Abstract
Quantitative real-time PCR (qRT-PCR) has been emerged as an effective method to explore the gene function and regulatory mechanisms. However, selecting appropriate reference gene (s) is a prerequisite for obtaining accurate qRT-PCR results. Peach is one of important fruit in Rosaceae and is widely cultivated worldwide. In this study, to explore reliable reference gene (s) in peach with different types during fruit ripening and softening (S1–S4), nine candidate reference genes (EF-1α, GAPDH, TBP, UBC, eIF-4α, TUB-A, TUB-B, ACTIN, and HIS) were selected from the whole-genome data. Then, the expression levels of the nine selected genes were detected using qRT-PCR in three peach types, including ‘Hakuho’ (melting type), ‘Xiacui’ (stony hard type), ‘Fantasia’ and ‘NJC108’ (non-melting type) cultivars were detected using qRT-PCR. Four software (geNorm, NormFinder, BestKeeper and RefFinder) were applied to evaluate the expression stability of these candidate reference genes. Gene expression was characterized in different peach types during fruit ripening and softening stages. The overall performance of each candidate in all samples was evaluated. The Actin gene (ACTIN) was a suitable reference gene and displayed excellent stability in ‘Total’ set, ‘Hakuho’ samples, S3 and S4 fruit developmental stages. Ubiquitin C gene (UBC) showed the best stability in most independent samples, including ‘Fantasia’, ‘NJC108’, S2 sets. Elongation factor-1α gene (EF-1α) was the most unstable gene across the set of all samples, ‘NJC108’ and S2 sets, while showed the highest stability in ‘Xiacui’ samples. The stability of candidate reference genes was further verified by analyzing the relative expression level of ethylene synthase gene of Prunus persica (PpACS1) in fruit ripening and softening periods of ‘Hakuho’. Taken together, the results from this study provide a basis for future research on the mining of important functional genes, expression patterns and regulatory mechanisms in peach.
Subject terms: Plant biotechnology, Plant development, Plant molecular biology
Introduction
Quantitative real-time PCR (qRT-PCR) is widely used for expression analysis of genes due to its fast, sensitive, specific and accurate characteristics1,2. However, the results of qRT-PCR are often affected by many factors, such as RNA concentration, reverse transcription efficiency, amplification efficiency, and experimental process3,4. A suitable internal reference gene can eliminate these errors, so the internal reference gene is usually introduced in the qRT-PCR analysis for data correction and standardization5. In fact, there is no generality of reference genes in all experiments, and the expression levels of the reference genes in different tissues will change under different treatments6,7. Blind use of reference genes to standardize qRT-PCR data will lead to unreliable results, therefore, it is necessary to select appropriate internal reference genes to minimize the distractions.
Previous studies showed that reference genes are always expressed all the time to maintain the basic life activities of cells and their expression levels are less affected by environmental factors2,8. Most of the traditional reference genes are the basic components of the cytoskeleton or genes involved in the basic metabolic regulation of the organism9. For example, ACTIN encodes a cytoskeleton structural protein, TUB (β-Tubulin) is mainly involved in cell growth, and EF-1α (eukaryotic elongation factor-1α) is involved in transcriptional extension. Due to the importance of reference genes in gene expression analysis, many studies on the screening of reference genes with stable expression in higher plants have been conducted and the results showed different reference genes have been applied in different experimental materials and conditions. ACTINT7 and TBP (TATA-box binding protein) are the best reference genes in tripterygium and kumquat10,11, GAPDH (Glyceraldehyde-3-phosphate dehydrogenase) is the most stable reference gene in all samples in Primula forbesii12. In Arabidopsis, UBC (Ubiquitin-conjugating enzyme) is the stable reference gene in seeds, while UBQ5 (ubiquitin5), APT1 (adenine phosphoribosyl-transferase 1) and EF-1α were the stable reference genes in different tissues13. The most stable reference gene of rice in different tissues was CYC (cyclophilin)14, while in different types of rice samples under drought stress were UBQ and GAPDH15. PP2A (protein phosphatase 2A) is the most stable gene under abiotic stress in sorghum14. UBC is the best reference gene for all samples and different cultivars in Osmanthus fragrans, while ACTIN is the best reference gene for different flower development stages and different temperature treatments16. All these studies showed that the reference genes should be screened, evaluated and verified accurately according to the plant cultivars, different tissues and conditions.
As one popular stone fruit, peach (Prunus persica (L.) Batsch) has been known as one of the most common and economically important species worldwide. Recent studies have been payed more attention on the physiological and metabolites changes of peach during fruit development17–19, and exogenous treatments on peach ripening and quality in postharvest20–22. However, the mechanism of different peach cultivars on peach fruit ripening and softening is still unclear23. Biochemical processes occur in a well-defined order under the control of a number of ripening/softening-related genes leading to considerable changes in texture, pigmentation, taste and aroma24. Screening suitable reference genes in different peach cultivars at different developmental periods can provide effective help for exploring gene function at molecular level. To our knowledge, only a few reports have been conducted on the suitability of reference gens for normalization of gene expression in peach25–27. Limited to the traditional ones published previously in other higher plants, the selected reference genes have been proven to vary significantly across different experimental conditions. Therefore, it is necessary to accurately select the appropriate reference genes in different flesh texture cultivars during peach fruit ripening and softening.
In this study, to validate appropriate reference genes in different cultivars, ‘Hakuho’ (a melting flesh peach genotype, M), ‘Xiacui’ and ‘Fantasia’ (a stony hard flesh peach genotype, SH), and ‘NJC108’ (a non-melting flesh peach genotype, NM) were selected. Nine candidate reference genes: EF-1α, GAPDH, TBP, UBC, eIF-4α (eukaryotic translation initiation factor 4), TUB-A (α-Tubulin), TUB-B, ACTIN, and HIS (Histone) were used to identify the most stable reference genes for normalization from peach whole-genome data. Four software (geNorm, NormFinder, BestKeeper and RefFinder) were used to comprehensively analyze the stability of expression level for these selected reference genes. The results would enrich the reference gene selection in peach, which further improved the stability, repeatability and accuracy of peach gene expression analysis. The suitable reference genes obtained in this study provided a theoretical basis for further exploring the gene expression and regulation mechanisms in peach.
Results
Assessment of amplification efficiency and specificity of nine candidate reference genes of peach
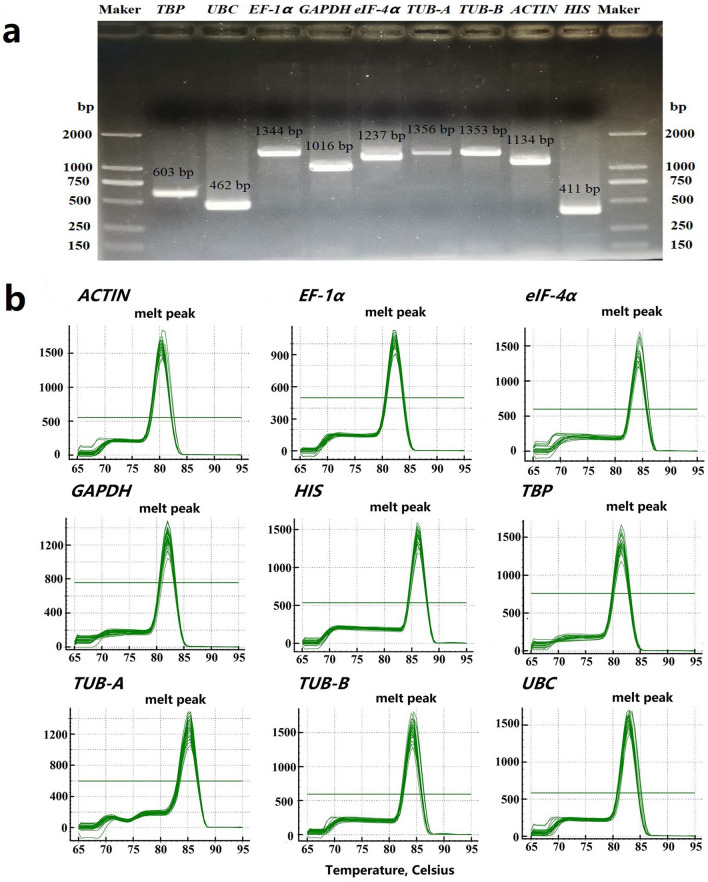
According to the sequences in peach genome, cloning primer pairs of the selected nine candidate reference genes were appropriately designed for PCR amplification (Table S1). The nine candidate genes were successfully cloned and the sequences were confirmed in subsequent experiments (Fig. 1a). Subsequently, qRT-PCR specific primer pairs of the nine genes were further designed, with the amplicon length ranging from 144 to 320 bp. Melting curve analysis of qRT-PCR showed that all the nine selected genes had a single peak, indicating that the amplified products did not have apparent primer dimer or nonspecific amplification (Fig. 1b). The amplification efficiencies (E%) varied from 90.5% to 108% and correlation coefficient (R2) ranged from 0.992 to 0.998 (Table 1, Fig. S1), which met the standard level (90 < E% < 110; R2 > 0.99)28.
Figure 1.
The specificity of primers for PCR amplification. (a) Amplification length of nine reference genes in peach by RT-PCR. (b) Melting curves generated for nine candidate reference genes by qRT-PCR.
Table 1.
Descriptions of candidate reference genes and amplification characteristics for qPCR.
| Gene symbol | Gene name | Primer sequence (5′-3′) | Amplicon size (bp) | E% | R2 |
|---|---|---|---|---|---|
| ACTIN | Actin gene | AGCAGAGCGATTCCGTTGTCC/CCTCCACTCAGCACTATGTTACCAT | 153 | 91.6 | 0.997 |
| EF-1a | Elongation factor -1α gene | GACCAACTGCCTTGCTCCTCTT/CTTGATGAAGTCACGATGTCCAGGT | 176 | 107.5 | 0.997 |
| GAPDH | Glyceraldehyde-3-phosphate gene | GACCAACTGCCTTGCTCCTCTT/GCTCTTCCACCTCTCCAATCCTTAG | 144 | 108 | 0.994 |
| TBP | TATA-box binding protein gene | CCCTTCTGGAATTGTCCCTACTCTC/GCAGTCGTCTTCGGTTCTCTTATTC | 162 | 98.0 | 0.998 |
| eIF-4α | Eukaryotic translation initiation factor 4 | TTGGCACAGCAGATTGAGAAGGTT/TCAGGTGGCATTGTAGCAGAGAAC | 320 | 93.1 | 0.995 |
| TUB-A | α-Tubulin gene | GGCTGGTATTCAGGTCGGCAATG/GGTGGAAGAGTTGGCGGTATGTC | 233 | 91.5 | 0.992 |
| TUB-B | β-Tubulin gene | GAGCGAGCAGTTCACAGCCATG/GTTCTCTTCAGCACCGTCCTCCT | 193 | 101.5 | 0.997 |
| UBC | Ubiquitin C gene | GAGTCCTGCTCTCCAGATACGAACT/CGGGTCCATTCCTTTGCTGTTTCA | 150 | 90.5 | 0.996 |
| HIS | Histone gene | GAGTCAAGAAGCCTCACCGTTACC/CGCCTAGCAAGCTGAATGTCCTT | 286 | 95.4 | 0.993 |
Expression levels and variation of the candidate reference genes of peach
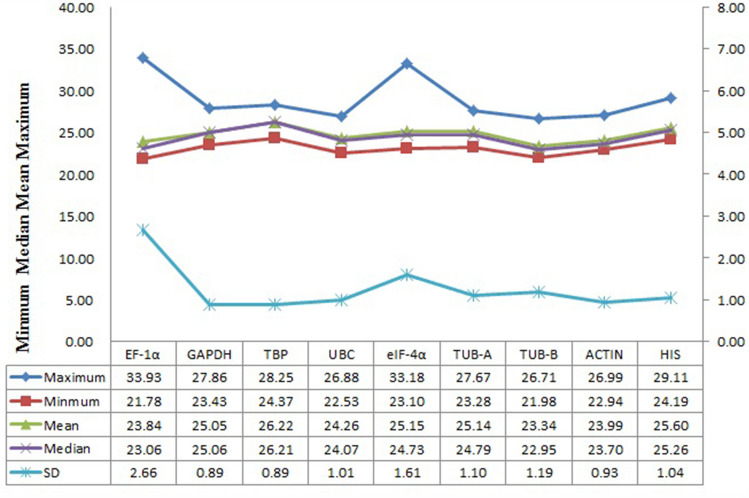
The expression levels of the nine candidate genes were displayed as Cq values and the raw data were listed in Table S2. A lower Cq value indicates a higher transcriptional expression level of the gene, while a higher Cq value indicates a lower expression level. The nine genes showed different expression levels with great changes of Cq values under experimental conditions (Fig. 2). In general, Cq values of the nine candidate genes ranged from 21.78 (EF-1α) to 33.18 (eIF-4α), and most Cq values of all test samples were concentrated between 22 and 28. EF-1α and TUB-B showed higher expression levels with lower Cq values, while TBP and TUB-A showed lower expression levels with higher Cq values. EF-1α had the largest variance (SD = 2.66), GAPDH and TBP had the smallest variance (SD = 0.89). The Cq value distribution of ACTIN was more concentrated than other candidate reference genes.
Figure 2.
Distribution of the Cq values of nine candidate reference genes across all samples in qRT-PCR analysis. The two ends of the graph represent the maximum and minimum Cq values, respectively. The red line across the graph indicates the mean value.
Evaluation of expression stability of the reference genes of peach
In the present study, four statistical approaches, geNorm, Normfinder, BestKeeper, and RefFinder were used to evaluate the expression stability of the nine reference genes. The analysis is based on three different data sets: (1) ‘total’, including all experimental samples; (2) samples of different cultivars; (3) samples of different fruit ripening and softening stages.
geNorm
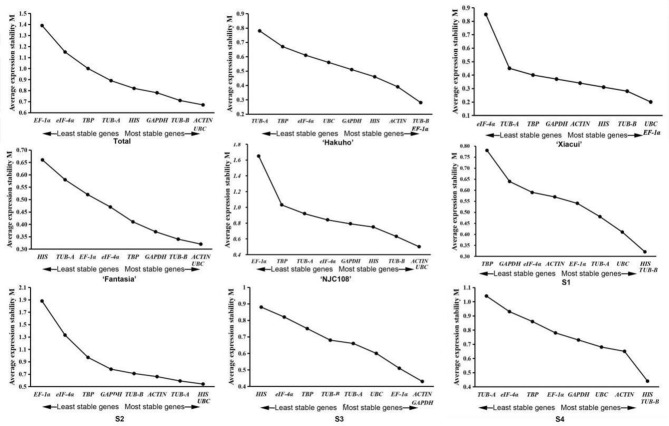
The geNorm software evaluates the stability of the reference gene by comparing the calculated average expression stability value (M). M-value is negatively correlated with expression stability. Reference gene with the lowest M-value has the highest stability. The software defaults to 1.5 as the critical value. As shown in Fig. 3, the M-values of the nine candidate reference genes were all less than 1.5 in different cultivars and different fruit ripening periods. These results indicated that these genes generally had good stabilities. However, the most stable reference gene (s) is (are) varied in different cultivars and periods. Among the ‘Total’, ‘Fantasia’ and ‘NJC108’, UBC and ACTIN were the most suitable reference genes; the genes EF-1 α and UBC were the most stable reference genes in ‘Xiacui’; the genes EF-1 α and TUB-B were recommended in ‘Hakuho’. During the periods in peach fruit ripening and softening, TUB-B and HIS were the most stable genes in S1 and S4 stages, while HIS and UBC were the most stable genes in S2 and the genes GAPDH and ACTIN were most stable in S3, respectively.
Figure 3.
geNorm expression stability with the M values of the nine candidate genes in all sample sets. A lower M value indicates greater stability and the largest value indicates the least stable reference gene.
NormFinder
The NormFinder software ranks the candidate reference genes by the stability value based on both intra- and inter-group variation. The gene with the lowest stability value has the highest stability. In the ‘Total’ group, the stability of the reference genes was ranked as follows: ACTIN > UBC > TUB-B > GAPDH > HIS > TUB-A > TBP > eIF-4α > EF-1α (Table 2). Gene stabilities were also analyzed by different cultivars (Table 3) and different fruit ripening and softening stages (Table 4). The ACTIN has the highest stability in S4 and ranks secondly in S1; UBC showed the best stability in ‘Xiacui’, ‘Fantasia’, ‘NJC108’ and S2; the gene EF-1α ranked first in the S3 but ranked last in the ‘Total’, ‘NJC108’ and S2. According to the analysis of NormFinder, the genes ACTIN and UBC showed good stability, while the gene EF-1α displayed unstable characteristic.
Table 2.
The stability ranking of candidate reference genes in “Total” analysis by geNorm, NormFinder, BestKeeper and RefFinder.
| Rank | geNorm | NormFinder | BestKeeper | RefFinder | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability value | Gene | SD [± Cq] | CV [%Cq] | Gene | Stability | ||
| Total | 1 | UBC | 0.67 | ACTIN | 0.23 | ACTIN | 0.66 | 2.76 | ACTIN | 1.00 |
| 2 | ACTIN | 0.67 | UBC | 0.42 | GAPDH | 0.67 | 2.68 | UBC | 1.86 | |
| 3 | TUB-B | 0.71 | TUB-B | 0.44 | UBC | 0.74 | 3.05 | GAPDH | 3.36 | |
| 4 | GAPDH | 0.78 | GAPDH | 0.64 | TBP | 0.75 | 2.86 | TUB-B | 3.57 | |
| 5 | HIS | 0.82 | HIS | 0.74 | HIS | 0.80 | 3.14 | HIS | 5.00 | |
| 6 | TUB-A | 0.89 | TUB-A | 0.96 | TUB-B | 0.91 | 3.90 | TBP | 6.09 | |
| 7 | TBP | 1.00 | TBP | 1.17 | TUB-A | 0.94 | 3.72 | TUB-A | 6.24 | |
| 8 | eIF-4α | 1.15 | eIF-4α | 1.46 | eIF-4α | 1.09 | 4.33 | eIF-4α | 8.00 | |
| 9 | EF-1α | 1.39 | EF-1α | 2.06 | EF-1α | 1.56 | 6.53 | EF-1α | 9.00 | |
Table 3.
The stability ranking of candidate reference genes in different variety of samples by geNorm, NormFinder, BestKeeper and RefFinder.
| Variety | Rank | geNorm | NormFinder | BestKeeper | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability value | Gene | SD [± Cq] | CV [%Cq] | Gene | Stability | ||
| ‘Hakuho’ | 1 | EF-1α | 0.28 | ACTIN | 0.18 | HIS | 0.18 | 0.70 | ACTIN | 1.57 |
| 2 | TUB-B | 0.28 | TUB-B | 0.20 | ACTIN | 0.31 | 1.31 | TUB-B | 1.86 | |
| 3 | ACTIN | 0.39 | EF-1α | 0.33 | TUB-B | 0.33 | 1.46 | EF-1α | 2.59 | |
| 4 | HIS | 0.46 | HIS | 0.40 | GAPDH | 0.37 | 1.45 | HIS | 2.83 | |
| 5 | GAPDH | 0.51 | GAPDH | 0.43 | EF-1α | 0.40 | 1.69 | GAPDH | 4.73 | |
| 6 | UBC | 0.56 | UBC | 0.59 | UBC | 0.50 | 2.13 | UBC | 6.00 | |
| 7 | eIF-4α | 0.61 | eIF-4α | 0.65 | eIF-4α | 0.58 | 2.37 | eIF-4α | 7.00 | |
| 8 | TBP | 0.67 | TBP | 0.79 | TBP | 0.61 | 2.36 | TBP | 8.00 | |
| 9 | TUB-A | 0.78 | TUB-A | 1.10 | TUB-A | 0.86 | 3.39 | TUB-A | 9.00 | |
| ‘Xiacui’ | 1 | EF-1α | 0.20 | UBC | 0.11 | TUB-B | 0.17 | 0.76 | EF-1α | 1.41 |
| 2 | UBC | 0.20 | EF-1α | 0.14 | EF-1α | 0.18 | 0.81 | UBC | 1.57 | |
| 3 | TUB-B | 0.28 | HIS | 0.27 | UBC | 0.23 | 0.98 | TUB-B | 2.45 | |
| 4 | HIS | 0.31 | TUB-B | 0.28 | HIS | 0.29 | 1.17 | HIS | 3.72 | |
| 5 | ACTIN | 0.34 | GAPDH | 0.28 | ACTIN | 0.30 | 1.28 | ACTIN | 5.23 | |
| 6 | GAPDH | 0.37 | ACTIN | 0.33 | TBP | 0.36 | 1.35 | GAPDH | 6.19 | |
| 7 | TBP | 0.40 | TBP | 0.40 | GAPDH | 0.38 | 1.58 | TBP | 6.48 | |
| 8 | TUB-A | 0.45 | TUB-A | 0.60 | TUB-A | 0.47 | 1.96 | TUB-A | 8.00 | |
| 9 | eIF-4α | 0.85 | eIF-4α | 2.21 | eIF-4α | 1.38 | 5.38 | eIF-4α | 9.00 | |
| ‘Fantasia’ | 1 | UBC | 0.32 | UBC | 0.27 | GAPDH | 0.61 | 2.44 | UBC | 1.32 |
| 2 | ACTIN | 0.32 | TUB-B | 0.30 | TUB-A | 0.72 | 2.85 | TUB-B | 2.78 | |
| 3 | TUB-B | 0.34 | TBP | 0.32 | UBC | 0.73 | 2.92 | GAPDH | 2.99 | |
| 4 | GAPDH | 0.37 | GAPDH | 0.35 | eIF-4α | 0.86 | 3.33 | ACTIN | 3.08 | |
| 5 | TBP | 0.41 | ACTIN | 0.35 | ACTIN | 0.88 | 3.66 | TBP | 4.53 | |
| 6 | eIF-4α | 0.47 | eIF-4α | 0.39 | TUB-B | 0.88 | 3.68 | eIF-4α | 5.42 | |
| 7 | EF-1α | 0.52 | EF-1α | 0.62 | TBP | 0.99 | 3.85 | TUB-A | 5.66 | |
| 8 | TUB-A | 0.58 | TUB-A | 0.64 | HIS | 1.01 | 3.78 | EF-1α | 7.45 | |
| 9 | HIS | 0.66 | HIS | 0.85 | EF-1α | 1.20 | 4.98 | HIS | 8.74 | |
| ‘NJC108’ | 1 | UBC | 0.50 | UBC | 0.25 | HIS | 0.50 | 1.91 | UBC | 1.78 |
| 2 | ACTIN | 0.50 | ACTIN | 0.25 | TBP | 0.59 | 2.24 | ACTIN | 1.93 | |
| 3 | TUB-B | 0.63 | TUB-B | 0.34 | GAPDH | 0.72 | 2.82 | HIS | 3.13 | |
| 4 | HIS | 0.75 | eIF-4α | 0.42 | TUB-A | 0.84 | 3.24 | TUB-B | 3.57 | |
| 5 | GAPDH | 0.79 | GAPDH | 0.71 | UBC | 0.97 | 3.96 | GAPDH | 4.4 | |
| 6 | eIF-4α | 0.84 | HIS | 0.78 | TUB-B | 1.10 | 4.51 | TBP | 5.66 | |
| 7 | TUB-A | 0.92 | TUB-A | 1.14 | ACTIN | 1.12 | 4.58 | eIF-4α | 5.83 | |
| 8 | TBP | 1.03 | TBP | 1.50 | eIF-4α | 1.24 | 5.01 | TUB-A | 6.09 | |
| 9 | EF-1α | 1.65 | EF-1α | 3.74 | EF-1α | 3.92 | 15.34 | EF-1α | 9.00 | |
Table 4.
The stability ranking of candidate reference genes in different stage of samples by geNorm, NormFinder, BestKeeper and RefFinder.
| Stage | Rank | geNorm | NormFinder | BestKeeper | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability value | Gene | SD [± Cq] | CV [%Cq] | Gene | Stability | ||
| S1 | 1 | TUB-B | 0.32 | TUB-A | 0.27 | TUB-A | 0.33 | 1.36 | TUB-A | 1.41 |
| 2 | HIS | 0.32 | ACTIN | 0.38 | UBC | 0.36 | 1.47 | TUB-B | 2.38 | |
| 3 | UBC | 0.41 | EF-1α | 0.38 | eIF-4α | 0.44 | 1.76 | UBC | 3.50 | |
| 4 | TUB-A | 0.48 | TUB-B | 0.40 | TUB-B | 0.45 | 1.96 | ACTIN | 3.94 | |
| 5 | EF-1α | 0.54 | UBC | 0.41 | ACTIN | 0.48 | 2.01 | HIS | 4.14 | |
| 6 | ACTIN | 0.57 | eIF-4α | 0.50 | HIS | 0.48 | 1.92 | EF-1α | 4.21 | |
| 7 | eIF-4α | 0.59 | HIS | 0.60 | EF-1α | 0.51 | 2.23 | eIF-4α | 5.24 | |
| 8 | GAPDH | 0.64 | GAPDH | 0.70 | GAPDH | 0.70 | 2.81 | GAPDH | 8.00 | |
| 9 | TBP | 0.78 | TBP | 1.15 | TBP | 0.77 | 2.90 | TBP | 9.00 | |
| S2 | 1 | UBC | 0.54 | UBC | 0.25 | HIS | 0.64 | 2.49 | UBC | 1.78 |
| 2 | HIS | 0.54 | TUB-B | 0.34 | GAPDH | 0.78 | 3.10 | HIS | 1.86 | |
| 3 | TUB-A | 0.59 | ACTIN | 0.34 | TBP | 0.88 | 3.39 | ACTIN | 2.91 | |
| 4 | ACTIN | 0.66 | HIS | 0.52 | TUB-A | 0.91 | 3.62 | TUB-A | 3.94 | |
| 5 | TUB-B | 0.71 | TUB-A | 0.56 | UBC | 0.99 | 4.03 | TUB-B | 4.33 | |
| 6 | GAPDH | 0.78 | GAPDH | 0.75 | ACTIN | 1.06 | 4.35 | GAPDH | 4.56 | |
| 7 | TBP | 0.97 | TBP | 1.50 | TUB-B | 1.43 | 6.08 | TBP | 5.66 | |
| 8 | eIF-4α | 1.33 | eIF-4α | 2.40 | eIF-4α | 1.70 | 6.50 | eIF-4α | 8.00 | |
| 9 | EF-1α | 1.88 | EF-1α | 3.68 | EF-1α | 3.91 | 15.33 | EF-1α | 9.00 | |
| S3 | 1 | GAPDH | 0.43 | EF-1α | 0.36 | ACTIN | 0.28 | 1.18 | ACTIN | 1.73 |
| 2 | ACTIN | 0.43 | UBC | 0.38 | TBP | 0.36 | 1.38 | EF-1α | 1.86 | |
| 3 | EF-1α | 0.51 | ACTIN | 0.48 | GAPDH | 0.44 | 1.78 | GAPDH | 2.63 | |
| 4 | UBC | 0.60 | GAPDH | 0.49 | EF-1α | 0.55 | 2.41 | UBC | 2.99 | |
| 5 | TUB-A | 0.66 | TUB-B | 0.62 | UBC | 0.64 | 2.67 | TBP | 5.12 | |
| 6 | TUB-B | 0.68 | TUB-A | 0.63 | eIF-4α | 0.67 | 2.71 | TUB-B | 5.69 | |
| 7 | TBP | 0.75 | TBP | 0.80 | TUB-B | 0.78 | 3.35 | TUB-A | 6.16 | |
| 8 | eIF-4α | 0.82 | eIF-4α | 0.86 | TUB-A | 0.88 | 3.54 | eIF-4α | 7.44 | |
| 9 | HIS | 0.88 | HIS | 0.89 | HIS | 0.89 | 3.46 | HIS | 9.00 | |
| S4 | 1 | TUB-B | 0.44 | ACTIN | 0.23 | TUB-A | 0.70 | 2.67 | ACTIN | 1.73 |
| 2 | HIS | 0.44 | TUB-B | 0.41 | GAPDH | 0.72 | 2.85 | TUB-B | 2.11 | |
| 3 | ACTIN | 0.65 | UBC | 0.52 | ACTIN | 0.78 | 3.22 | HIS | 3.25 | |
| 4 | UBC | 0.68 | HIS | 0.61 | TBP | 0.90 | 3.40 | UBC | 3.83 | |
| 5 | GAPDH | 0.73 | GAPDH | 0.62 | TUB-B | 0.96 | 4.04 | GAPDH | 3.98 | |
| 6 | EF-1α | 0.78 | EF-1α | 0.77 | UBC | 1.08 | 4.46 | TUB-A | 5.20 | |
| 7 | TBP | 0.86 | TBP | 0.82 | HIS | 1.11 | 4.32 | TBP | 6.09 | |
| 8 | eIF-4α | 0.93 | eIF-4α | 1.12 | EF-1α | 1.28 | 5.33 | EF-1α | 6.45 | |
| 9 | TUB-A | 1.04 | TUB-A | 1.30 | eIF-4α | 1.35 | 5.40 | eIF-4α | 8.24 | |
BestKeeper
The BestKeeper software evaluates the expression stability of reference genes with SD and CV of Cq values, and the smaller SD and CV value suggest the more stable results. For the three sets (Tables 2, 3, 4), the order of the stability of the reference genes in ‘Total’ is as follows: ACTIN > GAPDH > UBC > TBP > HIS > TUB-B > TUB-A > eIF-4α > EF-1α; the gene HIS ranks the first in ‘Hakuho’ and ‘NJC108’, but ranks the fourth in ‘Xiacui’ and the eighth in ‘Fantasia’; the gene TUB-A ranks the first in S1 and S4, but the seventh and eighth in S2 and S3, respectively. According to the analysis of BestKeeper, the genes ACTIN and GAPDH have good stability, while EF-1α has the worst stability.
RefFinder
The stability ranking calculated by the three software geNorm, NormFinder and BestKeeper were not entirely consistent. RefFinder is used to integrate the results to get the comprehensive index ranking. As shown in Tables 2, 3 and 4, the genes ACTIN and UBC have good stability, while eIF-4α and EF-1α have poor stability in ‘Total’ set. The order of stability was: ACTIN > UBC > GAPDH > TUB-B > HIS > TBP > TUB-A > eIF-4α > EF-1α. The genes ACTIN and EF-1α rank first in ‘Hakuho’ and ‘Xiacui’, respectively. The gene UBC ranks first in ‘Fantasia’, ‘NJC108’, and ranks the second in ‘Xiacui’, while ranks the sixth in ‘Hakuho’. The gene ACTIN shows the highest stability in S2, S3 and S4, while ranks the fourth in S1.
Analysis of the optimal number of reference genes of peach
The geNorm software was used to determine the optimal number of reference genes by analyzing the pairwise variation (Vn/n + 1) between normalization factors (NFn and NFn + 1, n ≥ 2). When Vn/(n + 1) < 0.15, it indicates that an extra reference gene is not necessary, whereas Vn/(n + 1) ≥ 0.15 means that at least n + 1 genes should be required in qRT-PCR analysis. As shown in Fig. 4, in the ‘Total’ set, only the value of V5/6 is less than 0.15, indicating that five reference genes (ACTIN, UBC, GAPDH, TUB-B and HIS) were proposed to be used. For ‘Hakuho’ (ACTIN and TUB-B), ‘Xiacui’ (TUB-B and EF-1α), ‘Fantasia’ (UBC and TUB-B) and S1 (TUB-A and TUB-B), two reference genes were enough with the V2/3 less than 0.15, while four genes were needed in ‘NJC108’ (UBC, ACTIN, HIS and TUB-B), S2 (UBC, HIS, ACTIN and TUB-A) and S3 (ACTIN, EF-1α, GAPDH and UBC) with the V4/5 dropping to 0.15. For S4, three genes (ACTIN, TUB-B and HIS) needs to be introduced.
Figure 4.
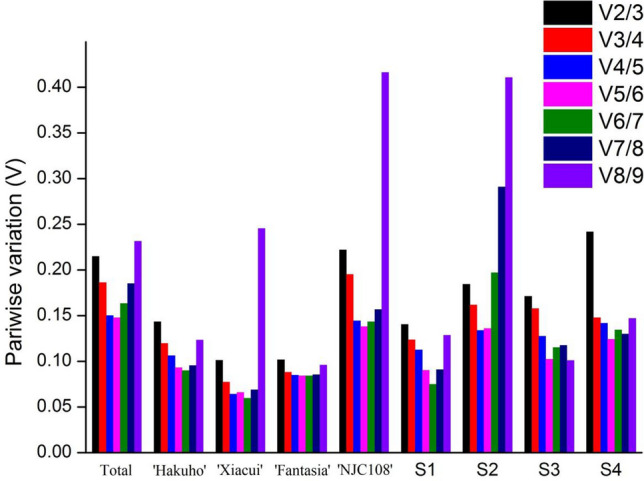
Pairwise variation analysis to determination of the optimal number of the nine candidate reference genes. All pairwise variation (Vn/Vn + 1) were calculated via geNorm, and the values determined the minimum number of reference genes for accurate normalization in each experimental set. The Vn/Vn + 1 values below 0.15 indicate that an additional reference genes are not necessary for gene expression normalization.
Determination of flesh firmness and validation of the selected reference genes of peach
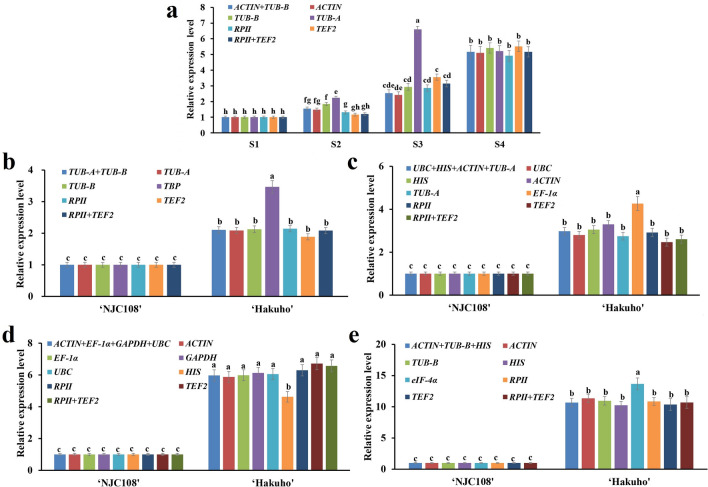
Verifying the expression stability of candidate reference genes in different sets will greatly improve the reliability of gene expression data. In this study, the sampling stages and the flesh firmness of ‘Hakuho’ during four fruit ripening stages were displayed (Fig. 5). The relative expression level of PpACS1, encoding 1-aminocyclopropane-1-carboxylic acid synthase in ‘Hakuho’ samples during fruit development were evaluated using the several top ranking reference genes, as recommended by RefFinder, alone or with a combination for data normalization. Combined with the optimal number of reference genes recommended by geNorm, the reference genes including the optimal reference genes of ACTIN and TUB-B in ‘Hakuho’, TUB-A and TUB-B in S1, UBC, HIS, ACTIN and TUB-A in S2, ACTIN, EF-1α, GAPDH and UBC in S3, ACTIN, TUB-B and HIS in S4 (combined Fig. 4, Tables 3, 4), two stable reference genes PpRPII (encoding RNA polymeraseII) and PpTEF2 (encoding translation elongation factor 2) have been reported in previous study27, and the least stable reference genes (TUB-A, TBP, EF-1α, HIS and eIF-4α) from the sample set, respectively. As illustrated in Fig. 6a, the PpACS1 expression level showed the similar (but not identical) trend when using single or a combination of reference genes ACTIN, TUB-B and RPII, TEF2. The expression level of PpACS1 increased gradually from S1 to S3, and increased rapidly from S3 to S4. There is no significant difference among the expression level of PpACS1 normalized by most stable genes ACTIN , TUB-B, and those normalized by control internal reference genes RPII and TEF2, individually and in combination, in S1 and S4 stages (Fig. 6a). However, PpACS1 expression level normalized by TUB-B showed significant difference with the expression level of PpACS1 when RPII and TEF2, individually and in combination, were used as internal control genes in S2. In addition, PpACS1 expression level normalized by ACTIN showed significant difference with the expression level of PpACS1 when TEF2 was used as internal control genes in S3. PpACS1 expression level when normalized with the least stable gene TUB-A showed significant difference with PpACS1 expression level when ACTIN, TUB-B and RPII TEF2, individually and in combination, were used as internal control genes from S1 to S4 stage. Furthermore, alone or the combination of two (TUB-A and TUB-B) or two (RPII and TEF2) reference genes were used to normalize qRT-PCR data showed the same expression patterns of PpACS1 in S1. However, using the least stable gene TBP as internal control, the expression level of PpACS1 showed significant difference with the data when normalized using other optimal reference genes individually or in combination (Fig. 6b) and the results were in accordance with expression level of PpACS1 when the least stable reference genes: EF-1α, HIS and eIF-4α were used for normalization in S2, S3 and S4 stage, respectively (Fig. 6c–e). Taken together, these results indicate that using inappropriate internal reference genes for normalization may lead to in accuracy qRT-PCR results. Therefore, select appropriate and reliable control genes is essential for gene expression analysis.
Figure 5.
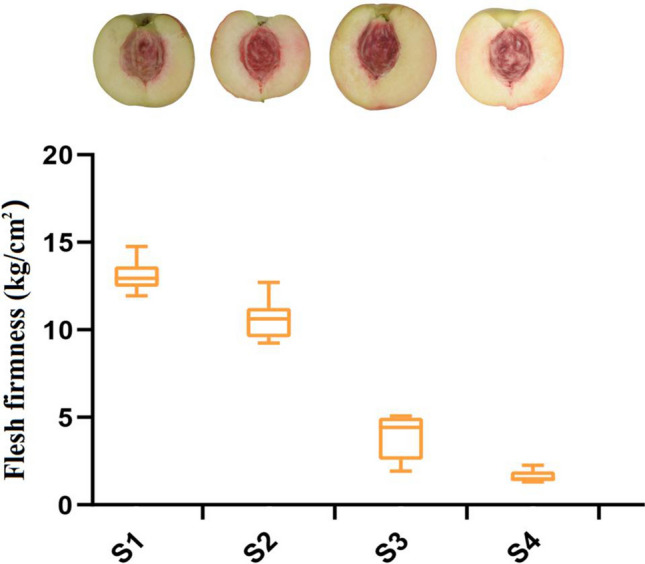
The fruit sampling stages and the flesh firmness of ‘Hakuho’ during fruit ripening and softening. The samples were randomly collected from trees at 4-day intervals for four times, starting from nearly 85 days after full bloom (DAFB) through to ripening, and designated as S1, S2, S3 and S4, respectively.
Figure 6.
Relative expression level of PpACS1 gene in ‘Hakuho’ peach cultivar from S1 to S4 stage. (a) Genes were normalized to ACTIN, TUB-B, TUB-A, RPIIand TEF2, individually and in combination, in ‘Hakuho’ peach cultivar. (b) Genes were normalized to TUB-A, TUB-B, TBP, RPIIand TEF2, individually and in combination, in S1. (c) Genes were normalized to UBC, HIS, ACTIN, TUB-A, EF-1α, RPII and TEF2, individually and in combination, in S2. (d) Genes were normalized to ACTIN, EF-1α, GAPDH, UBC, HIS, RPII and TEF2, individually and in combination, in S3. (e) Genes were normalized to ACTIN, TUB-B, HIS, eIF-4α, RPII and TEF2, individually and in combination, in S4. The statistical significance was determined by Duncan’s multiple comparison tests. Different letters indicate a significant difference (P < 0.05).
Discussion
To date, high throughput sequencing29, microarray30 and qRT-PCR31 approaches have been applied to detect gene expression, qRT-PCR has become a powerful tool and the most frequently used approach for normalization because of its sensitivity and specificity8. Accurate results of qRT-PCR are closely related to the normalization of certain suitable reference gene, and using of inappropriate reference gene will lead to deviated analysis and even wrong conclusions32–34. A good reference gene should have consistent expression levels under any experimental conditions and be independent of organs, tissues, stages of development and various treatments, etc. However, none of the reference genes has a constant expression profile under all experimental conditions. Therefore, screening and evaluating the best stable reference genes as well as establishing an effective evaluation system of multiple reference genes in specific research models are important for improving the accuracy of qRT-PCR experiments and clarifying qRT-PCR results-represented scientific questions in those models.
According to the textural changes during ripening, peach fruits are mainly classified into three types, melting, non-melting and stony hard35. Although there have been some reports on the selection of reliable reference genes in peach based on geNorm, NormFinder and/or BestKeeper software, and TEF2, UBQ10 and RPII25–27; eIF-1A, MUB625 and miR5059, miR507226 were found to be the most suitable reference genes across the special set of all samples. In this study evaluated the expression stabilities of 9 reference genes in 16 experimental samples (4 peach cultivars and 4 peach ripening and softening stages) were evaluated using qRT-PCR to determine the most stable reference genes. The present study is the first and systematical survey focused on the stability analysis of reference genes in the three types of peach fruit during its ripening and softening, geNorm, NormFinder, BestKeeper and RefFinder were applied.
Different rankings for the 9 candidate reference genes were developed after comparison to the ranking of these candidates generated by the four algorithms (Tables 2, 3, 4). Based on calculations by geNorm, 5 optimal reference genes were selected, and UBC, ACTIN expressed the most stably in the ‘Total’ sample, and they were considered to be the most suitable reference candidate genes under situations that different cultivars and ripening stages are blended. The NormFinder software showed nearly the same results in the ‘Total’ sample as geNorm. Based on the discrepancies of the statistical algorithms between the two software, geNorm and NormFinder, the reference gene stability ranking varied in the different cultivar and stage setups between these two programs (Tables 3, 4)36–38. The geNorm is highly dependent on the assumption that none of the genes is co-regulated as this would lead to an erroneous choice of optimum normalizer pair25,27,37, The geNorm provide more reliable results than NormFinder in terms of the best reference pairs for each experimental setup (Tables 2, 3, 4). In BestKeeper analysis, it is found that ACTIN and GAPDH had relatively good stability, EF-1α has the worst performance in the ‘Total’ setup. In view of the discrepancies among the analysis results of the above three software. RefFinder was further introduced to integrate all the results to avoid one sidedness of single software analysis and make the screening results more reliable. Comprehensive analysis by RefFinder software, the stability ranking turned out to be ACTIN > UBC > GAPDH > TUB-B > HIS > TBP > TUB-A > eIF-4α > EF-1α. Therefore, ACTIN and UBC are recommended as the most suitable reference genes, while ACTIN, UBC, GAPDH, TUB-B and HIS are the optimal reference genes when research model involves total analysis of different cultivars and ripening stages. In fact, the stability of the reference gene is not the same across plants. For example, ACTIN, the third mainly used reference gene, has been widely used as reference gene in gene expression studies in many organisms27, and ACTIN is stably expressed in this experiment, however ACTIN performed unsatisfactory in wheat39, maize40 and garlic41. This partly attributed to the fact that ACTIN as one of the major components of cytoplasmic microfilaments in eukaryotic cells, not only supports the cell and determines its shape but also participates in other cellular functions42. GAPDH has only moderate stability in the ‘Total’, ‘Fantasia’ and ‘S3’ experimental setups. And some researches have showed that GAPDH had a good performance in grape43 and Primula forbesii12, but not in wheat39. EF-1 α can be used as an appropriate reference gene in zucchini44, wolfberry45, and wheat39, but our analysis showed that EF-1 α was not the reliable gene for comparative expression in our peach model. UBC is a reference gene with good expression stability in all samples in different development stages of pearl millet46, which also behaves well in current peach model, but the expression stability is the worst in different tissues and fruit stages of pitaya fruit47.
Fruit ripening and softening is a complex process involving major traditions in fruit development and metabolism, and this developmental transition involves coordinated changes in a number of biochemical pathways. Ethylene regulates at least part of this developmental transition, ethylene biosynthesis, ethylene responses, and ethylene-regulated gene expression have been extensively studied in ripening fruit18. ACS is a key rate-limiting enzyme that controls ethylene synthesis in plants and plays an important regulatory role in plant development, fruit maturation48,49. The expressions of ACS homologous genes are differentially regulated by plant developmental and hormonal signals50 and PpACS gene expression is closely related to fruit ripening in peach23. Six PpACS genes (PpACS1-6) have been identified in nectarine (Prunus persica var. nectarina) and the expression level of the three genes (PpACS1, PpACS4 and PpACS5) showed dynamically changed during fruit ripening and softening in peach23. Peach, a climatic fruit, undergoes textural changes that lead to loss of tissue firmness during fruit ripening and softening and is accompanied by an increase in ethylene evolution. The expression level of PpACS1 which induces peach ripening, is related to ethylene production51,52.
To test the reliablity of the reference genes, the expression profiles of PpACS1 were assayed in ‘Hakuho’. Usually, only one reference gene was used for qRT-PCR data normalization in most reported gene expression studies53. However, single reference gene is insufficient sometimes54. RefFinder, which considers the optimal reference gene results of three algorithms (geNorm, NormFinder and BestKeeper) together. Therefore, PpACS1 expression level of ‘Hakuho’ during S1 to S4 were analyzed using several most stable reference genes recommended by RefFinder and the stable genes (TEF2 and RP II) reported in previous study27 individually and in combination to confirm the importance of selecting appropriate reference genes in experimental design. Combined the firmness of ‘Hakuho’ during peach ripening and softening and the expression level of PpACS1 which was normalized by ACTIN, TUB-B, TUB-A, RPIIand TEF2 genes alone or a combination, the firmness of ‘Hakuho’ was decreased from 13.67 kg/cm2 to 1.61 kg/cm2 during fruit ripening and softening stage (from S1 to S4) (Fig. 5). The PpACS1 expression level when normalized by RPII and TEF2 in S4 was 3.9 and 4.5 times higher than that in S1, respectively, while the expression level of PpACS1 which was normalized by the most stable reference genes ACTIN and TUB-B in S4 was 4.1 and 4.4 times higher than that in S1. In addition, we found that PpACS1 expression level normalized by ACTIN showed no significant difference with the expression level of PpACS1 normalized by RPII and TEF2 from S1 to S4 (Fig. 6a). And this results just confirmed that ACTIN was the best stable reference gene in ‘Hakuho’ (Table 4). PpACS1 expression showed the same trend from S1 to S4 when ACTIN, TUB-B, RPII and TEF2 were used as internal genes. While the most unstable gene TUB-A was used for normalization, PpACS1 expression level was severely overestimated during S3 stage in ‘Hakuho’ (Fig. 6a).
Generally speaking, flesh texture types of peach including the non-melting type and the stony hard type in addition to the melting type55. Mature peach fruit of melting and non-melting flesh types produce ethylene during ripening and this character is controlled by the related genes such as PpACS1, however, stony hard fruit texture is characterized by the absence of both ethylene production and post-harvest softening in mature fruit56. The melting type is characterized by rapid softening of the fruit flesh, while the non-melting type is characterized by a more limited softening. To validate the candidate reference genes at different ripening stages (S1: TUB-A and TUB-B, S2: UBC, HIS, ACTIN and TUB-A, S3: ACTIN, EF-1α, GAPDH and UBC, S4: ACTIN, TUB-B and HIS) of ‘Hakuho’, these multiple reference genes were used as internal controls, individually and in combination, to normalize the expression level of PpACS1, while the non-melting peach cultivar ‘NJC108’ was used as a control (Fig. 6b–e). The results with the optimal reference genes alone or a combination as internal controls were in consistent with the reduction of the peach fruit firmness and the expression characteristics of PpACS1 from S1 to S4 stage in ‘Hakuho’ (Figs. 5, 6a), while the PpACS1 expression level with the least stable reference genes (S1: TBP, S2: EF-1α, S3: HIS and S4: eIF-4α) (Fig. 6b–e) showed discrepancy with the expression trend of PpACS1 from S1 to S4 stage in ‘Hakuho’ (Fig. 6a). All the results fully showed that different reference genes were used to normalize in qRT-PCR analysis may lead different conclusions. Especially using an unstable reference gene, the results may be biased.
In conclusion, the selection of a suitable internal reference gene is a key step in gene expression analysis. The results from this study not only provide a favorable basis for selection of suitable reference genes in different cultivars and fruit maturation of peach, but also can be used as a reference for the exploration of related functional genes, expression patterns and regulatory mechanism in peach.
Materials and methods
Plant materials and growth conditions
Samples were collected from 8-year-old peach trees of ‘Hakuho’ (a melting flesh peach genotype), ‘NJC108’ (a non-melting flesh peach genotype), ‘Xiacui’ and ‘Fantasia’ (a stony hard flesh peach genotype) cultivar. All these four peach cultivars were growing in the National Peach Germplasm Repository of Zhengzhou Fruit Research Institute (Henan, China) under the same common field conditions. The collection of these peach cultivars was permitted by Zhengzhou Fruit Research Institute and it complies with local and national guidelines and legislation. Peach fruits with no visible defects, at a stage equivalent to commercial ripeness, were randomly collected from trees at 4-day intervals for four times, starting from nearly 85 days after full bloom (DAFB) through to ripening and softening, and designated as S1, S2, S3 and S4, respectively. Samples were collected and were immediately frozen in liquid nitrogen and then stored at − 80 °C until RNA extraction. Three biological replicates were performed for each sample.
Total RNA isolation and cDNA synthesis
Total RNA of all samples were extracted by using an RNA kit (Tiangen, Beijing, China) in accordance with the manufacturer’s method. The quality and purity of total RNA were further assessed with spectrophotometer NanoDrop 2000C (Thermo, USA) and 1% agarose gel electrophoresis. Approximately 1000 ng of total RNA was reverse transcribed into cDNA using the Prime Script RT Reagent Kit (TransGene, Beijing, China). The cDNA was subjected to tenfold serial dilutions (10×, 102×, 103×, 104×, 105×) for determining the amplification efficiency (E) and coefficient of correlation (R2) analysis; and 20-fold diluted for PCR amplification.
Gene selection, primer design and gene cloning
Nine candidate reference genes, EF-1α, GAPDH, TBP, UBC, eIF-4α, TUB-A, TUB-B, ACTIN and HIS, were selected from the whole-genome of peach. Primer Premier 6 was used to design the specific primers according to the gene sequences. The nine candidate reference genes were cloned from peach with the primer sequences listed in Table S1.
qRT-PCR, and statistical analysis of gene expression stability
The qRT-PCR was performed with a Bio-Rad real-time PCR System (Bio-Rad, CA, USA), with a SYBR qRT-PCR Mix (TsingKe, Beijing, China). The amplification procedure was conducted as follows: 95 °C for 1 min pre-denaturation, followed by 40 cycles at 95 °C for 5 s for denaturation, 60 °C for 30 s for annealing and extension, and melting curve analysis (61 cycles) at 65 °C for 10 s. The primer sequences of nine reference genes for qRT-PCR were listed in Table 1. Each assay contained a standard curve based on different dilutions of cDNA template to test the amplification efficiency (E, E% = (− 1 + 10 [−1/slope]) × 100%) of primer pair of each gene. Controls without template were also included in each run.
Expression levels of the nine candidate reference genes of peach were quantified by the number of amplification cycles (Cq). Four software, geNorm57, NormFinder58, BestKeeper59, and RefFinder60 were used to evaluate the gene expression stability for all samples. For geNorm and NormFinder software, Cq values would be converted into relative quantities according to the formula 2−ΔCq, ΔCq = the corresponding Cq—minimum Cq. geNorm evaluates the stability of the reference gene expression by comparing the pairwise variation (M). Besides, the optimal number of reference genes in qRT-PCR normalization were determined by the pairwise variation (Vn/n + 1) among normalization factors (NFn and NFn + 1, n ≥ 2) in geNorm. NormFinder compares the intra- and inter-group variation and combines the variation into a stability value for each gene. BestKeeper could analyze the raw Cq value to rank the stability with standard deviation (SD) and coefficient of variation (CV). RefFinder calculates a comprehensive stability ranking by integrating the three computational programs (geNorm, Normfinder, and BestKeeper).
Determination of flesh firmness and validation of reference genes of peach
Peach flesh firmness was determined with a fruit pressure tester (GY-4-J; TOP, China) equipped with an 11-mm diameter probe. The probe was pressed into the tissue of peach surface to 10 mm depth in every single smooth motion, and three disks were removed from opposite sides of each peach. Three peaches per sample were measured.
ACS1, a gene encoding an ethylene synthase and involved into fruit maturation, was selected to validate the suitable reference genes. The primer sequence of PpACS1 was 5′-TGCGTGGAGCCTGGTTGGTT-3′ for forward and 5′-CGAACGAGAGGAGAGTGAGGAGAC-3′ for reverse. The optimal reference genes of ACTIN and TUB-B in ‘Hakuho’, two stable genes (RPII, TEF2) reported in previous study27, the least stable gene (TUB-A) and multiple reference gene combinations at different ripening stages in peach: S1 (TUB-A and TUB-B), S2 (UBC, HIS, ACTIN and TUB-A), S3 (ACTIN, EF-1α, GAPDH and UBC), S4 (ACTIN, TUB-B and HIS) were used as an internal standard to normalize PpACS1 expression, respectively. The relative expression level of PpACS1 gene in ‘Hakuho’ was calculated using 2−ΔΔCt method59.
Supplementary Information
Acknowledgements
This study was supported by the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2019-ZFRI-01), Sci-Tech innovation special project for social undertakings and people’s livelihood guarantee of Chongqing science and technology bureau (cstc2019jscx-gksbX0138) and the Excellent Variety Innovation Program for characteristic benefit of Chongqing Academy of Agricultural Science (NKY-2019AB017, NKY-2020AB004). And thanks Professor Li of Connecticut university for providing language checking.
Author contributions
L.R.W., designed the experiments. S.H.Y. analyzed the data, performed the manuscripts and prepared the manuscript. K.C., Y.L. and J.L.W. checked the manuscript, approved the final draft. C.W.C. took the photos of the sample and prepared RNA samples. G.R.Z., W.C. F. and X.W.W. prepared the materials.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shuanghong You and Ke Cao.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86755-5.
References
- 1.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 2.Chapman JR, Waldenström J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE. 2015;10:1–18. doi: 10.1371/journal.pone.0141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dheda K, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005;344:141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Nestorov J, Matic G, Elakovic I, Tanic N. Gene expression studies: How to obtain accurate and reliable data by quantitative real-time RT-PCR. J. Med. Biochem. 2013;32:325–338. doi: 10.2478/jomb-2014-0001. [DOI] [Google Scholar]
- 5.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper -Excel-based tool using pairwise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 6.Govindaraj R, et al. Hepatoprotective effect of tricholoma giganteum (Agaricomycetes) in a nonalcoholic fatty liver disease rat model. Int. J. Med. Mushrooms. 2016;18:661–669. doi: 10.1615/IntJMedMushrooms.v18.i8.20. [DOI] [PubMed] [Google Scholar]
- 7.Selvey S, et al. β-actin-an unsuitable internal control for RT-PCR. Mol. Cell Probes. 2001;15:307–311. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- 8.Artico S, Nardeli SM, Brilhante O, Grossi-de-Sa MF, Alves-Ferreira M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010;10:1471–2229. doi: 10.1186/1471-2229-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozera B, Rapacz M. Reference genes in real-time PCR. J. Appl. Genet. 2013;54:391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, et al. Selection of reference genes for real-time quantitative PCR studies of kumquat in various tissues and under abiotic stress. Sci. Hortic. 2014;174:207–216. doi: 10.1016/j.scienta.2013.12.003. [DOI] [Google Scholar]
- 11.Zhang J. Validation of suitable reference genes for quantitative gene expression analysis in Tripterygium wilfordii. Mol. Biol. Rep. 2019;46:4161–4174. doi: 10.1007/s11033-019-04867-8. [DOI] [PubMed] [Google Scholar]
- 12.Jia Y, et al. Reference gene selection and validation by qRT-PCR during flower development and in different organs of Primula forbesii. J. Hortic. Sci. Biotechnol. 2019;25:1–12. [Google Scholar]
- 13.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palakolanu SR, et al. Evaluation of sorghum [Sorghum bicolor (L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Front Plant Sci. 2016;7:529. doi: 10.3389/fpls.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bevitori R, et al. Selection of optimized candidate reference genes for qRT-PCR normalization in rice (Oryza sativa L.) during Magnaporthe oryzae infection and drought. Genet. Mol. Res. 2014;13:9795–9805. doi: 10.4238/2014.November.27.7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Fu J, Wang Y, Bao Z, Zhao H. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCRanalysis of sweet osmanthus (Osmanthus fragransLour.) PLoS ONE. 2015;10:e0136355. doi: 10.1371/journal.pone.0136355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao K, et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016;7:13246. doi: 10.1038/ncomms13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer RL, Bennett AB. Role of cell wall hydrolases in fruit ripening. Plant Mol. Biol. 1991;42:675–703. [Google Scholar]
- 19.Guo J, et al. Comparative transcriptome and microscopy analyses provide insights into flat shape formation in Peach (Prunus persica) Front. Plant Sci. 2018;8:2215. doi: 10.3389/fpls.2017.02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lurie S, Crisosto CH. Chilling injury in peach and nectarine. Postharvest. Biol. Technol. 2005;37:195–208. doi: 10.1016/j.postharvbio.2005.04.012. [DOI] [Google Scholar]
- 21.Spadoni A, Guidarelli M, Sanzani SM, Ippolito A, Mari M. Influence of hot water treatment on brown rot of peach and rapid fruit response to heat stress. Postharvest. Biol. Technol. 2014;94:66–73. doi: 10.1016/j.postharvbio.2014.03.006. [DOI] [Google Scholar]
- 22.Xi WP, et al. Postharvest temperature influences volatile lactone production via regulation of acyl-CoA oxidases in peach fruit. Plant Cell Environ. 2012;35:534–545. doi: 10.1111/j.1365-3040.2011.02433.x. [DOI] [PubMed] [Google Scholar]
- 23.Zeng W, et al. Characterization of 1-aminocycl-opropane-1-carboxylic acid synthase (ACS) genes during nectarine fruit development and ripening. Tree Genet. Genomes. 2015;11:18. doi: 10.1007/s11295-015-0833-6. [DOI] [Google Scholar]
- 24.Fonseca S, et al. Monitoring gene expression along pear fruit development, ripening and sene-scence using cDNA microarrays. Plant Sci. 2004;167:457–469. doi: 10.1016/j.plantsci.2004.03.033. [DOI] [Google Scholar]
- 25.Kou X, Zhang L, Yang S, Li G, Ye J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experiment conditions. Sci. Hortic.-Amst. 2017;225:195–203. doi: 10.1016/j.scienta.2017.07.004. [DOI] [Google Scholar]
- 26.Luo X, et al. Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci. Hortic. 2014;165:281–287. doi: 10.1016/j.scienta.2013.10.030. [DOI] [Google Scholar]
- 27.Tong Z, Gao Z, Wang F, Zhou J, Zhang Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009;10:7. doi: 10.1186/1471-2199-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 29.Jin P, et al. Selection and validation of endogenous reference genes using a high throughput approach. BMC Genomics. 2004;5:55. doi: 10.1186/1471-2164-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paolacci AR, Tanzarella OA, Porceddu E, Ciaffifi M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, et al. A genome-wide characterization of microRNA genes in maize. Plos Genet. 2009;5:e1000716. doi: 10.1371/journal.pgen.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez L, et al. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 33.Guenin S, et al. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009;60:487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, He LL, Fu QT, Xu ZF. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using real-time quantitative PCR. Int. J. Mol. Sci. 2013;14:24338–24354. doi: 10.3390/ijms141224338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haji T, Yaegaki H, Yamaguchi M. Inheritance and expression of fruit texture melting, non-melting and stony hard in peach. Sci. Hortic.-Amst. 2005;105:241–248. doi: 10.1016/j.scienta.2005.01.017. [DOI] [Google Scholar]
- 36.Fan C, et al. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis) PLoS ONE. 2013;8:e56573. doi: 10.1371/journal.pone.0056573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, et al. Selection and validation of suitable reference genes for mRNA qRT-PCR analysis using somatic embryogenic cultures, floral and vegetative tissues in citrus. Plant Cell Tissue Organ Cult. 2013;113:469–481. doi: 10.1007/s11240-013-0288-0. [DOI] [Google Scholar]
- 38.Mafra V, et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS ONE. 2012;7:e31263. doi: 10.1371/journal.pone.0031263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long XY, et al. Genome-wide identification and evaluation of novel internal control genes for q-PCR based transcript normalization in wheat. Plant Mol. Biol. 2010;74:307–311. doi: 10.1007/s11103-010-9666-8. [DOI] [PubMed] [Google Scholar]
- 40.Galli V, Messias RDS, Silva SDDA, Rombaldi CV. Selection of reliable reference genes for quantitative real-time polymerase chain reaction studies in maize grains. Plant Cell. 2013;32:1869–1877. doi: 10.1007/s00299-013-1499-x. [DOI] [PubMed] [Google Scholar]
- 41.Liu M, Wu Z, Jiang F. Selection and validation of garlic reference genes for quantitative real-time PCR normalization. Plant Cell Tissue Organ. Cult. 2015;22:435. doi: 10.1007/s11240-015-0780-9. [DOI] [Google Scholar]
- 42.Stürzenbaum SR, Kille P. Control genes in quantitative molecular biological techniques: The variability of invariance. Comp. Biochem. Phys. B Biochem. Mol. Biol. 2001;130:281–289. doi: 10.1016/S1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- 43.Selim M, et al. Identification of suitable reference genes for real-time RT-PCR normalization in the grapevine-downy mildew pathosystem. Plant Cell Rep. 2012;31:205–216. doi: 10.1007/s00299-011-1156-1. [DOI] [PubMed] [Google Scholar]
- 44.Obrero A, et al. Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J. Agric. Food Chem. 2011;59:5402–5411. doi: 10.1021/jf200689r. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Wang Y, Zhou P. Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiol. Biochem. 2013;70:304–310. doi: 10.1016/j.plaphy.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 46.Shivhare R, Lata C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 2016;6:23036. doi: 10.1038/srep23036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods. 2019;15:70. doi: 10.1186/s13007-019-0455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haji T, Yaegaki H, Yamaguchi M. Softening of stony hard peach by ethylene and the induction of endogenous ethylene by 1-aminocyclopropane-1-carboxylic acid (ACC) J. Jpn. Soc. Hortic. Sci. 2003;72:212–217. doi: 10.2503/jjshs.72.212. [DOI] [Google Scholar]
- 49.Hayama H, Ito A, Moriguchi T, Kashimura Y. Identification of a new expansin gene closely associated with peach fruit softening. Postharvest Biol. Tech. 2003;29:1–10. doi: 10.1016/S0925-5214(02)00216-8. [DOI] [Google Scholar]
- 50.Hayama H, Shimada T, Fujii H, Ito A, Kashimura Y. Ethylene-regulation of fruit softening and softening-related genes in peach. J. Exp. Bot. 2006;57:4071–4077. doi: 10.1093/jxb/erl178. [DOI] [PubMed] [Google Scholar]
- 51.Hayama H, Tatsuki M, Ito A, Kashimura Y. Ethylene and fruit softening in the stony hard mutation in peach. Postharvest. Biol. Technol. 2006;41:16–21. doi: 10.1016/j.postharvbio.2006.03.006. [DOI] [Google Scholar]
- 52.Tatsuki M, Mori H. Rapid and transient expression of 1-aminocyclopropane-1-carboxylate synthase isogenes by touch and wound stimuli in tomato. Plant Cell Physiol. 1999;40:709–715. doi: 10.1093/oxfordjournals.pcp.a029597. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 54.Veazey KJ, Golding MC. Selection of stable reference genes for quantitative RT-PCR comparisons of mouse embryonic and extra-embryonic stem cells. PLoS ONE. 2011;6:e27592. doi: 10.1371/journal.pone.0027592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haji T, Yaegaki H, Yamaguchi M. Changes in ethylene production and flesh firmness of melting, nonmelting and stony hard peaches after harvest. J. Jpn. Soc. Hortic. Sci. 2001;70:458–459. doi: 10.2503/jjshs.70.458. [DOI] [Google Scholar]
- 56.Tatsuki M, Haji T, Yamaguchi M. The involvement of 1-aminocyclopropane-1-carboxylic acid synthase isogene, Pp-ACS1, in peach fruit softening. J. Exp. Bot. 2006;57:1281–1289. doi: 10.1093/jxb/erj097. [DOI] [PubMed] [Google Scholar]
- 57.Vandesompele J, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 59.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie F, Peng X, Chen D, Xu L, Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.