD Millipore), and the mPC chambers were seeded at a density of 106 cells cm−2. To assure complete filling of each chamber, 50 μL of cell suspension was used. Control HIE monolayers were also formed in 96-well plates at the same density, but with larger media volumes (100 μL/well). After 24 hours, the cells were nearly confluent on the collagen-IV coated glass coverslip base of the mPC chamber; cells did not adhere to the PDMS walls of the chamber. The media was then changed to differentiation media and replaced every 48 hours. Cells were differentiated for 5 days after seeding prior to experiments. To increase the volume of media available to the cells in the devices, temporary wells were formed by placing strips of UV-sterilized PDMS with circular holes over the inlet and outlet channels (Figure 1A). These wells, formed by making 3 mm holes in a strip of PDMS using a biopsy punch, increased the capacity of the chambers so that equivalent volumes of media could be used in the chambers and the 96 well plates (100 μL of media each). The two PDMS surfaces (at the top of the mPC and at the bottom of the temporary well strip) readily adhered together with a temporary bond that was sufficient for this purpose. The mPCs were stored in humidified 35 mm petri dishes (Corning) to minimize media evaporation.
After the HIEs were seeded and differentiated, the media and temporary wells were removed from the mPCs, and syringes filled with differentiation media were connected to the mPC ports with Tygon microbore tubing (0.02” inner diameter, 0.06” outer diameter, Cole Parmer). The chambers were primed with media and flow was started at 200 μL hr−1 with a syringe pump (PhD Ultra, Harvard Apparatus). To prevent air bubbles, the differentiation media was equilibrated in a cell culture incubator overnight prior to loading into the syringes. HIE monolayers cultured in static conditions also had media exchanged.
Characterization of HIE phenotype by RT-qPCR
Differentiated HIE monolayers were cultured in 3 conditions: in 96-well plates, in the mPC without flow, and in the mPC with flow (200 μL hr−1) as described above. After 24 hours, the cells were washed 2x with PBS and lysed with Trizol reagent (Ambion). RNA was isolated with the Direct-Zol RNA MiniPrep Kit (Zymo Research) following manufacturer’s instructions. 2-3 wells were pooled for one biological sample. qScript XLT 1-step RT-qPCR ToughMix with ROX master mix (Quantabio) and Taqman primer-probe mixes (ThermoScientific) were used to perform RT-qPCR (StepOne Plus, Applied Biosystems). Probe information is in Supplemental Table 1. Gene expression levels were normalized to GAPDH and fold change was calculated with the 2−ΔΔCt method.
Characterization of mucin expression by HIEs
Differentiated HIE monolayers were cultured with and without flow in the mPCs for 24 hours as described above. Afterwards, the HIEs were fixed with Carnoy’s solution (60% ethanol, 30% chloroform, 10% acetic acid) for 30 minutes and then stained with Alcian blue for 30 minutes. The samples were gently washed 3x with PBS before brightfield microscopy imaging (Eclipse TE300, Nikon).
Human Norovirus (HuNoV) infections of HIEs in mPCs
Differentiated HIE monolayers were infected with HuNoV as described previously.5 Briefly, the HIE monolayers (the secretor positive J2 line, which is susceptible to HuNoV infection5) were infected with 1.8 x 105 genome equivalents of GII.4 Sydney 2012 (TCH12-580) HuNoV for 1 hour. 500 μM of the bile acid GCDCA was added to facilitate noroviral entry to the HIEs. After one hour, the cells were washed twice to remove unbound virus, and flow through the mPCs was initiated at 200 μL hr−1 for 23 hours. Wells not subjected to flow (mPC and 96-well plate) were refilled with fresh differentiation media (with 500 μM GCDCA). HuNoV genome equivalents were harvested from the HIEs at 1 and 24 hours post-infection, measured by RT-qPCR, and compared to a standard curve of noroviral RNA. All work with HuNoV was conducted under BSL-2 conditions in a biological safety cabinet with appropriate airflow, and with the use of proper personal protective equipment.
Bacterial infection of HIEs in mPCs
All bacterial infection experiments followed the same generic approach (adapted from previously reported methods22). Duodenal HIE monolayers (D109 line) were formed in the mPCs. Bacteria were expanded in tryptic soy broth at 37°C overnight (one day before the experiment), quantified via optical density, and diluted in differentiation media to the final multiplicity of infection (MOI). This bacteria-containing media was added to the HIEs in the mPC chambers for 1 hour at 37°C to initiate infection, and then the media and any non-adherent bacteria were removed from the well. The wells were then either refilled with fresh differentiation media (no flow) or connected to syringes filled with differentiation media, which was flowed through the wells at 400 μL hr−1 for 7 hours. Afterwards, the wells were washed with PBS and prepared for downstream analysis. All work with EAEC was conducted under BSL-2 conditions in a biological safety cabinet with appropriate airflow, and with the use of proper personal protective equipment.
To evaluate the effects of flow on wildtype EAEC infections, two strains of EAEC were used: EAEC 042 (serotype O44:H18), the prototype strain; and EAEC A2A, a clinical isolate. Both strains were added to the HIEs at a MOI of 5, and the infections were fixed and evaluated with Wright-Giemsa staining using the Hema3 fixative and solutions (Fisher Scientific) per manufacturer’s recommendations. The mPC chambers were imaged using an upright microscope with a 100x oil-immersion objective (Ci-L, Nikon). Imaged cultures were evaluated to ascertain the general morphology of the HIEs and the aggregative adherence behavior of the EAECs, such as clustering into aggregates or biofilms. Aggregates are groups of 20-100 bacteria in which individual bacteria can be seen. In contrast, biofilms are a lawn of aggregately clustered colonies with bacteria numbering in the 1000s, and in which most bacteria cannot be identified individually.
To investigate the role of the fimbrial subunit aafA in EAEC adherence and aggregation, EAEC 042ΔaafA, an isogenic mutant of EAEC 042 that does not express the adhesion protein aafA, the major structural component of the AAF virulence factor,18 was compared to wildtype EAEC 042. Both bacteria were added at a MOI of 0.01. Following the application of flow, the wells were fixed, stained, and imaged as described above for the wildtype-only infections.
To evaluate EAEC infections with and without commensal E. coli, HIE monolayers were infected with either EAEC 042 expressing GFP, the non-pathogenic commensal E. coli K12 strain expressing RFP, or both. Each bacterial strain was added to the HIEs at a MOI of 5, with the combined EAEC 042/E. coli K12 condition receiving a total MOI of 10. The monolayers were fixed in 4% PFA overnight, the nuclei were stained with DAPI, and each well was imaged with confocal microscopy (SP8, Leica)
Data Analysis
All experiments were conducted at least three times with the exception of the aafA study, which was a single proof-of-concept experiment, and the alcian blue staining which was performed in triplicate but for one experimental run only. All statistical analyses were performed using R (The R Foundation). One-way ANOVA with post-hoc Tukey’s HSD testing was performed to compare (i) viral replication and (ii) HIE phenotypic differences between conditions. Significance was accepted as p<0.05.
Results
Fabrication and characterization of the mPCs
The mPCs were prepared from PDMS using 3D-printed molds affixed to coverglass (Figure 2A–B). The completed mPCs were rectangular (46 x 52 x 4 mm3, Figure 1; all dimensions are L x W x H). Each cassette contained 6 wells, each with a 3 x 7 x 1.5 mm3 central cell chamber. Triangular transition zones, 3 mm long, connected the cell chambers to inlet and outlet channels (4 x 1 x 0.5 mm3). The cell growth area was equivalent to that of a standard 96-well plate (~ 0.33 cm2), and the volume of each well was ~45 μL. The PDMS layer of the mPCs was plasma-bonded to #2 coverglass. The mPCs were treated with 3-aminopropyl(triethoxy)silane to provide an adsorptive surface for basement membrane ECM (in this case, collagen IV), which promoted the successful formation of HIE monolayers.
Figure 2: Fabrication and operation of the millifluidic perfusion cassette (mPC).
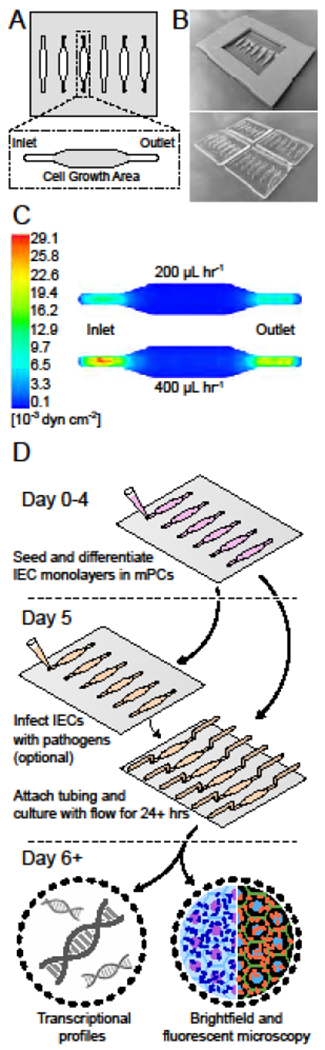
(A) Schematic of the mPC and its wells. (B) Example images of the master molds for the mPCs created through 3D printing (top) and the mPCs (bottom). (C) Computational fluid dynamics model of shear stresses present within the well. (D) Illustration of workflow for a sample experiment with the mPC including HIE culture, application of flow, infection with a pathogen, and molecular analysis.
Computational fluid dynamics simulations showed that the average shear stress in the mPC cell growth areas was 2.4x10−3 dyn cm−2 and 4.7x10−3 dyn cm−2 for flow rates of 200 and 400 μL hr−1, respectively (Figure 2C). These values fell within the estimated range of shear stresses in the human intestine (2.0x10−3 – 8.0x10−2).10,17
Operation of the mPC is flexible and user-friendly
HIE monolayers could be formed, differentiated, and inoculated with bacteria and viruses in the mPCs using minor adaptations of established protocols for 96-well plates and Transwell inserts.30 After seeding, the cells became confluent after 24 hours, and were subsequently differentiated for 5 days before flow experiments were initiated, with or without co-cultured microbial species (Figure 2D). The inlet and outlet ports easily accommodated micropipette tips during cell seeding, media exchange, and microbial inoculation steps, and the chamber size and geometry minimized air bubble formation.
Flow was applied to the mPCs by connecting a syringe pump to the chamber ports with microbore tubing (Figure 1B). HIE monolayers survived throughout the 6-day workflow, from initial culture through experiments, and were then characterized with microscopy and transcriptional analysis. Our experiments were run for twenty-four hours, but longer duration studies are possible.
Culture in the mPCs with or without flow does not significantly affect HIE phenotype
To assess the effect of culture in the mPC, we compared HIEs grown in 96-well plates to those grown in the mPCs under static conditions or under flow at 200 μL hr−1. HIEs in all three conditions expressed CD44, Lgr5, Wnt3A, and Ki67, which are markers for undifferentiated or proliferative crypt cells (Figure 3A). The cells also expressed markers of differentiated intestinal epithelium (Figure 3B): SI for enterocytes, Muc2 for goblet cells, and Lyz for Paneth cells. Expression of all markers relative to GAPDH, both differentiated and undifferentiated, was consistent with previous reports.1 Furthermore, there was no statistically significant difference between the three culture conditions for all markers evaluated, indicating that neither the mPCs nor flow had a significant effect on the HIE phenotype. There was also no observable effect of flow on the alignment of the HIEs.
Figure 3: Culture in the mPC does not alter the phenotype of human intestinal enteroids (HIEs).
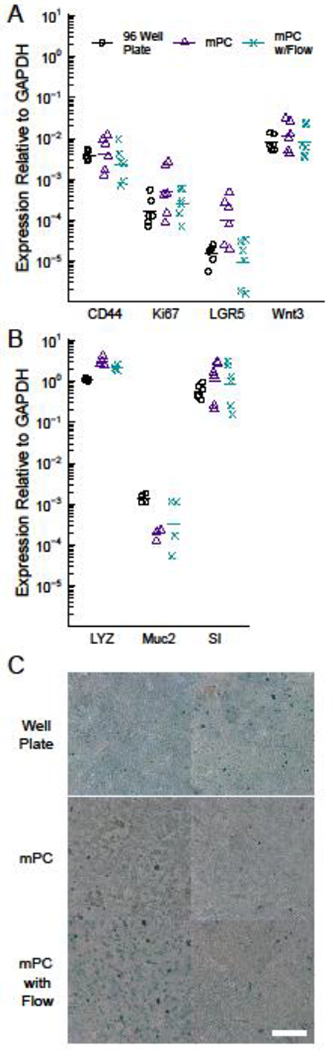
RT-qPCR quantification of markers for proliferative, stem-like cells (A) and terminally differentiated (B) intestinal epithelial cells reveal no statistically significant differences between HIEs cultured in 96-well plates, static mPCs, and dynamic mPCs. Data are compiled from three independent experiments. (C) Alcian blue staining reveals no difference in mucin expression between the three experimental conditions, scale = 50 μm. Two independent, representative samples are shown for each experimental condition. The slightly brighter color of the blue stain in the 96-well plate cultures is likely an artifact of the illumination light focused and reflected by the tissue culture plastic well base and walls when imaging these cultures using a dissecting microscope.
Because intestinal mucus strongly influences interactions between luminal microbes and the mucosal surface,2 we further investigated mucin expression of the HIE monolayers through an alcian blue stain. The stain appeared as a light blue wash across the cultures, but otherwise revealed no qualitative differences in protein-level mucin expression between the three experimental conditions (Figure 3C), consistent with the gene expression of Muc2.
Enteroaggregative E. coli form biofilms on HIE monolayers when exposed to flow
Next, we investigated if flow would improve models of the pathogenesis of EAEC infections. HIEs derived from the duodenum were used for these experiments since EAEC has a predilection for this intestinal segment in human infections. We conducted preliminary experiments (not shown) to adapt EAEC adherence assays for the mPC.22 Decreasing the MOI from 10 to 5 extended the survival of HIE monolayers from 3-4 hours to 8 hours, and HIE survival was more consistent with flow rates of 400 μL hr−1 vs. 200 μL hr−1. Co-cultured HIEs did not survive infections of 16 – 24 hours, even when using MOIs as low as 1. However, it is likely that MOI, flow rates, and infection time can be optimized further to improve HIE survival.
Wright-Giemsa staining was used to evaluate adherence of EAEC 042 (the prototype strain) and EAEC A2A (a clinical isolate) to the HIEs in the presence and absence of flow (Figure 4). Without flow, both strains of EAECs displayed a sheet-like adherence pattern consisting of aggregative clusters of bacteria diffusely scattered over the apical surface, resembling previous reports;22 individual bacteria could be identified within each cluster. With the addition of flow, both EAEC 042 and A2A formed thick biofilms characterized by large, dense, 3D colonies in which individual bacteria were rarely identifiable. Despite the heavy bacterial load present with flow, the underlying HIE monolayer remained intact.
Figure 4: Flow in the mPC promotes formation of EAEC biofilms.
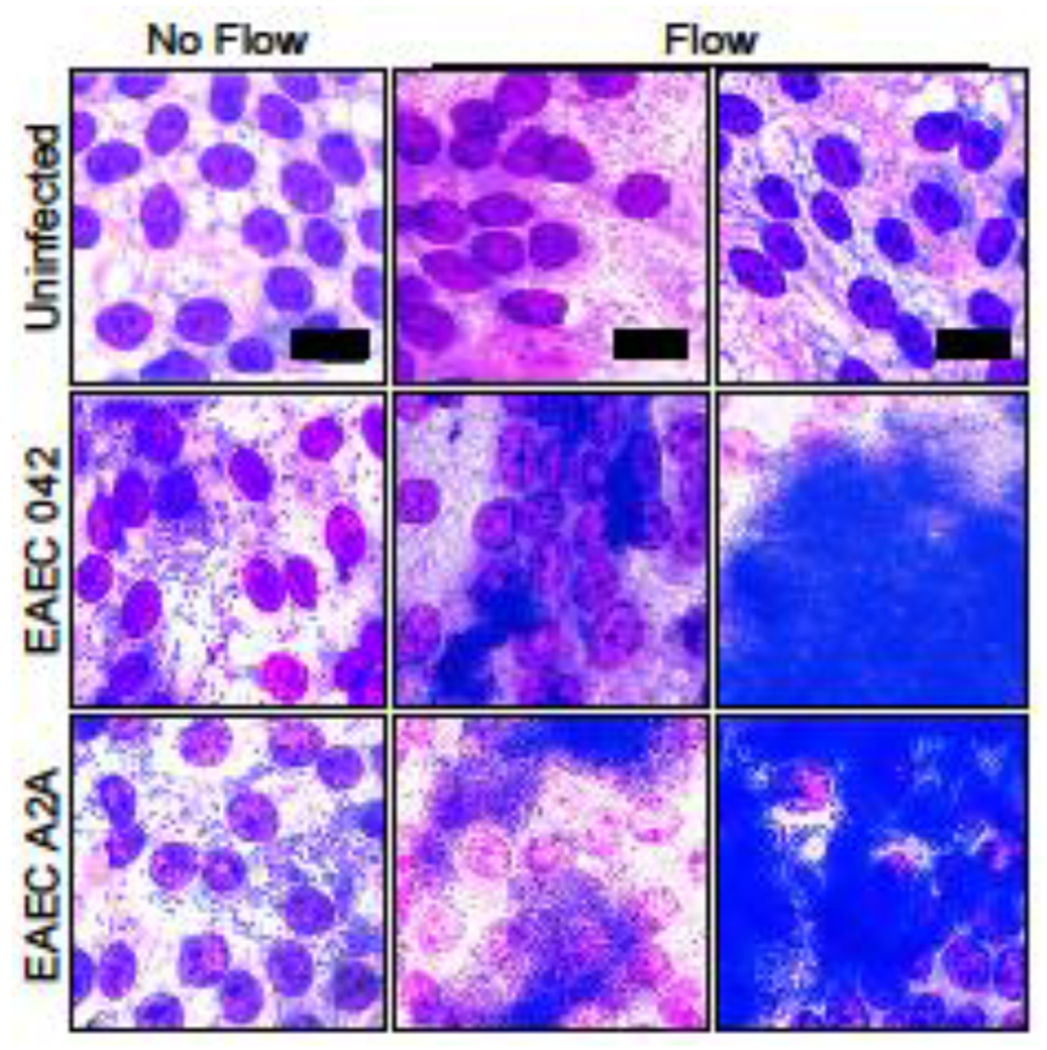
Wright Giemsa stain of HIE monolayers infected with EAEC 042 (prototype strain) and EAEC A2A (clinical isolate) with and without flow in the mPCs (8 hours post-infection). EAEC in the cultures without flow demonstrate aggregative adherence into small clusters (20-100 bacteria) whereas they formed biofilms in the cultures with flow (>1000 bacteria forming a lawn of aggregates). Cell nuclei appear purple and bacteria appear dark blue, scale = 30 μm. Images shown are representative of results from five independent experimental runs.
Expression of aggregative adherence virulence factors is required for the formation of EAEC biofilms
We next investigated the role of the aggregative adherence fimbriae (AAF), an important virulence factor expressed by most pathogenic strains of EAEC. Although AAF regulates bacterial adhesion, it has not yet been evaluated in a dynamic mechanical environment. Here, we infected HIEs with either wildtype EAEC or EAEC 042ΔaafA, which lacks aafA, a major structural subunit of AAF. We used a low MOI of 0.01 to ensure that the number of bacteria bound to the HIE surface represented AAF-epithelial interactions, as opposed to bacterial recruitment via paracrine signaling.
Without flow, both wildtype EAEC 042 and EAEC 042ΔaafA demonstrated comparable adhesion and aggregation with the HIE monolayer (Figure 5). After exposure to flow, their adherence patterns diverged significantly. Consistent with the observations in Figure 4, wildtype EAEC 042 in the flow condition formed large, 3D colonies, albeit smaller ones than before due to the lower MOI. In contrast, the density of EAEC 042ΔaafA bound to the epithelial cells decreased significantly with flow.
Figure 5: EAEC expression of aafA is required for biofilm formation.
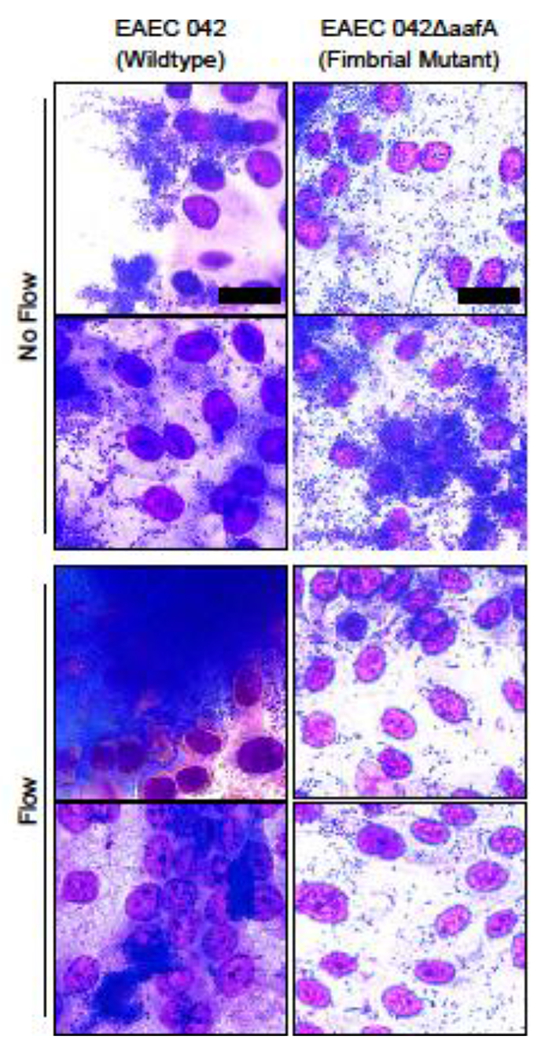
Wright Giemsa stain of HIE monolayers infected with wildtype EAEC 042 and EAEC 042ΔaafA, mutants with defective aggregative adherence fimbriae (AAF), with and without flow. EAECs only form large aggregative biofilms when they possess functional AAF. Images shown are representative of results from a proof-of-concept experiment. Scale = 20 μm.
mPCs allow co-administration of commensal organisms alongside pathological bacteria
Because the intestinal microbiome can impact host-pathogen interactions,21 we exposed HIE monolayers to EAEC 042, a commensal strain of E. coli (K12), or both. In the static mPCs, each of the three bacterial conditions caused complete destruction of the HIE monolayer (Figure 6), whereas under the flow condition, the monolayers remained intact for all three cases: EAEC 042, E. coli K12, and both, as shown by the DAPI stain. With flow, the pathological and commensal bacteria had different effects. Flow removed almost all of the commensal E. coli, with very few bacteria visible on the HIE monolayer afterwards. In contrast, the bacteria in the EAEC-only condition formed the thick biofilms when subjected to flow as seen earlier (Figure 4). The wells that received both commensals and EAECs did not have biofilms, but instead displayed dense aggregates of EAECs with small amounts of commensal E. coli.
Figure 6: Flow promotes the survival of HIE monolayers infected with both commensal and pathogenic bacteria.
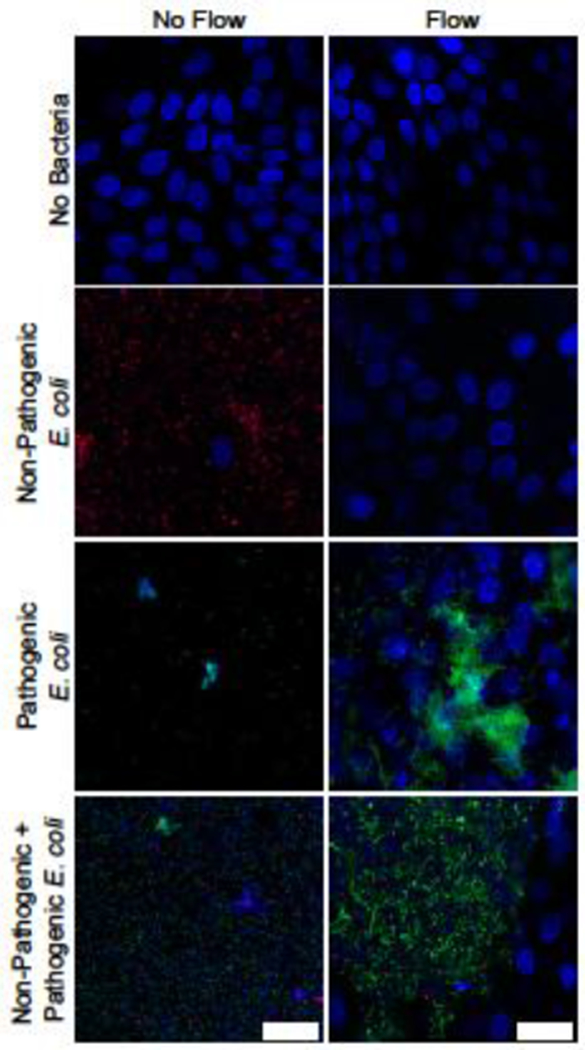
DAPI-stained HIE monolayers infected with RFP-expressing E. coli K12 (red), a commensal bacteria, and GFP-expressing EAEC 042 (green) with and without flow. Bacteria and HIE nuclei appear blue, scale = 25 μm. Both strains of bacteria destroy the HIE monolayer in static culture, but do not in the flow condition (top three panels). Co-infection with commensal bacteria reduces EAEC biofilm formation (comparison of the bottom two panels). Images shown are representative of results from three independent experimental runs.
mPCs permit human norovirus infections of HIE monolayers, and flow enhances viral replication
Finally, we infected jejunal HIEs with human norovirus to establish whether the mPCs were suitable for modeling viral causes of enteric disease; the jejunal response to noroviral infection is the most studied.5 In the 96-well plates, a 1.8 log10 (58-fold) increase in viral replication was seen after 24 hours (Figure S1). In the mPCs, 2.3 log10 (213-fold) and 2.6 log10 (386-fold) increases were observed after 24 hours for the wells cultured statically and under flow at 200 μL hr−1, respectively. While there was a trend towards increased viral replication in the static mPCs compared to 96-well plates, and the mPCs with flow compared to those without flow, a significant difference in norovirus genome equivalents was only observed between the 96-well plate and the mPCs with flow.
Discussion
Physical forces in the intestine, such as luminal mixing and fluid shear, significantly alter how pathogens interact with the intestinal epithelium,33 but how these factors influence enteric infectious disease is not well understood. Gaining more insight regarding the role of mechanics in this pathogenesis will require new experimental models. Adoption of these models, however, will depend on their appeal to a wide range of investigators regardless of their expertise with microfluidics or their access to expensive equipment for microfabrication. For these reasons, we developed a millifluidic perfusion cassette (mPC) that can model infections of intestinal epithelial cells with common viral and bacterial pathogens under flow.
The mPC is advantageously sized between microfluidic devices and typical parallel plate systems. This size niche allows for fabrication via 3D printed master molds instead of requiring specialized photolithography. Because the mPC chambers are close in size to wells of 96-well plates, cell culture protocols can be readily adapted for use with the device. This advantage allowed the HIEs to be seeded in the mPCs at higher densities than some previous reports,12 allowing confluent monolayers to form within 24 hours. As a result, the mPCs were able to successfully and reproducibly culture HIE monolayers, which can be fastidious and challenging. Furthermore, the mPC is transparent and compatible with various microscopy setups. Protein, DNA, or RNA (as shown here) can also be easily collected with micropipettes at the chamber inlet and outlet ports. The HIE monolayers produced in the mPC were flat and did not contain villi-like structures. Although we did not demonstrate the presence of apical microvilli in this work, their presence has been confirmed by prior work by our research group using both electron microscopy and immunofluorescence staining for actin and villin.5 Thus, the mPC is easy to fabricate and operate, especially for non-bioengineers, has moderate experimental throughput and media/cell requirements, and is compatible with common microscopic and biochemical analyses.
Using the mPC, we found that EAEC infections of intestinal epithelial cells conducted with flow produced biofilms, unlike those without flow. Until now, this pathophysiological hallmark has been difficult to reproduce in vitro, only succeeding when growing the bacteria in isolation (i.e., without intestinal epithelial cells).18 Previous static culture studies of EAEC infection of intestinal epithelial cells have been limited to adherence assays of 3–4 hours duration, since the bacteria would quickly overgrow and kill the intestinal epithelial cells. The ability to drive biofilm formation within the mPCs will allow investigators to develop an improved understanding of how this bacteria interacts with human hosts over a longer time frame, including the effect of the pathogen on the epithelial cell phenotype.
Our ability to establish biofilms in co-culture with intestinal epithelial cells allowed us to demonstrate in a proof-of-concept study that AAF, a virulence factor previously shown to mediate EAEC adherence and aggregation, is required for the formation of these biofilms. While such a role had previously been supported indirectly by a number of in vitro and in vivo studies,20,23 it could not be verified empirically due to the challenge of forming biofilms in co-culture. Using flow will enable investigation of how AAF engages with epithelial mucins22 in the mechanical environment of the intestinal lumen.
Similarly, the addition of flow to commensal-HIE co-cultures improved HIE survival, likely by replenishing the oxygen and nutrients in the mPC chambers. Studying how the complex intestinal microbiomes affect host-pathogen interactions has been hindered by the lack of experimental models allowing intestinal epithelial cells to be exposed to both commensals and pathogens. Indeed, in our experiments HIE monolayers exposed to commensal microorganisms in traditional static culture did not survive to the experimental endpoint. With flow, we showed significant differences between these pathogenic and commensal organisms that are consistent with their effects in vivo. We also noted that EAEC infections performed in the presence of commensal E. coli exhibited less extensive aggregation. This finding may have been due to competition between the two bacterial strains, or decreased EAEC virulence due to pathogen-commensal crosstalk. In-depth molecular analysis will be needed to assess these possibilities. Overall, these results indicate that the mPC enables studying how pathogenic bacteria behave in the presence of commensal organisms, and underline the importance of studying infections under flow conditions. While significant work remains to elucidate the mechanisms underlying these effects, the mPC is an effective tool that can be readily adopted by research groups to create more realistic in vitro models of enteric disease.
We speculate that flow improves the capabilities of these culture models by preventing the rapid bacterial overgrowth often found in static in vitro bacterial/eukaryotic co-cultures.13 By removing non-adherent bacteria and continually providing fresh media to the cell growth chamber, flow increased the availability of nutrients and oxygen to both the HIEs and the EAECs. This action would improve HIE survival and delay the stationary phase of the EAEC growth curve, revealing the pathophysiology of EAEC infection. Mechanobiological cues could also contribute to the flow-driven biofilm production: bacteria can transduce mechanical signals from their environment.19 Furthermore, fluid shear can increase E. coli adhesion through the catch-bond mechanism of the FimH fimbriae,29 although this effect reportedly requires shear forces at least 100 times greater than what we calculated for the mPC. This flow-driven biofilm formation may be linked to dispersin, a soluble factor produced by EAECs that sterically masks bacterial adhesins and inhibits EAEC aggregation.31 In static environments, dispersin collects near the bound EAECs, inhibiting biofilm formation. With flow, however, shear forces may push dispersin away from the EAECs, thereby lowering the local concentration, unmasking adhesins, and promoting biofilm formation.
It appears unlikely that biofilm formation was due to flow-induced changes in HIE phenotype, given that flow did not increase the production of epithelial mucins (MUC2), the major target of the EAEC adhesins that promote colonization.20 Expression of the Paneth cell marker LYZ was similarly unaffected, suggesting that flow did not promote proliferation of Paneth cells, which secrete antimicrobial compounds. This absence of overt changes in cellular behavior allows the effects of flow to be studied directly, without confounding changes in HIEs susceptibility to EAEC infection. Additionally, the shear stresses used in our experiments were chosen to minimize the potential for cellular changes, as they fall at the low end of the shear stresses estimated for the human intestine.10,17 For endothelial cells, shear stress is also known to induce a range of functional changes, including realignment of the cells, although the shear stresses applied in endothelial cell studies is commonly on the order of tens of dyn cm−2.4,9 We speculate that the range of shear stress applied in this work was too low to induce alignment of the HIEs. It was reported that even tenfold-higher shear stresses of 0.02 – 1 dyn cm−2 did not cause alignment of renal epithelial cells when they were cultured on a flat surface.7 The shear stresses applied in our mPCs were also approximately tenfold less than those in previous experiments reporting shear-induced phenotypic changes in intestinal cells.12,14 Moreover, those studies simultaneously applied cyclic strain alongside shear stress, making it difficult to attribute the changes to a specific mechanical stimuli. Finally, in contrast to prior work,12 we only applied mechanical stimulation to terminally differentiated HIEs, which have greatly limited phenotypic plasticity. Therefore, we do not believe that the biofilm formation in this work was due to shear stress-induced phenotypic changes in the HIEs. However, it will be of great interest to use the mPC in future investigations of HIE phenotype that apply flow at higher rates and during the HIE differentiation period, and that examine known mechanosensitive targets in more detail.
Unlike bacteria, viruses do not have independent metabolism or mechanosensory apparatuses when they are outside of a host cell. Therefore, the most likely way for flow to affect viral replication is through the secondhand effects of flow on infected intestinal epithelial cells. Norovirus binds to histo-blood group antigens present in the glycocalyx of the intestinal epithelium of secretor-positive persons,27 and there is considerable evidence that endothelial glycocalyces are altered by exposure to shear stress.32 It is possible that increased histo-blood group antigen expression drives the increased viral replication seen with flow, however, this needs experimental confirmation. Another possibility is that flow improves the overall health and/or metabolism of the HIEs, allowing improved viral replication. Finally, presentation of the virus particles to the HIEs may be enhanced by more rapid mass transport due to flow and/or shorter chamber height in the mPCs compared to 96-well plates.
In conclusion, we have engineered a millifluidic perfusion cassette that is easy to produce, requires minimal adaptation of traditional cell culture techniques, and is compatible with a wide array of microscopic and biochemical analysis. Using this simple system, we investigated the pathogenesis of enteric infections and found important differences between infections performed under static conditions and infections in the presence of flow. These results provide compelling motivation to investigate the mechanisms driving flow-induced alterations of bacterial and viral infection. The ability to form EAEC biofilms in co-culture with HIEs, in particular, opens up many possible lines of inquiry. Taken together, these initial findings demonstrate the utility of our millifluidic culture system, and underscore the importance of studying these pathological processes with in vitro models that mimic the dynamic intestinal environment.
Supplementary Material
Figure S1: RT-qPCR quantification of viral replication in HIE monolayers in 96-well plates and mPCs with and without flow. Viral RNA was measured at 1 hour and 24 hours post-infection. The fold change in viral genome equivalents between the two time points is shown for each condition. *** p < 0.005. Data are compiled from three separate experiments. Each data point represents three individual wells pooled together.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (U19 AH 16497 and F30 DK108541) and CPRIT RP160283 – Baylor College of Medicine Comprehensive Cancer Training Program. The authors thank Drs. Noah Shroyer and Sue Crawford for helpful discussions and insights, Dr. Jim Broughman and Xi-Lei Zheng for technical assistance with the flow experiments, and Dr. Jennifer Connell for editorial assistance.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- 1.Blutt SE, Broughman JR, Zou W, Zeng XL, Karandikar UC, In J, Zachos NC, Kovbasnjuk O, Donowitz M, and Estes MK. Gastrointestinal microphysiological systems. Exp. Biol. Med 242:1633–1642, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson GP, Lee SM, and Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 14:20–32, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donowitz M, Alpers DH, Binder HJ, Brewer T, Carrington J, and Grey MJ. Translational approaches for pharmacotherapy development for acute diarrhea. Gastroenterology 142:e1–e9, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebong EE, V Lopez-Quintero S, Rizzo V, Spray DC, and Tarbell JM. Shear-induced endothelial NOS activation and remodeling via heparan sulfate, glypican-1, and syndecan-1. Integr. Biol 6:338–347, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, and Estes MK. Replication of human noroviruses in stem cell-derived human enteroids. Science. 353:1387–1393, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, Hyser JM, Zeng X-L, Crawford SE, Broughman JR, Estes MK, and Donowitz M. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp. Biol. Med 239:1124–34, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frohlich EM, Zhang X, and Charest JL. The use of controlled surface topography and flow-induced shear stress to influence renal epithelial cell function. Integr. Biol 4:75–83, 2012. [DOI] [PubMed] [Google Scholar]
- 8.Gayer CP, and Basson MD. The effects of mechanical forces on intestinal physiology and pathology. Cell. Signal 21:1237–44, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang RB, and Eniola-Adefeso O. Shear stress modulation of IL-1β-induced E-selectin expression in human endothelial cells. PLoS One 7:e31874, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa T, Sato T, Mohit G, Imai Y, and Yamaguchi T. Transport phenomena of microbial flora in the small intestine with peristalsis. J. Theor. Biol 279:63–73, 2011. [DOI] [PubMed] [Google Scholar]
- 11.James BD, Montoya N, and Allen JB. MechanoBioTester: A decoupled multistimulus cell culture device for studying complex microenvironments in vitro. ACS Biomater. Sci. Eng 6:3673–3689, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, and Ingber DE. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep 8:2871, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HJ, Huh D, Hamilton GA, and Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12:2165–74, 2012. [DOI] [PubMed] [Google Scholar]
- 14.Kim HJ, Li H, Collins JJ, and Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci 201522193, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotloff KL et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 382:209–222, 2013. [DOI] [PubMed] [Google Scholar]
- 16.Kovbasnjuk Ο, Zachos NC, In J, Foulke-Abel J, Ettayebi K, Hyser JM, Broughman JR, Zeng X-L, Middendorp S, de Jonge HR, Estes MK, and Donowitz M. Human enteroids: preclinical models of non-inflammatory diarrhea. Stem Cell Res. Ther 4:S3, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lentle RG, and Janssen PWM. Physical characteristics of digesta and their influence on flow and mixing in the mammalian intestine: A review. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol 178:673–690, 2008. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed JA, Huang DB, Jiang ZD, DuPont HL, Nataro JP, Belkind-Gerson J, and Okhuysen PC. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J. Clin. Microbiol 45:121–126, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, and Stone HA. The mechanical world of bacteria. Cell 161:988–997, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Philipson CW, Bassaganya-Riera J, and Hontecillas R. Animal models of enteroaggregative Escherichia coli infection. Gut Microbes 4:281–291, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preidis GA, Hill C, Guerrant RL, Ramakrishna BS, Tannock GW, and Versalovic J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology 140:8–14.e9, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan A, Vela L, Zeng X, Yu X, Shroyer N, Blutt SE, Poole NM, Carlin LG, Nataro JP, Estes MK, Okhuysen PC, and Maresso AW. Novel segment- and host-specific patterns of enteroaggregative escherichia coli adherence to human intestinal enteroids. MBio 9:1–15, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz-Perez F, Sheikh J, Davis S, Boedeker EC, and Nataro JP. Use of a continuous-flow anaerobic culture to characterize enteric virulence gene expression. Infect. Immun 72:3793–3802, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato T, and Clevers HC. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340:1190–4, 2013. [DOI] [PubMed] [Google Scholar]
- 25.Sato T, Vries RGJ, Snipped HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, and Clevers HC. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–5, 2009. [DOI] [PubMed] [Google Scholar]
- 26.Saxena K, Blutt SE, Ettayebi K, Zeng X, Broughman JR, Crawford SE, Karandikar UC, Sastri NP, Conner ME, Opekun AR, Graham DY, Qureshi W, Sherman V, Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Donowitz M, and Estes K. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol 90:43–56, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroten H, Hanisch FG, and Hansman GS. Human norovirus interactions with histo-blood group antigens and human milk oligosaccharides. J. Virol 90:5855–5859, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah P, V Fritz J, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jager C, Seguin-Devaux C, Zenhausern F, and Wilmes P. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun 7:, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas WE, Nilsson LM, Forero M, Sokurenko EV, and Vogel V. Shear-dependent “stick-and-roll” adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol 53:1545–1557, 2004. [DOI] [PubMed] [Google Scholar]
- 30.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, and Stappenbeck TS. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64:911–20, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velarde JJ, Varney KM, Inman KG, Farfan M, Dudley E, Fletcher J, Weber DJ, and Nataro JP. Solution structure of the novel dispersin protein of enteroaggregative Escherichia coli. Mol. Microbiol 66:1123–1135, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Tiemeier GL, van den Berg BM, and Rabelink TJ. Endothelial glycocalyx hyaluronan: regulation and role in prevention of diabetic complications. Am. J. Pathol 190:781–790, 2020. [DOI] [PubMed] [Google Scholar]
- 33.De Weirdt R, and Van De Wiele T. Micromanagement in the gut: Microenvironmental factors govern colon mucosal biofilm structure and functionality, npj Biofilms Microbiomes 1:1–6, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: RT-qPCR quantification of viral replication in HIE monolayers in 96-well plates and mPCs with and without flow. Viral RNA was measured at 1 hour and 24 hours post-infection. The fold change in viral genome equivalents between the two time points is shown for each condition. *** p < 0.005. Data are compiled from three separate experiments. Each data point represents three individual wells pooled together.


