Abstract
Grafting is an ancient technique used for plant propagation and improvement in horticultural crops for at least 1,500 years. Citrus plants, with a seed-to-seed cycle of 5–15 years, are among the fruit crops that were probably domesticated by grafting. Poncirus trifoliata, a widely used citrus rootstock, can promote early flowering, strengthen stress tolerance, and improve fruit quality via scion–rootstock interactions. Here, we report its genome assembly using PacBio sequencing. We obtained a final genome of 303 Mb with a contig N50 size of 1.17 Mb and annotated 25,680 protein-coding genes. DNA methylome and transcriptome analyses indicated that the strong adaptability of P. trifoliata is likely attributable to its special epigenetic modification and expression pattern of resistance-related genes. Heterografting by using sweet orange as scion and P. trifoliata as rootstock and autografting using sweet orange as both scion and rootstock were performed to investigate the genetic effects of the rootstock. Single-base methylome analysis indicated that P. trifoliata as a rootstock caused DNA demethylation and a reduction in 24-nt small RNAs (sRNAs) in scions compared to the level observed with autografting, implying the involvement of sRNA-mediated graft-transmissible epigenetic modifications in citrus grafting. Taken together, the assembled genome for the citrus rootstock and the analysis of graft-induced epigenetic modifications provide global insights into the genetic effects of rootstock–scion interactions and grafting biology.
Subject terms: Genomics, Epigenomics, Plant breeding
Introduction
Plant grafting, as a traditional method of asexual propagation, is accomplished most commonly by connecting two plant segments, namely, a shoot piece called the ‘scion’ and a root piece known as the ‘rootstock’ (Fig. 1). This technique has been practiced in agriculture for over 2500 years1. Grafting has been widely used in modern production of many horticultural crops and some forest trees, such as citrus2,3, pear4,5, grape6, cassava7, and cedar8. Plant grafting is an ancient agricultural practice for propagation of uniform seedlings for commercial fruit species and to avoid a juvenile state, as an adult scion grafted onto a juvenile rootstock will maintain its adult state and ability to bear fruit9. Moreover, grafting can modulate plant growth10, improve the yield and quality of crops11,12, and enhance crop resistance to abiotic and biotic stresses10,13–15 in the form of scion–rootstock combinations.
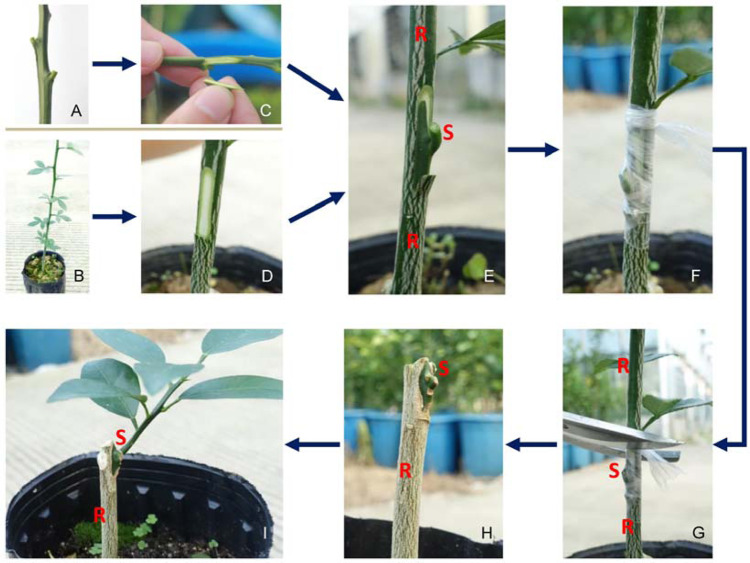
Fig. 1. Schematic diagram of the citrus grafting process.
A Mature sweet orange branch with full buds, which can be used as scion for grafting. B Annual seedling of Poncirus trifoliata, which can be used as rootstock for grafting. C Cutting the bud. D Cutting the rootstock. E Putting the bud on rootstock. F Binding the graft union with plastic film. G Cutting out the rootstock shoot above the graft union after graft union healing. H The live bud grafted on the rootstock. I New shoot coming out from the grafted bud. S scion, R rootstock
Recently, increasing effort has been made to dissect the molecular and physiological mechanisms underlying grafting. The long-distance transport of signaling molecules, such as mobile proteins, mRNAs, small RNAs, and small molecules, between the scion and rootstock has been proven to play pivotal roles in grafting physiology16. Additionally, heterografting-induced DNA methylation polymorphisms have been detected in Hevea brasiliensis17, Solanaceae plants18, and Cucurbitaceae plants19. Some evidence also suggests that epigenetic modification of DNA methylation patterns may account for certain graft transformation phenomena20–22. Currently, advances in genomic resources and molecular techniques provide important opportunities for improving the understanding of scion–rootstock interactions.
Citrus crops are among the most important fruit tree crops in the world, with global production exceeding 147 million tons in 2017 (FAO, 2017). Due to their long juvenility (time to bearing) and high heterozygosity, citrus plants are generally reproduced by grafting to maintain the fine properties of the cultivar and reduce juvenility23. In the citrus industry, the use of suitable rootstocks plays a very important role in commercial citrus production. The rootstock has a significant impact on plant vigor, yield, fruit quality, and disease resistance24–27. Additionally, the rootstock can also affect the metabolome of citrus fruit juice, which determines the flavor and nutrition of the fruit3,28. Moreover, the rootstock can modulate the metabolic response to Candidatus Liberibacter asiaticus in grafted sweet orange29.
With a deeper understanding of the interaction between the rootstock and scion, the breeding of excellent citrus rootstocks is becoming one of the most important ways to improve the efficiency of citrus production and cope with the increasingly harsh planting environment and climate. In long-term citrus production practices, some suitable rootstocks have been widely used in different citrus growth regions, such as Troyer citrange, Carrizo citrange, and Swingle citrimelo in America30, Rangpur lemon in Brazil, sour orange in Italy and Mexico, Palestine sweet lemon and lime in Israel, and rough lemon in India31. The common advantages of these rootstock species are improved tree potential and enhanced tolerance to environmental stress or plant diseases. However, each rootstock still has some disadvantages that prevent it from meeting production demands. For example, Troyer citrange and Carrizo citrange are sensitive to Citrus exocotis viroid32, and the rough lemon as a rootstock produces poor fruit quality33. Therefore, breeding excellent rootstocks for the citrus industry is still ongoing.
Poncirus trifoliata, a wild species closely related to Citrus belonging to the Aurantioideae subfamily of the Rutaceae family, is a popular rootstock for the citrus industry in China. It is diploid and has the same number of chromosomes (2n = 18) as the Citrus genus34. It shows good grafting compatibility with most citrus varieties and exhibits favorable adaptation to a variety of environmental conditions, such as cold hardiness and tolerance to biotic stress factors, including the devastating Huanglongbing35–37. Poncirus seeds are highly polyembryonic and can produce uniform seedlings for ease of grafting and nursery management. Additionally, P. trifoliata is also a valuable parent for rootstock breeding because of its favorable characteristics. Crossing of P. trifoliata with orange gives Carrizo and Troyer citrange, which are used as the top commercial rootstocks in many citrus production areas30. Mining the excellent genetic resources of P. trifoliata and exploring rootstock–scion interactions may promote the improvement of citrus rootstocks and the development of the citrus industry.
In the present study, we aimed to understand the genetic basis of P. trifoliata as a citrus rootstock. We de novo assembled a high-quality genome of P. trifoliata by single-molecule sequencing, and whole-genome DNA methylation maps of P. trifoliata and sweet orange were drawn. Heterografting-induced changes in whole genome DNA methylation and sRNA abundance were evaluated. This study provides an important citrus rootstock genome for understanding the unique biology of grafting and should facilitate better application of grafting in the citrus industry.
Results
Citrus rootstock genome
Screening of 169 P. trifoliata accessions collected from Hubei, Henan, and Shanxi provinces in China indicated that the accession of ZK8 showed the lowest heterozygosity38 (Supplementary Table 1). This genotype was sequenced for genome assembly by using 91× coverage of third-generation long reads generated from the PacBio RS II platform (Supplementary Table 2). In addition, 38× Illumina-sequencing data were used to correct sequencing errors (Supplementary Fig. 1 and Supplementary Table 3). The sequence reads were assembled by Falcon39, resulting in 707 contigs. The total contig sequence length (303 Mb) covered 90.4% of the estimated P. trifoliata genome, and the contig N50 of the final assembly was 1.17 Mb (Table 1). To verify the quality of the assembly, the Illumina sequences were mapped to the assembled genome with a mapping rate of 96.6%, and the error rate of assembly was <0.01%, as estimated by the heterozygous SNP rate. BUSCO40 was also used to assess assembly completeness, and 97.4% of the eukaryotic gene sets were classified as complete. To construct pseudochromosomes, we mapped the contigs to 1934 markers with known sequences in a genetic linkage groups41, and 231 contigs (each >10 kb) were anchored, accounting for 231 Mb of the assembled P. trifoliata genome (Supplementary Table 4). Furthermore, 111 contigs that accounted for the majority of the length of the anchored scaffolds (164.5 Mb, or 71% of total anchored contigs) were in matched orientation within the genetic map, suggesting high alignment accordance between the anchored genetic markers and the sequence contigs.
Table 1.
Statistics for the genome assembly of Poncirus trifoliata
| Assembly feature | Statistic |
|---|---|
| Estimated genome size (by k-mer analysis) | 335 Mb |
| Assembled size | 303 Mb |
| Chromosome size | 231 Mb |
| Number of contigs | 707 |
| Contig N50 | 1.17 Mb |
| Contig N90 | 189 kb |
| TE percentage | 46.5 |
| Annotated protein-coding genes | 25,680 |
| Average coding sequence length | 1297 bp |
| Average exons per gene | 6.4 |
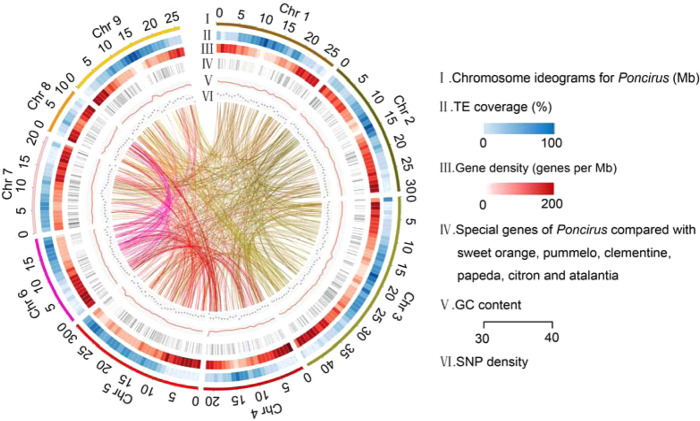
Ab initio gene predictions, homology searches, and RNA-seq analysis were integrated to predict gene models. In total, 25,680 genes with 39,675 transcripts were identified, with an average coding sequence length of 1297 bp and an average of 6.4 exons per gene. In addition, more than 99% of the protein-coding genes could be functionally annotated by GO terms, motifs, domains, and associated pathways. On the basis of homology searches and de novo methods, we identified a total of 140.9 Mb of repetitive elements, representing 46.5% of the genomic assembly (Table 1). Among the repetitive sequences, long terminal retrotransposons (LTRs) were the most abundant, accounting for 23.9% of the assembly (Supplementary Table 5). An overview of the gene density, repetitive elements, SNPs, and all detected syntenic blocks is presented in Fig. 2.
Fig. 2. Characterization of the Poncirus trifoliata genome.
The designation of each track is listed on the right. The lines in the center of the circle indicate pairs of homologous genes on the different chromosomes of P. trifoliata
Genome comparison among Poncirus and other Citrinae genomes
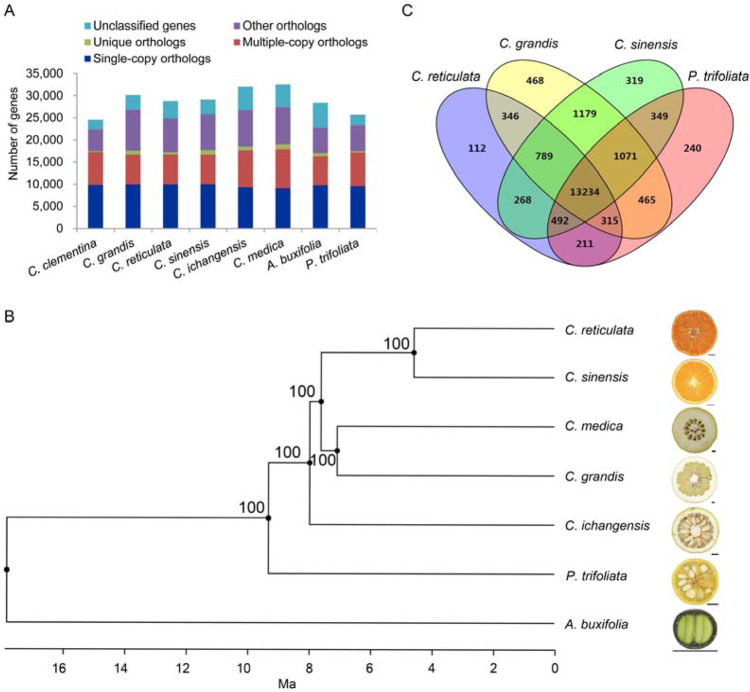
Pairwise comparisons of putative orthologs and paralogs were analyzed among P. trifoliata and seven other Citrinae group members, including Citrus grandis, Citrus reticulata, Citrus sinensis, Citrus clementina, Citrus medica, Citrus ichangensis, and Atalantia buxifolia (Fig. 3A and Supplementary Table 6)42–45. Based on the analysis of gene family clustering, we identified 25,888 gene families, of which 11,860 were shared by all eight species, and 7261 of these shared families were single-copy gene families. The sequences of these single-copy orthologous genes were retrieved from the eight Citrinae species, and alignments were performed based on these sequences. We combined all the alignments to produce an alignment matrix for the construction of a phylogenetic tree (Fig. 3B). P. trifoliata is located between Atalantia and the cultivated citrus species, indicating its closer relationship to the cultivated citrus species than to Atalantia. These results also support the better grafting and sexual compatibility of P. trifoliata with cultivated citrus species than with Atalantia46,47.
Fig. 3. Protein orthology comparison among genomes of eight Citrinae species.
A Clusters of orthologous and paralogous gene families in the eight Citrinae species identified by OrthoMCL. B Phylogenetic status of P. trifoliata in the Citrinae group. A maximum-likelihood phylogenetic tree was constructed by using all single-copy orthologs shared by the eight Citrinae species. The numbers near tree nodes are bootstrap values. Divergence times are indicated by bars below the phylogenetic tree scaled as million years ago (Ma). C Overlap of gene families in P. trifoliata and three widely cultivated citrus species
To gain clues regarding the genes specific to P. trifoliata, we compared the gene families among P. trifoliata and three widely cultivated species. As shown in Fig. 3C, 240 gene families were specific to P. trifoliata, and 13,234 gene families were shared by P. trifoliata, C. grandis, C. sinensis, and C. clementina. GO studies based on the 240 P. trifoliata-specific gene families showed enrichment of genes encoding “multicellular organismal homeostasis”, “response to temperature stimulus”, and “tachykinin receptor signaling pathway”, suggesting that some of these genes may be related to the special cold resistance of P. trifoliata (Supplementary Table 7).
Gene expression and DNA methylation variation between roots and shoots of P. trifoliata
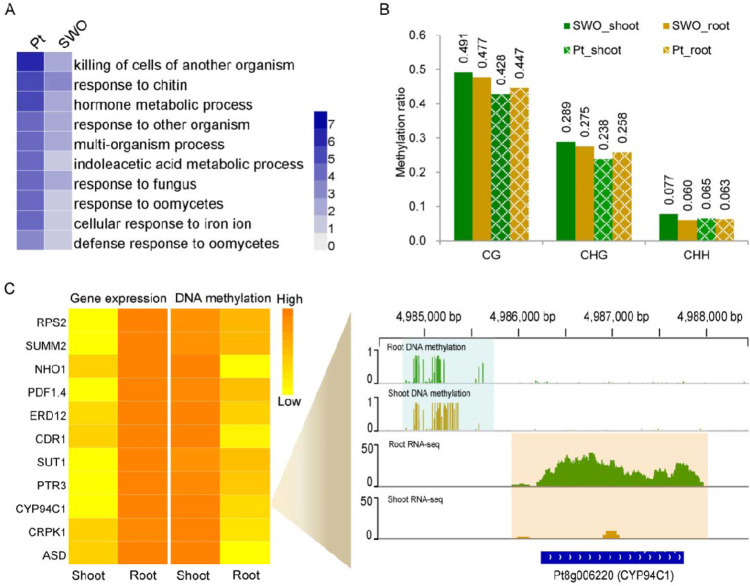
To dissect the transcriptomic characteristics of P. trifoliata roots, we collected roots (representative of underground tissue) and shoots (representative of aboveground tissue) from 2-month-old seedlings of P. trifoliata and sweet orange to isolate RNA for transcriptome sequencing. The shoot and root transcriptomes were compared within each species, namely, P. trifoliata and sweet orange, separately. In sweet orange, the number of genes highly expressed in shoots (4057) was larger than that in roots (3581). In P. trifoliata, more genes were highly expressed in roots (3372) than in shoots (3290), suggesting that transcriptomic events are more frequent in the roots of P. trifoliata than in those of sweet orange. GO analysis indicated that the genes highly expressed in the roots of P. trifoliata and sweet orange were both significantly enriched (P-value < 0.05, FDR < 0.05) in secondary metabolic process, phenylpropanoid metabolic process, and regulation of root development (Supplementary Tables 8 and 9). Notably, we found that the genes highly expressed in P. trifoliata roots were specifically enriched (P-value < 0.05, FDR < 0.05) in 34 GO categories, including killing of cells of another organism, response to chitin, response to fungus, and defense response to oomycetes (Fig. 3A and Supplementary Table 8). These enriched genes may contribute to the strong environmental adaptability of P. trifoliata as a rootstock.
In citrus, DNA methylation is dynamic and shows tissue specificity48. To investigate the DNA methylation changes between roots and shoots, whole-genome bisulfite sequencing was performed on the same set of materials used for transcriptome analysis. In total, 282,798,908, 291,934,320, 260,251,696, and 242,702,062 raw reads were generated for the roots of P. trifoliata (Pt_root), shoots of P. trifoliata (Pt_shoot), roots of sweet orange (SWO_root), and shoots of sweet orange (SWO_shoot), respectively (Supplementary Fig. 2 and Supplementary Table 10). The genome sequences of P. trifoliata and sweet orange were used as the reference for data analysis separately. Approximately 80% of cytosines were covered by at least one uniquely mapped read. Overall, the DNA methylation level of sweet orange was higher than that of P. trifoliata in the CG and CHG contexts (Fig. 4B). In P. trifoliata, the CG and CHG methylation levels in roots were slightly higher than those in shoots, while the opposite situation was observed in sweet orange (Fig. 4B). On each chromosome, methylcytosine densities in all contexts were not evenly spread, and DNA methylation was enriched predominantly in the pericentromeric regions (Supplementary Fig. 3). A chromosome-scale view of the DNA methylation levels and the densities of genes and transposable elements (TEs) showed that DNA methylation sites were most likely enriched in the regions containing numerous TEs but few genes. Similarly, the relative abundance of sRNAs was plotted along each chromosome, which showed a higher density of sRNAs in TE-rich regions, suggesting a role for these sRNAs in TE methylation (Supplementary Fig. 3).
Fig. 4. Differentially expressed genes and DNA methylation variation between roots and shoots.
A Selected GO terms specifically enriched for the highly expressed genes in P. trifoliata roots. A darker blue color indicates a more significant P-value. All GO terms have a P-value < 0.05. B DNA methylation levels in the shoots and roots of P. trifoliata and sweet orange. C Heatmaps showing the expression levels and promoter methylation levels of selected resistance-related genes. The genome browser snapshot in the panel on the right shows the methylation level and expression level of CYP94C1 in each developmental stage. The differentially methylated regions and expressed transcripts are shadowed with green and yellow boxes, respectively
To further evaluate the genes affected by DNA methylation, we identified differentially methylated regions (DMRs) between Pt_root and Pt_shoot. In total, we identified 13,331 high-confidence (FDR < 0.01) DMRs in all contexts. Regarding the DMR-associated genes, we identified 1705 genes that contained DMRs in their promoter region—1097 genes showed increased amounts of DNA methylation in roots, whereas the remaining 608 genes showed increased amounts of DNA methylation in shoots. Among the DMR-associated genes, we identified some genes with important roles in the defense response, such as CDR1, RPS2, and PDF1.4. The expression levels of these genes were upregulated in the roots of P. trifoliata, which may be associated with DNA hypomethylation occurring in their promoter regions (Fig. 4C).
P. trifoliata as a rootstock induced DNA demethylation in the scion
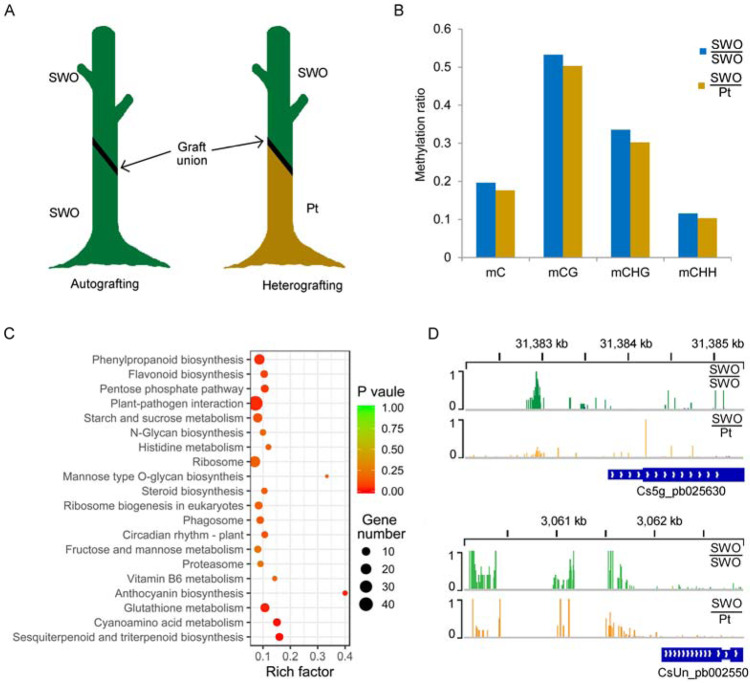
Recent studies have suggested that modifications of DNA methylation may be an important cause of grafting-induced variations18,19,21. Considering the special DNA methylation pattern observed in the root of P. trifoliata as mentioned above, we conducted whole-genome bisulfite sequencing to explore DNA methylation variation between leaves of sweet orange autografted on sweet orange (SWO/SWO) and heterografted on P. trifoliata (SWO/Pt) (Fig. 5A). In total, an average of 158,550,092 reads were generated for each replicate, yielding ~23.8 Gb of data representing >60× of the sweet orange genome (Supplementary Fig. 4 and Supplementary Table 11). Whole-genome methylation level comparison revealed that the levels of CG and CHG methylation both decreased by ~3% at the whole-genome level in the heterografting combination relative to the autografted plant (Fig. 5B), suggesting that demethylation was caused by heterografting. We examined the expression of all the DNA methylase genes (MET, CMT2, CMT3, DRM, and DDM) and demethylase genes (ROS1, DME, and DML3) by q-PCR. The expression of the DNA methylase genes showed no significant difference between SWO/SWO and SWO/Pt, except the expression of CMT2. The three DNA demethylase genes (ROS1, DME, and DML3) were upregulated when P. trifoliata was used as rootstock, which is consistent with hypomethylation being detected when P. trifoliata was used as rootstock (Supplementary Fig. 5). We investigated DNA methylation patterns throughout the gene and TE regions in scions of SWO/SWO and SWO/Pt and found that both grafting combinations showed similar patterns of CG, CHG, and CHH methylation in gene regions (Supplementary Fig. 6). In the TE and flanking regions, the methylation levels of all three contexts in the scion of SWO/Pt were ~2% lower than those in the scion of SWO/SWO (Supplementary Fig. 6), possibly accounting for the major DNA methylation difference caused by heterografting.
Fig. 5. Poncirus as a rootstock induced DNA demethylation in the scion.
A Schematic diagram showing autografting (SWO/SWO) and heterografting (SWO/Pt). SWO, sweet orange; Pt Poncirus trifoliata. The black lines represent the graft junction. B Genome-wide methylation levels in scions of two grafting combinations. C KEGG pathway-enrichment analysis of differentially methylated genes (DMGs) between scions of heterografted and autografted plants. D Genome browser snapshot showing DNA methylation levels of two disease resistance-related genes (Cs5g_pb025630 and CsUn_pb002550) in scions of two grafting combinations
To further investigate changes in DNA methylation, DMRs were identified between SWO/SWO and SWO/Pt. In total, we identified 9027 DMRs in SWO/Pt compared to SWO/SWO, among which 4464 were hypermethylated and 4563 were hypomethylated. To assess how DNA methylation contributes to rootstock–scion interactions, 1537 genes that contained DMRs in their promoter region were identified (Supplementary Table 12). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of these genes revealed that many distinctive biological pathways were affected, such as phenylpropanoid biosynthesis, flavonoid biosynthesis, pentose phosphate pathway, and plant–pathogen interaction (Fig. 5C and Supplementary Table 13). Notably, we found 47 genes with DMRs in their promoter region that were related to plant–pathogen interaction, such as the disease resistance genes RPP8 and RPS2 and the transcription factors MYB108 and MYB44 (Fig. 5D and Supplementary Table 13). Additionally, some genes involved in these biological pathways were upregulated in SWO/Pt (Supplementary Table 14). These results indicated that heterografting-induced DNA methylation variation may be responsible for the strong adaptability of citrus cultivars when using P. trifoliata as rootstock.
Heterografting reduced root-to-shoot mobile sRNAs
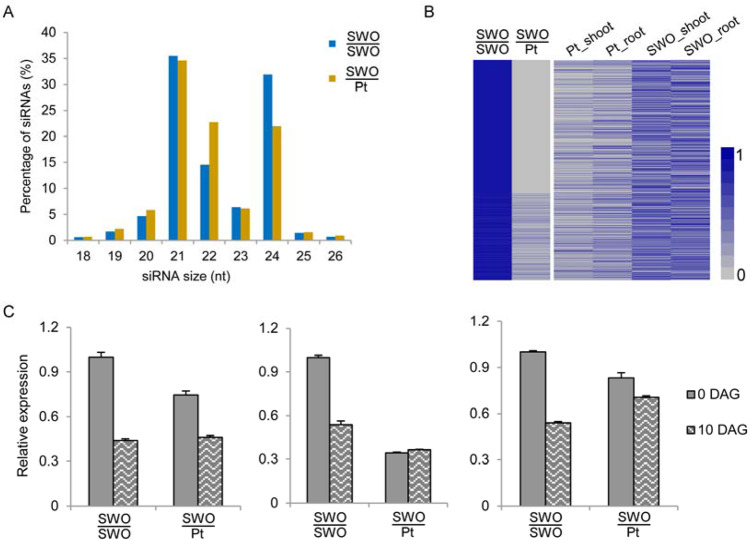
To test the link between changes in DNA methylation and small RNAs, we generated small RNA-seq data from SWO/SWO and SWO/Pt leaves (Supplementary Table 15). The data showed that the percentage of 24-nt sRNAs was much lower in SWO/Pt than in SWO/SWO, which was consistent with the trend of DNA demethylation caused by heterografting (Fig. 6A, Supplementary Fig. 7A). Furthermore, we found that the methylation level of the regions overlapping with sRNAs was significantly higher than that of random genomic regions, suggesting that heterografting-induced DNA demethylation was significantly associated with sRNAs (P < 0.01) (Supplementary Fig. 7B).
Fig. 6. Heterografting reduced root-to-shoot mobile sRNAs.
A Percentages of unique sRNAs in scions of two grafting combinations. B Heatmaps showing the expression pattern of DNA demethylation-associated 24-nt sRNAs in scions of two grafting combinations of roots and shoots of sweet orange and P. trifoliata. C qPCR detection of the expression levels of three selected 24-nt sRNAs in scions of the two grafting combinations before and 10 days after girdling (DAG). The three sRNAs from left to right are AAATGGATTAGGTATCCCATACCT, ATACCAACATTCTTTTCCAAGATT, and AACTATTACGCCTATTGAGCGATC. The values in each column are the means of three biological replicates. Error bars indicate the SD
The decreased 24-nt sRNAs in the scion of heterografted plants could be caused by repression of the sRNA biogenesis pathway or asymmetric movement of sRNAs between the rootstock and scion. To investigate these two possibilities, we first examined the expression levels of the key genes involved in 24-nt sRNA biogenesis in the scions of SWO/SWO and SWO/Pt. For the canonical RNA-directed DNA methylation (RdDM) pathway in Arabidopsis, five key genes are involved in 24-nt sRNA biogenesis, namely, SHH1, NRPD1, CLSY1, RDR2, and DCL349. In the sweet orange genome, we identified the orthologs of these genes using BLASTP and found that none of the five genes were differentially expressed between the scions of SWO/SWO and SWO/Pt (Supplementary Fig. 8), indicating that the decreased 24-nt sRNAs in the scion of heterografted plants cannot be attributed to sRNA biogenesis. Then, we analyzed the expression levels (miRNA read counts were normalized to reads per million, RPM) of all the detected 24-nt sRNAs and the DNA methylation levels of their target loci. Among the 24-nt sRNAs with reduced DNA methylation in their target loci in the scion of SWO/Pt, 548 were SWO/SWO specific (RPM > 0 in SWO/SWO, not detected in SWO/Pt) and 356 were highly expressed in SWO/SWO (≥1.5-fold) (Table 2 and Supplementary Table 16). Further investigation of these 903 24-nt sRNAs in the roots of sweet orange and P. trifoliata revealed 534 24-nt sRNAs that were highly (≥1.5-fold) expressed in sweet orange roots (Fig. 6B). Therefore, we speculate that the higher abundance of the 24-nt sRNAs in sweet orange roots may have led to more 24-nt sRNAs moving to the scion when sweet orange was used as a rootstock for grafting. To verify the root-to-shoot movement of the 24-nt sRNAs, girdling treatment above the grafting union was applied for the two grafting combinations. qRT-PCR analyses of three selected 24-nt sRNAs revealed that in the scion of SWO/Pt, none of the 24-nt sRNAs showed significant changes in transcript levels before and after girdling (Fig. 6C). In the scion of SWO/SWO, the expression levels of the 24-nt sRNAs were obviously reduced after girdling (Fig. 6C), indicating that the phloem-mobile 24-nt sRNAs from roots to shoots were blocked by girdling.
Table 2.
Expression of decreased 24-nt sRNAs in the scion of the heterografted plant and methylation level of the matched DMRs
| siRNA sequence | Expression level | Methylation level | ||
|---|---|---|---|---|
| SWO/SWO | SWO/Pt | SWO/SWO | SWO/Pt | |
| TGAGATAGTGGGGGAGCCTGGGTC | 1.10 | 0.08 | 0.33 | 0.13 |
| TACTCCGTGACTTCTTAAGTCGGT | 1.02 | 0 | 0.28 | 0.13 |
| GATCTACAAGAGAAAAAGAGGAGT | 0.90 | 0.05 | 0.36 | 0.15 |
| TAGCAATAAGCTGGTTGGGGGGTG | 0.85 | 0.10 | 0.27 | 0.12 |
| TTGGTTCCGGTGGAACATCCACAC | 0.81 | 0 | 0.48 | 0.23 |
| GGTCACATCATTTCGAGCAAAGGT | 0.76 | 0 | 0.33 | 0.15 |
| ACGATCCTGTTGGGCTAAAATTCT | 0.75 | 0 | 0.38 | 0.14 |
| TTTGTGGCTGTATCATTACTCTTT | 0.75 | 0.05 | 0.27 | 0.04 |
| TTGACTTTTGAAGTTTGACCAGCC | 0.70 | 0.07 | 0.16 | 0.06 |
| CTTTAAAATGCAAAGACCCAGGCT | 0.69 | 0 | 0.33 | 0.13 |
| TGCCACGTCACCATCAACAGTACA | 0.59 | 0.04 | 0.33 | 0.16 |
| AGATACCCAAGTACGTCATTTCAA | 0.57 | 0.04 | 0.46 | 0.22 |
| AACACACGCTTTTCTCCCCAAATT | 0.54 | 0.04 | 0.42 | 0.18 |
| TTTTCAAGGTACGATTTCTAGAAT | 0.54 | 0.05 | 0.61 | 0.29 |
| GAGACGACTAACTCTTCTCCAGTC | 0.52 | 0 | 0.41 | 0.14 |
| GGCTTCATACCCGGGCCGGGTAAT | 0.51 | 0 | 0.32 | 0.15 |
| AATGAGCTCCACGCCTGCAAAACA | 0.51 | 0 | 0.26 | 0.08 |
| GCTTGATGACATGACCAGTGTTCT | 0.49 | 0 | 0.25 | 0.09 |
| TCTAATGATCATTTTCAACATACT | 0.48 | 0 | 0.81 | 0.52 |
| CTGCTAATGAGCTACTTGATATGT | 0.46 | 0.07 | 0.29 | 0.13 |
Discussion
P. trifoliata is a Chinese deciduous species with trifoliolate leaves that has been used as a rootstock for citrus cultivars for a long time. It is resistant to Phytophthora, nematodes, and tristeza virus and can withstand a cold temperature of −26 °C50,51. Ancient people observed that the graft union of mandarin and P. trifoliata grown in South China was mandarin, while in North China, the graft union grown was trifoliate orange. This is now easy to explain, as only the rootstock trifoliate orange is highly resistant to cold temperatures and can survive in the winter in North China, while the scion mandarin with good fruit quality is vulnerable to cold temperatures. P. trifoliata is also sexually compatible with the Citrus genus, and the most widely grown sexual hybrid is Troyer citrange, which is also a very important citrus rootstock in many countries23. In this study, we sequenced and assembled a high-quality genome of a landrace of P. trifoliata from Shanxi Province based on genetic evaluation of a set of 169 accessions. Thus, this genotype represents an original type of P. trifoliata relative to the narrow genetic background in the United States; moreover, this genome is more complete than the recently published genome52 (Supplementary Table 6). Both genomes provide valuable genomic resource for citrus rootstock genetics and breeding improvement. This genome together with the genomes of cultivated citrus plants (as scions) should facilitate a deep understanding of scion–rootstock interactions at the genomic level. In addition, the genomic information may also be valuable for further investigation of the basis of medical uses and some other unique biological traits of trifoliate orange, such as cold hardiness, trifoliate leaf character, and resistance to citrus tristeza virus.
Grafting has been widely used to improve the performance of horticultural plants for thousands of years. Although increasing physiological evidence indicates the existence of rootstock–scion interactions in plants53, molecular evidence at the genetic and epigenetic levels revealing the influence of rootstock–scion interactions is scarce. Recently, the discovery of mobile genetic elements such as DNA, RNA, and proteins has gradually revealed the molecular mechanisms underlying several agronomic traits affected by rootstock–scion interactions16. For example, microRNA399 was identified as a long-distance signal for the regulation of plant phosphate homeostasis in rapeseed and pumpkin54. Epigenetic modification may also play important roles in creating heritable phenotypic variation by grafting. In this study, we found that the DNA methylation level in the scion grafted on P. trifoliata decreased by ~3% (Fig. 5). A previous study reported that DNA methylation levels in grafted cucumber and melon were significantly increased when pumpkin was used as rootstock, while there was no significant change in grafted watermelon19. Grafting between plant species of Solanaceae caused extensive DNA methylation variation, and some variation could be stably passed on to offspring18. Thus, we can conclude that grafting does cause variations in DNA methylation, but the degree and trend of DNA methylation variation may vary based on species and graft combinations. The effect of grafting on DNA methylation may be a regulatory mechanism for intercell interactions between scions and rootstocks21.
Our observation that the DNA demethylation pattern in the scions of heterografted plants is concomitant with reduced abundance of 24-nt sRNAs implies a possible mechanism of graft-induced alteration in DNA methylation. sRNA-mediated graft-transmissible epigenetic modifications have been confirmed in Arabidopsis thaliana by grafting experiments. Molnar et al. demonstrated that the movement of transgene-derived and endogenous sRNAs from shoot to root across graft unions can cause epigenetic changes in rootstock cells55. A subsequent study showed that mobile sRNAs originating in the shoots guided RdDM at thousands of loci in the roots56. Our finding that the sRNAs are associated with the methylation levels of the sRNA-overlapping regions suggests that the DNA demethylation in the scion of heterografted plants may be attributed to the reduction in sRNAs leading to reduced RdDM. As the genotypes of both the rootstock and scion in autografted plants are the same, it is difficult to determine whether the highly expressed sRNAs in the scions of autografted plants are from the rootstock. However, the higher expression of the sRNAs in the roots of sweet orange than in those of Poncirus, combined with the downregulation of the highly expressed sRNAs in the scions of autografted plants after griding treatment, suggested the possibility that the reduction in the rootstock-to-scion movement of sRNAs leads to decreased sRNA abundance in the scions of heterografted plants. Based on the above finding, the DNA demethylation in the scion of SWO/Pt was possibly caused by the reduced graft-transmissible epigenetic modifications mediated by rootstock-to-scion movement of sRNAs.
The high-quality citrus rootstock genome provides an important basis for future studies on rootstock genetic improvement, and our multiomic analysis of P. trifoliata may promote a deeper understanding of graft biology, not only in citrus plants but also in other horticultural crops. Given the important biological roles played by DNA methylation in plants, it is reasonable to suspect that graft-transmissible epigenetic modifications may have functional consequences. In the future, the use of transgenic plants as rootstocks for grafting will further enhance the opportunity to improve practical nontransgenic cultivars in the field.
Methods
Plant materials and sequencing
The P. trifoliata used for genome sequencing was collected from Hanzhong, Shanxi Province, China. For whole-genome bisulfite sequencing, the seeds of sweet orange and P. trifoliata were germinated and cultured in an artificial climate incubator (28 °C, 16 h light and 8 h darkness) for 2 months, and then, the shoots and roots of the seedlings were collected separately. Two biological repeats were performed for each tissue, and 8–10 seedlings were mixed for each biological repeat. The same materials were used to perform RNA and small-RNA sequencing, and three biological repeats were performed.
The grafting experiment was performed at the National Citrus Breeding Center, Huazhong Agricultural University, Wuhan, China. Healthy and uniform annual branches were selected from one adult sweet orange tree as scions, and biennial seed-germinated P. trifoliata and sweet orange seedlings with uniform growth were selected as rootstocks. Two grafting combinations were constructed: one with sweet orange (SWO/SWO) grafted as rootstock and one with P. trifoliata (SWO/Pt) grafted as rootstock. Each grafting combination was used for at least eight plants. All seedlings were put into the same greenhouse with the same water, fertilizer, and pest management conditions. Five months after grafting, gene expression and DNA methylation analyses were carried out on the 5th–8th leaves above the graft union. Every three grafted seedlings were combined as one biological repeat. Two biological repeats were performed for bisulfite sequencing and small-RNA sequencing.
Genome assembly and annotation
Illumina reads were used to estimate the P. trifoliata genome features by GCE software57. The genome size was 335 Mb, and the heterozygosity was 1.02% based on the k-mer depth distribution. Approximately 20-kb SMRT libraries were prepared according to the released protocol for the PacBio sequel platform. This generated a total sequence length of 30.49 Gb. We used Falcon/Falcon_unzip39 to assemble these SMRT sequences. Subsequently, the draft-assembled contigs were polished with Quiver. Finally, Pilon v.1.1.858 was utilized to perform the second round of error correction with Illumina reads. The genetic map with 1934 markers was used for anchoring the assembled contigs41. For TE annotation, a de novo repeat library was first constructed by RepeatModeler v.1.0.1159 (http://www.repeatmasker.org). Then, the library was integrated with the repBase60 plant repeat database. Finally, RepeatMasker v.4.0.761 was used to mask repeat elements.
Ab initio gene predictions, homology searches and RNA-seq analysis were integrated to predict gene models. Ab initio gene prediction and annotation were performed by Augustus v.3.2.262 and GlimmerHMM v.3.0.463. Then, the gene structure was further confirmed based on the published proteins and expressed sequence tags (ESTs) of citrus species by using the AAT package64 and Exonerate v.2.2.065. For RNA-seq analysis, transcriptomes from shoot, leaf, and flower material were aligned to the genome by TopHat2 v.2.1.166. Then, these sequences were subjected to genome-guided and de novo assembly by Trinity v.2.3.267. The assemblies were further refined by using PASA68. All the predicted gene structures above were integrated by EVM69. Finally, the gene models were generated after annotating the UTR and alternative splicing isoforms using PASA pipeline v.2.3.368. For functional annotation of protein-coding genes, nucleotide sequences of high-confidence genes were searched against the SwissProt and TrEMBL databases. The motifs and domains within gene models were identified by InterProScan v.5.32.7170.
Phylogenetic tree construction
An all-vs.-all BLASTP search was first performed using the corresponding protein sequences of P. trifoliata and seven other citrus species. Then, gene family clustering was conducted by OrthoMCL v.2.0.971. The single-copy orthologous genes were retrieved from the eight Citrinae species and aligned by Muscle v.3.8.42572. The poorly aligned sequences were eliminated using Gblocks73 with default parameters. RAxML v.8.2.1274 was finally used to construct the maximum-likelihood phylogenetic tree with 1000 bootstraps.
Whole-genome bisulfite analysis
High-quality whole-genome bisulfite reads were mapped to the assembled P. trifoliata genome using Bismark v.0.18.175. The unique mapped reads were used to identify differentially methylated cytosines and regions using the methylKit76 package. Bases that had coverage below 4× and had more than the 99.9th percentile of coverage were discarded. For each treatment, methylation call files corresponding to the three methylation sequence contexts were generated. The methylation levels of annotated features, including genes, promoter regions (2 kb upstream of transcription start site), and TEs, were calculated by a customized Perl script. The DMRs were identified by MethylKit package76. Hyper-DMR and hypo-DMR in the CG, CHG, and CHH contexts were identified using a 1000-bp window. Regions with a minimum methylation difference of 25% or for which the fold change in the methylation level was ≤0.5 and ≥2 were regarded as DMRs, and regions containing <4 methylated cytosines were removed. The DMRs were allocated to gene bodies and promoter regions. KEGG (http://www.genome.jp/kegg/) was used to understand the pathway enrichment of DMR-related genes77. In addition, the conversion rate of WGBS was assessed by using lambda phage DNA samples, and the conversion rate ranged from 99.54% to 99.61%.
RNA extraction and transcriptome analysis
Total RNA from different tissues was extracted using an RNA extraction kit (RNAiso Plus, TaKaRa) following the manufacturer’s instructions. RNA-seq libraries were constructed and sequenced on the Illumina Genome Analyzer platform. The clean RNA-seq reads were mapped to the reference genome by Hisat2 v.2.0.478. The correlation coefficients of biological replicates were calculated by the cor function in R. The Ballgown package79 was utilized to estimate gene expression levels. The Cuffdiff v.2.2.180 procedure was followed to identify differentially expressed genes (FDR < 0.05). DEGs were assigned to GO terms, and GO enrichment was performed by the agriGO database81.
Small RNA-seq and data analysis
sRNA sequencing raw reads were filtered first by removing the low-quality reads, filtering the contaminants, and trimming the adaptor sequences. The filtered reads were then mapped to the reference genome by Bowtie2 v.2.1.082 with no mismatch. For the mapped reads, only those of 20–24 nt with counts ≥2 were retained. Then, the retained reads were further filtered for tRNAs, rRNAs, snRNAs, and snoRNAs based on their alignment to the Rfam database83. The remaining small RNAs were further processed to assess their positions in the chromosome and their overlap with methylated regions. For comparison of expression levels, the sRNA read counts were normalized to RPM.
Girdling experiments
One year after the graft union completely healed, both grafting combinations (SWO/SWO and SWO/Pt) were used for the girdling experiments. The removed bark was located 3–5 cm above the graft union, and a 5-mm-wide section of bark was removed down to the xylem. Leaf samples were collected from the scion (above the girdling position) before and 10 days after girdling. Leaves from three grafted plants were combined as one biological repeat, and three biological repeats were prepared for each graft combination.
Quantitative PCR analysis
Total RNA from all tissues was extracted using TRIzol reagent (Takara). cDNA was synthesized using 1 μg of total RNA and HiScript II QRT SuperMix for qPCR (Vazyme, R223-01). qRT-PCR was performed on an LC480 instrument (Roche) using SYBR Green PCR Mastermix according to the manufacturer’s instructions (Kapa, RR420). The cycling conditions included incubation for 5 min at 95 °C followed by 40 cycles of amplification (95 °C for 5 s and 60 °C for 35 s). Using the citrus β-actin gene as the internal reference gene, relative gene expression values were calculated using the 2−ΔΔCt method84. miRNA expression was detected by stem-loop qRT-PCR85. Three independent biological replicates and at least three technical replicates were performed. All the primers used in qRT-PCR are listed in Supplementary Table 17.
Data access
The assembled genome sequences of P. trifoliata have been deposited at DDBJ/ENA/GenBank under accession number VKKW00000000 and can also be downloaded at our website http://citrus.hzau.edu.cn/orange/download/index.php. Whole-genome sequencing data, transcriptome data, whole-genome bisulfite sequencing data, and sRNA data have also been deposited in the NCBI database, and the accession numbers of each sample are recorded in Supplementary Tables 10, 11,14, 15, 18, and 19.
Supplementary information
Acknowledgements
This work was funded by the National Key Research and Development Program of China (2018YFD1000101), the National Natural Science Foundation of China (31925034 and 31872052) and the National Postdoctoral Program for Innovative Talents (BX20200146). We are grateful to Mr. Dekuan Ding from the Citrus experimental station in Shannan of the MATS program of the Ministry of Agriculture of China (CARS-27) for providing the P. trifoliata samples.
Author contributions
Q.X. conceived and designed the project and the strategy. Y.H. assembled the P. trifoliata genome, performed bioinformatics analyses and managed the database. Y.X. carried out the grafting and girdling experiment and prepared all the plant samples. X.J., H.Y., Y.H., M.J.R., and C.T. participated in the sample preparation and grafting experiments. H.J. and G.H. assisted in the data analyses. Y.X. wrote the manuscript with contributions from X.D., Y.H., and Q.X. All authors read and approved the final manuscript.
Conflict of interest
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yue Huang, Yuantao Xu
Supplementary information
The online version contains supplementary material available at 10.1038/s41438-021-00505-2.
References
- 1.Melnyk CW, Meyerowitz EM. Plant grafting. Curr. Biol. 2015;25:R183–R188. doi: 10.1016/j.cub.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 2.He W, et al. Dissection of the mechanism for compatible and incompatible graft combinations of Citrus grandis (L.) Osbeck (‘Hongmian Miyou’) Int. J. Mol. Sci. 2018;19:505. doi: 10.3390/ijms19020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tietel Z, et al. Impact of scion/rootstock reciprocal effects on metabolomics of fruit juice and phloem sap in grafted Citrus reticulata. PLoS ONE. 2020;15:e0227192. doi: 10.1371/journal.pone.0227192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, et al. Differential expression analysis of genes related to graft union healing in Pyrus ussuriensis Maxim by cDNA-AFLP. Sci. Hortic. 2017;225:700–706. doi: 10.1016/j.scienta.2017.07.028. [DOI] [Google Scholar]
- 5.Hudina M, Orazem P, Jakopic J, Stampar F. The phenolic content and its involvement in the graft incompatibility process of various pear rootstocks (Pyrus communis L.) J. Plant Physiol. 2014;171:76–84. doi: 10.1016/j.jplph.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Assuncao M, Santos C, Brazao J, Eiras-Dias JE, Fevereiro P. Understanding the molecular mechanisms underlying graft success in grapevine. BMC Plant Biol. 2019;19:396. doi: 10.1186/s12870-019-1967-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gakpetor PM, Mohammed H, Moreti D, Nassar NMA. Periclinal chimera technique: new plant breeding approach. Genet. Mol. Res. 2017;16:gmr16039790. doi: 10.4238/gmr16039790. [DOI] [PubMed] [Google Scholar]
- 8.Robert ML, Juarez-Gomez J, Chaires-Pacheco M, Pena-Ramirez YJ. Successive grafting confers juvenility traits to adult Spanish red cedar (Cedrela odorata Linnaeus): a tool for the rescue of selected materials. N. For. 2019;51:335–347. [Google Scholar]
- 9.Mudge K, Janick J, Scofield S, Goldschmidt EE. A history of grafting. Hortic. Rev. 2009;35:437–493. doi: 10.1002/9780470593776.ch9. [DOI] [Google Scholar]
- 10.Zhang L, Marguerit E, Rossdeutsch L, Ollat N, Gambetta GA. The influence of grapevine rootstocks on scion growth and drought resistance. Theor. Exp. Plant Physiol. 2016;28:143–157. doi: 10.1007/s40626-016-0070-x. [DOI] [Google Scholar]
- 11.Huang Y, Tang R, Cao Q, Bie Z. Improving the fruit yield and quality of cucumber by grafting onto the salt tolerant rootstock under NaCl stress. Sci. Hortic. 2009;122:26–31. doi: 10.1016/j.scienta.2009.04.004. [DOI] [Google Scholar]
- 12.Rouphael Y, Schwarz D, Krumbein A, Colla G. Impact of grafting on product quality of fruit vegetables. Sci. Hortic. 2010;127:172–179. doi: 10.1016/j.scienta.2010.09.001. [DOI] [Google Scholar]
- 13.Tramontini S, Vitali M, Centioni L, Schubert A, Lovisolo C. Rootstock control of scion response to water stress in grapevine. Environ. Exp. Bot. 2013;93:20–26. doi: 10.1016/j.envexpbot.2013.04.001. [DOI] [Google Scholar]
- 14.Agut B, Gamir J, Jaques JA, Flors V. Systemic resistance in citrus to Tetranychus urticae induced by conspecifics is transmitted by grafting and mediated by mobile amino acids. J. Exp. Bot. 2016;67:5711–5723. doi: 10.1093/jxb/erw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Rouphael Y, Cardarelli M, Colla G. Vegetable grafting as a tool to improve drought resistance and water use efficiency. Front. Plant Sci. 2017;8:1130. doi: 10.3389/fpls.2017.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Jiang L, Wu R. Plant grafting: how genetic exchange promotes vascular reconnection. N. Phytol. 2017;214:56–65. doi: 10.1111/nph.14383. [DOI] [PubMed] [Google Scholar]
- 17.Uthup TK, Karumamkandathil R, Ravindran M, Saha T. Heterografting induced DNA methylation polymorphisms in Hevea brasiliensis. Planta. 2018;248:579–589. doi: 10.1007/s00425-018-2918-6. [DOI] [PubMed] [Google Scholar]
- 18.Wu R, et al. Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants. PLoS ONE. 2013;8:e61995. doi: 10.1371/journal.pone.0061995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avramidou E, et al. Global DNA methylation changes in Cucurbitaceae inter-species grafting. Crop Breed. Appl. Biot. 2015;15:112–116. doi: 10.1590/1984-70332015v15n2n20. [DOI] [Google Scholar]
- 20.Cao L, et al. Heritability and reversibility of DNA methylation induced by in vitro grafting between Brassica juncea and B. oleracea. Sci. Rep. 2016;6:27233. doi: 10.1038/srep27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu N, et al. Maintenance of grafting-induced epigenetic variations in the asexual progeny of Brassica oleracea and B. juncea chimera. Plant J. 2018;96:22–38. doi: 10.1111/tpj.14058. [DOI] [PubMed] [Google Scholar]
- 22.Goldschmidt EE. Plant grafting: new mechanisms, evolutionary implications. Front. Plant Sci. 2014;5:727. doi: 10.3389/fpls.2014.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castle WS. A career perspective on citrus rootstocks, their development, and commercialization. Hortic. Sci. 2010;45:11–15. [Google Scholar]
- 24.Stover E, Inch S, Richardson ML, Hall DG. Conventional citrus of some scion/ rootstock combinations show field tolerance under high huanglongbing disease pressure. Hortic. Sci. 2016;51:127–132. [Google Scholar]
- 25.Benjamin G, Tietel Z, Porat R. Effects of rootstock/scion combinations on the flavor of citrus fruit. J. Agric. Food Chem. 2013;61:11286–11294. doi: 10.1021/jf402892p. [DOI] [PubMed] [Google Scholar]
- 26.Laino P, et al. Rootstock–scion interaction affecting citrus response to CTV infection: a proteomic view. Physiol. Plant. 2016;156:444–467. doi: 10.1111/ppl.12395. [DOI] [PubMed] [Google Scholar]
- 27.Souza JD, et al. Different adaptation strategies of two citrus scion/rootstock combinations in response to drought stress. PLoS ONE. 2017;12:e0177993. doi: 10.1371/journal.pone.0177993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini MK, Capalash N, Kaur C, Singh SP. Comprehensive metabolic profiling to decipher the influence of rootstocks on fruit juice metabolome of Kinnow (C. nobilis × C. deliciosa) Sci. Horticul. 2019;257:108673. doi: 10.1016/j.scienta.2019.108673. [DOI] [Google Scholar]
- 29.Albrecht U, Tripathi I, Bowman KD. Rootstock influences the metabolic response to Candidatus Liberibacter asiaticus in grafted sweet orange trees. Trees. 2019;34:405–431. doi: 10.1007/s00468-019-01925-3. [DOI] [Google Scholar]
- 30.Castle W. S., Tucker, D. P. H., Krezdom, A. H. & Youtsey, C. O. Rootstocks for Florida citrus. Institute of Food and Agriculture Science/University of Florida. 1-22 (1989).
- 31.Rom R. C. & Carlson, R. F. Rootstocks for Fruit Crops. 384–385 (A Wiley-Interscience Publication, 1987).
- 32.Ghobakhloo M, Dizadji A, Yamchi A. A real-time PCR assay for detection and absolute quantitation of Citrus exocortis viroid in two sensitive and tolerant rootstocks. Crop Prot. 2019;115:27–30. doi: 10.1016/j.cropro.2018.09.006. [DOI] [Google Scholar]
- 33.Al-Jaleel A, Zekri M, Hammam Y. Yield, fruit quality, and tree health of ‘Allen Eureka’ lemon on seven rootstocks in Saudi Arabia. Sci. Hortic. 2005;105:457–465. doi: 10.1016/j.scienta.2005.02.008. [DOI] [Google Scholar]
- 34.Befu M, Kitajima A, Ling YX, Hasegawa K. Classification of ‘Tosa-Buntan’ pummelo (Citrus grandis [L.] Osb.), ‘Washington’ navel orange (C. sinensis [L.] Osb.) and trifoliate orange (Poncirus trifoliata [L.] Raf.) chromosomes using young leaves. J. Jpn. Soc. Hortic. Sci. 2008;69:22–28. doi: 10.2503/jjshs.69.22. [DOI] [Google Scholar]
- 35.Gong XQ, Liu JH. Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tissue Org. 2012;113:137–147. doi: 10.1007/s11240-012-0267-x. [DOI] [Google Scholar]
- 36.Boava LP, et al. Global gene expression of Poncirus trifoliata, Citrus sunki and their hybrids under infection of Phytophthora parasitica. BMC Genom. 2011;12:39. doi: 10.1186/1471-2164-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.George J, Lapointe S. Host plant resistance associated with Poncirus trifoliata influence oviposition, development and adult emergence of Diaphorina citri (Hemiptera: Liviidae) Pest Manag. Sci. 2018;75:279–285. doi: 10.1002/ps.5113. [DOI] [PubMed] [Google Scholar]
- 38.Hu, Y. Studies on the Genetic Diversity of Trifoliate Orange (Poncirus trifoliata [L.] Raf) and Genetics of its Hybrids with Red Tangerine (Citrus reticulata Blanco). MD dissertation, Huazhong Agricultural University (2015).
- 39.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 41.Zheng X, et al. SLAF-based construction of a high-density genetic map and its application in QTL mapping of carotenoids content in citrus fruit. J. Agric. Food Chem. 2019;67:994–1002. doi: 10.1021/acs.jafc.8b05176. [DOI] [PubMed] [Google Scholar]
- 42.Wu GA, et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014;32:656–662. doi: 10.1038/nbt.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017;49:765–772. doi: 10.1038/ng.3839. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, et al. Genome of wild mandarin and domestication history of mandarin. Mol. Plant. 2018;11:1024–1037. doi: 10.1016/j.molp.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 46.Grosser JW, Gmitter FGJ, Chandler JL. Intergeneric somatic hybrid plants from sexually incompatible woody species: Citrus sinensis and Severinia disticha. Theor. Appl. Genet. 1998;75:397–401. doi: 10.1007/BF00276741. [DOI] [Google Scholar]
- 47.Hussain S, Luro F, Costantino G, Ollitrault P, Morillon R. Physiological analysis of salt stress behaviour of citrus species and genera: low chloride accumulation as an indicator of salt tolerance. S. Afr. J. Bot. 2012;81:103–112. doi: 10.1016/j.sajb.2012.06.004. [DOI] [Google Scholar]
- 48.Xu J, Xu H, Xu Q, Deng X. Characterization of dna methylation variations during fruit development and ripening of sweet orange. Plant Mol. Biol. Rep. 2014;33:1–11. doi: 10.1007/s11105-014-0732-2. [DOI] [Google Scholar]
- 49.Matzke MA, Mosher RA, RNA-directed DNA. methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 50.Saunt J. Citrus Varieties of the World. Sinclair International Limited; 1990. [Google Scholar]
- 51.Yelenosky G. Cold hardiness in citrus. Hortic. Rev. 1985;7:201–238. [Google Scholar]
- 52.Peng, Z. et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 10.1111/tpj.14993 (2020). [DOI] [PMC free article] [PubMed]
- 53.Koepke T, Dhingra A. Rootstock scion somatogenetic interactions in perennial composite plants. Plant Cell Rep. 2013;32:1321–1337. doi: 10.1007/s00299-013-1471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53:731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molnar Attila, et al. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 56.Lewsey MG, et al. Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl Acad. Sci. USA. 2016;113:E801–E810. doi: 10.1073/pnas.1515072113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu, B. et al. Estimation of genomic characteristics by analyzing kmer frequency in de novo genome projects. Preprint at http://arxiv.org/abs/1308 (2013).
- 58.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smit, A. F. A. & Hubley, R. RepeatModeler Open-1.0 http://www.repeatmasker.org (2008–2015).
- 60.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0. http://www.repeatmasker.org (1996–2015).
- 62.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 63.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 64.Huang X, Adams MD, Zhou H, Kerlavage AR. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- 65.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinforma. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas BJ, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 74.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akalin A, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian T, et al. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Griffiths-Jones S, et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–D124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 85.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.