Abstract
Dandruff is a recurrent chronic scalp disorder, affecting majority of the population worldwide. Recently a metagenomic study of the Indian scalp microbiome described an imperative role of bacterial commensals in providing essential vitamins and amino acids to the scalp. Coconut oil and its formulations are commonly applied on the scalp in several parts of the world to maintain scalp health. Thus, in this study we examined the effect of topical application of coconut oil on the scalp microbiome (bacterial and fungal) at the taxonomic and functional levels and their correlation with scalp physiological parameters. A 16-weeks-long time-course study was performed including 12-weeks of treatment and 4-weeks of relapse phase on a cohort of 140 (70 healthy and 70 dandruff) Indian women, resulting in ~ 900 metagenomic samples. After the treatment phase, an increase in the abundance of Cutibacterium acnes and Malassezia globosa in dandruff scalp was observed, which were negatively correlated to dandruff parameters. At the functional level, an enrichment of healthy scalp-related bacterial pathways, such as biotin metabolism and decrease in the fungal pathogenesis pathways was observed. The study provides novel insights on the effect of coconut oil in maintaining a healthy scalp and in modulating the scalp microbiome.
Subject terms: Clinical microbiology, Microbiology
Introduction
The human skin including the scalp surface, serves as the body’s first line of defence as well as a host to a myriad of microorganisms, which includes both bacteria and fungi1. The application of high-throughput next-generation sequencing and robust computational analysis has led to an in-depth understanding of the scalp microbiome in the recent years2–4, providing novel clues on the pathophysiology of scalp-related disorders such as dandruff and seborrheic dermatitis in different countries5–9. Dandruff is one of the most common scalp condition affecting majority of the population worldwide10. It is a recurrent, chronic, sub-inflammatory disorder, which is characterized by scaly patches and sometimes itching10,11. Various environmental and intrinsic factors are reported to be linked to the development of dandruff, such as the sebum composition, host susceptibility, scalp microbiome, and a combined interaction between all of these.
Global studies have revealed that the scalp microbiome is characterized by a rather low bacterial diversity, as compared to the other body sites12,13, and is dominated by Cutibacterium acnes (formerly Propionibacterium acnes), Staphylococcus epidermidis and Malassezia spp.4–7. Staphylococcus epidermidis and Cutibacterium acnes are found to be the key bacterial players, where dandruff is commonly marked with an increased abundance of S. epidermidis on the scalp5–7. Among the fungal microbiota, different species of Malassezia, specifically M. restricta and M. globosa have shown varying proportions in populations of different countries5,14–16. Specific strains of M. restricta have been identified in the dandruff patients at the genotypic level17. The ability of Malassezia sp. to metabolize and oxidize sebum-derived lipids (triglycerides, squalene, fatty acids, etc.) is an additional source of potential inflammatory compounds18. M. restricta is also known to induce cytotoxicity to skin cells in vitro, suggesting an active role in the acceleration of dandruff19. A strong association of uncharacterised Malassezia species with dandruff is also observed in a few recent studies4,7. Further, scalp bacteria have been reported to have a stronger association with scaling severity than fungi suggesting that bacteria could have an implication in the clinical symptoms14,15. However, whether the scalp microbiome variation is a cause or a consequence of the unhealthy condition of the scalp remains unclear. S. epidermidis and C. acnes are also a part of the commensal microbiota and reported to have beneficial activities on the skin through immune response modulation and protection against pathogens20–22. In our recent metagenomic study carried out on the scalp microflora of the Indian population, we have observed enrichment of bacterial pathways related to the synthesis and metabolism of amino acids, biotin and B-vitamins in healthy scalp compared to dandruff, revealing a new potential role of bacterial commensals in maintaining the scalp nutrient homeostasis4.
Current anti-dandruff therapies involve topical antifungal agents such as azoles, the clinical efficacy of which is accompanied by a reduction in the proportion of Malassezia spp. on the scalp23–26. However, it is usually observed that at the cessation of the treatment, the relapse restores the initial symptoms24,27. Scalp-related products such as oils, shampoo and other cosmetics are also used worldwide to maintain scalp health and hygiene28,29. Among which, coconut oil is the most widely used product in African and Asian countries, including India, to ameliorate scalp health and hair growth29–31. Not much is known about the mode of action of coconut oil. Firstly, the antifungal activity of lauric acid, the major fatty acid contained in the oil, is suspected to prevent the proliferation of pathogens29,31–34. Secondly, coconut oil is known to have a biophysical action on the skin barrier function, since it helps to decrease the TEWL (trans-epidermal water loss) on long-term application35,36. However, no study has yet systematically examined the effect of topical application of coconut oil on the scalp microbiome.
Due to the recently established role of the microbiome on skin and scalp health, a few studies have investigated the effect of emollients 37,38 and topical medication39,40 on the skin microbiome. Here, we carried out a 16-weeks-long time-course study to understand the impact of coconut oil application on the scalp microbiome (bacterial and fungal) of 140 individuals with healthy and dandruff scalp. A treatment phase was carried out for 12-weeks followed by 4-weeks of the relapse phase in which no application of oil was performed. The scalp clinical parameters were also recorded throughout the study and correlated with the taxonomic and functional profile of the scalp microbiome. The present study aims to provide insights on the potential effect of coconut oil on the scalp fungal and bacterial microbiome.
Results
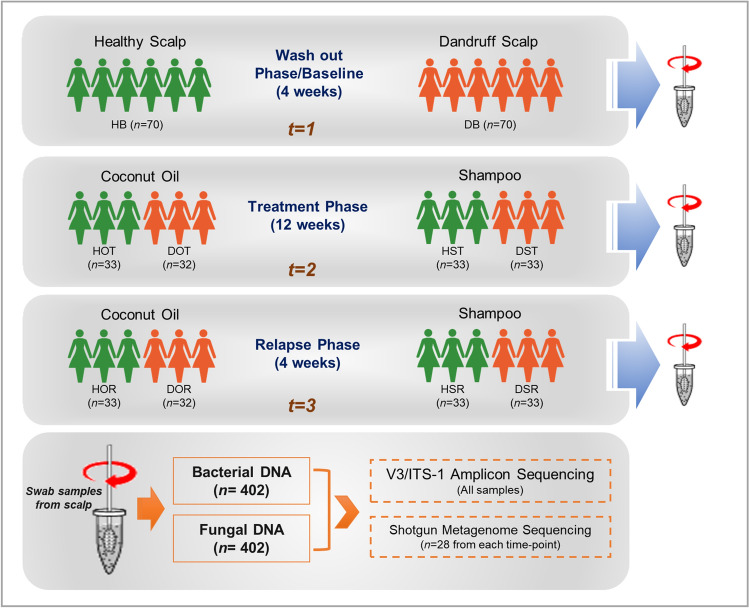
Recently, we reported the functional role of scalp microbiome (bacterial and fungal) in a cohort of 140 Indian individuals consisting of 70 individuals with healthy and 70 with dandruff scalp4. In the current study, we have used the same cohort to perform a time-course study to understand the impact of coconut oil application on the scalp. Amplicon and shotgun metagenomic analysis revealed the major microbial species and their functional pathways in the healthy and dandruff scalp microbiome at the baseline. From the amplicon analysis, the core scalp microbiome was defined as the taxonomic groups with ≥ 1% abundance in at least 80% of the samples, which represented the stable and consistent microbial population in the microbiome associated with the scalp environment. To examine the effect of coconut oil (O) on the scalp microbiome, the changes in the relative abundance of the core microbiome and their associated functional pathways were analysed and compared with a ‘neutral shampoo’ (S) after 12-weeks of treatment phase (T) followed by four weeks of relapse phase (R). The study design is presented in Fig. 1 and details on the nomenclature of groups and samples is provided in supplementary methods.
Figure 1.
Study Design. Swab samples were collected at three phases, baseline (t = 1), treatment phase (t = 2) and relapse phase (t = 3) from healthy and dandruff scalps. Bacterial and fungal DNA was extracted from the collected swab samples, and amplicon (bacterial 16S rRNA V3 and fungal ITS1 region) and shotgun metagenomic sequencing were performed to carry out the taxonomic and functional analysis. In the figure, H = healthy scalp, D = dandruff scalp, O = oil-treatment, S = shampoo-treatment, and B, T and R = the three phases or time-points i.e. Baseline, Treatment and Relapse phase, and n = number of subjects in each group.
Taxonomic variations in the fungal microbiome after the treatment and relapse phases
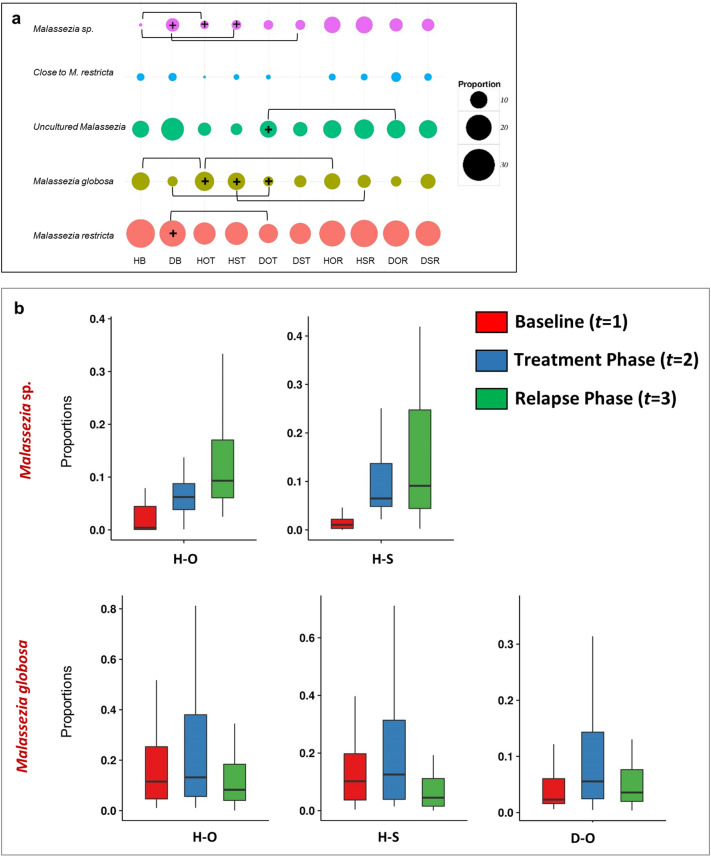
Taxonomic analysis of the baseline microflora showed the alpha-diversity of the fungal population to be significantly lower (p ≤ 0.001) in the healthy scalp (HB: Healthy scalp Baseline) compared to the dandruff scalp (DB: Dandruff scalp Baseline) (Fig. S1a). A high abundance of M. globosa (p ≤ 0.0001) was observed in HB (16.23%) compared to DB (6.41%) (Fig. S1b–d). A strikingly high proportion of OTUs corresponding to uncharacterized Malassezia spp. was observed. One of these OTUs belonged to uncultured species of Malassezia (> 95% identity with Uncultured Malassezia, Genbank ID—KC785585.1) and others belonged to unknown Malassezia species (also at > 95% identity), of which, six OTUs (sequences provided in Supplementary Text) showed an identity of ≥ 85% with M. restricta, and were assigned to a subgroup of Malassezia (i.e. species close to M. restricta). Therefore, the uncharacterized Malassezia sequences formed three subgroups: (1) uncultured Malassezia, (2) Malassezia sp., and (3) species close to M. restricta, as described previously4. The uncultured Malassezia was significantly abundant (p ≤ 0.0001) in DB (25.26%) compared to HB (14.44%). Malassezia sp. and species close to M. restricta were also significantly (p ≤ 0.01) higher in DB compared to HB (Fig. S1d).
The alpha-diversity (Shannon index and number of observed species) of the fungal microbiome increased significantly (p ≤ 0.0001) after oil treatment (HOT) and after-shampoo application (HST) in the healthy scalp compared to the baseline (HB) (Fig. S1a). It also showed a significant increase (p ≤ 0.0001) in oil-treated healthy scalp at the relapse phase (HOR) as compared to baseline (HB) and treatment phase (HOT). However, there was no significant effect seen on the alpha-diversity of the dandruff groups.
Proportions of M. restricta and M. globosa along with the Malassezia subgroups (1, 2 and 3), constituting the fungal core of the scalp, were examined in the three phases. After oil-treatment, the abundance of M. restricta reduced significantly (p = 0.03) in DOT (26.55%) compared to DB (33%) (Fig. 2a and Fig. S2a–e). The abundance of M. globosa increased significantly in both healthy (HOT = 23.23%, p = 0.02) and dandruff scalp (DOT = 9.74%, p ≤ 0.001) compared to the baseline, i.e. HB (16.23%) and DB (6.41%), respectively. It also increased significantly (p ≤ 0.05) in DST (14.32%) compared to DB (6.41%), while there was no significant difference between HST and HB. The abundance of Malassezia sp. increased significantly (p ≤ 0.0001) in HOT (8.86%) and HST (10%) compared to HB (2.37%). It also increased significantly (p ≤ 0.05) in DST (12.28%) compared to DB (10.12%). Additionally, the ratio of M. restricta to M. globosa, which was observed to be significantly higher (p = 0.004) in the dandruff scalp compared to the healthy scalp, decreased significantly (p = 0.001) in DOT compared to DB (Fig. S1e).
Figure 2.
Comparison of fungal population at the three phases. (a) Bubble plots representing the top five fungal species across all the groups. The bubble size indicates mean relative abundance of species within each group. Square brackets indicate the groups between which a significant difference in the species abundance was observed (p ≤ 0.05, Wilcoxon test, + indicates the group with the higher abundance among the two). (b) Core fungal species showing significant variations across the three phases (FDR adjusted p ≤ 0.05, repeated measures ANOVA).
Group-wise analysis using repeated measures ANOVA confirmed the significant variations (FDR adj. p ≤ 0.05) in M. globosa after oil-treatment in both healthy and dandruff scalp, and after shampoo-application in the healthy scalp (Fig. 2b and Table S1a). A similar trend was shown by Malassezia sp. in oil and shampoo-treated healthy scalp. The abundance of M. globosa decreased significantly (p ≤ 0.01) in HOR (13.41%) and HSR (9.44%) compared to HOT (23.23%) and HST (19.32%), respectively (Fig. S3a–e). A significant decrease (p = 0.03) in the abundance of uncultured Malassezia was also observed in DOR (18.37%) compared to DOT (23.60%) (Fig. 2a). These results suggest that the effect of oil-treatment on the changes in the fungal community was not sustained for dandruff group after the relapse phase, in contrast to the healthy group.
Taxonomic variations in the bacterial microbiome after the treatment and relapse phases
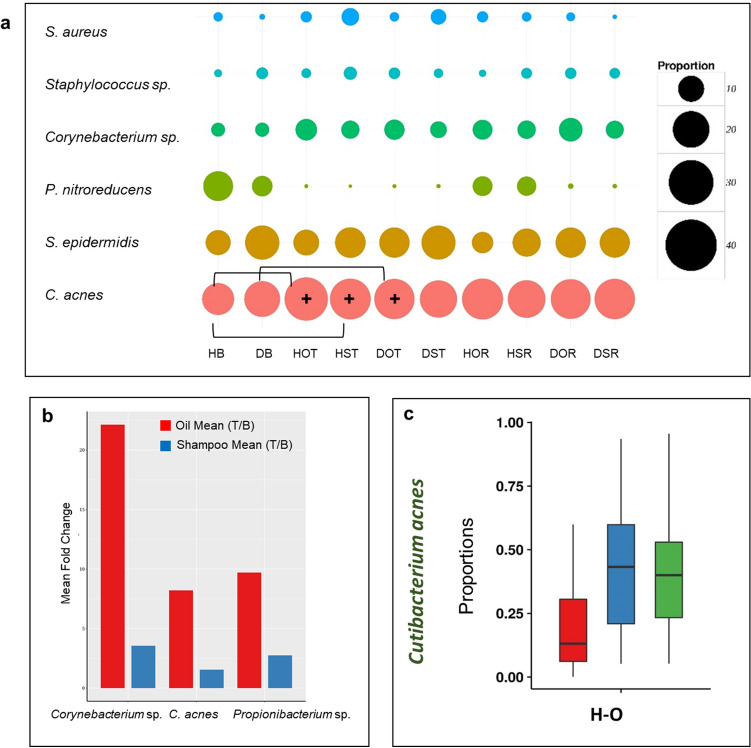
There was no significant difference observed in the alpha-diversity of the bacterial microbiome between healthy (HB) and dandruff (DB) group at the baseline (Fig. S4a). The weighted UniFrac distances does not show a significant difference between healthy (HB) and dandruff (DB) group at the baseline (Fig. S4b). At the species level, the abundance of S. epidermidis was observed to be significantly higher (p = 0.0002) in dandruff (28.11%) than in healthy (14.83%) scalp (Fig. S4c–e). The abundance of C. acnes did not vary significantly (p ≥ 0.05) between dandruff (30.83%) and healthy (24.42%) scalp.
After the treatment phase, there was a significant increase (p ≤ 0.01) in the bacterial diversity (Shannon index) in the healthy scalp after oil-treatment (HOT) and not with shampoo-application (HST) compared to baseline (Fig. S4a). The weighted UniFrac distances showed a significant decrease in within-sample distances in HOT compared to HB, while it was not significant for DOT compared to DB (Fig. S4b). Interestingly, oil-treated groups (HOT and DOT) showed lower UniFrac distances compared to the shampoo-treated group (HST and DST respectively), suggesting a distinct effect of coconut oil on the scalp bacterial communities.
At the taxonomic level, we have focussed on the variations in the abundance of the two main species C. acnes and S. epidermidis that constitute the core microbiome. The proportion of C. acnes increased significantly (p ≤ 0.0001) in HOT (45.29%) compared to HB (24.42%) (Fig. 3a and Fig. S5a–e). Although less than HOT, it also showed a significant increase (p ≤ 0.0001) in HST (40.09%) compared to HB. The ratio of C. acnes to S. epidermidis, which was significantly higher (p = 0.01) in the healthy scalp compared to the dandruff scalp, was significantly higher (p = 0.008) in HOT compared to HB (Fig. S5f). Significant fold-change differences were observed in the abundance of bacterial species between oil-treatment and shampoo-application (Fig. 3b). Interestingly, C. acnes and Propionibacterium sp. showed significant (p ≤ 0.05) fold-changes after oil-treatment compared to shampoo-application. Fold change analysis was applied to both bacterial and fungal population, however the results were found to be significant (p ≤ 0.05) only for the bacterial microbiome. We did not find any fungal species to show significant fold-changes between the groups and hence, the results only from the bacterial microbiome are included here. Among the other species, Corynebacterium sp., showed the maximum level of fold-change (from < 5 to > 20 times, p ≤ 0.05) after oil-treatment. Repeated measures ANOVA (FDR adj. p ≤ 0.05) confirmed an increase in the abundance of C. acnes in healthy subjects after oil-treatment (Fig. 3c, Table S1b). Changes in the proportion of C. acnes and S. epidermidis were retained (p ≥ 0.05) after the relapse phase (Fig. 3c and Fig. S6). These results suggest an apparent beneficial effect of coconut oil on the core bacterial species after 12 weeks treatment, which appears to be retained at the relapse phase.
Figure 3.
Comparison of bacterial population at the three phases. (a) Bubble plots representing the top five bacterial species across all the groups. The bubble size indicates mean relative abundance of species within each group. Square brackets indicate the groups between which a significant difference in the species abundance was observed (p ≤ 0.05, Wilcoxon test, + indicates the group with the higher abundance among the two). (b) Significant fold-change differences (p ≤ 0.05) observed in bacterial species abundance between oil-treatment and shampoo-application. No significant difference was observed in the fungal species between the two treatment groups. (c) Core bacterial species showing significant variations across the three phases (FDR adjusted p ≤ 0.05, repeated measures ANOVA).
Correlation of microbial species with host physiological parameters
Dandruff scalp is characterized by in increased TEWL indicating an altered barrier function, and a few studies have described the variation in the host-associated physiological parameters in the healthy and dandruff scalp8,41. Therefore, a systematic investigation of the host physiological parameters was carried out at all the three phases of the study and correlated with the scalp microbiome (Table S2).
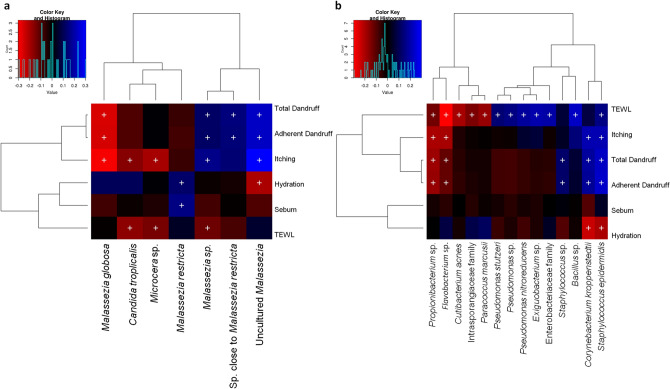
The three Malassezia subgroups showed significant positive correlation (FDR adj. p ≤ 0.05) with dandruff scores and itching, whereas, M. globosa was negatively correlated with these parameters (Fig. 4a). The taxa corresponding to unknown Malassezia sp. showed a significant negative correlation with TEWL. Uncultured Malassezia showed a negative correlation with hydration, and M. restricta showed a positive correlation with sebum level and hydration.
Figure 4.
Spearman’s correlation between microbial taxa and host physiological parameters. (a) Fungal and (b) bacterial taxa showing significant correlations (+ , FDR adjusted p ≤ 0.05) with any of the parameters are plotted as a heatmap.
Among the bacterial population, S. epidermidis displayed a significant positive correlation (FDR adj. p ≤ 0.05) with dandruff scores, TEWL and itching. However, Propionibacterium sp. correlated negatively with these parameters, and C. acnes showed a significant negative correlation with TEWL (Fig. 4b). Although being low in abundance, Flavobacterium sp. and C. kroppenstedtii showed a pattern similar to Propionibacterium sp. and S. epidermidis, respectively.
The host physiological parameters were also compared in the oil and shampoo-treated groups across the three phases using repeated measures ANOVA. The results showed a reduction in TEWL and dandruff scores in both healthy and dandruff groups after the coconut oil treatment phase (Table S1c). It is to be noted that the results for coconut oil treatment are more concordant when studied at taxonomic level compared to the host physiological parameters, which are influenced by many other factors beyond the scope of this study.
Functional variations in the scalp microbiome
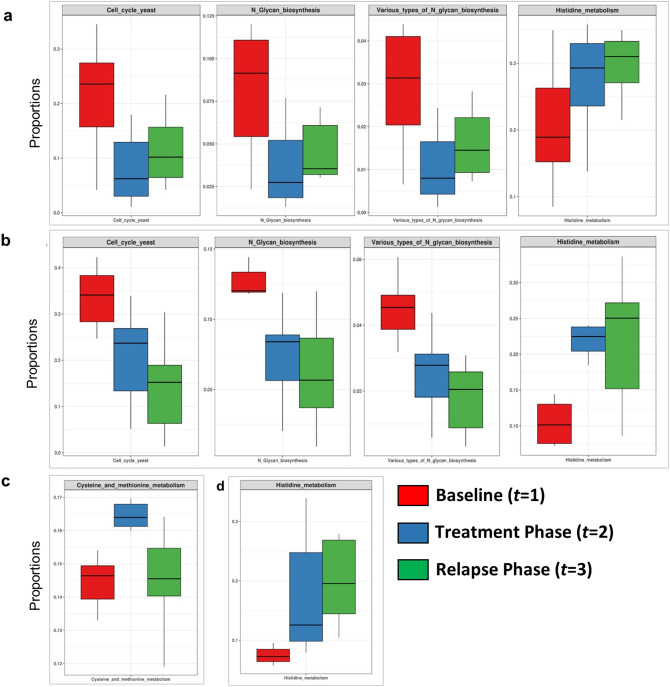
The shotgun metagenomic analysis showed several fungal KEGG pathways to vary significantly (p ≤ 0.05, Wilcoxon test) between the healthy and dandruff scalp at the baseline as reported earlier (Fig. S7a)4. In brief, the amino acid metabolism pathways (histidine, cysteine and methionine metabolism) and lipoic acid metabolism pathway were more abundant in healthy scalp than in dandruff scalp. Further, pathways for N-glycan biosynthesis, which are implicated in cell-host interaction, were enriched in the dandruff scalp42. Several fungal pathways also showed significant variations in their proportions after the treatment phase, which were statistically tested at all three phases using repeated measures ANOVA (Table S1d; Fig. 5). After oil-treatment, histidine metabolism pathway showed a significant increase in both healthy and dandruff groups, however, it was also observed in the shampoo group suggesting that it is not linked to the oil application. In contrast, the proportion of N-glycan biosynthesis and cell cycle pathways decreased in both healthy and dandruff scalps in the oil treated group and not in the shampoo group compared to baseline (Fig. 5). This effect was not sustained at the relapse phase, since there was a significant increase at the relapse phase (t = 3) compared to the treatment phase (t = 2). However, the initial baseline levels were not recovered.
Figure 5.
Functional analysis of fungal microbiome. The proportions of fungal pathways showing significant variations across all the three phases in the following groups (FDR adjusted p ≤ 0.05, repeated measures ANOVA) are shown in the box plots: (a) Healthy oil-treated group, (b) dandruff oil-treated group, (c) healthy shampoo-treated group and (d) dandruff shampoo-treated group.
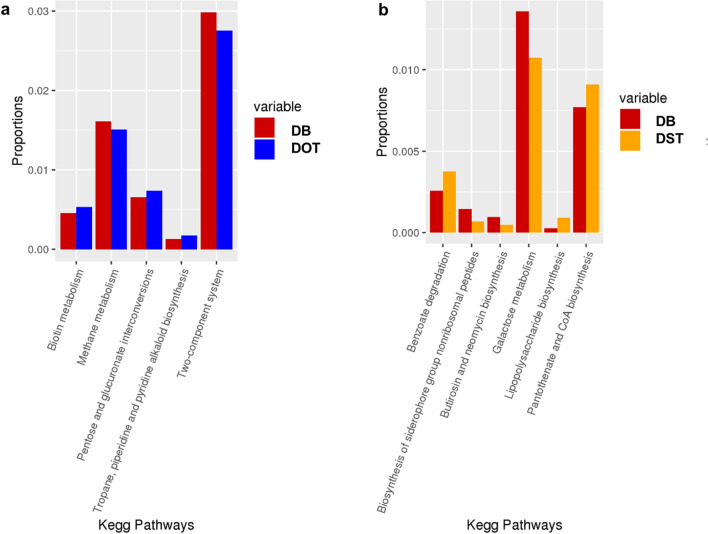
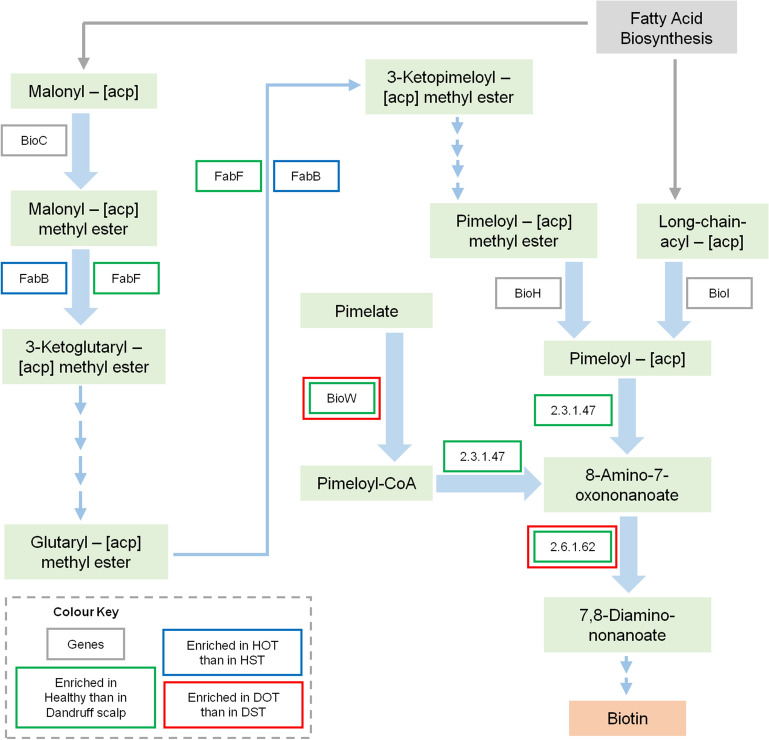
Functional analysis of the bacterial microbiome showed the pathways related to vitamins and cofactors (biotin, porphyrin and chlorophyll, vitamin-B6, nicotinate and nicotinamide metabolism, ubiquinone and other terpenoid-quinone biosynthesis), and amino acids (alanine, aspartate, arginine, glutamate and proline and lysine metabolism and biosynthesis) to be significantly higher in healthy scalp than dandruff scalp at the baseline, as reported earlier (Fig. S7b)4. Results from the previous study have shown these pathways to be positively correlated with C. acnes, suggesting it to be the major contributor for biotin and other B-vitamins on the scalp surface4. Bacterial KEGG pathways showed significant variations after oil-treatment compared to shampoo-application in the dandruff scalp (Fig. 6). The biotin metabolism pathway showed a substantial increase in DOT compared to DB. The variations in pathway abundance were also tested statistically at the three phases using repeated measures ANOVA (Table S1e). The abundance of KOs related to biotin metabolism and biotin transport pathways also increased after oil-treatment compared to shampoo-application in the healthy and dandruff scalp (Fig. 7, Fig. S7c and d).
Figure 6.
Functional analysis of bacterial microbiome. Differentially abundant bacterial KEGG pathways after (a) oil-treatment and (b) shampoo application in the dandruff group (which showed p ≤ 0.05, Wilcoxon test) are shown in the bar graphs. None of the pathways showed significant differences in the healthy group.
Figure 7.
Schematic representation of biotin metabolism pathway describing the significantly enriched KOs (p ≤ 0.05) in the dataset.
Discussion
This study reports the effect of topical application of coconut oil on the scalp microbiome in a cohort of 140 Indian women using high-throughput sequencing and computational analysis. Since, bacteria and fungi are the major scalp microbial species, we examined both using bacterial 16S rRNA and fungal ITS1 amplicon sequencing, respectively, followed by the shotgun metagenomic sequencing, which helped to understand the impact of coconut oil on the scalp microbiome.
Coconut oil is commonly used in several parts of the world to maintain scalp health and to moisturise the skin in addition to repair hair damage, through a direct or indirect mode of action29,31. Supporting studies have demonstrated the inhibitory role of coconut oil and its major component, lauric acid, on the growth and invasion of dermatophytes on the skin more efficiently than other hair oils commonly used in India such as mustard oil, cantharidine oil, amla oil, etc.32–34. It is also known to decrease the TEWL on long-term application on the skin surface, and increases the hydration levels36, which could play an important role in shaping the microbiome of the scalp surface. Thus, it seems reasonable to speculate a potent effect of coconut oil application on the scalp physiology, which in turn modifies the scalp microbiome4,8.
In our previous study of the scalp microbiome in the Indian cohort, we have obtained the baseline microbial composition of both fungal and bacterial communities, which has been reported and reanalysed for the current study4. The determination of baseline microflora helped to elucidate the potential beneficial role of microbiome by comparing their compositional changes after the treatment phase. Cutibacterium acnes and Staphylococcus epidermidis emerged as major bacterial colonizers, and Malassezia restricta and Malassezia globosa as the major fungal colonisers. Similar association of these species with the healthy and dandruff scalp has also been observed in other studies from different population such as, from France, China, Brazil, etc.5–8. A noteworthy observation at the baseline was the high abundance of uncharacterised Malassezia sp. in the dandruff scalp compared to the healthy scalp, and its significant positive correlation with dandruff-associated parameters. A high proportion of uncultured Malassezia (> 37% of the total fungal population) has also been observed recently in the Brazilian population7. The results also showed M. globosa to be negatively correlated with dandruff score and itching, and S. epidermidis and uncharacterised Malassezia sp. to be positively correlated with these parameters. Similar variation in the proportion of M. globosa was also reported in a Chinese cohort using the Illumina sequencing technology8. However, a few other studies have reported contrasting results, which could be a result of lower cohort size, differences in the country of origin of populations and usage of conventional sequencing technologies5,7,9,43.
The above taxonomic results were confirmed in this study by recording the host physiological parameters at all the three time-points. The results also showed the beneficial role of bacterial scalp microbiome in supplying essential vitamins and amino acids to the host as observed in the previous study, where a positive correlation of C. acnes was also observed with the pathways related to the metabolism of biotin and other B-vitamins that are essential for maintaining a healthy scalp4.
The time-course study revealed an apparent beneficial effect of coconut oil application on the fungal scalp microbiome in enhancing the core fungal species associated with a healthy microflora. The abundance of M. globosa, which was correlated with the healthy scalp, increased after the application of coconut oil in the healthy and dandruff scalp. The abundance of M. restricta reduced significantly in the dandruff scalp after oil-treatment as compared to the baseline. Further, a significant decrease in the ratio of M. restricta to M. globosa was observed in dandruff subjects after the application of coconut oil. This ratio was similar to the baseline healthy composition reported in this cohort and other populations, which points towards a probable role of coconut oil in maintaining a healthy fungal scalp microbiome7. The pathways related to pathogenesis, survival and adhesion (N-glycan biosynthesis and cell cycle pathways) showed a significant reduction after the application of coconut oil, suggesting its role in lowering the abundance of fungal pathogenic species. The effect of coconut oil was not sustained for dandruff group, mainly in the fungal community. This observation could be explained by a limited effect of the oil on the fungal microbiome in the dandruff scalp. The fungal taxa found on the dandruff scalp were significantly different compared to healthy scalp and could be more resistant to lauric acid.
The beneficial effects of coconut oil were more prominently evident on the bacterial microbiome compared to the fungal microbiome. It was observed that C. acnes and Propionibacterium sp., which are reportedly associated with a healthy scalp, showed a significant increase after oil-treatment in both the healthy and dandruff scalp4–7. Further, the ratio of C. acnes to S. epidermidis was also notably increased after the oil-treatment. These results suggest that oil helps in enhancing the beneficial bacterial species in both healthy and dandruff scalp. This was further confirmed by the significant increase in the biotin metabolism pathway after oil-treatment, which was majorly contributed by C. acnes and Propionibacterium sp. Biotin and other B-vitamins are crucial precursors for different enzymes required for vital biochemical reactions in the living cells, and are also essential for a healthy skin and scalp surface44. Biotin is also reported to reduce cellular inflammation and improve skin barrier quality, scalp health and hair growth35,36. Since, humans and other mammals cannot synthesize biotin, they obtain it through exogenous sources by import mechanism of the Na-dependent multivitamin transporters (SMVT) and biotin-specific transport components45,46. Thus, it is tempting to speculate the beneficial effect of coconut oil on the scalp microbiome primarily via creating physiological conditions that favour the beneficial microbial community involved in the biosynthesis of nutrients essential for scalp nutrition.
This study demonstrates a positive effect of coconut oil on the scalp microbial communities and their functional potential. We could speculate that microbiome changes are the first step towards the restoration of a healthy scalp that will lead to perceptible benefits to host much later than the time-lines included in this study, and thus provide a long-term benefit compared to short-term benefit as observed in the case of the neutral shampoo. Another significant outcome of the study is that beyond antifungal agents, other approaches could be considered for dandruff treatment targeting both fungal and bacterial microbiome. However, further studies are needed to understand the underlying mechanisms and nutritional significance of coconut oil for the scalp microbiome. A pre-emptive approach aimed at reducing the susceptibility to dandruff by maintaining/reinstating a healthy scalp microbiome, in addition to improving scalp barrier functions, seems a novel opportunity to achieve long-lasting effects.
Materials and methods
Ethics approval and consent to participate
The research protocol was approved by the Independent Ethics Committee for Evaluation of Protocols for Clinical Research, CLINICOM, Bengaluru, India (Study number ARC/COSB/1444) and was conducted in accordance with the principles expressed in the World Medical Association Declaration of Helsinki. A written informed consent was obtained from all subjects prior to any study-related procedures.
Subject recruitment and study design
The study was carried out on a previously reported cohort of 140 Indian women4. Firstly, 184 female volunteers were screened for the study, out of which 70 individuals with healthy and 70 with dandruff scalp (aged between 20–45 years, mean age 34.6) were enrolled with the association of MS Clinical research (Bengaluru, India), who had used coconut oil occasionally in the past one year. The volunteers were non-smokers, free from any cutaneous diseases, did not use anti-hair-loss treatment at least for three months prior to sampling, did not use anti-dandruff shampoos and hair-related products (such as for bleaching, straightening, dyeing, permanent waving, etc.) on scalp and hair for at least three weeks prior to sampling, and did not consume antibiotics or apply systemic antifungals for one month prior to sampling.
Dandruff grading and clinical evaluation of scalp physiological parameters
Dandruff level was scored according to a grading scale as previously described5. Scalp physiological parameters were measured using appropriate devices and protocols. Sebumeter (SM815, Courage & Khazaka, Germany) was used to measure the sebum level of the scalp following manufacturer’s instructions. VapoMeter (Delfin Technologies, Finland) was used to measure the TEWL (trans-epidermal water loss) that measures the integrity of barrier function of the scalp by following the manufacturer’s instructions47. All the measurements were performed in triplicates and values presenting coefficient of variation < 15% were considered as relevant. Corneometer (CK Electronic GmbH, Germany) measurement was performed on the scalp surface to check the hydration levels following the manufacturer’s instructions. The above measurements were performed on a shaved trimmed mini-area before the oil-treatment and shampoo wash at the investigational site by a dedicated operator. Following clinical or physiological parameters were recorded for each subject across the three phases: adherent dandruff score (ADS), total dandruff score (TDS) i.e. adherent and non-adherent, hydration, sebum level, TEWL, erythema and itching (Table S2).
Treatment
The volunteers were asked to use a bland or neutral shampoo (L’Oréal India Pvt. Ltd.) two times a week for a period of four weeks prior to the beginning of the study to standardize the scalp condition (baseline, Day-0 or t = 1). Four subjects from the healthy group and five from the dandruff group did not continue after the baseline sampling, and therefore, the remaining 66 subjects with healthy scalp and 65 with dandruff scalp were followed-up throughout the course of the study. During the treatment phase (Day-84 or t = 2), 32 subjects with dandruff scalp and 33 with healthy scalp received controlled oil-treatment twice a week for 12 weeks at the center (MS Clinical Research, Bengaluru, India). The treatment consisted of 20 min of scalp massage with 10 ml of pure coconut oil (100% refined coconut oil, Cargill, India) followed by two hours leave-on, and then a bland shampoo wash (20 ml). The remaining 33 subjects with dandruff scalp and 33 with healthy scalp were subjected to only the shampoo wash twice a week at the center (MS Clinical research, Bengaluru, India) for 12 weeks with no application of coconut oil. During the relapse phase (Day-112 or t = 3), all the 131 subjects received a topical application of only the bland shampoo on the scalp twice a week for four weeks. Subjects were advised not to use any hair or scalp products other than the study products.
Sampling of the scalp microbiome
The volunteers were asked not to perform scalp wash for two days prior to the sampling procedure. Samples from the scalp (vertex or crown of the head) were obtained at the baseline (t = 1), at the end of 12 weeks of treatment phase (t = 2) and after the relapse phase (t = 3). Sampling was conducted as previously described with minor modifications5. A sterile cotton swab soaked in a solution containing collection solution (0.15 M NaCl and 0.1% Tween 20) was rubbed onto the scalp surface (between the hairs) under a zig-zag pattern, to cover a total surface of 4 cm2 in a non-overlapping manner. At the end of the procedure, the head of each swab was cut from the handle and placed into a tube containing 5 ml of collection buffer. As described previously5, swabs were stored at 4 °C and processed for DNA isolation within 24 h. In addition, a few sterile cotton swabs (as negative controls) were cut from the handle and placed in the collection buffer, and further processed using identical procedure.
High-throughput sequencing DNA extraction
The DNA extraction method was validated, as described above4. Since, the DNA extraction strategy can influence the microbiota community profiling, the DNA extraction method was developed using different bacterial fungal species and ensured that maximum quantity of DNA was recovered by the optimized protocols.
Two identical samples of 2 ml each were generated from one collection tube. The microbial cell suspension from each tube was pelleted by centrifugation at 10,000 g for 30 min, at 4 °C. For bacterial DNA extraction, the cells were re-suspended in 180 ml of lysis buffer (20 mM Tris–HCl, 2 mM EDTA, 1.2% TritonX100 (w/v); pH 8.0) and incubated for 30 min at 37 °C. Further 25 µl of proteinase K and 200 µl buffer AL (Qiagen, MD, USA) were added to the mixture and incubated for 30 min at 56 °C.
For fungal DNA extraction, the cells were re-suspended in 600 ml of lysis buffer (1 M Sorbitol, 100 mM EDTA, 14 mM β-mercaptoethanol) and incubated with Zymolyase-T20 (200 U) for 30 min at 30 °C. The resulting spheroplasts were centrifuged for 10 min at 300 × g and the supernatant was discarded. The spheroplasts were re-suspended in 180 µl of Buffer ATL and incubated with 20 µl of proteinase K (Qiagen, MD, USA) for 15 min at 56 °C.
The remaining steps were performed according to the manufacturer’s protocol and the extracted DNA was stored at -20 °C. DNA concentration was measured using Qubit ds DNA HS kit on Qubit 2.0 fluorometer (Life technologies, Carlsbard, CA, USA).
PCR amplification and high-throughput sequencing
The PCR amplification and high-throughput sequencing was performed as explained in a previous study4. Equal concentration of bacterial and fungal DNA was used (~ 1 ng) for PCR amplification of 16S rRNA V3 hypervariable region and ITS1 region, respectively (see supplementary methods for details on primers and protocol used). Four bacterial samples and two fungal samples did not show any amplification, and therefore were not included in the study. After evaluating the amplified products on 2% w/v agarose gel, the products were purified using Ampure XP kit (Beckman Coulter, Brea, CA USA). Amplicon libraries were prepared using primers for V3 and ITS1 regions by following the Illumina 16S metagenomic library preparation guide. The metagenomic libraries were prepared using Illumina Nextera XT sample preparation kit (Illumina Inc., USA).
Based on the minimal DNA concentration (> 0.2 ng/µl) required to carry out the library preparation for shotgun sequencing, 14 subjects with healthy scalp and 14 with dandruff scalp were selected from each time-point for shotgun metagenome sequencing of their bacterial and fungal DNA. Thus, a total of 398 bacterial and 400 fungal amplicon samples, and 84 bacterial and 84 fungal metagenomic samples were sequenced in this study (Table S2).
Both the amplicon and shotgun metagenome libraries were evaluated on 2100 Bioanalyzer using DNA1000 kit for amplicon, and High Sensitivity DNA kit for metagenome (Agilent Technologies, Santa Clara, CA, USA) to estimate the library size. The libraries were further quantified on a Qubit 2.0 fluorometer using Qubit dsDNA HS kit (Life technologies, USA) and by qPCR using KAPA SYBR FAST qPCR Master mix and Illumina standards and primer premix (KAPA Biosystems, Wilmington, MA, USA) following the Illumina suggested protocol. Libraries in equal concentrations were loaded on Illumina NextSeq 500 platform using NextSeq 500/550 v2 sequencing reagent kit (Illumina Inc., USA) and 150 bp paired-end sequencing was performed for both types of libraries at the Next-Generation Sequencing (NGS) Facility, IISER Bhopal, India.
Assignment of bacterial 16S rRNA (V3) and fungal ITS1 amplicon reads
The raw sequence data was subjected to quality trimming and ambiguity filtering using NGSQC toolkit and the paired-end reads were assembled for each amplicon sequence using FLASH48,49. Quality filtration of fastq reads was carried out using NGSQC toolkit and all the reads with 80% of bases ≥ Q30 quality scores were selected for further analysis. Primers from the reads were removed using Cutadapt50 and reads without the primer sequences were discarded. Clustering was carried out using closed-reference OTU picking and de novo OTU picking protocol of QIIME v1.951 at ≥ 97% identity. The custom ITS1 database prepared previously4 and Greengenes database v13_5 were used as a reference for fungal and bacterial taxonomic assignment, respectively52.
α-diversity was calculated using the Shannon index metrics and observed species after rarefying from 100 sequences at a step size of 6,000 for V3 as well as for ITS1 amplicons using QIIME v1.9. Pielou’s evenness was calculated to identify the distribution of species with respect to their proportion in each sample groups using the R-package53. Weighted UniFrac distances were measured for the bacterial population, and not for fungal samples, due to the highly variable nature of ITS1 sequences, which makes them difficult to interpret informative and meaningful phylogenetic information54,55.
For the taxonomic assignment of de novo OTUs, sequences were aligned against the respective databases using BLAT, and the assignment was performed using Lowest Common Ancestor (LCA) algorithm52,56. The negative control samples showed a high abundance of fungal genus Pachysolen and bacterial genus Actinotalea, which are commonly found in environments such as air and soil, and not associated with the skin or scalp57,58. Hence, the OTUs from these genera were excluded from the analysis.
Shotgun metagenomic data analysis
Metagenomic reads with 60% bases above Q25 were considered for the analysis49. For fungal metagenome, the bacterial contaminant reads were removed by alignment against the bacterial reference genomes retrieved from NCBI, and the human contaminant reads were removed using BMTagger59. The remaining fungal reads from each sample were assembled independently into contigs using SOAPdenovo at a k-mer size of 75 bp.
For bacterial metagenome, the human and fungal contaminant reads were removed by aligning the sequences using BLAT against human HG19 assembly and custom fungal genome database, respectively60. These fungal genes and genomes were downloaded from Aspergillus database, FungiDB (release-30), Fungal Genome Initiative-Broad Institute, Fungi Ensembl, Saccharomyces Genome Database, Candida Genome Database and NCBI to construct a custom fungal genome database56,61–65. If any fungal gene sequences were not available in these databases, the gene sequences were extracted from the genome sequences based on the gtf information using SAMtools66. The high-quality reads were assembled at a k-mer length of 47 (the k-mer length was estimated using kmerGenie67) using SOAPdenovo, and the genes were predicted from the assembled contigs using MetaGeneMark68,69.
The fungal contigs with ≥ 300 bp length were selected and fungal genes were predicted from scaffolds using AUGUSTUS1,68,70. In order to increase the coverage of genes from fungal genomes found in our dataset, a total of 2,421,207 CDS were added from 303 fungal genomes from Ensembl fungi database. For bacteria, an additional parameter of identity > 50% with ≥ 60% coverage or aligned length > 300 was used. The relative abundance of each KO was calculated by adding up the abundance of genes mapping to the same KO ID, which was then used to calculate the relative abundance of KO ID in each sample. A similar approach was used to calculate the relative abundance of eggNOGs in each sample.
Non-redundant bacterial gene catalogue was generated by using CD-HIT, which consisted of 19,729,749 bacterial genes. A total of 207,763 genes were used as a reference fungal gene set consisting of non-redundant 27,937 genes from assembled metagenomes combined with CDS obtained from Ensembl Fungi genomes database71. In total, 587,400 bacterial and 81,395 fungal non-redundant genes were identified in the dataset (see supplementary methods for details on gene quantification).
Taxonomic assignment of reads from metagenomic data
The fungal reference genomes were retrieved from National Centre for Biotechnology Information (NCBI). The archaeal and bacterial genomes were retrieved from NCBI and Ensembl database. The metagenomic reads were aligned to the reference fungal and bacterial genomes and the mapped reads were considered for further analysis72,73.
KEGG assignment of genes
The KEGG v60 was updated by retrieving new sequences for KO IDs from the KEGG server74. The bacterial and fungal genes were annotated by alignment against KEGG and eggNOG v4.0 databases75–78. Protein sequences were assigned to eggNOG and KEGG orthologous groups based on the highest scoring hit containing at least one HSP (highest-scoring segment pair) above 60 bits and E-value ≤ 10–6 as described previously4. In total, 4,064 and 3,532 KO IDs were obtained from the combined fungal and bacterial gene catalogue, respectively. Pathway abundance was calculated by adding the relative abundance of each KO ID belonging to a particular pathway.
Statistical analysis
The species abundance and KEGG pathway composition were compared between different groups using Wilcoxon test to identify significant (p ≤ 0.05) variations. All the comparisons were performed pairwise for each group using R software79. In this study, same subjects were sampled at three different phases, therefore, to identify the species and pathways that presented significant (p ≤ 0.05) variations across the three phases, repeated measures ANOVA was performed. The species that showed a relative abundance of ≥ 1% in at least 20 samples were considered in this analysis. To identify the significantly varying fold-change in species abundance due to treatment, the fold-changes in species abundance were calculated as t = 2/t = 1 for each subject and compared using Wilcoxon test.
The Spearman’s Rank Correlation Coefficients with FDR adj. p-value were calculated to correlate clinical parameters with species. Species with ≥ 1% proportion in at least 20 samples were selected for the correlation analysis. Correlations were tested across all the phases to obtain the largest set of values for each parameter. Hierarchical clustering algorithm was used for clustering the highly-correlated pathways and species in the samples.
Figures 2b, 3b, 5, 6, S7a and S7b were created using ‘ggplot2’ package in R version 3.080. Figure 4 was created with ‘heatmap2’ function using ‘gplots’ package in R79. The Figures S4a and S4b were created using the ‘boxplot’ function from ‘graphics’ package in R79.
Supplementary Information
Acknowledgements
Authors would like to thank Dr. Mukta Sachdev, Medical Director, MS Clinical Research for her support during the clinical study. We also acknowledge Francois Pradier, Director, L’Oréal Research and Innovation, India for his support for the study and Dr. Audrey Gueniche for her help in interpreting the clinical results. We thank Subarna Saha, Mahesh M Veeranagaiah and Prashant Hegde for participating in preparing the clinical protocol and carrying out the clinical sampling. We thank the NGS facility and HPC facility at IISER Bhopal for providing the infrastructure to carry out the sequencing experiments and computational analysis, respectively. RS and DBD acknowledge DST-INSPIRE and UGC (Govt. of India), respectively for providing research fellowships.
Author contributions
C.C., N.M. and V.K.S. conceived the idea. C.C., N.R., L.B., N.M. and V.K.S. designed the study. R.S., C.C., N.M. and V.K.S. designed the experiments. R.S. carried out the experimental processing and sequencing of the DNA samples. P.M. performed the computational analysis of amplicon data. P.M. and D.B.D. performed the computational analysis of the metagenomic data. P.M., D.B.D. and R.S. carried out the statistical analysis of amplicon and metagenomic data and its correlation with the clinical parameters. R.S., P.M., C.C., D.B.D., N.M. and V.K.S. interpreted the results and prepared the figures and tables. R.S., P.M., C.C., D.B.D., N.R., L.B., N.M. and V.K.S. prepared the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by L’Oréal Research and Innovation (Project no. CONS/BIO/2015050) and intramural funding from IISER Bhopal.
Data availability
The high-throughput sequence data generated from this study have been deposited under the project number PRJNA415710 in the NCBI BioProject database and will be made publicly available on publication or on request at the time of peer review.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rituja Saxena, Parul Mittal and Cecile Clavaud.
Contributor Information
Namita Misra, Email: namita.misra@rd.loreal.com.
Vineet K. Sharma, Email: vineetks@iiserb.ac.in
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86454-1.
References
- 1.Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA, Program NCS. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena R, Sharma VK. A metagenomic insight into the human microbiome: its implications in health and disease. In: Kumar D, Antonarakis S, editors. Medical and Health Genomics. Mica Haley; 2016. pp. 107–119. [Google Scholar]
- 4.Saxena R, Mittal P, Clavaud C, Dhakan DB, Hegde P, Veeranagaiah MM, Saha S, Souverain L, Roy N, Breton L. Comparison of healthy and dandruff scalp microbiome reveals the role of commensals in scalp health. Front. Cell. Infect. Microbiol. 2018;8:346. doi: 10.3389/fcimb.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clavaud C, Jourdain R, Bar-Hen A, Tichit M, Bouchier C, Pouradier F, El Rawadi C, Guillot J, Menard-Szczebara F, Breton L, Latge JP, Mouyna I. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE. 2013;8(3):e58203. doi: 10.1371/journal.pone.0058203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Clavaud C, Bar-Hen A, Cui M, Gao J, Liu Y, Liu C, Shibagaki N, Gueniche A, Jourdain R, Lan K, Zhang C, Altmeyer R, Breton L. Characterization of the major bacterial-fungal populations colonizing dandruff scalps in Shanghai, China, shows microbial disequilibrium. Exp. Dermatol. 2015;24(5):398–400. doi: 10.1111/exd.12684. [DOI] [PubMed] [Google Scholar]
- 7.Soares RC, Camargo-Penna PH, de Moraes VCS, De Vecchi R, Clavaud C, Breton L, Braz ASK, Paulino LC. Dysbiotic bacterial and fungal communities not restricted to clinically affected skin sites in dandruff. Front. Cell. Infect. Microbiol. 2016;6:157. doi: 10.3389/fcimb.2016.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Wang Z, Yuan C, Liu X, Yang F, Wang T, Wang J, Manabe K, Qin O, Wang X, Zhang Y, Zhang M. Dandruff is associated with the conjoined interactions between host and microorganisms. Sci. Rep. 2016;6:24877. doi: 10.1038/srep24877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park HK, Ha MH, Park SG, Kim MN, Kim BJ, Kim W. Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS ONE. 2012;7(2):e32847. doi: 10.1371/journal.pone.0032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borda LJ, Wikramanayake TC. Seborrheic dermatitis and dandruff: a comprehensive review. J. Clin. Investig. Dermatol. 2015;3:2. doi: 10.13188/2373-1044.1000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay R. Malassezia, dandruff and seborrhoeic dermatitis: an overview. Br. J. Dermatol. 2011;165:2–8. doi: 10.1111/j.1365-2133.2011.10570.x. [DOI] [PubMed] [Google Scholar]
- 12.Perez GIP, Gao Z, Jourdain R, Ramirez J, Gany F, Clavaud C, Demaude J, Breton L, Blaser MJ. Body site is a more determinant factor than human population diversity in the healthy skin microbiome. PLoS ONE. 2016;11(4):e0151990. doi: 10.1371/journal.pone.0151990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibagaki N, Suda W, Clavaud C, Bastien P, Takayasu L, Iioka E, Kurokawa R, Yamashita N, Hattori Y, Shindo C. Aging-related changes in the diversity of women’s skin microbiomes associated with oral bacteria. Sci. Rep. 2017;7(1):10567. doi: 10.1038/s41598-017-10834-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka A, Cho O, Saito C, Saito M, Tsuboi R, Sugita T. Comprehensive pyrosequencing analysis of the bacterial microbiota of the skin of patients with seborrheic dermatitis. Microbiol. Immunol. 2016;60(8):521–526. doi: 10.1111/1348-0421.12398. [DOI] [PubMed] [Google Scholar]
- 15.Park T, Kim HJ, Myeong NR, Lee HG, Kwack I, Lee J, Kim BJ, Sul WJ, An S. Collapse of human scalp microbiome network in dandruff and seborrhoeic dermatitis. Exp. Dermatol. 2017;26(9):835–838. doi: 10.1111/exd.13293. [DOI] [PubMed] [Google Scholar]
- 16.Soares RC, Zani MB, Arruda ACBB, de Arruda LHF, Paulino LC. Malassezia intra-specific diversity and potentially new species in the skin microbiota from Brazilian healthy subjects and seborrheic dermatitis patients. PLoS ONE. 2015;10(2):e0117921. doi: 10.1371/journal.pone.0117921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiruma M, Cho O, Hiruma M, Kurakado S, Sugita T, Ikeda S. Genotype analyses of human commensal scalp fungi, Malassezia globosa, and Malassezia restricta on the scalps of patients with dandruff and healthy subjects. Mycopathologia. 2014;177(5–6):263–269. doi: 10.1007/s11046-014-9748-2. [DOI] [PubMed] [Google Scholar]
- 18.Jourdain R, Moga A, Vingler P, El Rawadi C, Pouradier F, Souverain L, Bastien P, Amalric N, Breton L. Exploration of scalp surface lipids reveals squalene peroxide as a potential actor in dandruff condition. Arch. Dermatol. Res. 2016;308(3):153–163. doi: 10.1007/s00403-016-1623-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donnarumma G, Perfetto B, Paoletti I, Oliviero G, Clavaud C, Del Bufalo A, Guéniche A, Jourdain R, Tufano MA, Breton L. Analysis of the response of human keratinocytes to Malassezia globosa and restricta strains. Arch. Dermatol. Res. 2014;306(8):763–768. doi: 10.1007/s00403-014-1479-1. [DOI] [PubMed] [Google Scholar]
- 20.Meisel JS, Sfyroera G, Bartow-McKenney C, Gimblet C, Bugayev J, Horwinski J, Kim B, Brestoff JR, Tyldsley AS, Zheng Q, Hodkinson BP, Artis D, Grice EA. Commensal microbiota modulate gene expression in the skin. Microbiome. 2018;6(1):20. doi: 10.1186/s40168-018-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo RL, Nakatsuji T. Microbial symbiosis with the innate immune defense system of the skin. J. Invest. Dermatol. 2011;131(10):1974–1980. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat. Rev. Microbiol. 2018;16(3):143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 23.Pierard-Franchimont C, Pierard G, Arrese J, De Doncker P. Effect of ketoconazole 1% and 2% shampoos on severe dandruff and seborrhoeic dermatitis: clinical, squamometric and mycological assessments. Dermatology. 2001;202(2):171–176. doi: 10.1159/000051628. [DOI] [PubMed] [Google Scholar]
- 24.Pierard G, Arrese J, Piérard-Franchimont C, De Doncker P. Prolonged effects of antidandruff shampoos-time to recurrence of Malassezia ovalis colonization of skin. Int. J. Cosmet. Sci. 1997;19(3):111–117. doi: 10.1111/j.1467-2494.1997.tb00174.x. [DOI] [PubMed] [Google Scholar]
- 25.Kamamoto C, Nishikaku A, Gompertz O, Melo A, Hassun K, Bagatin E. Cutaneous fungal microbiome: Malassezia yeasts in seborrheic dermatitis scalp in a randomized, comparative and therapeutic trial. Dermato-endocrinology. 2017;9(1):e1361573. doi: 10.1080/19381980.2017.1361573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zani M, Soares R, Arruda A, de Arruda L, Paulino L. Ketoconazole does not decrease fungal amount in patients with Seborrhoeic dermatitis. Br. J. Dermatol. 2016;175(2):417–421. doi: 10.1111/bjd.14501. [DOI] [PubMed] [Google Scholar]
- 27.Ortonne JP, Nikkels A, Reich K, Ponce Olivera R, Lee J, Kerrouche N, Sidou F, Faergemann J. Efficacious and safe management of moderate to severe scalp seborrhoeic dermatitis using clobetasol propionate shampoo 0–05% combined with ketoconazole shampoo 2%: a randomized, controlled study. Br. J. Dermatol. 2011;165(1):171–176. doi: 10.1111/j.1365-2133.2011.10269.x. [DOI] [PubMed] [Google Scholar]
- 28.Chiu C-H, Huang S-H, Wang H-MD. A review: hair health, concerns of shampoo ingredients and scalp nourishing treatments. Curr. Pharmaceut. Biotechnol. 2015;16(12):1045–1052. doi: 10.2174/1389201016666150817094447. [DOI] [PubMed] [Google Scholar]
- 29.Gavazzoni Dias MF. Hair cosmetics: an overview. Int. J. Trichol. 2015;7(1):2–15. doi: 10.4103/0974-7753.153450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorni, C., Sharma, P., Saikia, G., & Longvah, T. Fatty acid profile of edible oils and fats consumed in India. Food Chemistry (2017). [DOI] [PubMed]
- 31.Young F. Palm kernel and coconut oils: analytical characteristics, process technology and uses. J. Am. Oil Chem. Soc. 1983;60(2):374–379. doi: 10.1007/BF02543521. [DOI] [Google Scholar]
- 32.Garg A, Miiller J. Inhibition of growth of dermatophytes by Indian hair oils. Mycoses. 1992;35(11–12):363–369. doi: 10.1111/j.1439-0507.1992.tb00895.x. [DOI] [PubMed] [Google Scholar]
- 33.Hajini G, Kandhari K, Mohapatra L, Bhutani L. Effect of hair oils and fatty acids on the growth of dermatophytes and their in vitro penetration of human scalp hair. Sabouraudia J. Med. Vet. Mycol. 1970;8(3):174–176. doi: 10.1080/00362177085190851. [DOI] [PubMed] [Google Scholar]
- 34.Ohk SO, Park HG, Lee H, Kwon YJ, Kim BJ, Kim D, Chun YJ. Heterologous expression and characterization of CYP61A1 from dandruff-causing Malassezia globosa. Protein Exp. Purif. 2015;114:89–94. doi: 10.1016/j.pep.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Boemeke L, Marcadenti A, Busnello FM, Gottschall CBA. Effects of coconut oil on human health. Open J. Endocrine Metabolic Dis. 2015;5(07):84. doi: 10.4236/ojemd.2015.57011. [DOI] [Google Scholar]
- 36.Evangelista MT, Abad-Casintahan F, Lopez-Villafuerte L. The effect of topical virgin coconut oil on SCORAD index, transepidermal water loss, and skin capacitance in mild to moderate pediatric atopic dermatitis: a randomized, double-blind, clinical trial. Int. J. Dermatol. 2014;53(1):100–108. doi: 10.1111/ijd.12339. [DOI] [PubMed] [Google Scholar]
- 37.Lee HJ, Jeong SE, Lee S, Kim S, Han H, Jeon CO. Effects of cosmetics on the skin microbiome of facial cheeks with different hydration levels. MicrobiologyOpen. 2018;7(2):e00557. doi: 10.1002/mbo3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynde CW, Andriessen A, Bertucci V, McCuaig C, Skotnicki S, Weinstein M, Wiseman M, Zip C. The skin microbiome in atopic dermatitis and its relationship to emollients. J. Cutan. Med. Surg. 2016;20(1):21–28. doi: 10.1177/1203475415605498. [DOI] [PubMed] [Google Scholar]
- 39.Gonzalez ME, Schaffer JV, Orlow SJ, Gao Z, Li H, Alekseyenko AV, Blaser MJ. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J. Am. Acad. Dermatol. 2016;75(3):481–493. doi: 10.1016/j.jaad.2016.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seité S, Zelenkova H, Martin R. Clinical efficacy of emollients in atopic dermatitis patients–relationship with the skin microbiota modification. Clin. Cosmet. Investig. Dermatol. 2017;10:25. doi: 10.2147/CCID.S121910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouradier F, Liu C, Wares J, Yokoyama E, Collaudin C, Panhard S, Saint-Léger D, Loussouarn G. The worldwide diversity of scalp seborrhoea, as daily experienced by seven human ethnic groups. Int. J. Cosmet. Sci. 2017;39:629–636. doi: 10.1111/ics.12425. [DOI] [PubMed] [Google Scholar]
- 42.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007;71(2):282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gemmer CM, DeAngelis YM, Theelen B, Boekhout T, Dawson TL., Jr Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J. Clin. Microbiol. 2002;40(9):3350–3357. doi: 10.1128/JCM.40.9.3350-3357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 2013;24(2):160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Grafe F, Wohlrab W, Neubert RH, Brandsch M. Transport of biotin in human keratinocytes. J. Invest. Dermatol. 2003;120(3):428–433. doi: 10.1046/j.1523-1747.2003.12058.x. [DOI] [PubMed] [Google Scholar]
- 46.Uchida Y, Ito K, Ohtsuki S, Kubo Y, Suzuki T, Terasaki T. Major involvement of Na(+) -dependent multivitamin transporter (SLC5A6/SMVT) in uptake of biotin and pantothenic acid by human brain capillary endothelial cells. J. Neurochem. 2015;134(1):97–112. doi: 10.1111/jnc.13092. [DOI] [PubMed] [Google Scholar]
- 47.De Paepe K, Houben E, Adam R, Wiesemann F, Rogiers V. Validation of the VapoMeter, a closed unventilated chamber system to assess transepidermal water loss vs the open chamber Tewameter. Skin Res. Technol. 2005;11(1):61–69. doi: 10.1111/j.1600-0846.2005.00101.x. [DOI] [PubMed] [Google Scholar]
- 48.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 51.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Simpson GL, Solymos P, Stevens MHH, Wagner H. Package ‘vegan’. Commun. Ecol. Pack. Vers. 2013;2:9. [Google Scholar]
- 54.Leung MH, Chan KC, Lee PK. Skin fungal community and its correlation with bacterial community of urban Chinese individuals. Microbiome. 2016;4(1):46. doi: 10.1186/s40168-016-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjoller R, Koljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. Fungal community analysis by high-throughput sequencing of amplified markers—a user's guide. New Phytol. 2013;199(1):288–299. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yi H, Schumann P, Chun J. Demequina aestuarii gen. nov., sp. nov., a novel actinomycete of the suborder Micrococcineae, and reclassification of Cellulomonas fermentans Bagnara et al. 1985 as Actinotalea fermentans gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2007;57(1):151–156. doi: 10.1099/ijs.0.64525-0. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Kaas RS, Jensen PR, Workman M. Draft genome sequence of the yeast Pachysolen tannophilus CBS 4044/NRRL Y-2460. Eukaryot Cell. 2012;11(6):827–827. doi: 10.1128/EC.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotmistrovsky, K., & Agarwala, R. BMTagger: Best Match Tagger for removing human reads from metagenomics datasets (2011).
- 60.Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M. The UCSC genome browser database: 2015 update. Nucleic Acids Res. 2014;43(D1):D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J, Shah P. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2013;42(D1):D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2011;40(D1):D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44(D1):D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2011;40(D1):D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 2017;45(D1):D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2013;30(1):31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 68.Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132–e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:465–467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 72.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 74.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao Y, Tang H, Ye Y. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics. 2011;28(1):125–126. doi: 10.1093/bioinformatics/btr595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42:231–239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12(1):59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 79.Team, R. C. R: A language and environment for statistical computing (2013).
- 80.Wickham H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011;3(2):180–185. doi: 10.1002/wics.147. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequence data generated from this study have been deposited under the project number PRJNA415710 in the NCBI BioProject database and will be made publicly available on publication or on request at the time of peer review.