Abstract
Light plays a critical role in plant growth and development, but the mechanisms through which light regulates fruit ripening and nutritional quality in horticultural crops remain largely unknown. Here, we found that ELONGATED HYPOCOTYL 5 (HY5), a master regulator in the light signaling pathway, is required for normal fruit ripening in tomato (Solanum lycopersicum). Loss of function of tomato HY5 (SlHY5) impairs pigment accumulation and ethylene biosynthesis. Transcriptome profiling identified 2948 differentially expressed genes, which included 1424 downregulated and 1524 upregulated genes, in the Slhy5 mutants. In addition, genes involved in carotenoid and anthocyanin biosynthesis and ethylene signaling were revealed as direct targets of SlHY5 by chromatin immunoprecipitation. Surprisingly, the expression of a large proportion of genes encoding ribosomal proteins was downregulated in the Slhy5 mutants, and this downregulation pattern was accompanied by a decrease in the abundance of ribosomal proteins. Further analysis demonstrated that SlHY5 affected the translation efficiency of numerous ripening-related genes. These data indicate that SlHY5 regulates fruit ripening both at the transcriptional level by targeting specific molecular pathways and at the translational level by affecting the protein translation machinery. Our findings unravel the regulatory mechanisms of SlHY5 in controlling fruit ripening and nutritional quality and uncover the multifaceted regulation of gene expression by transcription factors.
Subject terms: Plant physiology, Plant molecular biology
Introduction
Light not only serves as a source of energy for plant photosynthesis, but also represents an important cue that regulates plant growth and development, including seed germination1, seedling growth2, flowering3, and eventually senescence4. To sense light, plants have evolved several classes of photoreceptors, among which cryptochromes and phototropins monitor blue light, phytochromes monitor red/far-red light, and UVR8 monitors UV-B light5. Upon light perception, the photoreceptors interpret and transduce light signals to the downstream core signaling networks, which leads to remodeling of the transcriptome and changes in growth and development6.
Transcriptional regulation plays critical roles in light-regulated growth and developmental processes. Nearly 20–35% of plant genes display massive transcriptional reprogramming under light7. Several transcription factors, such as PHYTOCHROME-INTERACTING FACTORs (PIFs) and ELONGATED HYPOCOTYL 5 (HY5), which exhibit antagonistic actions, have been identified as the key components in the light signaling pathway8. PIFs belong to the family of basic helix-loop-helix transcription factors, whereas HY5 is a member of the basic leucine zipper (bZIP) transcription factor family9,10. The stability of both PIFs and HY5 is regulated by CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ubiquitin ligase that mediates protein ubiquitination and degradation by the 26S proteasome11. COP1 directly targets HY5 for degradation but positively affects the PIF protein levels via an indirect effect12. In the presence of light, COP1 is repressed by photoreceptor phytochromes and cryptochromes, which results in instable PIFs and the accumulation of HY5 and thereby in the promotion of photomorphogenesis13.
As a central positive regulator in the light signaling pathway, HY5 is involved in fundamental developmental processes in plants, including cell proliferation, cell elongation, and chloroplast development14. Interestingly, in tomato (Solanum lycopersicum) fruit, HY5 regulates the accumulation of carotenoids15, a group of 40-carbon isoprenoid compounds that contribute to fruit color and nutritional quality in various horticultural crops. HY5 might act in concert with other components in the light signaling pathway, including PIF1a16, DAMAGED DNA BINDING PROTEIN 1/HIGH PIGMENT 1 (DDB1/HP1)17, DEETIOLATED 1/HIGH PIGMENT 2 (DET1/HP2)18, CULLIN 4 (CUL4)17, and COP1-like15, to modulate fruit carotenoid accumulation in tomato. However, the direct targets modulated by HY5 in horticultural crops and the mechanisms underlying its transcriptional regulation remain largely unknown. Furthermore, whether HY5 modulates other physiological aspects of fruit development remains unclear. Such information could be highly useful for unraveling the regulatory cascade controlled by HY5 and for understanding the connections with other regulatory networks that control fruit quality.
In the present study, to characterize the biological function of tomato HY5 (SlHY5) and its regulatory mechanisms, we generated Slhy5 mutants using CRISPR/Cas9 genome editing technology. We found that the SlHY5 mutation impaired the normal fruit ripening process through reduced pigment accumulation and ethylene release. A transcriptome analysis coupled with chromatin immunoprecipitation (ChIP) indicated that SlHY5 directly bound to promoters of genes involved in carotenoid and anthocyanin biosynthesis, and genes associated with ethylene generation. Surprisingly, we found that SlHY5 influenced the protein translation efficiency of numerous ripening genes by targeting ribosomal protein genes. Our findings suggest that SlHY5 regulates fruit ripening both at the transcriptional level by targeting specific molecular pathways and at the translational level by affecting the protein translation machinery.
Results
CRISPR/Cas9-engineered mutations in SlHY5 cause abnormal ripening in tomato fruit
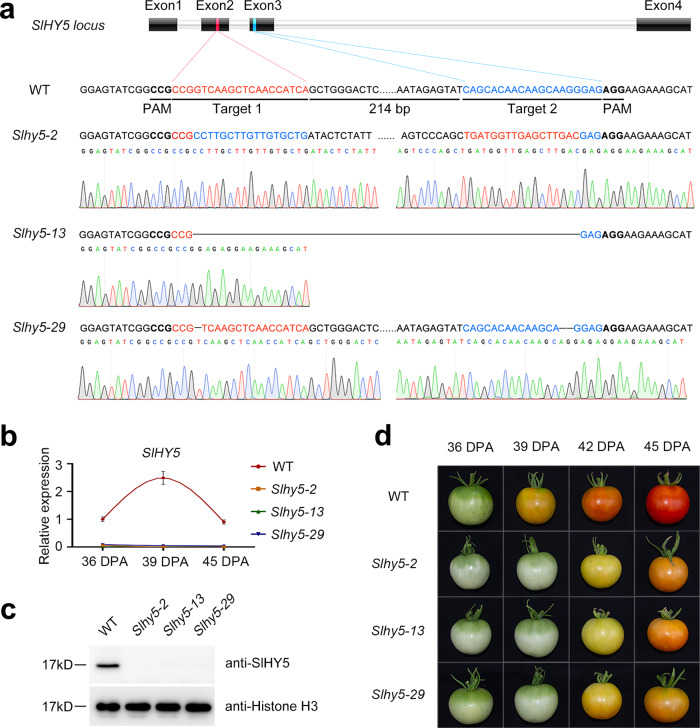
In recent years, CRISPR/Cas9 gene-editing technology has been widely used in studies of plant gene function. To better understand the biological function of SlHY5 (Solyc08g061130), the only HY5 gene in tomato, we generate mutations in the SlHY5 locus using the CRISPR/Cas9 gene-editing system. Two specific target sites were designed in the second and third exons (Fig. 1a). We ultimately obtained 31 independent transgenic lines of the T0 generation through Agrobacterium tumefaciens-mediated transformation and the mutation of SlHY5 was verified by sequencing genomic regions flanking the target sites. We isolated three distinct homozygous mutant lines (Slhy5-2, Slhy5-13, and Slhy5-29) of the T1 generation and used these for further analysis. A large 248-bp inversion between the two targets was found at the SlHY5 locus in the Slhy5-2 line, whereas a deletion of 248 bp appeared in the Slhy5-13 line, and the Slhy5-29 line harbored a 1-bp deletion in the first target and a 2-bp deletion in the second target (Fig. 1a). The three mutants were predicted to produce nonfunctional truncated SlHY5 proteins. We performed an off-target analysis and found that no genome editing occurred at the most likely off-target sites, which were predicted using CRISPR-P, in the three mutagenesis lines (Supplementary Fig. 1). Quantitative real-time PCR and western blotting analyses were performed to determine the expression of SlHY5 in the Slhy5 mutants at both the mRNA and protein levels. The SlHY5 transcript was almost completely silent in the Slhy5 mutants, and the SlHY5 protein was also completely undetectable (Fig. 1b, c). These results confirmed that SlHY5 was successfully mutated.
Fig. 1. SlHY5 is required for the normal ripening of tomato fruit.
a Genotype of mutations in the SlHY5 locus generated by the CRISPR/Cas9 genome editing system. Two target sequences were designed to specifically target the second and third exons with a 214-bp interval. The red and blue letters indicate the sequences of targets 1 and 2, respectively. The protospacer adjacent motif (PAM) is indicated. The mutations in the transgenic plants were confirmed by sequencing the genomic regions flanking the target sites. The sequences of the wild-type (WT) plants and three homozygous mutant lines (Slhy5-2, Slhy5-13, and Slhy5-29) are shown. b Transcript levels of SlHY5 in wild-type and Slhy5 mutant fruits determined by quantitative real-time PCR. c SlHY5 protein levels in wild-type and Slhy5 mutant fruits determined by western blotting. The histone H3 protein levels were used as a loading control. d Ripening phenotype of Slhy5 mutants. Wild-type and Slhy5 mutant fruits at 36, 39, 42, and 45 days post anthesis (DPA) are shown
The fruit ripening progress was markedly delayed in the Slhy5-2, Slhy5-13, and Slhy5-29 mutant lines compared with the wild-type plants (Fig. 1d). The onset of ripening in the wild-type fruit occurred at 39 days post anthesis (DPA), whereas a visible color change started to be observed in the fruit of Slhy5 mutants at 42 DPA. At 45 DPA, the wild-type fruit was fully ripe and showed a red color, whereas the fruits of the Slhy5 mutants remained orange. Even at the final stage of fruit ripening, the fruits of the Slhy5 mutants could not turn completely red and showed a light-red color (Supplementary Fig. 2). In addition, the green fruits of the Slhy5 mutants were whiter than those of the wild-type plants. The phenotype of delayed fruit ripening was also observed in the T2 generation, which further confirmed that SlHY5 regulates the ripening of tomato.
Seedlings of the Slhy5 mutants growing under standard conditions displayed a significantly increased hypocotyl growth compared with the wild-type seedlings. After the transplantation of 4-week-old seedlings into the field, the Slhy5 mutants exhibited a higher mortality rate than the wild-type plants. Moreover, the Slhy5 mutant plants showed pale green leaves and pale yellow flowers (data not shown).
The Slhy5 mutants exhibit impaired expression of ripening-related genes
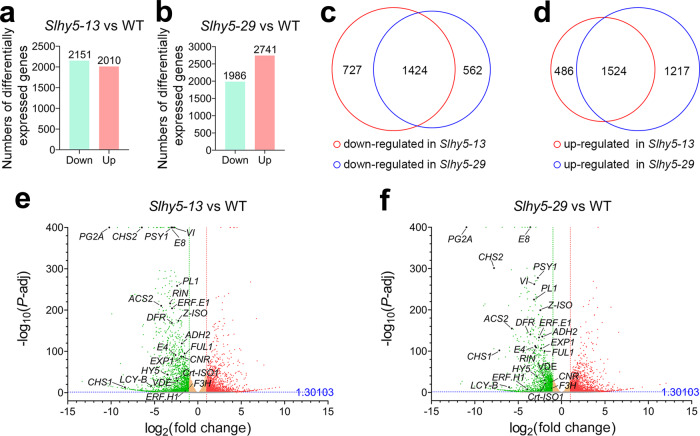
To determine how SlHY5 affects tomato fruit ripening, we performed a comparative transcriptome analysis between the wild-type plants and the Slhy5 mutants (Slhy5-13 and Slhy5-29) at 39 DPA based on RNA sequencing (RNA-seq). In total, we obtained >492 million clean reads, and >95% of these reads could be mapped to unique loci in the reference genome. A bioinformatics analysis identified 4161 differentially expressed genes, which included 2151 downregulated [log2(fold change) < −1, P-adj < 0.05] and 2010 upregulated [log2(fold change) > 1, P-adj < 0.05] genes in the Slhy5-13 line (Fig. 2a and Supplementary Table 1). Moreover, 1986 downregulated and 2741 upregulated genes were differentially expressed in the Slhy5-29 line (Fig. 2b and Supplementary Table 2). Overall, 1424 downregulated and 1524 upregulated differentially expressed genes were found in both mutants (Fig. 2c, d and Supplementary Table 3). A high Pearson correlation coefficient was found between the two mutants, which represented the high similarity between them (Supplementary Fig. 3). Notably, the SlHY5 mRNA level was significantly decreased in both lines (Fig. 2e, f and Supplementary Table 3), which suggested that genome editing might cause RNA instability due to frameshift mutations. Among the differentially expressed genes, genes involved in anthocyanin biosynthesis (CHS1, CHS2, CHI, F3H, F3’H, DFR, and ANS), ethylene biosynthesis and response (ACS2, E4, E8, ERF.E1, and ERF.H1), carotenoid metabolism (PSY1, Z-ISO, Crt-ISO1, LCY-B, and VDE), volatile aroma compound biosynthesis (ADH2), sucrose metabolism (VI), and cell wall degradation (PG2A, PL1, and EXP1) were substantially downregulated. The expression of some well-known ripening-related transcription factors, such as RIN, CNR, and FUL1, was also decreased in the Slhy5 mutants (Fig. 2e, f).
Fig. 2. Expression of numerous ripening-related genes is decreased in the Slhy5 mutants.
a, b Number of differentially expressed genes that were downregulated or upregulated in the Slhy5-13 (a) and Slhy5-29 (b) mutants versus wild-type (WT) plants at 39 days post anthesis (DPA). c, d Venn diagrams showing the overlapping downregulated (c) and upregulated (d) genes that were differentially expressed in the Slhy5-13 and Slhy5-29 mutants. e, f Volcano diagrams revealing the differentially expressed genes in the Slhy5-13 (e) and Slhy5-29 (f) mutant versus WT plants. The colored spots indicate P-adj < 0.05, which indicates that the differences in gene expression are significant. The green and red spots show downregulated and upregulated genes with more than twofold changes, respectively. Some of the ripening-related genes are marked by black spots. ACS2 1-aminocyclopropane-1-carboxylic acid synthase 2, ADH2 alcohol dehydrogenase 2, ANS anthocyanidin synthase, CHI chalcone-flavonone isomerase, CHS1 chalcone synthase 1, CHS2 chalcone synthase 2, CNR colorless non-ripening, Crt-ISO1 carotene isomerase 1, DFR dihydroflavonol-4-reductase, E4 ethylene response gene 4, E8 ethylene response gene 8, ERF. E1 ethylene response factor E1, ERF. H1 ethylene response factor H1, EXP1 expansin 1, F3H flavanone 3-hydroxylase, F3′H flavonoid 3′-hydroxylase, FUL1 FRUITFULL 1, HY5 ELONGATED HYPOCOTYL5, LCY-B lycopene beta-cyclase, PG2A polygalacturonase 2A, PL1 pectate lyase 1, PSY1 phytoene synthase 1, RIN ripening inhibitor, VDE violaxanthin de-epoxidase, VI vacuolar invertase, Z-ISO 15-cis-zeta-carotene isomerase
Carotenoid biosynthesis is inhibited in the Slhy5 mutants
The accumulation of carotenoids appears to be a typical feature during tomato fruit ripening. The RNA-seq assay revealed that eight genes (PSY1, Z-ISO, Crt-ISO1, LCY-E, LCY-B, VDE, NCED1, and NCED2) involved in carotenoid metabolism were differentially expressed in the Slhy5 mutants (Fig. 3a). PSY is the rate-limiting enzyme in the carotenoid biosynthetic pathway (Fig. 3b). The tomato genome contains three PSY genes, and only PSY1 is expressed specifically during fruit ripening and contributes to carotenoid accumulation in tomato fruit19. A ChIP-qPCR analysis was performed to identify whether genes (PSY1, Z-ISO, Crt-ISO1, LCY-E, LCY-B, and VDE) involved in the metabolism of lycopene, β-carotene, and phytoene, which are the main carotenoids in tomato fruit, were directly regulated by SlHY5.
Fig. 3. Carotenoid biosynthesis is regulated by SlHY5.
a Heatmap showing the differentially expressed genes involved in carotenoid biosynthesis identified from the comparison of the Slhy5 mutant (lines Slhy5-13 and Slhy5-29) versus wild-type (WT) fruits at 39 days post anthesis (DPA) based on RNA-seq analysis. b Sketch diagram displaying the core carotenoid metabolism pathway. c ChIP-qPCR assays showing that SlHY5 binds to the promoters of genes involved in carotenoid biosynthesis. The promoter structures of the SlHY5 target genes are shown. The blue boxes represent ACGT-containing elements, and the numbers indicate the positions of these motifs relative to the translational start site. The black lines with uppercase letters represent the regions used for ChIP-qPCR. The values show the percentages of DNA fragments that coimmunoprecipitated with anti-SlHY5 antibodies relative to the input DNAs. PSY1 phytoene synthase 1, Z-ISO 15-cis-zeta-carotene isomerase, Crt-ISO1 carotene isomerase 1, LCY-E lycopene epsilon-cyclase, LCY-B lycopene beta-cyclase, VDE violaxanthin de-epoxidase. d Carotenoid accumulation in Slhy5 mutants at 39 and 45 DPA. The error bars represent the standard deviations from three independent experiments. The asterisks indicate significant differences (*P < 0.05)
We scanned the presence of ACGT-containing elements, which have been elucidated as the binding sites of HY5 (refs. 20,21), in the promoter regions of the candidate genes and found one to five ACGT-containing elements in the 2-kb region upstream from the start codon (ATG). For the ChIP-qPCR assay, cross-linked DNA–protein complexes were immunoprecipitated using affinity-purified anti-SlHY5 polyclonal antibodies. Specific primers were designed to amplify the promoter regions surrounding the ACGT-containing elements from the immunoprecipitated DNA. The binding of SlHY5 to the promoters was expressed as the relative amount of immunoprecipitated DNA fragments versus the input DNA fragments. As shown in Fig. 3c, SlHY5 bound to the promoters of all the candidate genes involved in carotenoid biosynthesis, which indicated that SlHY5 directly regulates these genes.
We next determined the contents of lycopene, β-carotene, and phytoene in tomato fruit (Fig. 3d). At 39 DPA, only β-carotene could be detected, and higher levels of carotenoid were found in the wild-type fruits than in the Slhy5 mutants. At 45 DPA, phytoene was not detected in the Slhy5 mutants but appeared in the wild-type fruits. Although lycopene and β-carotene could be detected in the Slhy5 mutants at 45 DPA, their contents were significantly lower in the mutants than in the wild-type fruits. Taken together, these results indicated that SlHY5 affects carotenoid accumulation by targeting genes involved in the carotenoid biosynthetic pathway.
Anthocyanin accumulation is suppressed in the Slhy5 mutants
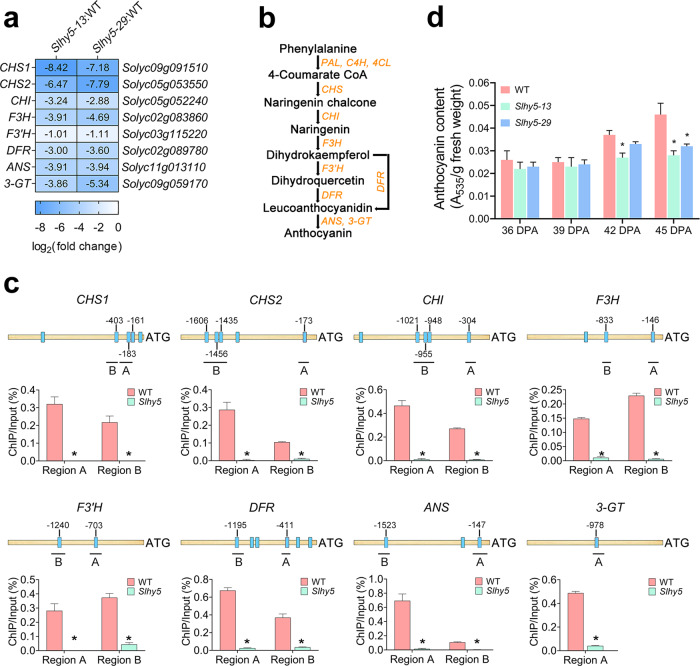
HY5 has been demonstrated to regulate anthocyanin biosynthesis in tomato and Arabidopsis22,23. Anthocyanin biosynthesis via the phenylpropanoid pathway is a complicated secondary metabolic process24, and chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H) contribute to the early steps of anthocyanin biosynthesis. Flavonoid 3′-hydroxylase (F3′H), a P450 enzyme, catalyzes the hydroxylation of dihydrokaempferol to form dihydroquercetin. Dihydrokaempferol and dihydroquercetin are then reduced to the corresponding leucoanthocyanidins by dihydroflavonol-4-reductase (DFR). Anthocyanins are converted from leucoanthocyanins by anthocyanidin synthase (ANS) and UDP-glucose:flavonoid-O-glycosyltransferase (3-GT). Our RNA-seq data showed that crucial genes involved in anthocyanin biosynthesis (CHS1, CHS2, CHI, F3H, F3′H, DFR, ANS, and 3-GT) were markedly reduced in the Slhy5 mutants (Fig. 4a, b), and a ChIP-qPCR analysis indicated that all of these genes were directly regulated by SlHY5 (Fig. 4c).
Fig. 4. Anthocyanin biosynthesis is regulated by SlHY5.
a Heatmap showing the differentially expressed genes involved in anthocyanin synthesis that were identified from the comparison of the Slhy5 mutant (lines Slhy5-13 and Slhy5-29) versus wild-type (WT) fruits at 39 days post anthesis (DPA) based on RNA-seq analysis. b Sketch diagram exhibiting the core anthocyanin biosynthesis pathway. c ChIP-qPCR assays showing that SlHY5 binds to the promoters of genes involved in anthocyanin synthesis. The promoter structures of the SlHY5 target genes are shown. The blue boxes represent ACGT-containing elements, and the numbers indicate the positions of these motifs relative to the translational start site. The black lines with uppercase letters represent the regions used for ChIP-qPCR. The values are the percentages of DNA fragments that coimmunoprecipitated with anti-SlHY5 antibodies relative to the input DNAs. ANS anthocyanidin synthase, CHI chalcone-flavonone isomerase, CHS1 chalcone synthase 1, CHS2 chalcone synthase 2, DFR dihydroflavonol-4-reductase, F3H flavanone 3-hydroxylase, F3′H flavonoid 3′-hydroxylase, 3-GT flavonol-3-glucosyltransferase. d Anthocyanin accumulation in Slhy5 mutants during fruit ripening. The error bars represent the standard deviations from three independent experiments. The asterisks indicate significant differences (*P < 0.05)
We then measured anthocyanin content in the wild-type and Slhy5 mutant fruits at various ripening stages. During the early ripening stages (36 DPA and 39 DPA), no significant difference in the anthocyanin content was found between the wild-type and Slhy5 mutants. However, at the later ripening stage (45 DPA), the accumulation of anthocyanin in the Slhy5 mutants was distinctly lower than that in the wild type (Fig. 4d). These results confirmed that SlHY5 regulates anthocyanin biosynthesis directly.
SlHY5 mutation influences ethylene production and response
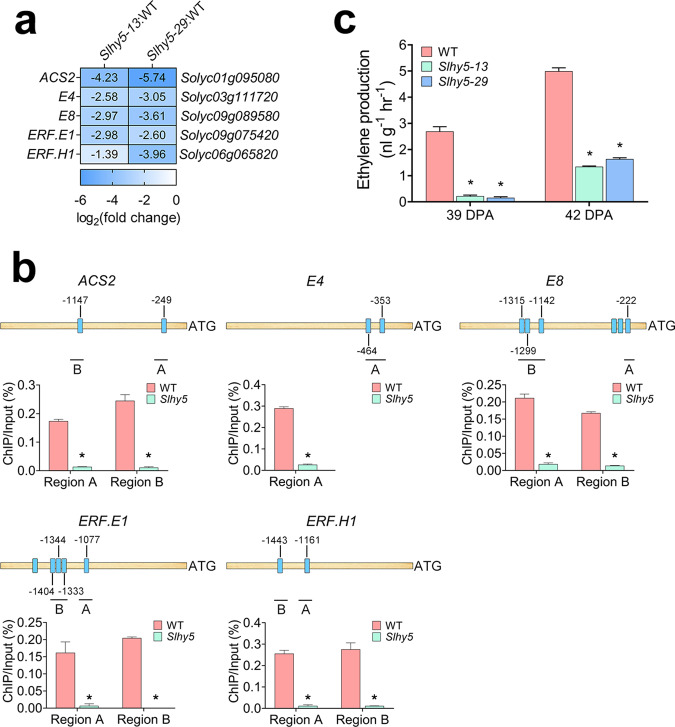
The RNA-seq analysis showed that a number of genes involved in ethylene biosynthesis and response were downregulated in the Slhy5 mutants (Fig. 5a and Supplementary Fig. 4). Five of these genes (ACS2, E4, E8, ERF.E1, and ERF.H1) were selected for the ChIP-qPCR assay due to their critical roles in ethylene signaling. ACS encodes 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, which is the rate-limiting enzyme in ethylene biosynthesis. Fourteen ACS isoforms have been identified in tomato25. ACS2 represents the most abundant ACS isoform in ripening fruit, and the inhibition of ACS2 by antisense RNA blocks fruit ripening26. A ChIP-qPCR assay showed that SlHY5 directly bound to the promoter of ACS2 (Fig. 5b).
Fig. 5. Ethylene production and response are regulated by SlHY5.
a Heatmap showing the differentially expressed genes involved in ethylene production and response identified from the comparison of the Slhy5 mutant (Slhy5-13 and Slhy5-29 lines) versus wild-type (WT) fruits at 39 days post anthesis (DPA) based on RNA-seq analysis. b ChIP-qPCR assays showing that SlHY5 binds to the promoters of genes involved in ethylene production and response. The promoter structures of the SlHY5 target genes are shown. The blue boxes represent ACGT-containing elements, and the numbers indicate the positions of these motifs relative to the translational start site. The black lines with uppercase letters represent the regions used for ChIP-qPCR. The values are the percentages of DNA fragments that coimmunoprecipitated with anti-SlHY5 antibodies relative to the input DNAs. ACS2 1-aminocyclopropane-1-carboxylic acid synthase 2, E4 ethylene response gene 4, E8 ethylene response gene 8, ERF.E1 ethylene response factor E1, ERF.H1 ethylene response factor H1. c Ethylene release in Slhy5 mutants at 39 and 42 DPA. The error bars represent the standard deviations from three independent experiments. The asterisks indicate significant differences (*P < 0.05)
E4 encodes a putative methionine sulfoxide reductase, and E8 encodes a 1-aminocyclopropane-1-carboxylate oxidase homolog without catalytic activity to convert ACC to ethylene27,28. Although the molecular functions of E4 and E8 remain unclear, they are considered typical ripening-related genes29,30. As shown in Fig. 5b, both E4 and E8 were directly targeted by SlHY5.
SlERF.E1, which was formerly known as LeERF2 (ref. 31), encodes an ethylene response factor that displays a ripening-related expression pattern32, whereas SlERF.H1, previously named LeERF1, affects tomato fruit ripening and softening33. ChIP-qPCR assays indicated that SlHY5 directly bound to the promoters of SlERF.E1 and SlERF.H1 (Fig. 5b).
We subsequently analyzed ethylene synthesis and found that the Slhy5 mutants produced appreciably decreased levels of ethylene than the wild-type fruits at 39 DPA and 42 DPA, when the ethylene burst arose (Fig. 5c). These data demonstrated that SlHY5 directly regulates ethylene production and response.
Translation is partially impaired in the Slhy5 mutants
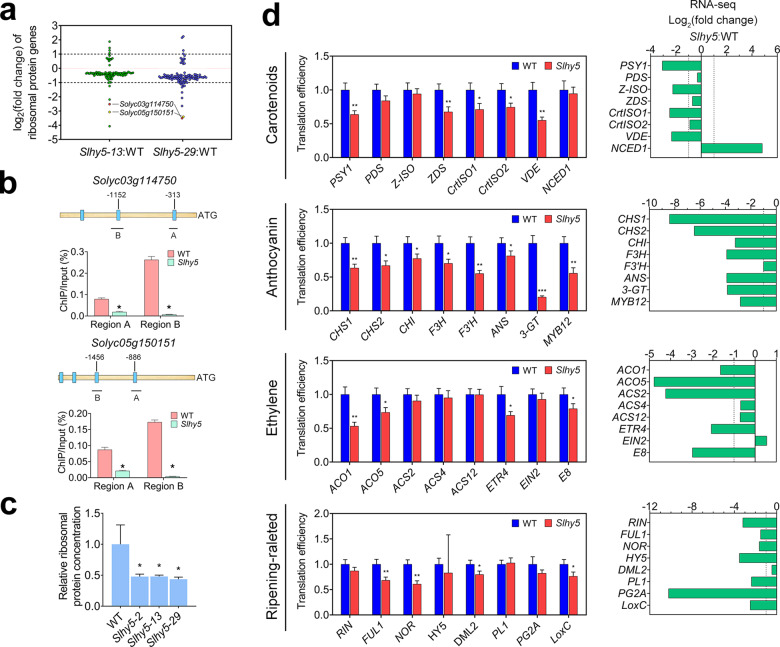
According to International Tomato Annotation Group version 4.0 (ITAG4.0), 544 annotated genes encode ribosomal proteins. Interestingly, in our RNA-seq analysis, we identified 107 genes encoding ribosomal proteins (129 identified in Slhy5-13 and 219 identified in Slhy5-29) whose expression was significantly altered (P-adj < 0.05) in both Slhy5 mutants (Fig. 6a and Supplementary Table 3). Most of these genes (86.0%) showed decreased expression in the mutants. We selected six genes with a fold change > 2 for the ChIP-qPCR assay, and the results indicated that two of these genes (Solyc03g114750 and Solyc05g150151) are direct targets of SlHY5 (Fig. 6b).
Fig. 6. Protein translation is affected in the Slhy5 mutant.
a Differential expression of ribosomal protein genes in the Slhy5 mutant (Slhy5-13 and Slhy5-29) versus wild-type (WT) fruits at 39 days post anthesis (DPA) based on RNA-seq data. b ChIP-qPCR assays showing that SlHY5 binds to the promoters of two ribosomal protein genes. The promoter structures of the SlHY5 target genes are shown. The blue boxes represent ACGT-containing elements, and the numbers indicate the positions of these motifs relative to the translational start site. The black lines with uppercase letters represent the regions used for ChIP-qPCR. The values are the percentages of DNA fragments that coimmunoprecipitated with anti-SlHY5 antibodies relative to the input DNAs. c Relative ribosomal protein concentration in Slhy5 mutant compared with WT fruits at 39 DPA. The error bars represent the standard deviations from three independent experiments. The asterisks indicate significant differences (*P < 0.05). d Translation efficiency of specific genes was altered in the Slhy5 mutants. PSY1 phytoene synthase 1, PDS phytoene desaturase, Z-ISO 15-cis-zeta-carotene isomerase, ZDS zeta-carotene desaturase, CrtISO1 carotene isomerase 1, CrtISO2 carotene isomerase 2, VDE violaxanthin de-epoxidase, NCED1 9-cis-epoxycarotenoid dioxygenase 1, ANS anthocyanidin synthase, CHI chalcone-flavonone isomerase, CHS1 chalcone synthase 1, CHS2 chalcone synthase 2, F3H flavanone 3-hydroxylase, F3′H flavonoid 3′-hydroxylase, 3-GT flavonol-3-glucosyltransferase, MYB12 MYB transcription factor 12, ACO1 1-aminocyclopropane-1-carboxylate oxidase 1, ACO5 1-aminocyclopropane-1-carboxylate oxidase 5, ACS2 1-aminocyclopropane-1-carboxylic acid synthase 2, ACS4 1-aminocyclopropane-1-carboxylic acid synthase 4, ACS12 1-aminocyclopropane-1-carboxylic acid synthase 12, ETR4 ethylene receptor 4, EIN2 ethylene insensitive 2, E8 ethylene response gene 8, RIN ripening inhibitor, FUL1 FRUITFULL 1, NOR non-ripening, HY5 ELONGATED HYPOCOTYL5, DML2 DEMETER-like DNA demethylase 2, PL1 pectate lyase 1, PG2A polygalacturonase 2A, LoxC lipoxygenase C. The error bars represent the standard deviations from three independent experiments. The asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001; Student’s t test). The corresponding transcriptional levels of these genes are listed in the right panel
A decrease in the expression of genes encoding ribosomal proteins might influence the pool of ribosomal proteins. We subsequently compared the abundances of ribosomal proteins, which were calculated from purified polysomes in the wild-type and Slhy5 mutant fruits. The concentration of total soluble proteins prior to polysome enrichment was used as a control. More ribosomal proteins were enriched in the wild-type than in the Slhy5 mutant fruits (Fig. 6c), which indicated that loss of function of SlHY5 affects the accumulation of ribosomal proteins in ripening fruit.
Since ribosomal proteins are core components of the ribosome, which is a protein translation machinery, we speculate that SlHY5 might affect protein translation. We next determined the translation efficiency by calculating the ratio of the expression level of specific genes in polysomal to total RNA. Genes involved in various metabolic processes, including carotenoid metabolism, anthocyanin accumulation, ethylene biosynthesis and response, and some well-known ripening-related genes were evaluated. As shown in Fig. 6d, the translation efficiency of all the detected genes associated with anthocyanin accumulation was decreased in the Slhy5 mutants. Approximately half of the tested genes involved in carotenoid metabolism and ethylene biosynthesis and response exhibited a reduced translation efficiency in the mutants. Four of the ripening-related genes, i.e., FUL1, NOR, DML2, and LoxC, displayed a reduced translation efficiency in the Slhy5 mutants. It is worth noting that some genes (CHS1, CHS2, CHI, F3H, F3’H, ANS, 3-GT, MYB12, ACO1, ACO5, ETR4, E8, PSY1, CrtISO1, VDE, FUL1, NOR, and LoxC) were regulated by SlHY5 at both the transcriptional and translational levels, whereas the others were regulated only at the transcriptional level (ACS2, Z-ISO, NCED1, RIN, PL1, and PG2A) or translational level (ZDS, CrtISO2, and DML2). These results suggest that SlHY5 has the ability to regulate gene expression not only at the transcriptional level, but also at the translational level during fruit ripening.
Discussion
SlHY5 regulates tomato fruit ripening at the transcriptional level
HY5 is well known for its function in the light signaling pathway. HY5 was first identified in Arabidopsis as a transcriptional regulator with a bZIP motif9. HY5 can bind up to eight types of cis-elements present in the promoters of the target genes, particularly G-box (CACGTG) and ACE-box (ACGT). Large-scale identifications of direct targets of HY5 have revealed that HY5 affects the expression of one-third of genes in the Arabidopsis genome and directly controls ~3000 of these genes20,21. In contrast to the understanding of HY5 in the model plant Arabidopsis, the functions and regulatory mechanisms of HY5 in the physiological processes of horticultural crops remain largely uncharacterized. In this study, we found that ~3000 genes were differentially expressed in the fruit of the Slhy5 mutants (Fig. 2), which suggested that SlHY5 plays a global regulatory role in the process of tomato fruit ripening. ChIP-qPCR assays showed that genes involved in carotenoid metabolism and anthocyanin biosynthesis were directly regulated by SlHY5 (Figs. 3 and 4). All of these genes were downregulated in the Slhy5 mutants, consistent with the results from the phenotypic analysis, which showed that Slhy5 mutants contain lower levels of carotenoids and anthocyanins than wild-type fruits.
Intriguingly, genes involved in ethylene production and response were identified as direct targets of SlHY5 in tomato (Fig. 5). Consistent with the downregulation of these genes, ethylene production was substantially reduced in Slhy5 mutants at 39 and 42 DPA (Fig. 5), which demonstrated that SlHY5 positively affects ethylene biosynthesis. Ethylene is necessary for climacteric fruit ripening. As a classic climacteric fruit, tomato shows a burst of ethylene at the onset of fruit ripening34. These data indicated that SlHY5 is necessary for normal fruit ripening in tomato. SlHY5 might influence fruit ripening directly by binding to the promoters of ripening-related genes, as observed with carotenoid biosynthetic genes, or indirectly by affecting ethylene signaling. Notably, Arabidopsis seedlings with hy5 mutation showed 1.5-fold higher ethylene emissions than wild-type seedlings, which implies that HY5 exerts a negative effect on ethylene production35. A similar antagonistic relationship between HY5 and ethylene generation has also been observed on hypocotyl growth in Arabidopsis, and exogenous ethylene can promote HY5 degradation36. The discrepancy could be explained by the difference between seedlings/hypocotyls and fruit, in which ethylene plays different roles.
As a programmed developmental process, fruit ripening involves epigenetic regulations and the coordination of plant hormones and transcription factors29,34,37–40. A number of transcription factors, including RIN, NOR, CNR, TAGL1, and FUL1/FUL2, have been characterized to be involved in fruit ripening29,37,38,41. In the present study, we found that the expression of RIN, CNR, and FUL1 was reduced in the Slhy5 mutants. Further ChIP-qPCR analysis revealed that SlHY5 bound to the promoters of RIN, CNR, and FUL1, which indicates that SlHY5 directly regulates these transcription factors (Supplementary Fig. 5). The interplay between SlHY5 and these ripening regulators deserves further investigation.
Notably, HY5 regulates various physiological processes of plants9,14,15, such as seedling growth, leaf development, and flowering, which may influence fruit physiology. SlHY5-mediated fruit ripening might be partly influenced by these developmental processes.
SlHY5 regulates the translation efficiency of numerous ripening-related genes
It has been elucidated that light modulates gene expression at multiple levels. In addition to the transcriptional regulation mediated by transcription factors, posttranscriptional regulation, such as alternative splicing and selected degradation of mRNA via small regulatory RNAs, has been demonstrated to attenuate the expression of light-responsive genes21,42. Light also regulates gene expression at the posttranslational level through the ubiquitin-dependent protein degradation of key components in the light signaling pathway or by affecting protein phosphorylation/dephosphorylation43. Furthermore, light regulates the translation of expressed genes in plants44. A global survey of transcripts under translational control during photomorphogenesis in Arabidopsis was recently performed45. Information about translational regulation offers knowledge on how an organism copes with intrinsic development cues and responds to external environmental stimuli45. Although a large number of genes are regulated at the translational level in plants45, the linkers connecting the light signals and the downstream translational regulation of gene expression have not been defined.
In the present study, we found that a large portion of genes encoding ribosomal proteins were downregulated in Slhy5 mutants (Fig. 6a), and several of these ribosomal protein genes were identified as direct targets of SlHY5 (Fig. 6b). Ribosomal proteins represent the core components of the translation machinery. A lack of ribosomal proteins causes an insufficient ribosome pool and impairs the translation of transcripts. We further demonstrated that the translation efficiency of multiple ripening-related genes was decreased in the Slhy5 mutants (Fig. 6d), which indicated that SlHY5 affects the translation of these genes. Interestingly, no significant changes in mRNA levels have been observed for some of these ripening-related genes (e.g., ZDS and DML2) in the Slhy5 mutants, which indicates that these genes are specifically regulated at translational level. Our data suggest that HY5 might serve as the linker in the light-regulated translational control of gene expression.
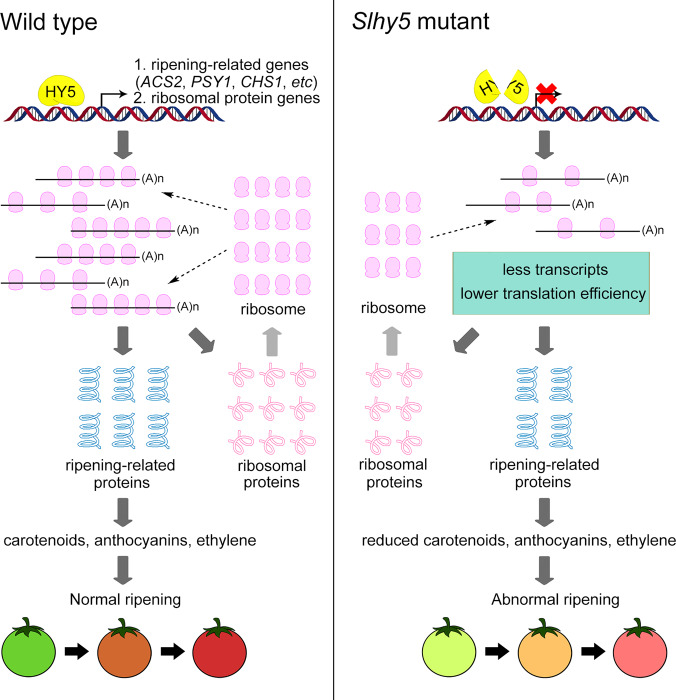
In summary, we demonstrated that SlHY5 participates in tomato fruit ripening through the transcriptional regulation of ripening-related genes, such as those involved in carotenoid biosynthesis and ethylene signaling. We also proved that SlHY5 affects the translation efficiency of a set of ripening-related genes by targeting ribosomal protein genes (Fig. 7). Our findings provide insight into the molecular regulatory network of fruit ripening and the multifaceted regulation of gene expression by transcription factors. Considering the conservation and vital role of HY5 in the light signaling pathway, we propose that the regulatory mechanisms described in this manuscript are also involved in the regulation of other light-controlled developmental processes.
Fig. 7. Model of the regulatory role of SlHY5 in tomato fruit ripening.
SlHY5 regulates fruit ripening at both the transcriptional and translational levels. SlHY5 can bind directly to the promoters of genes involved in pigment accumulation, ethylene production and response, and translation to modulate their expression at the transcriptional level. SlHY5 also regulates the translation efficiency of ripening-related genes by targeting ribosomal protein genes. Loss of function of SlHY5 influences the transcription and translation of ripening-related genes, which leads to delayed fruit ripening
Materials and methods
Plant materials
Wild-type tomato seeds (S. lycopersicum cv. Ailsa Craig) were kindly provided by Dr. James J. Giovonnoni (Boyce Thompson Institute for Plant Research, Cornell University, Ithaca, NY, USA). The tomato plants were grown in a greenhouse under standard culture conditions. Flowers were tagged on the day of anthesis to evaluate the ripening stages of the fruits. Wild-type fruits were harvested at the mature green (MG), breaker (Br), orange (Or), and red ripe (RR) stages, which occurred on average at 36, 39, 42, and 45 DPA. Fruits from the Slhy5 mutants were harvested at equivalent ripening stages, as determined by the number of DPA. Upon harvesting, the pericarps were collected, frozen immediately in liquid nitrogen, and stored at −80 °C until use.
Construction of Slhy5 mutants using the CRISPR/Cas9 system
A highly efficient CRISPR/Cas9 system improved for plant genome editing46 was used in this study. CRISPR-P (http://cbi.hzau.edu.cn/crispr/), an online tool, was used to generate a pool of target sequences, and we selected two target sites for SlHY5 based on previously reported principles46. Once the target sequences were selected, a PCR-based method was applied to rapidly generate multiple sgRNA expression cassettes that could be easily cloned into the pYLCRISPR/Cas9Pubi-H binary vector by Golden Gate ligation. The final construct pYLCRISPR/Cas9Pubi-H-SlHY5 was sequenced and then transformed into wild-type tomato through A. tumefaciens-mediated transformation.
After regeneration, tomato seedlings were transplanted into soil and cultured in a greenhouse. Genomic DNA of regeneration seedlings was extracted from fresh leaves using the FastPure Plant DNA Isolation Mini Kit (Vazyme, DC104). The presence of the transgene was verified by PCR to confirm the insertion of T-DNA. Subsequently, to validate the mutation of the transgenic plants, second-round PCR was performed using primers flanking the target sites. The PCR products were sequenced directly, and the superimposed sequencing chromatograms were decoded manually or using the automated web tool DSDecodeM (http://skl.scau.edu.cn/dsdecode/). Subsequently, the mutated and original sequences were aligned to show the mutagenesis.
RNA extraction and quantitative real-time PCR
Pericarps of fruits (at least eight fruits) from six plants were used to extract total RNA, following a previously described method47. One microgram of total RNA was used for first-strand cDNA synthesis with a HiScript III RT SuperMix for qPCR kit (Vazyme, R323-01). Quantitative real-time PCR was performed with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711), using the StepOnePlus Real-Time PCR System (Applied Biosystems). Gene expression levels were calculated using the 2−ΔΔCt analysis method, and the samples were normalized using ACTIN (Solyc11g005330). Three distinct biological replicates of each experiment were included.
RNA sequencing
For RNA-seq, pericarps of fruits (at least eight fruits) at 39 DPA from six wild-type and Slhy5 mutant (Slhy5-13 and Slhy5-29) plants were collected for total RNA isolation. RNA-seq libraries were constructed using the NEBNext® UltraTM RNA Library Prep Kit for Illumina (NEB) and sequenced using a HiSeq PE150 instrument (Illumina). In total, ~500 million raw reads were obtained, and all the data were uploaded to the BIG Data Center Genome Sequence Archive (https://bigd.big.ac.cn/gsub/) under accession number CRA003226. The raw reads were filtered using FastQC to generate clean data, which were aligned to the tomato reference genome SL4.0 using HISAT2 (version 2.0.5). FeatureCounts (version 1.5.0-p3) was used to count the numbers of reads mapped to each gene, and RPKM values were calculated based on the length of each gene and the read count mapped to the gene. Differential expression analysis between the wild-type and Slhy5 mutant plants was performed, using the DESeq2 R package (version 1.16.1). The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (the adjusted values are denoted P-adj hereafter). The fold changes were calculated using the formula FPKMSlhy5/FPKMWT. Transcripts satisfying the criteria |log2(fold change)| > 1 and P-adj < 0.05 were defined as differentially expressed.
Ethylene production assay
To measure ethylene release, at least six fruits from six wild-type and Slhy5 mutant plants (Slhy5-13 and Slhy5-29) were harvested at 39 and 42 DPA. After harvest, the fruits were placed at room temperature for 3 h to dissipate the field heat and wound ethylene. The fruits were then transferred to jars, and the jars were then sealed and incubated for another 2 h. The gas samples were analyzed using a gas chromatograph (Agilent 7820 A). The production of ethylene was calculated through a comparison with the standard sample and normalization of the fruit weight.
Determination of pigments
Total anthocyanins were extracted following a previously described method48 with some modifications. One gram of pericarp sample was ground into fine powder in liquid nitrogen and incubated in 3 ml of 1% HCl in methanol (v/v) overnight at room temperature. An additional 3 ml of chloroform was then added and mixed thoroughly to remove fat-soluble pigments. After centrifugation at 12,000 × g for 2 min, the supernatants were collected and filtered through a 0.45-μm filter. The content of anthocyanins was determined spectrophotometrically at 535 nm and expressed as A535 g−1 fresh weight. Average values were calculated from three independent replicates.
Carotenoids were extracted as described previously49. The carotenoid content was measured using a Waters ACQUITY UPC2 System (Milford, MA, USA) equipped with a C18 column, according to the manufacturer’s recommendations. The individual carotenoids (phytoene, lycopene, and β-carotene) were identified according to the retention time and quantified using standard curves.
Recombinant SlHY5 protein expression and specific antibody preparation
The full-length SlHY5 open reading frame was amplified from tomato cDNA using the primers HY5-F (CTTTAAGAAGGAGATATACATATGCACCATCATCATCATCATCAAGAGCAAGCGACGAGTTC) and HY5-R (GTGGTGGTGGTGGTGGTGCTCGAGCTACTTCCTCCCTTCCTGTG). The PCR product was inserted into the pET30a vector digested with Nde I and Xho I using a ClonExpress Ultra One Step Cloning Kit (Vazyme, C115) to generate pET30a-6×His-SlHY5. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) competent cells, and the expression of the recombinant protein was then induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside. The recombinant protein was purified using Ni-NTA His Bind Resin (Merck, 70666) following the manufacturer’s instructions, and rabbits were immunized with the purified recombinant protein at Beijing Protein Innovation Company (http://www.proteomics.org.cn/). Polyclonal antibody was affinity purified from antisera using AminoLink Plus Coupling Resin according to the manual (Thermo Scientific, 20501).
Nuclei enrichment
Nuclei were isolated from tomato fruit at 39 DPA according to a previously described method47. Briefly, the pericarp was ground into fine powder in liquid nitrogen and suspended in buffer 1 (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 5 mM β-mercaptoethanol, and 1 mM PMSF). The homogenates were filtered through two layers of miracloth and centrifuged at 3000 × g for 20 min. The pellets were suspended softly in buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and 1 mM PMSF) and set aside on ice for 10 min. After centrifugation at 12,000 × g for 10 min, the pellets were resuspended in 500 μl of buffer 3 (1.7 M sucrose, 10 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 0.15% Triton X-100, 5 mM β-mercaptoethanol, and 1 mM PMSF) and overlaid on top of another 500 μl of buffer 3. The nuclei were enriched by centrifugation at 16,000 × g for 45 min and stored at −80 °C until use.
Nuclear protein extraction and western blotting
The purified nuclei were thoroughly resuspended in extraction buffer (0.7 M Sucrose, 0.5 M Tris-HCl, pH 8.0, 0.5 M EDTA, 0.1 M KCl, 5 mM β-mercaptoethanol, and 1 mM PMSF) and broken by sonication on ice. The homogenate was mixed with an equal volume of Tris-HCl saturated-phenol (pH 7.4) and vortexed for 10 min at 4 °C. After centrifugation at 20,000 × g for 15 min, the upper-layer phenol phase was re-extracted with extraction buffer. The proteins in the phenol phase were precipitated by adding five volumes of methanol containing 0.1 M ammonium acetate and stored overnight at −20 °C. The protein pellets were collected by centrifugation at 20,000 × g for 15 min, and washed several times with ice-cold methanol and acetone. The nuclear proteins were air-dried and dissolved in 0.5 M Tris-HCl (pH 8.0) containing 1% SDS.
For immunoblot analysis, 15 μg of nuclear proteins was separated by 12% SDS–PAGE and then transferred to an Immobilon-P PVDF membrane (Millipore, IPVH00005) by semidry transfer (Amersham Biosciences, TE77 PWR). The membranes were blocked with 5% nonfat milk in TBS-T buffer for 2 h at room temperature, and incubated with an anti-SlHY5 (1:1000) or anti-histone H3 (1:10000) antibody for 1 h. The membranes were then washed four times with TBS-T buffer and incubated with the secondary antibody (1:5000) for 1 h. The membranes were washed four times with TBS-T buffer and visualized using a chemiluminescence detection kit (SuperSignal, Pierce Biotechnology).
Chromatin immunoprecipitation
ChIP assays were performed as described previously with some modifications47. Pericarp tissues of fruits (at least eight fruits) at 39 DPA from six wild-type and Slhy5-13 mutant plants were sliced and submerged in 1% formaldehyde under vacuum for 20 min for the cross-linking of genomic DNA and protein. The fixed materials were subjected to nuclear isolation as described above. The enriched nuclei were sonicated to shear gDNA to an average size of 300–800 bp. After centrifugation at 12,000 × g for 2 min, the supernatant, which contained the sheared chromatin, was collected. A portion of the supernatant was reverse cross-linked and used as the input DNA control. The affinity-purified anti-SlHY5 antibody was incubated with preblocked Magna ChIPTM Protein A + G Magnetic Beads (Millipore, 16-663) overnight at 4 °C, and the remaining chromatin sample was then added. After incubation for 4 h, the beads were washed successively with low-salt wash buffer, high-salt wash buffer, lithium chloride wash buffer, and TE buffer. The DNA–protein complexes were eluted from the beads by occasional rotation at 65 °C for 1 h. The cross-linking between immunoprecipitated DNA and SlHY5 protein was interrupted by adding NaCl to a final concentration of 0.2 M, and the eluate was incubated overnight at 65 °C. The immunoprecipitated DNA was purified using a TIANquick Mini Purification Kit (TIANGEN, DP203). The HY5-binding sites (ACGT-containing elements) in the promoters of the candidate genes were analyzed with PlantPAN 3.0 (http://plantpan.itps.ncku.edu.tw/TFsearch.php). The primers used for quantitative PCR were designed using Primer Premier 5 software and are listed in Supplementary Table 4.
Translation efficiency and ribosomal protein determination
The translation efficiency was assayed following a previously described method with some modifications50. All procedures were operated on ice or at 4 °C. Pericarps of fruits (at least eight fruits) at 39 DPA from six wild-type and Slhy5-13 mutant plants were ground into fine powder in liquid nitrogen. One gram of sample was used for total RNA extraction, and the remaining sample was suspended in 15 ml of polysome extraction buffer (200 mM Tris-HCl, pH 9.0, 200 mM KCl, 35 mM MgCl2, 25 mM EGTA, 1% Triton X-100, 1% IGEPAL CA-630, 5 mM DTT, 1 mM PMSF, 100 μg/ml cycloheximide, and 50 μg/ml chloramphenicol) and maintained on ice for 10 min. The extract was filtered through two-layer miracloth and then centrifuged at 16,000 × g for 15 min. Exactly 10 ml of the supernatant was loaded on a sucrose cushion (1.75 M sucrose, 400 mM Tris-HCl, pH 9.0, 200 mM KCl, 35 mM MgCl2, 5 mM EGTA, 5 mM DTT, 50 μg/ml cycloheximide, and 50 μg/ml chloramphenicol), and the remaining supernatant was reserved for total soluble protein determination. Polysomes were collected after ultracentrifugation at 190,000 × g for 4 h and resuspended in 500 μl of RNase-free ddH2O (50 μl was reserved for ribosomal protein determination). RNA in the polysomes were extracted using the hot phenol method. The relative expression of the target genes in total or polysomal RNA was calculated using the 2−ΔΔCt analysis method, and the samples were normalized using ACTIN. The translation efficiency was calculated as the relative expression in polysomal/total RNA followed by normalization against that in the wild-type fruits. The primers used for quantitative PCR were designed using Primer Premier 5 software and are listed in Supplementary Table 5.
The total soluble protein (TSP) and ribosomal protein (RP) concentrations were determined using a PierceTM BCA Protein Assay Kit (Thermo, 23225). The relative RP content was calculated using the formula Concentration(RP)/Concentration(TSP) followed by normalization against that of the wild-type plants.
Supplementary information
Supplementary Information (Fig. S1-S5, Table S4-S5)
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant Nos. 31801601, 31925035, and 31871855), the China Postdoctoral Science Foundation (2018T110153), and the Youth Innovation Promotion Association CAS (2019083).
Author contributions
G.Q. and W.W. conceived and designed the experiments; W.W., P.W., X.L., and Y.W. performed the experiments; and S.T. provided critical discussions. G.Q. and W.W. analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00523-0.
References
- 1.Yang L, Jiang Z, Jing Y, Lin R. PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J. Integr. Plant Biol. 2020;62:1372–1384. doi: 10.1111/jipb.12938. [DOI] [PubMed] [Google Scholar]
- 2.Zhou P, et al. Both PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2 promote seedling photomorphogenesis in multiple light signaling pathways. Plant Physiol. 2014;164:841–852. doi: 10.1104/pp.113.227231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouwer B, Gardeström P, Keech O. In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis. J. Exp. Bot. 2014;65:4037–4049. doi: 10.1093/jxb/eru060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casual JJ. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013;64:403–427. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- 6.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 7.Jiao Y, Ma L, Strickland E, Deng XW. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell. 2005;17:3239–3256. doi: 10.1105/tpc.105.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, et al. Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell. 2013;25:1657–1673. doi: 10.1105/tpc.112.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Gene. Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paik I, Kathare PK, Kim J, Huq E. Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant. 2017;10:1035–1046. doi: 10.1016/j.molp.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoecker U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 2017;37:63–69. doi: 10.1016/j.pbi.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Llorente B, Martinez-Garcia JF, Stange C, Rodriguez-Concepcion M. Illuminating colors: regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 2017;37:49–55. doi: 10.1016/j.pbi.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, et al. PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis. Plant Cell. 2014;26:1992–2006. doi: 10.1105/tpc.114.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 2016;9:1353–1365. doi: 10.1016/j.molp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, et al. Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl Acad. Sci. USA. 2004;101:9897–9902. doi: 10.1073/pnas.0400935101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llorente B, et al. Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J. 2016;85:107–119. doi: 10.1111/tpj.13094. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, et al. Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J. 2008;55:89–103. doi: 10.1111/j.1365-313X.2008.03489.x. [DOI] [PubMed] [Google Scholar]
- 18.Davuluri GR, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L, Shao Z, Zhang M, Wang Q. Regulation of carotenoid metabolism in tomato. Mol. Plant. 2015;8:28–39. doi: 10.1016/j.molp.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, et al. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and posttranscriptional regulation. Plant J. 2011;65:346–358. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 22.Shin DH, et al. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP transcription factor in Arabidopsis. FEBS Lett. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- 23.Qiu Z, et al. Identification of candidate HY5-dependent and-independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 2019;60:643–656. doi: 10.1093/pcp/pcy236. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiol. 2015;169:2380–2390. doi: 10.1104/pp.15.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oeller PW, Lu MW, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- 27.Deikman J, Kline R, Fischer RL. Organization of ripening and ethylene regulatory regions in a fruit-specific promoter from tomato (Lycopersicon esculentum) Plant Physiol. 1992;100:2013–2017. doi: 10.1104/pp.100.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery J, Goldman S, Deikman J, Margossian L, Fischer RL. Identification of an ethylene-responsive region in the promoter of a fruit ripening gene. Proc. Natl Acad. Sci. USA. 1993;90:5939–5943. doi: 10.1073/pnas.90.13.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vrebalov J, et al. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–3062. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai J, Qin G, Chen T, Tian S. The mode of action of remorin1 in regulating fruit ripening at transcriptional and post‐transcriptional levels. N. Phytol. 2018;219:1406–1420. doi: 10.1111/nph.15264. [DOI] [PubMed] [Google Scholar]
- 31.Tournier B, et al. New members of the tomato ERF family show specific expression pattern and diverse DNA‐binding capacity to the GCC box element. FEBS Lett. 2003;550:149–154. doi: 10.1016/S0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, et al. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016;170:1732–1744. doi: 10.1104/pp.15.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, et al. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 2007;26:1999–2008. doi: 10.1007/s00299-007-0394-8. [DOI] [PubMed] [Google Scholar]
- 34.Yokotani N, et al. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J. Exp. Bot. 2009;60:3433–3442. doi: 10.1093/jxb/erp185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Z. et al. The ethylene response factor AtERF11 that is transcriptionally modulated by the bZIP transcription factor HY5 is a crucial repressor for ethylene biosynthesis in Arabidopsis. Plant J. 68, 88–99 (2011). [DOI] [PubMed]
- 36.Yu, Y. et al. Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet.9, e1004025 (2013). [DOI] [PMC free article] [PubMed]
- 37.Vrebalov J, et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science. 2002;296:343–346. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 38.Bemer M, et al. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 2012;24:4437–4451. doi: 10.1105/tpc.112.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lü P, et al. Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants. 2018;4:784–791. doi: 10.1038/s41477-018-0249-z. [DOI] [PubMed] [Google Scholar]
- 40.Zhou, L., Tian, S. & Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol.20, 156 (2019). [DOI] [PMC free article] [PubMed]
- 41.Gao, Y. et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot.71, 3560–3574 (2020). [DOI] [PMC free article] [PubMed]
- 42.Godoy Herz, M. A. et al. Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell73, 1066–1074 (2019). [DOI] [PubMed]
- 43.Li, J. et al. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun.11, 1592 (2020). [DOI] [PMC free article] [PubMed]
- 44.Chen, G. H., Liu, M. J., Xiong, Y., Sheen, J. & Wu, S. H. TOR and RPS6 transmit light signals to enhance protein translation in de-etiolating Arabidopsis seedlings. Proc. Natl Acad. Sci. USA115, 12823–12828 (2018). [DOI] [PMC free article] [PubMed]
- 45.Liu, M. J. et al. Translational landscape of photomorphogenic Arabidopsis. Plant Cell25, 3699–3710 (2013). [DOI] [PMC free article] [PubMed]
- 46.Ma, X. et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant8, 1274–1284 (2015). [DOI] [PubMed]
- 47.Wang, Y. et al. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol.15, 548 (2014). [DOI] [PMC free article] [PubMed]
- 48.Solfanelli, C., Poggi, A., Loreti, E., Alpi, A. & Perata, P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol.140, 637–646 (2006). [DOI] [PMC free article] [PubMed]
- 49.Xiong, C. et al. A tomato B‐box protein SlBBX20 modulates carotenoid biosynthesis by directly activating PHYTOENE SYNTHASE 1, and is targeted for 26S proteasome‐mediated degradation. N. Phytol. 221, 279–294 (2019). [DOI] [PubMed]
- 50.Merchante, C. et al. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell163, 684–697 (2015). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (Fig. S1-S5, Table S4-S5)