Abstract
Recent advances in the generation, purification and cellular delivery of RNA have enabled development of RNA-based therapeutics for a broad array of applications. RNA therapeutics comprise a rapidly expanding category of drugs that will change the standard of care for many diseases and actualize personalized medicine. These drugs are cost effective, relatively simple to manufacture, and can target previously undruggable pathways. It is a disruptive therapeutic technology, as small biotech startups, as well as academic groups, can rapidly develop new and personalized RNA constructs. In this review we discuss general concepts of different classes of RNA-based therapeutics, including antisense oligonucleotides, aptamers, small interfering RNAs, microRNAs, and messenger RNA. Furthermore, we provide an overview of the RNA-based therapies that are currently being evaluated in clinical trials or have already received regulatory approval. The challenges and advantages associated with use of RNA-based drugs are also discussed along with various approaches for RNA delivery. In addition, we introduce a new concept of hospital-based RNA therapeutics and share our experience with establishing such a platform at Houston Methodist Hospital.
Keywords: RNA therapeutics, delivery of RNA therapeutics, hospital-based RNA therapeutics, messenger RNAs (mRNAs), self-amplifying mRNA
Introduction
RNA Therapeutics comprise a rapidly expanding category of drugs that will speed solutions to the clinic; will actualize personalized medicine; and will make the term “undruggable” obsolete. The first RNA drugs have been approved, and many more are in development. We are in the midst of a therapeutic revolution, the likes of which have not been seen since the advent of recombinant protein technology almost 50 years ago in Silicon Valley. Accordingly, we will review recent developments in RNA Therapeutics, and their promise to alter the landscape of the pharmaceutical industry.
Conventional drug strategy relies on the ability of small molecule drugs to target active sites of proteins so as to inhibit or alter their function. It is well known that only ∼1.5% of the human genome encodes proteins (Ezkurdia et al., 2014). Furthermore, only 10–14% of proteins have active binding sites that are “druggable” targets for small molecules (Hopkins and Groom, 2002). Thus the “druggable” targets for small molecule therapies is limited. This limitation was addressed in part by the revolution of recombinant protein technology which had its beginnings in Silicon Valley in the late 1970’s (Russo, 2003). The development of genetic engineering by Stanley Cohen at Stanford University and Herb Boyer at UCSF provided the platform for generating recombinant proteins. Recombinant protein technology has become a significant share of the pharmaceutical market. In 2019, the FDA Center for Drug Evaluation and Research (CDER) approved 48 new agents, of which 10 were recombinant proteins, peptides or drug-antibody conjugates (Mullard, 2020). The Center for Biologics Evaluation and Research (CBER) is primarily responsible for evaluation and approval of vaccines, allergenic products, blood and blood products, plasma derivatives, cellular and gene therapy products. In 2019, CBER approved 22 new applications, one of which was a recombinant protein, three were attenuated virus vaccines, and one was a DNA therapeutic, Zolgensma (FDA, 2019). Recombinant proteins have limitations as drugs, particularly due to size and stability issues (Antosova et al., 2009; Lam et al., 2015). Furthermore, they must be properly folded and often require post-translational modifications (Li et al., 2015) that complicate the synthetic process. By contrast, nucleic-acid based strategies avoid many of these limitations as they make use of the translational machinery of the mammalian cell. Because DNA therapeutics pre-dated RNA therapeutics, a brief discussion of these nucleic-acid based cousins of RNA is useful for historical and comparative purposes.
DNA Therapeutics
DNA drugs generate therapeutic proteins when delivered to the nuclei of the patient’s cells. DNA drugs may be delivered as plasmids or integrated into a viral vector.
DNA Plasmids
DNA plasmids are high molecular weight, double-stranded (ds), circular DNA molecules that encode a therapeutic protein. Such proteins could replace defective or missing proteins in the patient (Saraswat et al., 2009). DNA plasmids can be used in: (1) gene therapy, (2) vaccination, and (3) cell therapy. The plasmid DNA (pDNA) must penetrate the cytoplasmic and nuclear membranes to gain access to the nucleus. In the nucleus, the pDNA is transcribed into mRNA that encodes the desired protein in the patient’s body. As an example, the pDNA drug VM202 is a 7377 base pair plasmid DNA gene therapy that encodes both isoforms of human hepatocyte growth factor, HGF (Kessler et al., 2015; ClinicalTrials.gov, 2020d). This drug is in a Phase III clinical trial to assess its benefit in treating painful diabetic peripheral neuropathy (DPN).
Viral Vectors
DNA-based drugs may be directed toward replacing defective or missing protein(s) (Schmeer et al., 2017; Daley, 2019). However, concerns regarding integration of foreign DNA into the host chromosomes and disruption of normal gene function has led to a search for non-integrating strategies. Adeno-Associated virus (AAV) is a common viral vector for the delivery of DNA encoding a therapeutic protein with minimal risk for integration. The AAV is a small (25 nm) icosahedral human parvovirus that contains a linear single-stranded DNA (4.7 kb). After removing key AAV viral genes, the desired gene is inserted into the AAV DNA for expression. The AAV vector delivers the desired gene to the target cell, for example to restore normal protein function (Tratschin et al., 1984; Naso et al., 2017). Alternatively, the AAV vector can be utilized to deliver interference RNA to downregulate the expression of a specific gene. Tomar et al. (2003) demonstrated the use of AAV to express hairpin siRNA in HeLa S3 cells to downregulate expression of caspase 8 and p53.
The Chinese food and drug regulatory agency approved the first DNA therapy drug, Gendicine, in 2003 to treat head and neck squamous cancer. Gendicine is an adenoviral vector encoding the wild type (wt) p53 gene which restores the expression of this tumor suppressor function to treat cancer (Zhang et al., 2018). Approximately 60–80% of all cancers manifest a p53 deficiency. In 2012, Glybera became the first DNA therapy to be approved in Europe. Glybera was an adeno-associated virus (serotype 1) encoding the lipoprotein lipase (LPL) gene to reverse LPL deficiency. This is a rare genetic disease that increases the levels of fat in the blood and cause severe pancreatitis. Glybera was not commercially successful and is no longer available (Bryant et al., 2013). Luxturna is an AAV (serotype 2) therapy encoding the RPE65 gene which received FDA approval in December 2017 and European approval in 2018. It is indicated for patients for RPE65-mediated inherited retinal dystrophy and improves vision by restoring RPE65 protein levels (Maguire et al., 2019). Zolgensma is an AAV vector-based gene therapy that delivers a fully functional copy of the human SMN gene into the target motor neuron cells. A one-time intravenous administration results in expression of the SMN protein in a child’s motor neurons, which improves muscle movement and function and survival of children with spinal muscular atrophy, SMA (Mahajan, 2019).
DNA-based vaccines encode specific antigen(s) to induce a protective immune response (Daley, 2019). Imlygic is a genetically modified therapy approved in United States, China, Europe, and Australia to treat malignant tumors. In this DNA therapy, the ICP34.5 gene is deleted to attenuate the natural herpes simplex virus type 1, HSV-1, which diminishes infection of normal tissues, but which enhances preferential tumor killing. The drug inserts into tumor cells, replicating and expressing a protein which evokes a cytotoxic immune response to the cancer cells (Conry et al., 2018; Amgen, 2020). DNA-based vaccines may also target infectious agents. For example, Innovio has developed a DNA vaccine encoding the spike protein of SARS-CoV-2 for intramuscular injection to induce immunity to the virus (INOVIO, 2020). Theirs was the first DNA vaccine to enter clinical trials for COVID-19. Other DNA vaccine developers include Genexine Consortium; Osaka University with Takara Bio, Anges & Cytiva; and Zydus Cadila Healthcare Limited (CovidVax, 2020; Milkeninstitute, 2020).
DNA vectors can be used to generate novel cell therapies. Typically, the therapeutic DNA is transfected into the cells ex vivo to alter cell phenotype or function, and then these cells are expanded and delivered into the patient (Daley, 2019). Kymriah is CD19-targeting CAR T cells immunotherapy to treat leukemia, lymphoma, and pediatric cancer in a single dose (approved in United States and Europe in 2017 and 2018 respectively). Enriched T cells from patient’s peripheral blood mononuclear cells are transduced with a lentiviral vector encoding a chimeric antigen receptor (CAR) against CD19. Subsequently, the transduced T cells are expanded, formulated into a suspension, and cryopreserved for later delivery (Zheng et al., 2018; FDA, 2020). Yescarta is another CD19-targeting CAR immunotherapy to treat large B cell lymphoma. Yescarta uses a retroviral vector to insert the CD-19 specific CAR into T cells. The anti-CD19 CAR T-cells are infused back into the patient’s body to kill CD19-expressing target cells. This drug was approved in the United States (2017) and Europe (2018) (Fala, 2018; Zheng et al., 2018).
Strimvelis is a personalized DNA-based medicine approved in Europe in 2016 to treat patients with a very rare disease called Severe Combined Immunodeficiency due to Adenosine Deaminase deficiency (ADA-SCID). Strimvelis uses the patient’s own CD34+ cells generated from hematopoietic stem cell (HSCs). The CD34+ cells are then transduced with a gammaretrovirus vector carrying the gene for human adenosine deaminase (ADA), then reinfused into the patient. These cells home to the patient’s bone marrow where CD34+ cells replicate and generate normal ADA protein to correct the deficiency (South et al., 2018). Zyntelgo was approved in Europe in 2019 for the treatment of beta-thalassemia. Zyntelgo comprises a lentiviral vector to introduce the beta globin gene into autologous blood-derived CD34+ cells ex vivo. The genetically modified cells are then re-infused and home to the patient’s bone marrow to generate red blood cells with normally functioning hemoglobin (Schuessler-Lenz et al., 2020). A similar approach was taken by investigators at St. Jude Children’s Research Hospital, to treat severe combined immunodeficiency (SCID), also called bubble boy disease. This rare- life-threatening disorder is caused by mutations in the gene that encodes the common γ-chain (IL2RG), which is shared by multiple cytokine receptors. This protein is necessary for the development and function of lymphocytes, so children with this disease lack normal immune function. The St. Jude investigators used a lentiviral vector to transfect autologous blood-derived CD34+ cells ex vivo with the IL2RG gene, which cells are then re-infused (Stjude, 2020; St Jude Children’s Research Hospital, 2020). As described below, this gene therapy is effective at restoring immune function in SCID.
CRISPR-Associated Protein 9 (CRISPR-Cas9)
Genome editing tools are used to add, remove or alter the genetic material at particular locations in the genome. There are several genome editing methods, as typified by clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9, CRISPR-Cas9 (Jiang and Doudna, 2017). Ishino et al. (1987) were first to discover the CRISPR DNA sequence. The CRISPR-Cas9 is a component of the bacterial adaptive immune system. In bacteria, two RNA molecules and the protein Cas9 bind to a foreign target. One of these molecules, called trans-activating CRISPR RNA (tracrRNA), serves as a scaffold and binds to Cas9, a DNA endonuclease. The other molecule, called CRISPR RNA (crRNA), has sequence homology to the foreign DNA (Jiang and Doudna, 2017; Karlgren et al., 2018; Zhan et al., 2019) and ensures cleavage specificity. This native immune defense has been modified for genome editing. In one version of the modified CRISPR-Cas9 technology, both RNA molecules are linked into a single guide RNA (sgRNA). The target DNA sequence is followed by a protospacer adjacent motif (PAM), also known as “NGG” sequence, which is a short (2–6 bp) DNA sequence. Cas9 cleaves the target DNA sequence 3 bases upstream of the PAM and creates a double-strand break. The latter can be repaired by two mechanisms: non-homologous end joining (NHEJ), and homology-directed repair (HDR). NHEJ is an error-prone process that introduces small deletions or insertions (indels) and disrupts the targeted locus (gene knock-out). HDR is a more precise process where a short donor DNA sequence is used for the double-stranded break repair (Jiang and Doudna, 2017; Karlgren et al., 2018), facilitating a gene knock-in.
EDIT-101 (Editas Medicine) is an early stage clinical CRISPR medicine to treat patients suffering from Leber Congenital Amaurosis type 10 (LCA10). LCA10 is a genetic blindness caused by an A to G point mutation within intron 26 in the CEP290 gene involved in phototransduction. This mutation results in a splicing defect to include a 128 bp cryptic exon in CEP290 transcript, thereby leading to expression of non-functional mutant protein. EDIT-101 is designed to deliver Staph. aureus Cas9 and two CEP290-specific guide RNAs to target cells by subretinal injection using an adeno-associated viral vector. In EDIT-101, Cas9 is driven by photoreceptor cell-specific promoter, rhodopsin kinase, and the resulting break in DNA is repaired by NHEJ, resulting in normal CEP290 expression and restoration of photoreceptor function (Maeder et al., 2019; Research and Pipeline – Editas, 2020).
A few other CRISPR-Cas9-based drugs are also in clinical trials now. For instance, CTX001, CTX110, CTX120, and CTX130 from CRISPR Therapeutics (Pipeline – CRISPR, 2019c). CTX001 is designed to treat both sickle cell disease and beta thalassemia. It disrupts the erythroid lineage-specific enhancer of the BCL11A gene in autologous blood stem cells and thus increases production of γ-globin, a component of fetal hemoglobin (HbF). The latter can serve as an alternative to the patient’s defective adult hemoglobin (HbA), and its increased level is observed in the patient’s blood cells after treatment with CTX001(Hemoglobinopathies – CRISPR, 2019a). CTX 110, CTX120, and CTX130 are being developed as immunotherapies, which, respectively, create allogenic CAR-T cells against CD19 for B-cell malignancies, BCMA for multiple myeloma, and CD70 for both hematologic cancers (certain lymphomas) and solid tumors (renal cell carcinoma) (Immuno-Oncology – CRISPR, 2019b).
All DNA-based drugs must penetrate two membranes, the cytoplasmic and the nuclear membranes, to have their effect. Because they enter the nucleus, DNA drugs raise safety concerns due to potential integration into the host genome (Ledwith et al., 2000). These limitations can be addressed by RNA therapeutics which only need to reach the cell cytoplasm, and which represents no risk of chromosomal integration (Lundstrom, 2018a; Shin et al., 2018).
RNA Therapeutics
The development of RNA drugs has largely focused on two approaches: (1) antisense RNA (RNAi), where short oligonucleotides recognize and hybridize to complementary sequences in endogenous RNA transcripts and alter their processing (Stephenson and Zamecnik, 1978; Shen and Corey, 2018); and (2) message RNA (mRNA), where mRNAs encoding certain peptides or proteins elicit their transient expression in the cytoplasm (for instance, to replace defective proteins or present antigens for vaccination) (Wolff et al., 1990; Ulmer and Geall, 2016). The development of RNA therapeutics required that several major hurdles be overcome, specifically the (1) rapid degradation of exogenous RNA by RNases that are ubiquitous in the environment and tissues; (2) delivery of negatively charged RNA across hydrophobic cytoplasmic membrane; and (3) strong immunogenicity of exogenous RNA that caused cell toxicity and impaired translation into therapeutic proteins.
These hurdles have been substantially overcome with recent advancements in RNA biology, bioinformatics, separation science and nanotechnology thereby facilitating the recent rapid development of RNA therapeutics. Advantages of RNA-based drugs that are driving development include: (1) their ability to act on targets that are otherwise “undruggable” for a small molecule or a protein; (2) their rapid and cost effective development, by comparison to that of small molecules or recombinant proteins; (3) the ability to rapidly alter the sequence of the mRNA construct for personalized treatments or to adapt to an evolving pathogen. Below, we review the different classes of RNA therapeutics (see Figure 1), the challenges and advantages associated with their use, and provide an overview of the therapeutics under development as well as of those already available on the market (see Table 1). In addition, we introduce a new concept of hospital-based RNA therapeutics and share our experience with establishing such a platform at Houston Methodist Hospital.
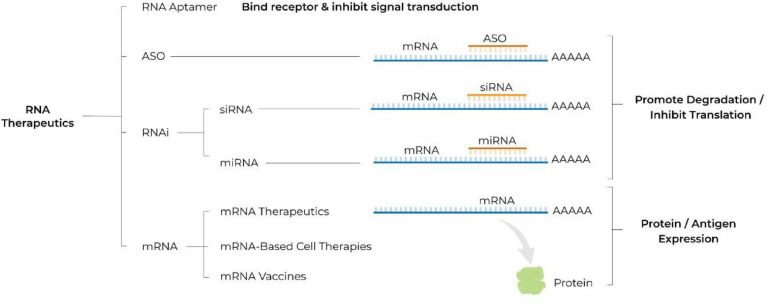
FIGURE 1.
Schematic illustrating different classes of RNA therapeutics. ASO, antisense oligonucleotide; RNA, ribonucleic acid; RNAi, RNA interference; siRNA, small interfering RNA; miRNA, microRNA; mRNA, messenger RNA; A, adenosine molecule; AAAAA, poly A tail.
TABLE 1.
RNA therapeutics approved for clinical use or undergoing clinical trials.
| Drug | Type of RNA | Company | Route of delivery | Condition/Disease | Status |
| Nusinersen (Spinraza) | ASO | Ionis | Intrathecal | Spinal muscular atrophy | FDA approval in 2016 |
| Eteplirsen (Exondys 51) | ASO | Sarepta | Intravenous | Duchenne muscular dystrophy | FDA approval in 2016 |
| Inotersen (Tegsedi) | ASO | Ionis | Subcutaneous | Familial amyloid polyneuropathy | FDA approval in 2018 |
| Volanesorsen (Waylivra) | ASO | Ionis | Subcutaneous | Familial chylomicronemia syndrome | EU approval in 2019 |
| Patisiran (Onpattro) | siRNA | Alnylam | Intravenous | Polyneuropathy | FDA approval in 2018 |
| Givosiran (Givlaari) | siRNA | Alnylam | Subcutaneous | Acute hepatic porphyria | FDA approval in 2019 |
| Cobomarsen (MRG-106) | miRNA | miRagen (Viridian) | Intravenous/subcutaneous | Blood cancers | Phase II |
| Remlarsen (MRG-201) | miRNA | miRagen (Viridian) | Intradermal | Keloids | Phase II |
| MRG-110 | miRNA | miRagen (Viridian) | Intradermal | Tissue Repair | Phase I |
| Pegaptanib (Macugen) | Aptamer (RNA) | Bausch + Lomb | Intravitreal | Macular Degeneration | FDA approval in 2014 |
| Emapticap pegol (NOX-E36) | Aptamer (RNA) | NOXXON | Intravenous/Subcutaneous | Diabetic nephropathy, lung and pancreatic cancer | Phase II |
| Olaptesed pegol (NOX-A12) | Aptamer (RNA) | NOXXON | Intravenous | Brain cancer | Phase I/II |
| BNT162b2 | mRNA | BioNTech and Pfizer | Intramuscular | COVID-19 | FDA authorization for emergency use in 2020 |
| mRNA-1273 | mRNA | Moderna | Intramuscular | COVID-19 | FDA authorization for emergency use in 2020 |
| CVnCoV | mRNA | CureVac | Intramuscular | COVID-19 | Phase III |
| AZD8601 | mRNA | Moderna/Astrazeneca | Epicardial | Ischemic heart disease | Phase II |
| mRNA-1647 | mRNA | Moderna | Intramuscular | Cytomegalovirus infection | Phase II |
| P-BCMA-101 | mRNA | Poseida | Intravenous | Multiple myeloma | Phase II |
| mRNA-4157 | mRNA | Moderna | Intramuscular | Cancer | Phase II |
| mRNA-3704 | mRNA | Moderna | Intravenous | Methylmalonic aciduria | Phase I/II |
| MRT5005 | mRNA | Translate Bio | Inhalation | Cystic Fibrosis | Phase I/II |
| mRNA-2416 | mRNA | Moderna | Intratumoral | Solid tumors/lymphoma/advanced ovarian carcinoma | Phase I/II |
| BNT131 (SAR441000) | mRNA | BioNTech/Sanofi/Genmab | Intratumoral | Advanced melanoma | Phase I/II |
| Descartes-08 | mRNA | Cartesian | Intravenous | Generalized myasthenia gravis | Phase I/II |
| BNT122 | mRNA | BioNTech | Intravenous | Solid tumors | Phase I/II |
| mRNA-2752 | mRNA | Moderna | Intratumoral | Solid tumors | Phase I |
| MEDI1191 | mRNA | Moderna | Intratumoral | Solid tumors | Phase I |
| mRNA-1944 | mRNA | Moderna | Intravenous | Chikungunya infection | Phase I |
| CV8102 | mRNA | CureVac | Intratumoral | Solid tumors | Phase I |
| ARCT-810 | mRNA | Arcturus | Intravenously | Urea disorder | Phase I |
| CV7202 | mRNA | CureVac | Intramuscular | Rabies | Phase I |
| mRNA-1893 | mRNA | Moderna | Intramuscular | Zika | Phase I |
| CV9202 | mRNA | CureVac | Intradermal | Non-small cell lung cancer | Phase I |
| mRNA-5671 | mRNA | Moderna | Intravenous | Cancer | Phase I |
| BNT111 | mRNA | BioNTech | Intravenous | Advanced Melanoma | Phase I |
The table summarizes information available at ClinicalTrials.gov (https://clinicaltrials.gov/) as of January 26, 2021. ASO, antisense oligonucleotide; siRNA, small interfering RNA; miRNA, microRNA; RNA, ribonucleic acid; mRNA, messenger RNA.
Antisense Oligonucleotide (ASO), Small Interfering Rna (siRNA), and microRNA as Therapeutics
Antisense oligonucleotides are short single-stranded DNA, phosphorothioate DNA, RNA analogs, conformationally restricted nucleosides (locked nucleic acids, LNA), or morpholino phosphorodiamidate oligonucleotides complementary to a certain region of RNA that they are meant to target. The modifications in backbone, and sugar molecules give antisense oligos more affinity and stability (Bennett and Swayze, 2010). Zamecnick and Stephenson pioneered the field of antisense oligonucleotides in late 1970s. They designed and synthesized tridecamer deoxynucleotides complementary to reiterated terminal sequences of Rous sarcoma virus (RSV) at 3′ and 5′ ends. To test its function, RSV-infected chick embryo fibroblasts (CSF) were transfected with the tridecamer which hybridized to its complementary sequence in the target RNA (RSV RNA), thereby inhibiting viral production and cell transformation (Stephenson and Zamecnik, 1978).
There are two classes of ASOs: RNase H-dependent ASO (Wu et al., 2004), and RNase H-independent (steric block) ASO (Baker et al., 1997). The former is more commonly used and is dependent on the endogenous RNase H enzyme that hydrolyzes the RNA strand of an RNA/DNA duplex. The RNase H-dependent ASOs are more efficient in knockdown of gene expression than RNase H-independent ASOs (Larrouy et al., 1992; Dean et al., 1994; Dias and Stein, 2002). Minshull and Hunt (1986) were the first to demonstrate the RNase H-dependent ASO mechanism in reticulocyte lysate. They added RNase H-treated DNA-mRNA hybrid complexes directly to the reticulocyte lysate for translation, and observed a loss of full length translation products of the hybridized mRNA (Minshull and Hunt, 1986). Giles et al. (1995) further demonstrated the RNase H-mediated antisense effect in human leukemia cells by identifying the RNase H-generated mRNA fragments with a reverse ligation-mediated PCR method.
Steric block ASOs physically inhibit or prevent translation or splicing, and can be engineered to either prevent polyadenylation (Vickers et al., 2001), inhibit (Baker et al., 1997) or enhance translation (Liang et al., 2016), or alter splicing (Hua et al., 2007). For instance, Baker et al. (1997) designed RNase H-independent 2′-O-2-methoxyethyl ASOs that were complementary to the 5′ cap region of the intercellular adhesion molecule 1 (ICAM-1) transcript in human umbilical vein endothelial cells. These ASOs inhibited protein expression of the targeted transcript by interfering with the formation of the 80S translation initiation complex (Baker et al., 1997).
To date, three ASO drugs have received FDA approval: (1) nusinersen (Ionis Pharmaceuticals) (Neil and Bisaccia, 2019); (2) eteplirsen (Sarepta Therapeutics) (Lim et al., 2017); and (3) inotersen (Ionis Pharmaceuticals and Akcea Therapeutics) (Mathew and Wang, 2019). Nusinersen (FDA approval in December 2016) is used to treat spinal muscular atrophy (SMA), which is caused by deletions or mutations in the survival motor neuron 1 (SMN1) gene. The mutated or deleted gene does not produce enough SMN protein. SMN2, a homolog gene, is different from SMN1 only in that it is spliced with exon 8, rather than exon 7. This results in expression of small amount of full length SMN protein. Nusinersen is a 2′-O-methoxyethyl-phosphorothioate-modified drug that modulates splicing of SMN2 so as to increase the transcript containing exon 7 (see Figure 2, Rigo et al., 2012), and, therefore enhances production of full length SMN protein (Wurster and Ludolph, 2018).
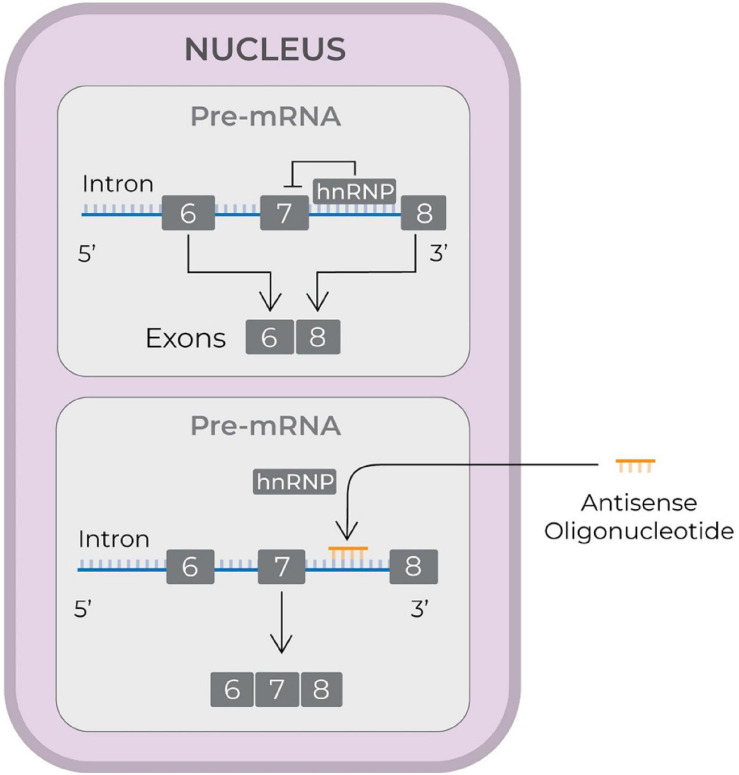
FIGURE 2.
Schematic illustrating the mode of action of an antisense drug to treat spinal muscular atrophy. The antisense drugs reach nucleus, displace hnRNP proteins and increase the synthesis of transcripts containing exon 7 and thereby generate full length SMN protein (Rigo et al., 2012). SMN, survival of motor neuron; hnRNP, heterogeneous nuclear ribonucleoprotein; pre-mRNA, precursor mRNA; mRNA, messenger RNA; RNA, ribonucleic acid.
Eteplirsen (Exondys 51, FDA approval in September 2016) is used to treat Duchenne muscular dystrophy (DMD). DMD is caused by mutations in the DMD gene that result in a premature stop codon and a non-functional dystrophin protein, with a mutation in exon 51 being most frequent single exon mutation (Bladen et al., 2015). Eteplirsen is 30-mer phosphomorpholidate oligonucleotide that recognizes and hybridizes to exon 51 of the DMD gene thereby modulating the pre-messenger RNA splicing process to exclude exon 51 from the mature mRNA. This restores the reading frame of the DMD gene and a shortened but functional dystrophin protein is produced (Lim et al., 2017).
Inotersen (Tegsedi, FDA approval in October 2018) is used to treat familial amyloid polyneuropathy caused by the mutation in the transthyretin (TTR) gene. The mutated gene leads to translation and accumulation of the abnormal TTR protein in different organs, including heart, kidney, eyes, and nerves. Tegsedi is a 20-mer chemically modified RNA molecule that hybridizes to TTR mRNA, mimicking a DNA/RNA complex, and thereby interferes with TTR protein production (Mathew and Wang, 2019).
Volanesorsen (Ionis Pharmaceuticals) was recently approved in Europe (Waylivra, EU conditional marketing authorization in May 2019). This is an antisense oligonucleotide indicated for treatment of familial chylomicronemia syndrome (Paik and Duggan, 2019). It recognizes and hybridizes to the complementary sequence in the 3′ untranslated region of apolipoprotein CIII mRNA to increase its degradation by RNase H1.
Small interfering RNAs (siRNAs) are small non-coding RNA duplexes that originate from precursor siRNAs. The latter are either transcribed or artificially introduced and range from 30 bp to more than 100 bp. The precursor siRNA duplex is processed by the endogenous Dicer enzyme into 20–30 bp long siRNA with two base overhangs in the 3′ region, which interacts with the endogenous RNA-induced silencing complex (RISC) to elicit RNA interference (RNAi). The endonuclease argonaute 2 (AGO2) component of the RISC cleaves the sense strand, leaving intact the antisense strand, which guides the active RISC to its target mRNA. Then AGO2 cleaves the phosphodiester backbone of the target mRNA. The antisense strand is usually fully complementary to the coding region of the target mRNA, therefore siRNA knocks down one specific target gene (Wittrup and Lieberman, 2015; Dana et al., 2017). Fire et al. (1998) were first to demonstrate RNAi by blocking the expression of unc-22, unc-54, fem1, and hlh-1 genes in C. elegans. They showed that dsRNA are more effective than ssRNA to artificially induce RNAi and destroy an mRNA target (Fire et al., 1998; Lam et al., 2015). Patisiran (Onpattro, Alnylam Pharmaceuticals, FDA approval in August 2018) was the first marketed siRNA-based drug (see Figure 3, Kristen et al., 2018). It is used to treat adult patients with polyneuropathy caused by hereditary TTR-mediated amyloidosis. Patisiran is a dsRNA that acts through RNAi and induces degradation of mRNA encoding TTR (Kristen et al., 2018). Recently, another siRNA drug, Givosiran (Givlaari, Alnylam Pharmaceuticals) received FDA approval in November 2019 (Alnylam, 2020b) for the treatment of acute hepatic porphyria. The drug targets aminolevulinate synthase 1 mRNA in liver, and reduces the levels of disease-causing neurotoxic intermediates aminolevulinic acid and porphobilinogen (Alnylam, 2020b; Givlaari, 2020).
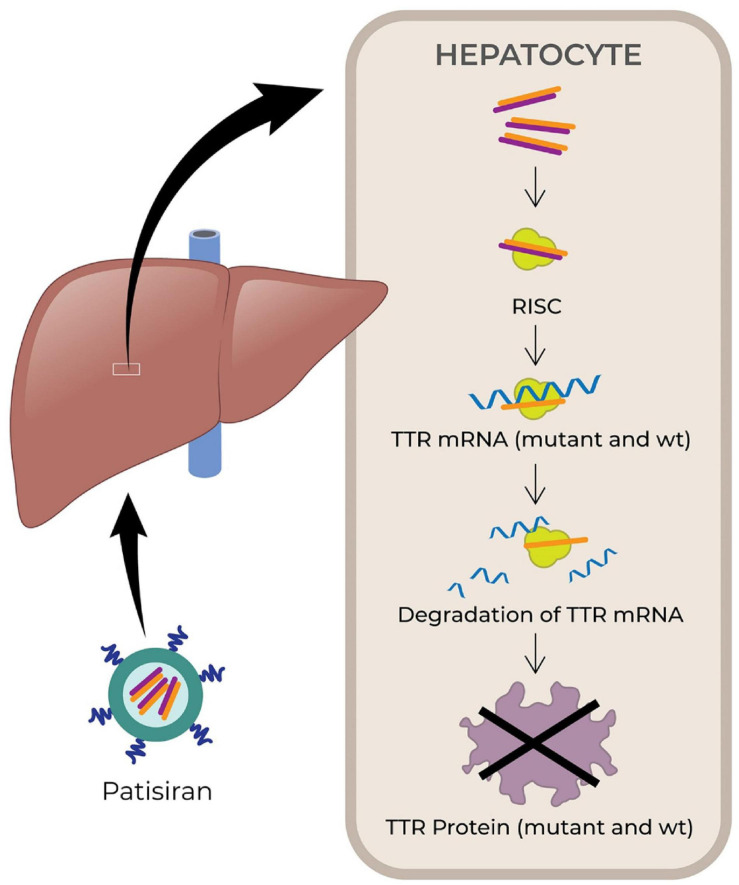
FIGURE 3.
Schematic illustrating the mechanism of action of small interfering RNA (siRNA) drug, patisiran. The drugs are encapsulated in lipid nanoparticles and administered intravenously. After administration, the drugs finally reach hepatocyte and released into the cytoplasm, where it is loaded onto the RISC. The antisense strand hybridizes with target mRNA to suppress the production of target protein (TTR) (Kristen et al., 2018). TTR, transthyretin; wt, wild type; RISC, RNA-induced silencing complex; mRNA, messenger RNA; RNA, ribonucleic acid.
Unlike siRNAs, microRNAs (miRNAs) are small non-coding RNA molecules that regulate the expression of multiple mRNAs by blocking translation or promoting degradation of the target mRNAs. They were first discovered in C. elegans (Lee et al., 1993). This class of non-coding RNAs are transcribed from genomic DNA as primary miRNAs (pri-miRNAs). The latter adopt a loop structure with interspersed mismatches and are cleaved by Drosha to a 70–100 bp precursor miRNAs (pre-miRNAs), before leaving the nucleus. Exportin 5 transports the pre-miRNAs to the cytoplasm, where Dicer processes them into 18–25 bp RNA duplexes with two base overhangs in the 3′ region. These structures are now referred to as miRNAs. The miRNA is then loaded into the RISC to form a miRISC complex. The miRNA duplex unwinds to release the sense strand. The antisense strand guides the miRISC. Hybridization usually occurs at 2–7 bases of the 5′ end of miRNA and the 3′ UTR of the target mRNA. The target mRNA is inhibited via translational repression, degradation or cleavage (Lam et al., 2015; Lou et al., 2018; O’Brien et al., 2018). The miRNA-based therapeutics could be categorized into two types: miRNAs mimics and miRNAs inhibitors. The former are double-stranded RNA molecules that mimic miRNAs, while the latter are single-stranded RNA oligos designed to interfere with miRNAs. Although there are no miRNA-based drugs on the market as of today, some promising candidates are currently in clinical trials. For instance, miRagen Therapeutics (Viridian Therapeutics) is developing cobomarsen (MRG-106), remlarsen (MRG-201), MRG-229 (miR-29 mimic; pre-clinical), and MRG-110 (miR-92 inhibitor)(Pipeline – miRagen, 2019; Pipeline – Viridian, 2021). Cobomarsen (MRG-106) is designed to treat patients with blood cancers by reducing miR-155 levels (Pipeline – miRagen, 2019; Pipeline – Viridian, 2021). Remlarsen (MRG-201) is designed to mimic miR-29 to treat keloids (ClinicalTrials.gov, 2019; Hanna et al., 2019; Pipeline – miRagen, 2019; Pipeline – Viridian, 2021), MRG-229 mimics miR-29 to treat pathologic fibrosis (Hanna et al., 2019; Pipeline – miRagen, 2019; Pipeline – Viridian, 2021), while MRG-110 inhibits miR-92 to accelerate tissue repair (Hanna et al., 2019; Pipeline – miRagen, 2019; Pipeline – Viridian, 2021). InteRNA Technologies has designed INT-1B3 to mimic tumor suppressor miRNA in order to modulate the immunosuppressive tumor microenvironment and treat solid cancers (Yahyanejad et al., 2018; Businesswire, 2019). Regulus Therapeutics’ product RG-012 inhibits miR-21 to treat Alport Syndrome (Regulus, 2019). Besides RG-012, Regulus has several other miRNA-based drugs in the pipeline to treat ADPKD (miR-17), glioblastoma multiforme (miR-10b), and others (Regulus, 2019). Miravirsen, which is under development by Roche Innovation Center Copenhagen, targets miR-122 and decreases viral load in patients with chronic hepatitis C (van der Ree et al., 2016). Another candidate, a TargomiR drug MesomiR-1 (EnGeneIC Limited) mimics tumor suppressor miR-16 to treat patients with mesothelioma (Reid et al., 2016). Overall, both siRNAs and miRNAs are attractive candidates as therapeutic agents and may overcome the major limitations of small molecule-based and protein-based drugs.
Aptamers as Therapeutics
Aptamers are short single-stranded nucleic acids that can bind to variety of targets, such as proteins, peptides, carbohydrates, and other molecules, by virtue of the tertiary structure of the aptamer, rather than its sequence. Aptamers were first described in 1990 independently by two groups, Ellington and Szostak (1990) and Tuerk and Gold (1990), through systematic evolution of ligand by exponential enrichment (SELEX) technique. Aptamers are evolved from highly diverse nucleic acid pools to bind to the targets with high specificity and affinity, and can serve as agonists (Dollins et al., 2008; McNamara et al., 2008), antagonists (Santulli-Marotto et al., 2003; Berezhnoy et al., 2012), bispecific aptamers (Schrand et al., 2014, 2017), and even carriers for other drugs (McNamara et al., 2006; Lozano et al., 2016).
Although aptamers hold promise as therapeutics, there is only one FDA-approved aptamer-based drug on market. Pegaptanib (Macugen, Bausch + Lomb Pharmaceutical Retina Portfolio, FDA approval in December 2004) is a 27 base RNA aptamer (see Figure 4 for predicted structure, Ng et al., 2006) selected against vascular endothelial growth factor (VEGF) and is used to treat age-related macular degeneration, a leading cause of irreversible blindness worldwide (Ruckman et al., 1998). Many other aptamer-based drugs are currently in clinical trials and have been recently reviewed elsewhere (Kaur et al., 2018). Emapticap pegol (NOXXON, 2020b; NOXXON Pharma), for instance, was evolved to specifically bind and inhibit the pro-inflammatory chemokine C-C motif-ligand 2 (CCL2) to treat diabetic nephropathy as well as lung and pancreatic cancer (NOXXON, 2020b). Similarly, Olaptesed pegol (NOXXON, 2020a; NOXXON Pharma) was evolved to target CXC chemokine ligand (CXCL12) for clinical treatment of brain cancer (glioblastoma/glioma). REG1 is an anticoagulation combination therapy that consists of pegnivacogin, an RNA aptamer inhibitor of coagulation factor IXa, and anivamersen, a complementary sequence to rapidly reverse the anticoagulant effect of pegnivacogin (Lincoff et al., 2016). The intent of this combination aptamer is that the first will induce anticoagulation as needed, whereas the second will reverse the anticoagulation as necessary to avoid excessive bleeding. However, in a trial comparing the aptamer therapy against the approved anticoagulant bivalirudin, in patients undergoing percutaneous coronary artery intervention, REG1 did not provide any greater benefit, and had a greater frequency of serious allergic reactions. However, the concept of having a second aptamer to act as an antidote to the first, so as to treat side effects or adverse reactions is an intriguing idea.
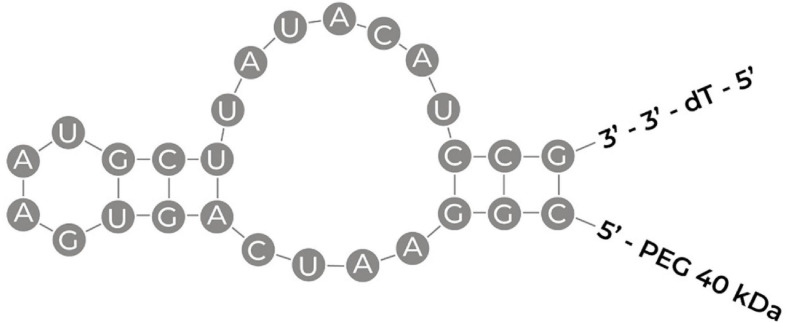
FIGURE 4.
Schematic illustrating the sequence of pegaptanib with its secondary structure. PEG, polyethylene glycol; dT, deoxthymidine (Ng et al., 2006; Amadio et al., 2016).
Aptamers have great potential to replace monoclonal antibodies in therapeutic and diagnostic (Damase et al., 2018; Bauer et al., 2019) applications because they can be produced via chemical synthesis, are more cost-effective in manufacturing, simpler to modify, and elicit little immunogenicity. However, despite the fact that aptamers have many advantages over antibodies, their clinical translation is challenging due to suboptimal pharmacokinetic properties (highly sensitive to nucleases, readily excreted by kidneys) and complexity of selection techniques (a time consuming process with low success rates) (Nimjee et al., 2017).
mRNA as Therapeutics
Messenger RNAs (mRNAs) serve as the intermediates between the coding genomic DNA and the encoded proteins (Gilbert, 1986). Essentially, mRNAs are transient blueprints of genes that are encoded in the genomic DNA. The mRNA transcripts of these genes deliver the genetic information to the translational machinery to generate the encoded proteins (Wolff et al., 1990; Conry et al., 1995). Wolff et al. (1990) were amongst the first to use exogenous mRNA to induce the expression of a protein in vivo. They injected synthetic mRNA encoding luciferase, chloramphenicol acetyltransferase, or beta-galactosidase into mouse skeletal muscle and detected proteins translated from these RNAs at the injection site. Importantly, the extent of expression from RNA was comparable to that obtained from injection of a DNA vector encoding the same proteins (Wolff et al., 1990). Jirikowski et al. (1992) demonstrated the therapeutic effect of mRNA in a rodent model of central diabetes insipidus. Brattleboro rats have a genetic deficiency of vasopressin. The investigators purified cytoplasmic mRNA from wild-type rat hypothalamus, or synthesized mRNA that encodes for vasopressin. Injection of either mRNA into the hypothalamus of Brattleboro rats induced the synthesis of vasopressin and transiently reversed diabetes insipidus (Jirikowski et al., 1992).
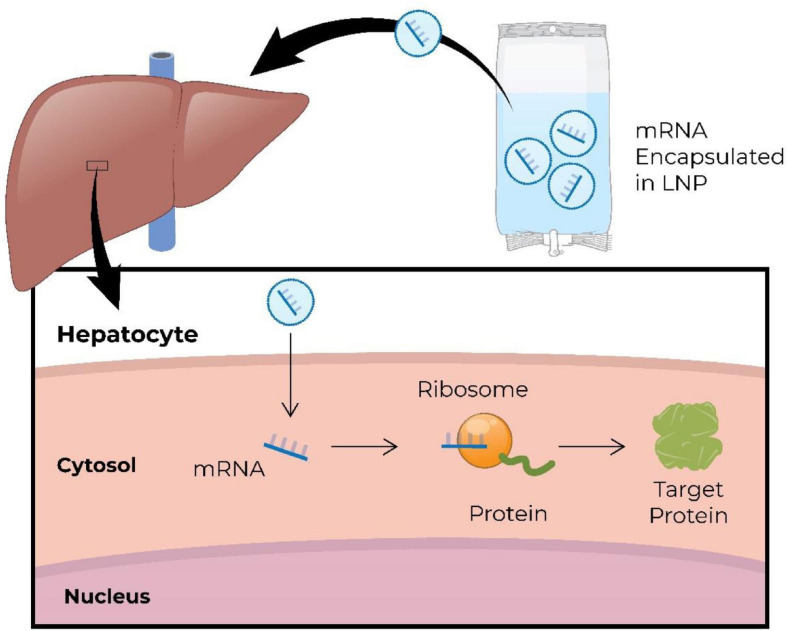
Synthetic or in vitro-translated mRNA is engineered to mimic naturally occurring mRNA (Jirikowski et al., 1992; Conry et al., 1995). It consists of a single-stranded open reading frame flanked by untranslated regions, and has a 5′ cap for translation, and a 3′ poly(A) tail for stability (Conry et al., 1995; Whisenand et al., 2017; Shin et al., 2018). Exogenous mRNA is translated into protein in the cytoplasm, and degrades within the cytoplasm typically within minutes to hours, posing no risk of integration into the genome. Figure 5 demonstrates systemically delivered mRNA expressing hepatic protein (An et al., 2017). Development and manufacturing of RNA therapeutics is relatively simple and a more cost-effective process comparing to that for recombinant proteins or small molecules. In addition, RNA sequences can be easily modified allowing for personalization of RNA therapy. There are several therapeutic modalities that utilize mRNA: (1) replacement therapy, where mRNA is administered to the patient to compensate for a defective gene/protein, or to supply therapeutic proteins; (2) vaccination, where mRNA encoding specific antigen(s) is administered to elicit protective immunity; (3) cell therapy, where mRNA is transfected into the cells ex vivo to alter cell phenotype or function, and then these cells are delivered into the patient.
FIGURE 5.
Schematic illustration of intravenous administration of mRNA encapsulated in lipid nanoparticles (LNP) to restore missing/defective protein in hepatic cells (An et al., 2017). mRNA, messenger RNA; RNA, ribonucleic acid.
mRNA as Replacement Therapy
Currently, there are three major biopharmaceutical companies that develop mRNA therapeutics: Moderna (Boston, MA, United States), CureVac (Tubingen, Germany), and BioNTech (Mainz, Germany). Together, they have a diverse portfolio of gene therapy products in the pipeline, covering metabolic diseases, heart diseases, and immunomodulators for applications in immuno-oncology. For instance, Moderna’s AZD8601 is a naked mRNA encoding vascular endothelial growth factor (VEGF-A) that is intended to be delivered through epicardial injection during coronary artery bypass surgery. The intent is to enhance local angiogenesis, thereby reducing myocardial ischemia and improving left ventricular systolic function (Carlsson et al., 2018) in patients with ischemic heart disease. The efficacy of this drug is being evaluated in a phase II trial conducted by AstraZeneca. Another therapeutic candidate, mRNA-3704 (Moderna) encodes methylmalonyl-CoA mutase and is designed to treat the deficiency of this enzyme that leads to methylmalonic aciduria (Modernatx, 2021a). MRT5005 (Translate Bio) is being developed to treat cystic fibrosis, an inherited disease caused by a mutation in the cystic fibrosis transmembrane conductance regulator (CFTR), a chloride channel. When it is disrupted in the epithelium, there is an accumulation of thickened mucous in different organs, including the pancreas and lungs. MRT5005 encodes fully functional CFTR and is delivered to lung epithelial cells through nebulization (TranslateBio, 2019). Other therapeutics being developed by Moderna include mRNA-2416, mRNA-2752, and MEDI1191, which are mRNA-based immunomodulators. The mRNA-2416 encodes OX40 Ligand (OX40L), which is a co-stimulatory membrane-bound protein that enhances the expansion, function and survival of T cells to mount an attack against cancer cells. When delivered by intratumoral injection (solid tumors/lymphoma/advanced ovarian carcinoma), cells in the tumor may express this ligand on their surfaces, leading to a stronger T cell attack against the tumor cells. Additionally, Moderna is investigating whether mRNA-2416 has the potential to elicit an abscopal effect in metastatic cancer, in which localized injection into a tumor, and cytolytic release of tumor antigens, would generate a secondary immune response and have an effect on metastases throughout the body (Moderna, 2020). Another therapeutic candidate, mRNA-2752, delivers OX40L into the tumors, as well as mRNAs encoding immunostimulatory cytokines IL-23 and IL-36γ to further promote T cell-mediated cytotoxicity. The mRNA drug MEDI1191 is indicated for the solid tumors as well, and encodes IL-12, one of the most potent cytokines in mediating antitumor activity (Tugues et al., 2015).
BioNTech, in collaboration with Sanofi, is evaluating an mRNA-based intratumoral immunomodulator BNT131 (SAR441000) as a monotherapy in patients with advanced melanoma. This immunomodulator is a combination of IL-12sc, IL-15sushi, IFNα, and GM-CSF mRNAs, increased concentration of which in the local tumor microenvironment may promote natural killer cell activation and induce cytotoxic T-cell responses, resulting in an immune-mediated destruction of tumor cells (National Cancer Institute, 2011; Pipeline – BioNTech, 2021). The mRNA drug CV8102 from CureVac is a TLR7/8/RIG1 agonist designed to induce a systemic immune response to the injected primary tumor as well as distant metastases of some solid tumors. This agent is targeted against advanced melanoma, cutaneous squamous cell carcinoma, and squamous cell carcinoma of head and neck (Pipeline – CureVac, 2020).
Although the aforementioned pharmaceutical companies were pioneers in commercialization of mRNA therapeutics, there are many other research groups and biotech start-ups worldwide entering the field. For instance, Connolly et al. (2018) have proposed mRNA-based therapy for α-1-antitrypsin deficiency. This genetic disorder is characterized by destruction of lung tissue by neutrophil elastase due to lack of its natural inhibitor α-1-antitrypsin and by liver damage due to deposition of defective α-1-antitrypsin. The latter is normally produced by the liver to be transported to the lungs, and is encoded by the SERPINA1 gene, a mutation in which leads to defects in transport and activity. Human SERPINA1-encoding mRNA systemically delivered to wild-type mice resulted in target protein expression in both liver and lungs (Connolly et al., 2018).
Cao et al. (2019) recently developed an mRNA drug for citrin deficiency. This disorder is caused by a mutation in the SLC25A13 gene, which encodes citrin, a mitochondrial membrane transport protein involved in the urea cycle. Citrin deficiency leads to hyperammonemia and neuropsychiatric disturbances. Intravenous administration of a human citrin-encoding mRNA in SLC25A13-knockout mice reduced hepatic citrulline and blood ammonia levels following an oral sucrose challenge, and reduced sucrose aversion, a hallmark of citrin deficiency (Cao et al., 2019). Another mRNA drug for a disorder of the urea cycle is being developed by Arcturus Therapeutics. Their mRNA-based therapeutic ARCT-810 encodes ornithine transcarbamylase (OTC) to treat the genetic deficiency of this enzyme and is now in early clinical trials (ClinicalTrials.gov, 2020b). Zhu et al. (2019) reported encouraging data from preclinical studies of an mRNA therapy for Fabry disease. The latter is caused by mutations in the GLA gene that encodes enzyme α-galactosidase, required for utilization of glycolipids. In Fabry disease, glycolipid derivates (globotriaosylceramide, globotriaosylsphingosine) accumulate over time in multiple tissues leading to range of clinical symptoms. A single dose of GLA mRNA intravenously administered in GLA-deficient mice significantly reduced accumulation of globotriaosylsphingosine in plasma and tissues. Of note, this beneficial effect of mRNA was observed for up to 6 weeks after administration (Zhu et al., 2019).
mRNA Vaccines
mRNAs encoding antigens and/or adjuvants can be used as vaccines to evoke protective immunity against infectious diseases (prophylactic vaccines), or to harness the immune system to fight cancer (therapeutic vaccines). The COVID-19 pandemic has showcased the utility and advantages of RNA technology for vaccination, as out of all COVID-19 vaccines under development, the first two to have received emergency use authorization by the FDA were RNA-based. The first authorized vaccine was developed by BioNTech in collaboration with Pfizer. Initial clinical trials conducted in Germany helped to identify two leading vaccine candidates, BNT162b1 and BNT162b2, which were then tested further in the US (Walsh et al., 2020a, b). BNT162b1 encodes secreted trimerized receptor binding domain of spike protein, whereas BNT162b2 encodes membrane-anchored full length spike protein modified by two proline mutations to stabilize the prefusion conformation. Both RNA candidates incorporate 1-methyl-pseudouridine, which dampens innate immune sensing and increases mRNA translation in vivo (Karikó et al., 2008) and are formulated in the lipid nanoparticles (refer to Delivery of RNA therapeutics section below for details). The safety and immunogenicity data of these two vaccine candidates in younger and older adults supported advancement of BNT162b2 into subsequent trials (Mulligan et al., 2020). A large Phase III clinical trial revealed extraordinarily high efficacy (with vaccination preventing about 90% of symptomatic infections) with excellent safety, leading to emergency use authorization by the FDA on December 11, 2020 (FDA, 2021b).
The Moderna RNA vaccine against COVID-19 received emergency use authorization shortly thereafter (December 18, 2020) (FDA, 2021a), based on a large Phase III trial showing at least equal efficacy and safety as BNT162b2. Similarly to BNT162b2, this vaccine encodes stabilized SARS-CoV-2 spike immunogen, and like the BNT mRNA, is delivered intramuscularly as a lipid nanoparticle-encapsulated mRNA (Corbett et al., 2020; Jackson et al., 2020). The development timeline of both vaccines was unprecedented. It was reported that it took merely 2 days after the SARS-CoV-2 genome sequence by Chinese scientists in January 2020 (Novel 2019 Coronavirus Genome, 2020; Wang et al., 2020, p. 2), to select the appropriate sequence for the Moderna vaccine candidate, 25 days to manufacture the first clinical batch of mRNA-1273, and another 35 days to dose the first participant (Jackson et al., 2020; Modernatx, 2021b). Other biotech companies are developing mRNA-based vaccines to address the COVID-19 pandemic. For instance, at the time of this writing, CureVac has initiated a Phase 3 clinical trial of its SARS-CoV-2 vaccine candidate CVnCoV (Pipeline – CureVac, 2020).
The biotechnology response to the COVID-19 pandemic has highlighted the speed and flexibility of mRNA vaccines (Vogel and Sarver, 1995), and reveals mRNA therapeutics to be a powerful tool to address epidemic outbreaks caused by newly emerging viruses. The relative simplicity of the development process and flexibility of the manufacturing platform can markedly accelerate clinical development (Liu, 2019). As such, mRNA-based vaccine technology has attracted a lot of attention during the COVID-19 pandemic (Le et al., 2020; Lurie et al., 2020). Unlike viral vector-based vaccines, mRNA vaccines are not confounded by pre-existing immunity against the vector (Ura et al., 2014; Condit et al., 2016). They also do not need to enter the nucleus for translation, unlike plasmid DNA vaccines that require nuclear localization to elicit their protective effects (Ulmer et al., 1993; Ledwith et al., 2000).
RNA vaccines can activate both cell-mediated and humoral immunity. For instance, Martinon et al. (1993) demonstrated induction of anti-influenza cytotoxic T lymphocytes in vivo by immunizing animals with subcutaneous injection of liposome-encapsulated mRNA, which encoded the nucleoprotein of the influenza virus. Petsch et al. (2012) evaluated effects of a multi-valent mRNA vaccine (hemagglutinin, neuraminidase, and nucleoprotein) against influenza A H1N1, H3N2, and H5N1 strains in mice, ferrets, and pigs. This intradermally injected mRNA vaccine induced antigen-specific neutralizing antibodies and protected the animals from influenza A virus (Petsch et al., 2012). Similarly, the intramuscularly administered vaccines against avian influenza A viruses H10N8 and H7N9 have been shown to be safe and immunogenic in first-in-man trials conducted by Moderna (Feldman et al., 2019). Moderna is currently evaluating vaccine candidates for other infectious respiratory diseases: respiratory syncytial virus (mRNA-1172, mRNA-1777) and metapneumovirus/parainfluenza 3 (mRNA-1653).
The protective effects of mRNA vaccines have been also proven useful beyond respiratory pathogens. For example, Schnee et al. (2016) demonstrated the potency of mRNA vaccines against rabies in rodents and pigs. This vaccine against a viral glycoprotein induced antigen-specific immune responses in vivo (Schnee et al., 2016). Notably, specific CD4+ T cells induced by the mRNA vaccine were higher than those induced by a licensed vaccine, and the titers of neutralizing antibody in mice remained stable for the entire observation period (up to 1 year). The safety, reactogenicity and immune response of this vaccine is currently being evaluated in a phase I trial conducted by CureVac (CV7202) (ClinicalTrials.gov, 2020c). Pardi et al. (2017) proposed a bi-valent modified mRNA vaccine that encodes pre-membrane and envelope glycoproteins of a Zika virus strain responsible for the outbreak in 2013. A single dose of this vaccine, encapsulated in lipid nanoparticles and delivered intradermally, was sufficient to protect mice against viral challenges at 2 weeks or 5 months after vaccination, and non-human primates at 5 weeks after vaccination (Pardi et al., 2017). Utilizing the same antigens, Moderna has been developing a non-modified encapsulated mRNA-1893 vaccine against Zika, which received fast track FDA designation and is currently in phase I trials (ClinicalTrials.gov, 2020f) to assess its safety, tolerability and immunogenicity. Importantly, mRNA-1893 had prevented congenital transmission of the virus in mouse models of congenital infection (Jagger et al., 2019). John et al. designed a cytomegalovirus (CMV) vaccine to prevent CMV infection and/or disease during pregnancy and in transplant patients. The vaccine consists of six modified mRNA constructs encoding CMV glycoproteins and pentameric complex and is delivered intramuscularly in lipid nanoparticles. Its single dose elicited robust immune response in mice and non-human primates (John et al., 2018), and is currently in clinical trials (mRNA-1647) sponsored by Moderna (ModernaTX Inc, 2019a).
In addition to mounting active immunity, mRNA can be also used for passive immunization. A drug candidate mRNA-1944 (Moderna) is a great example of mRNA therapeutics encoding human monoclonal neutralizing antibodies (mAb). This drug is designed to provide passive protection against chikungunya infection. The ultrapotent antibodies were isolated from the B cells of a survivor of natural infection, and their sequences were encoded into mRNA molecules, encapsulated in lipid nanoparticles and delivered by infusion into mice. After mRNA delivery, one human mAb, CHKV-24, was found to be expressed at immunologically relevant levels, and its protective capacity was evaluated in mouse models of chikungunya. Treatment with CHKV-24 mRNA reduced viremia to undetectable levels at 2 days after inoculation and protected mice from mortality. Further studies with non-human primates also demonstrated a long-lasting immunogenic effects of the drug (Kose et al., 2019). Overall, the pre-clinical data encouraged a first-in-man trial (ModernaTX Inc, 2019b).
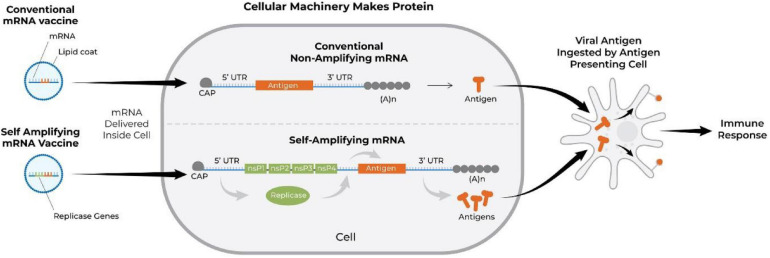
Another interesting RNA-based approach to vaccination is that with self-amplifying RNA vaccines. The backbone sequence for the latter is adapted from an alphavirus, a positive-sense single-stranded RNA virus with high capacity for replication (Lundstrom, 2018b; Vogel et al., 2018). Such mRNA vaccine contains an antigen-encoding sequence and viral RNA dependent RNA polymerase-encoding sequence along with other elements required for replication (Hyde et al., 2015). The advantage of the self-replicating approach is that significantly higher amount of antigen can be expressed with lower doses of mRNA (Vogel et al., 2018). Both types of RNA vaccines degrade after transient expression; however, the self-replicating RNA achieves longer term expression of the antigen (Ulmer and Geall, 2016). Figure 6 demonstrates the mode of action of conventional and self-amplifying mRNA vaccines (Kowalski et al., 2019). Harvey et al. (2003) were among the first to demonstrate the ability of self-amplifying mRNA vaccines to elicit antigen-specific immunity. They constructed vaccine vectors based on replicon RNA of the Australian flavivirus Kunjin that encoded HIV-1 Gag antigen and delivered them intramuscularly as either naked RNA or in the form of virus-like particles in BALB/c mice. Such immunization induced both Gag-specific antibody and protective Gag-specific CD8+ T-cell responses (Anraku et al., 2002; Harvey et al., 2003). Similarly, Moyo et al. (2018) engineered a vaccine against HIV-1 using an RNA replicon derived from Semliki Forest virus, which encoded 6 highly conserved regions of HIV-1 proteins. The self-replication approach was also used by McKay et al. (2020) (Imperial College London) to develop another mRNA COVID-19 vaccine which encodes the pre-fusion stabilized SARS-CoV-2 spike protein, which vaccine has now entered a phase I trial.
FIGURE 6.
Schematic showing the mechanism of action of conventional (BioNTech and Moderna COVID-19 vaccines) and self-amplifying mRNA vaccines. The mRNA vaccine is translated into protein, processed by antigen presenting cells, and subsequently activates immune responses. nsP, non-structural protein; Cap, N7-methylated guanosine structure covalently joined to the first nucleotide of the mRNA through a reverse 5′ to 5′ triphosphate linkage (Kowalski et al., 2019; Dolgin, 2021); A, adenosine molecule; (A)n, poly-A tail; UTR, untranslated region.
There are multiple RNA cancer vaccines in clinical trials such as CV9202 (CureVac), which is a self-adjuvating RNA vaccine targeting six antigens commonly expressed in non-small cell lung cancer (Sebastian et al., 2014). The mRNA-5671 (Moderna) targets KRAS and is being currently evaluated in patients with KRAS-mutant advanced or metastatic non-small cell lung cancer, colorectal cancer, or pancreatic adenocarcinoma (ClinicalTrials.gov, 2020e). The mRNA-4157 is another cancer vaccine from Moderna, but, unlike mRNA-5671, it is an individualized therapeutic vaccine against melanoma. In this approach, genetic sequencing and bioinformatic analysis is performed on a patient’s tumor to identify 20 patient-specific neoantigen epitopes which are encoded by an mRNA construct manufactured for a single patient (National Cancer Institute, 2011; ClinicalTrials.gov, 2020a; van Dülmen and Rentmeister, 2020). Another example of the individualized cancer vaccine in clinical trials is BNT122 (phase II) from BioNTech, which is designed for locally advanced or metastatic solid tumors (including melanoma, non-small cell lung cancer, bladder cancer, and others). BioNTech has many other anti-cancer vaccines in their pipeline, with the following ones having reached clinical studies: BNT111 for advanced melanoma; BNT112 for metastatic castration-resistant prostate cancer and high-risk localized prostate cancer; BNT113 against HPV16-derived oncoproteins E6 and E7 found in HPV16-positive solid cancers, such as head and neck squamous cell carcinoma; BNT114 for triple negative breast cancer; and BNT115 for ovarian cancer (Pipeline – BioNTech, 2021).
mRNA-Enhanced Cell Therapies
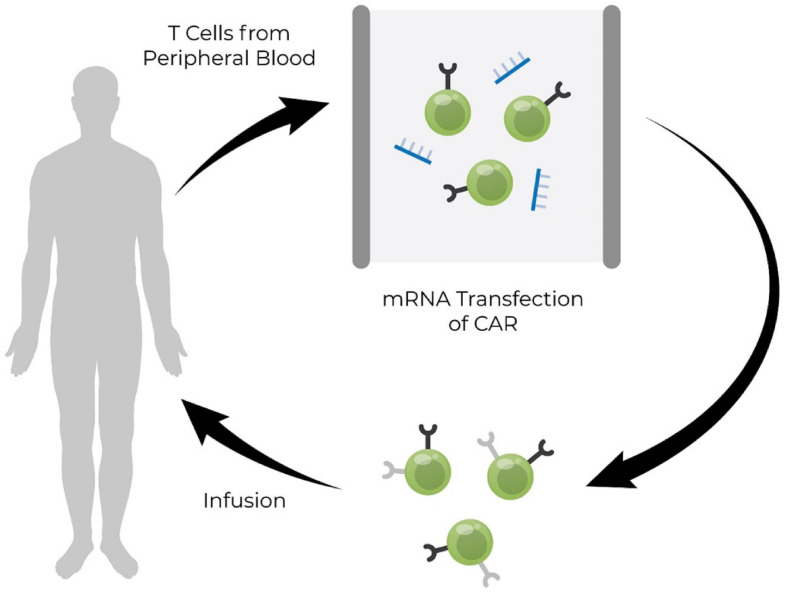
Cell therapy may be enhanced by mRNA. The cells may be obtained from the patient or the cell bank. Subsequently, they are therapeutically modified ex vivo with mRNA encoding the desired proteins (such as CARs, reprogramming or transdifferentiation factors, telomerase, etc.). Then the mRNA-enhanced cells are re-infused into the patient to treat the disease (see Figure 7, Bertoletti and Tan, 2020). Currently, there are several cell-based therapies that have reached clinical trials and employ mRNA. For example, TriMix-based immunotherapy (ECI-006) is a combination of mRNAs encoding DC-activating molecules (CD40L, CD70, and caTLR4) and melanoma-specific tumor-associated antigens (tyrosinase, gp100, MAGE-A3, MAGE-C2, and PRAME) (Arance Fernandez et al., 2019, p. 011) that are transfected into autologous DCs ex vivo. This therapy has demonstrated significant clinical activity in combination with ipilimumab without increasing regimen toxicity in metastatic melanoma (De Keersmaecker et al., 2020). MCY-M11 (MaxCyte) is an autologous CAR-T cell therapy for mesothelin-expressing solid tumors. The peripheral blood lymphocytes from a single leukapheresis are transfected with anti-human mesothelin mRNA CAR and cryopreserved as multiple cell aliquots for repeat administrations (Hung et al., 2018). Descartes (Cartesian Therapeutics) is an autologous cell therapy, where anti-BCMA CAR T-cells are engineered ex vivo by transient expression of mRNA, to treat myasthenia gravis and relapsed/refractory multiple myeloma (SparkCures, 2020). Although mRNA in aforementioned therapies is used to achieve transient expression of proteins, it can also be designed to serve as a gene editing tool to achieve stable expression of target proteins.
FIGURE 7.
Schematic illustrations of the use of mRNA in engineering cells ex vivo. T cells derived from peripheral blood of patients suffering from disease are modified ex vivo with mRNA expressing the chimeric antigen receptor (CAR) and then modified cells are reinfused into the patient (Bertoletti and Tan, 2020). mRNA, messenger RNA; RNA, ribonucleic acid.
It is also possible to combine mRNA and DNA into a therapeutic. In one approach, a transposon system is used to genetically modify cells for therapy. This system consists of a DNA plasmid which encodes a gene of interest flanked by a mirrored set of inverted repeats (transposon), together with mRNA encoding the transposase enzyme. The plasmid DNA encoding the transposon is co-delivered along with the mRNA transposase enzyme in a single electroporation reaction. The transposase that is translated from the mRNA then binds to the inverted repeats and cuts the DNA, to release the transposon. After that, the transposon binds a strand of genomic DNA with a TA dinucleotide, where the transposase creates a double-stranded break, allowing the transposon to integrate (Singh et al., 2014). Using this approach, Poseida Therapeutics has developed a cell therapy for multiple myeloma, P-BCMA-101, which is now in clinical trials. The company’s proprietary mRNA-based transposon system is used to integrate a transposon encoding the anti-BCMA CAR into resting T cells (Singh et al., 2014, p. 19; Ostertag, 2018).
Delivery of RNA Therapeutics
Targeted delivery is a major hurdle for effective RNA Therapeutics, a hurdle that must be overcome to broaden the application of clinical translation of this type of therapeutic. Because mRNA is inherently unstable it requires delivery vehicles that will protect the cargo from RNAase degradation. There is a need for novel delivery vehicles that will deliver the RNA drug to the site of therapeutic action facilitating the entry of the RNA drug into the cytoplasm where it may exert its effect. In the following paragraphs we provide a brief review of the advances made in RNA delivery vehicles.
Lipid-Based Nanoparticles
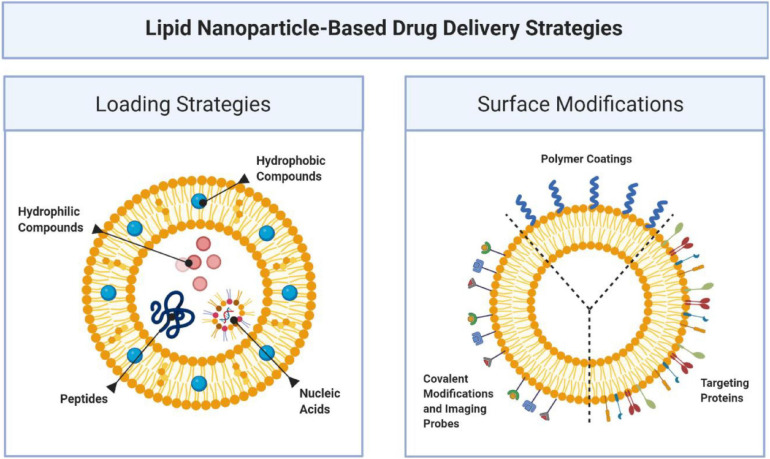
Liposomes are formed when materials containing polar head groups and non-polar tails (phospholipids) are dispersed in aqueous phase. They are spherical vesicles consisting of at least one phospholipid bilayer enclosing an aqueous core. They are a flexible drug delivery particle which may have various surface modifications (see Figure 8, Sushnitha et al., 2020; Zinger et al., 2020) capable of delivering a variety of therapeutic payloads (Torchilin, 2005). Most notably amongst these application is Doxil [Barenholz (Chezy), 2012] a liposome encapsulated formulation of doxorubicin for the treatment of cancer, that reduces heart toxicity related to the drug. This breakthrough treatment demonstrated the efficacy of the use of nanoparticles to change the biodistribution of drug and increase its safety.
FIGURE 8.
Lipid based nanoparticles accommodate a variety of therapeutic payloads including hydrophobic and hydrophilic small molecules, genetic materials, and proteins. One of the advantages of this platform is its ability to functionalize the surface with various proteins that can serve to target and localize nanoparticles over specific targets, imaging probes or covalent modification (Sushnitha et al., 2020; Zinger et al., 2020).
Liposomes have an inherent advantage in that they mimic cell membrane composition and can encapsulate mRNA when combined with cationic lipids. Positively charged lipids can electrostatically interact with negatively charged mRNA to form complexes of RNA and liposomes. In this way, RNA is encapsulated within liposomes. Cationic lipids such as, DOTMA (1,2-di-O-octadecenyl-3-trimethylammonium-propane) and DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) readily form complexes with negatively charged RNA. The use of cholesterol modified lipid makes the resulting complex more stable and improves transfection. Hattori et al. (2015) demonstrated the use of cationic cholesterol modified liposome to deliver siRNA into mouse liver. Subsequently, Kranz et al. (2016) demonstrated the use of liposomes for intravenous delivery of mRNA vaccines encoding four tumor antigens NY-ESO-1, MAGE-A3, tyrosinase and TPTE targeting dendritic cells (DCs) in patients with advanced malignant melanoma, effectively expressing the antigens in spleen cells. However, the liposome delivery systems have flaws; (1) liposomes are less stable, and may fuse or leak RNA resulting in low efficiency of delivery; (2) they entrap less RNA; (3) they can be harmful if oxidized; and (4) are not consistent in size. The heterogeneity of the particles increases batch to batch variability (Laouini et al., 2012; Sercombe et al., 2015; Shin et al., 2018).
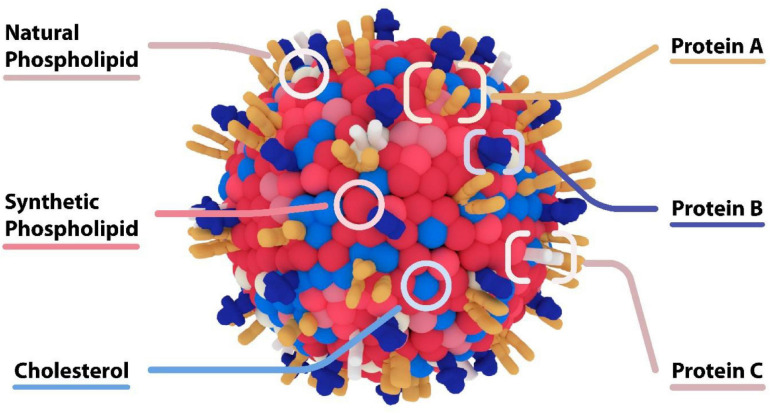
Some of the aforementioned hurdles have been addressed with advances in the surface modifications of liposomes. Lipid nanoparticles (LNP) made of cationic and other lipids, cholesterol, and polyethylene glycol (PEG) with a hydrophilic inner core retain the capacity to carry anionic RNA, protect it from degradation and prolong its circulation. An example of the efficacy of the LNP platform is patisiran (Onpattro, Alnylam Pharmaceuticals, FDA approval August 2018), the siRNA-based therapeutic against hereditary transthyretin-mediated amyloidosis discussed above. In patisiran, the dsRNA is encapsulated inside four lipid excipients (Kristen et al., 2018): DSPC [1,2-distearoyl-sn-glycero-3-phosphocholine], cholesterol (DLin-MC3-DMA) [(6Z,9Z,28Z,31Z)-heptatriaconta-6,9,28, 31-tetraen-19-yl-4-(dimethylamino) butanoate] and PEG2000-C-DMG [α-(3′-{[1,2-di(myristyloxy)proponoxy] carbonyl-amino}propyl)-ω-methoxy, polyoxyethylene] (Zhang et al., 2020). The PEG functionalization is crucial for this platform as it allows enough circulation time for the drug to be localized in the liver.
Another example is PEG-liposome mRNA vaccines developed by BioNTech and Moderna for the treatment of COVID-19. As described above, these vaccines encode the prefusion-stabilized SARS-CoV-2 spike protein. Minor concerns have been raised as the proprietary lipid is cleared slowly from target tissues and studies are ongoing if accumulation of this lipid could represent a safety concern. The promising results have been seen in non-human primates and human, and, at the time of the writing of this manuscript, this formulation has been approved by FDA for emergency use. Another concern was raised by the rare anaphylactic reactions to the RNA vaccines, which have been attributed to the PEG component of the LNPs (Castells and Phillips, 2021).
Beyond PEG-functionalized liposomes, more sophisticated surface modalities have emerged to create “biomimetic” nanoparticles that functionalized membrane proteins on the surface of nanoparticles (see Figure 9). We have generated “leukosomes” by incorporating leukocyte membrane proteins into the surface of liposomes (Molinaro et al., 2016). These particles have the virtue of localizing at sites of inflammation, binding to the activated endothelium via endothelial adhesion molecules such as LFA-1 and CD-45 (Molinaro et al., 2016). Furthermore, this biomimetic platform has shown intrinsic anti-inflammatory effects upon endothelium through its interaction with macrophages. In a lipopolysaccharide (LPS) induced murine model of sepsis, administration of biomimetic nanoparticles derived from macrophages decreased pro-inflammatory genes (IL-6, IL-1b, and TNF-α), and increased anti-inflammatory genes (IL-10 and TGF-β) (Molinaro et al., 2019). The intrinsic anti-inflammatory effects of these biomimetic leukosomes have been demonstrated in other disease models including inflammatory bowel disease (IBD) (Corbo et al., 2017), atherosclerosis (Martinez et al., 2018; Boada et al., 2020), and cancer (Martinez et al., 2018). Most recently, we have shown the capability of leukosomes to home to sites of vascular inflammation in the apo E deficient hypercholesterolemic mouse. In this model, the elevated levels of cholesterol cause the accumulation of lipid and macrophages in the aorta. Leukosomes were more efficient than standard LNPs at delivering rapamycin to the aorta, where the rapamycin inhibited macrophage proliferation and generation of inflammatory cytokines (Boada et al., 2020). While significant work remains to be done testing the efficacy of this platform in larger animal models, there is great promise for biomimetic nanoparticles to create new mRNA therapeutics that has the potential to selectively target all the inflammatory-based conditions.
FIGURE 9.
Biomimetic Nanoparticles – “Leukosome” Technology. Biomimetic nanoparticles developed by Molinaro et al. (2016) “Leukosomes.” The composition of these particles consists of a liposome together with proteins derived from leukocytes. These particles preferentially adhere to activated endothelium at sites of inflammation, and have intrinsic anti-inflammatory properties.
Polymer Nanomaterials
Polymer nanomaterials normally refer to synthetic compounds made of a handful of base units that come together to form complex structures. These materials usually include synthetic polymers such as PLGA [ploy(lactic-co-glycolic acid)], PLA (polylactic acid), chitosan, gelatin, polycaprolactone, and poly-alkyl-cyanoacrylates. These materials have the virtue of a long shelf life; the ability to encapsulate hydrophilic and hydrophobic compounds and proteins; and the capability for tuned delivery of therapeutic compounds (Devulapally et al., 2016; Amini et al., 2017). The flexibility inherent in polymeric nanomaterials is due to the fact that small chemical modifications of the basic polymeric units permits exquisite control over release profile. Polymers can be synthesized to create injectable nanoparticles that can be delivered as intravenous injections or administered as intramuscular, subdermal or intraperitoneal drug depots that degrade over a period of months or weeks (Molina et al., 2015).
The ease of encapsulation extends to nucleic acids. Cationic hydrophilic polymers with hydrophobic modification can self-assemble in aqueous phase to encapsulate RNA. An example of this was employed by Zhao et al. (2016), who developed cationic polyethyleneimine-stearic acid (PSA) copolymer to deliver HIV-1 gag encoding mRNA to dendritic cells (DC2.4 cells) and BALB/c mice. The optimum mass ratio of PSA: mRNA was 4:1, as assessed by transfection efficiency. They injected 6–8 weeks old female mice (BALB/c) subcutaneously with HIV-1 gag mRNA encapsulated in PSA and detected levels of anti-HIV-1 gag specific antibodies. They also demonstrated HIV gag specific CD8+ and CD4+ T cell responses after immunization with PSA/mRNA vaccine (Zhao et al., 2016). McCullough et al. (2014) used chitosan nanoparticles to deliver self-amplifying replicon RNA encoding influenza virus hemagglutinin and nucleoprotein to dendritic cells in mice and rabbit (McCullough et al., 2014). Andreozzi et al. (2017) explored the pH sensitivity of their nucleic acids carrier to trigger endosomal delivery and found that the carrier is stable at narrow pH range (7–9). They used poly(allylamine) phosphate supramolecular nanocarriers to deliver green fluorescence protein (GFP) siRNA in A549 cells to effectively silence the expression of GFP protein.
Consideration must also be given to the intrinsic tendency of polymers to induce inflammation within the immediate microenvironment, due either to lack of degradation such as in the case of PLA or through byproducts. In the case of PLGA, degradation into its basic monomers, lactic acid and glycolic acid leads to a decrease in pH at sites of degradation, promoting inflammation (Kumari et al., 2010). This is not an insurmountable problem. One popular and practical solution has been the use of lipid-polymer hybrids pioneered by Zhang et al. (2008). This type of material, a nanoparticle with a lipid surface and a polymer core, combines the best of both worlds and provides an exquisite control over release profile from its polymer core and lipid surface that better resembles the cell membrane, with possibility for modification that can further enhance targeting toward diseased tissues. Application of this material to deliver RNA therapeutics is promising based on pre-clinical results with xenograft tumors (Shi et al., 2011). The combination of lipid and polymer permits combination therapy. In a landmark study, (Desai et al., 2013) delivered anti-TNFα siRNA (siTNFα) together with capsaicin, in a psoriatic plaque murine model. Their polymer nanoparticle was able to penetrate the cutaneous barrier to deliver the therapeutic interference RNA. Given these pre-clinical results the use of polymer-based materials is extremely promising for delivery of RNA therapeutics (Hadinoto et al., 2013).
Silica Nanoparticles
Mesoporous silica nanoparticles (MSNPs) have gained more attention for their therapeutic applications. The nanoparticles consist of an amorphous silica (silicon dioxide) matrix with ordered porosity in the mesoporous range. The peculiar feature of this nanoparticle includes large surface areas with large pore volumes, ease of modification, and established silanol chemistry. The surface of the nanoparticles can be modified by positively charged moieties to transport negatively charged RNA. The MSNPs pore sizes are tunable with a broad range and can be quite uniform. The particles have high loading capacity for nucleic acids and efficient delivery. RNA can be loaded inside pores for their transport and the surface can be modified with cancer specific ligands and antibodies to deliver the RNA to the target site. Particle aggregation is a hurdle that must be overcome so that the product is safe for injection. Aggregation can cause thrombosis or induce tissue injury. However, PEGylation of the surface of the nanoparticles greatly decreases aggregation and tissue damage. These nanoparticles degrade into non-toxic products and are safely excreted (Möller et al., 2016; Paris and Vallet-Regí, 2020). Meka et al. (2016) developed the MSNPs with large pore and bicontinuous cubic mesostructure to deliver siRNA in human colon cancer cells (HCT116). They demonstrated the high efficiency of the silica particles in drug delivery and suppression of the tumor proteins in their work (Meka et al., 2016). Lee et al. (2018) developed hybrid particles with a positive charge structure with large pores to load anionic siRNA drug (knockdown B cell lymphoma 2, Bcl-2) and a negatively charged structure with small pores to load the anticancer drug, doxorubicin. They used their hybrid particles to deliver siRNA and doxorubicin to HeLa cells. Their work demonstrated the flexibility of this platform to provide dual pore hybrid silica nanoparticles to treat cancer with combination of both genetic and chemotherapeutic drugs simultaneously (Lee et al., 2018).
Carbon and Gold Nanomaterials
Gold nanoparticles, quantum dots, nanographene oxide, carbon nanotubes are each synthesized nanostructures that have the capacity to harbor RNA, protecting it from degradation and delivering it to the targeted disease site. Zheng et al. (2012) used gold nanoparticles for topical application of therapeutic nucleic acids (siRNA, knockdown EGFR) in mouse and human skin (Zheng et al., 2012). Li et al. (2011) used quantum dots-siRNA complexes to suppress a target gene (HPV18 E6 gene) in HeLa cells. Yang et al. (2014) utilized nanographene oxide modified with gadolinium(Gd-NGO) to deliver a small molecule anti-cancer drug, epirubicin together with Let-7g miRNA (tumor suppressors, decrease expression of the Ras oncogene family) to image and treat glioblastoma in mice (Yang et al., 2014). Kam et al. (2005) observed that functionalized carbon nanotubes could deliver siRNA against lamin A/C to suppress the expression of this protein in HeLa cells.
N-Acetylgalactosamine (GalNAc)
GalNAc is a trivalent ligand that binds to asialoglycoprotein (ASGPR) receptors in hepatocytes. Clinical studies suggest that GalNAc conjugated siRNAs are very efficient to knockdown gene expression in the liver. Zimmermann et al. (2017) performed the first clinical trial of GalNAc conjugated siRNA drugs (Revisuran) to treat transthyretin-mediated amyloidosis (ATTR) (Zimmermann et al., 2017). However, an imbalance in deaths of the Revisuran siRNA treated patients in phase III clinical trials terminated the study of this drug. Givosiran (Givlaari, developed by Alnylam Pharmaceuticals) is the world’s first ever GalNAc conjugated siRNA drug approved by FDA (November, 2019) (Alnylam, 2020b). As discussed above, Givosiran is used to prevent acute attacks of hepatic porphyria by silencing the expression of aminolevulinate synthase 1 mRNA in liver. This has the effect of reducing neurotoxic levels of aminolevulinic acid and porphobilinogen that can induce seizures, paralysis, respiratory failure, neurological damage and death (Balwani et al., 2020; Nature Biotechnology, 2020). There are few more GalNAc conjugated siRNA drugs (Fitusuran, Lumasiran, Vutrisiran, and Inclisiran) in Phase 3 clinical trials from Alnylam Pharmaceuticals (Alnylam, 2020a).
Hospital-Based RNA Therapeutics
We are witnessing the dawn of a new era of biopharmacotherapy. The promise of mRNA therapeutics has galvanized the pharmaceutical companies and investment sector, as evidenced by the current market caps of Biontech ($22.4B), Curevac ($18.6B), and Moderna ($52B), as well as the rapid proliferation of biotech start-ups in this field (FierceBiotech, 2020). Each of the three major companies has dozens of new mRNA therapeutics under development, some of which have been mentioned in this review. Inherent advantages of mRNA therapeutics have spurred this unprecedented investment. High-purity RNA constructs can be generated much faster than traditional small molecule drugs or recombinant proteins, and at lower costs; the manufacturing process is adaptable to any RNA sequence allowing for personalized RNA therapeutics; moreover, RNA has a superior safety profile and simpler regulatory roadmap compared to DNA-based gene therapy because it doesn’t integrate into the host genome (Sahin et al., 2014; Karikó, 2019). The rapid growth of mRNA therapeutics has been made possible by recent advances which have overcome key obstacles such as innate immune activation, RNA stability, and delivery.
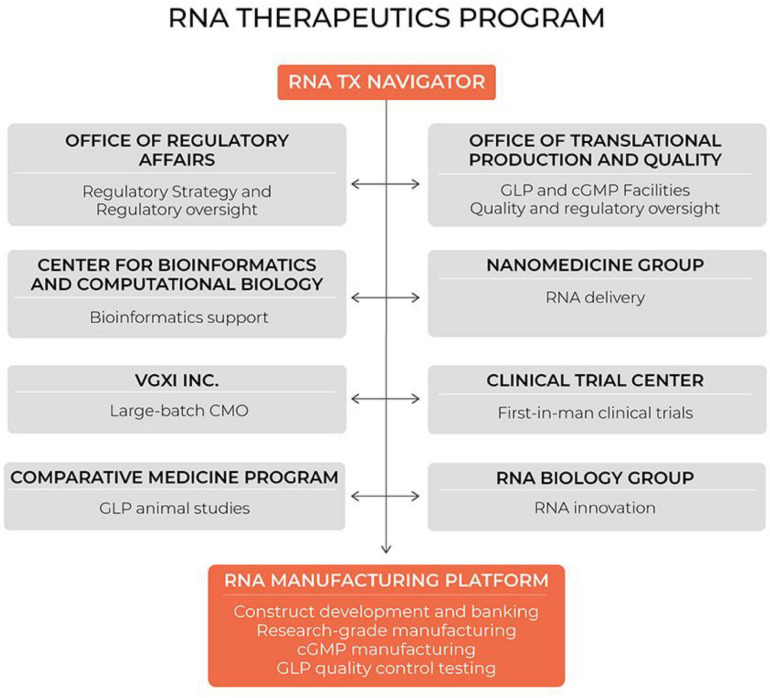
Notably, mRNA therapeutics are a disruptive therapeutic technology, as small biotech startups, as well as academic groups, can rapidly develop new and personalized mRNA constructs. Our group has long-standing expertise in designing and manufacturing RNA therapeutics for the scientific community. Initially we were funded by the NHLBI to serve as an mRNA core for stem cell investigators in the Progenitor Cell Biology Consortium. Subsequently, the Cancer Prevention and Research Institute of Texas funded us to address an unmet need for cancer researchers in Texas who use mRNA as a research tool or a therapeutic. The demand for our services has increased substantially within the last 5 years, while we strengthened our expertise through extensive collaboration with cancer biologists, immunologists, RNA biologists, bioinformaticians, and nanomedicine scientists. During this time we established our hospital-based cGMP-compliant manufacturing capability and quality control methods (see Figure 10). We licensed our proprietary processes to VGXI Inc, a local company with large-batch manufacturing capabilities for DNA-based gene therapies. With this collaboration, we built a seamless transition for academic groups and small companies to go from pre-clinical development and first-in-man clinical trials supported by our hospital-based program, to late-stage clinical trials and commercialization supported by our industry partner VGXI, Inc.
FIGURE 10.
Hospital-based RNA therapeutics (TX) program in Houston Methodist. CMO, contract manufacturing organization; GLP, good laboratory practice; cGMP, current good manufacturing practice.
During these years we learned that many small biotech companies and academic groups, who have innovative ideas for disruptive RNA therapeutics, lack key competencies to reach the clinic, such as manufacturing capabilities, delivery technologies, or regulatory expertise, while our group has been operating in the environment established to accelerate novel therapeutics from conception to the clinic. Houston Methodist Research Institute is adjacent to the Houston Methodist Hospital, which is ranked by US News and World Report as the No. 1 hospital in Texas. The Institute was established with the focus on clinical translation. As such, the Institute has the infrastructure necessary to support pre-clinical and clinical development of novel therapeutics. We have clean rooms and calibrated equipment for cGMP manufacturing of RNA constructs, as well as the equipment to synthesize and analyze lipid nanoparticles for RNA encapsulation. We have an Office of Translational Production and Quality that provides guidance and quality oversight for manufacturing, QC testing and release. In addition, our Office of Regulatory Affairs assists with regulatory roadmaps and translational research planning. Our Comparative Medicine Program supports GLP animal studies for IND applications, while our Cockrell Clinical Trial Center designs and implements first-in-man clinical trials. This infrastructure has facilitated our development of the first fully integrated hospital-based RNA therapeutics program.
Our RNA core is the central pillar, as the RNA core team develops new constructs and manufactures research or clinical grade RNA. The RNA core is complemented by RNA biologists, bioinformaticians, and nanomedicine experts. This program offers a single entry point with consultation to ensure a seamless transition between development stages. This system facilitates the development of RNA therapeutics by academic groups and small biotechs and dramatically shortens the time “from bench to bedside.” Furthermore, it promotes collaboration between our RNA faculty and clinician-scientists at the hospital to identify unmet clinical needs and develop mRNA-based solutions.
The success of hospital-based programs to generate novel nucleic acid drugs has already been demonstrated. For instance, the investment of Nationwide Children’s hospital into cGMP facilities and a critical mass of gene therapy experts led to the creation of transformative gene therapies for lethal childhood diseases: SMA (onasemnogene abeparvovec xioi; Zolgensma, reviewed in DNA therapies) and DMD (golodirsen; Vyondys 53). Golodirsen is a phosphorodiamidate morpholino oligomer that hybridizes to exon 53 of dystrophin pre-mRNA, and restores the mRNA reading frame in patients with confirmed DMD mutations amenable to exon 53 skipping (Frank et al., 2020). Besides golodirsen, the group is currently evaluating another treatment for DMD – a gene transfer of micro-dystrophin delivered by recombinant AAV and driven by a skeletal and cardiac muscle-specific promoter with enhanced cardiac expression (Mendell et al., 2020).
Another example of a successful hospital-based gene therapy program is that of St. Jude Children’s Research Hospital. As described above, they developed and generated material in house for first-in-man studies of a lentiviral gene therapy for X-linked SCID. Their lentiviral vector encoding IL2RG was designed to include insulators to block activation of genes adjacent to where IL2RG is inserted into the patients’ DNA. The goal was to prevent the gene therapy from inadvertently causing leukemia by switching on an oncogene in the patient’s blood stem cells. Vector production and gene therapy treatment were streamlined using a stable producer cell line and cryopreservation. Combined with low-exposure, targeted busulfan conditioning in infants with newly diagnosed SCID-X1, the therapy had only low-grade adverse effects and resulted in multilineage engraftment of transduced cells, reconstitution of functional T cells and B cells, and normalization of NK-cell counts during a median follow-up of 16 months (Mamcarz et al., 2019).
In addition to accelerating clinical translation of novel therapeutics, hospital-based programs facilitate development and implementation of personalized medicines, when relatively small quantities of a drug are required in treating a single patient. While manufacturing of the drug at such scale is rarely financially justified for a big pharma, it is quite reasonable for a hospital, can be done within a hospital’s cGMP facilities and then quickly delivered to the patient. An outstanding example of patient-customized treatment was recently demonstrated by Boston Children’s Hospital. In patient presenting with symptoms diagnostic of Batten’s disease, genetic testing for known mutations were not observed. Whole-genome sequencing revealed a previously unknown insertion that altered splicing of the MFSD8 gene and led to premature translational termination. Knowing this, the investigators designed a 22-mer ASO, milasen, with the same backbone and sugar chemistry modifications as nusinersen (reviewed in ASO section) to correct the misplicing and restore MFSD8 expression. Soon after pre-clinical studies to confirm efficacy and assess toxicity studies, the drug was administered to the patient with clinical improvement observed (Kim et al., 2019). Notably, the first injection of milasen was done within 1 year after first contact with the patient, a testament to the speed of this hospital-based gene therapy platform.
Conclusion and Future Perspectives
RNA Therapeutics is a rapidly emerging field of biotherapeutics. These therapies are based upon powerful and versatile platform which has nearly unlimited capacity to address unmet clinical needs. RNA Therapeutics are destined to change the standard of care for many diseases. The number of RNA drugs under development, and in clinical trials, is growing rapidly. The rapid growth of RNA therapeutics has been due to solving the problems of stability, delivery and immunogenicity. Whereas there is room for further improvement and innovation in each of these areas, the solutions have advanced to the point that RNA Therapeutics are now feasible. Although several dominating players in the RNA biopharma sector have emerged, new small biotech startups as well as academic groups with transformative ideas are propagating. In addition, hospital-based RNA therapeutics programs will facilitate RNA-based drug development and accelerate translation of transformative therapies from lab bench to patient’s bedside.
Author Contributions
TRD: first draft and illustrations. RS, CB, FT, and RIP: subsequent revisions of manuscript. JPC: revisions of the manuscript and funding of effort. All authors contributed to the article and approved the submitted version.
Conflict of Interest
Houston Methodist Hospital has been assigned intellectual property related to the synthesis, purification, validation, and delivery of nucleic acid therapeutics. JPC is an inventor on issued patents related to mRNA telomerase therapy, which have been assigned to Stanford University and licensed to his company. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Supported by funding from the National Institutes of Health (R01HL133254, R01HL148338, and R01GM125632); Cancer Prevention and Research Institute of Texas (RP150611 and RP200619); George and Angelina Kostas Research Center for Cardiovascular Medicine; as well as the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation.
References
- Alnylam (2020a). Alnylam® Development Pipeline of Investigational RNAi Therapeutics Alnylam. Available online at: https://www.alnylam.com/alnylam-rnai-pipeline/ (accessed January 3, 2020). [Google Scholar]
- Alnylam (2020b). Alnylam Announces Approval of GIVLAARITM (givosiran) by the U.S. Food and Drug Administration (FDA) Alnylam Pharm. Inc. Available online at: http://investors.alnylam.com/news-releases/news-release-details/alnylam-announces-approval-givlaaritm-givosiran-us-food-and-drug (accessed January 3, 2020). [Google Scholar]
- Amadio M., Govoni S., Pascale A. (2016). Targeting VEGF in eye neovascularization: What’s new?: a comprehensive review on current therapies and oligonucleotide-based interventions under development. Pharmacol. Res. 103 253–269. 10.1016/j.phrs.2015.11.027 [DOI] [PubMed] [Google Scholar]
- Amgen (2020). FDA Approves IMLYGIC Talimogene Laherparepvec As First Oncolytic Viral Therapy In The US Amgen Inc. Available online at: http://www.amgen.com/en/media/news-releases/2015/10/fda-approves-imlygic-talimogene-laherparepvec-as-first-oncolytic-viral-therapy-in-the-us (accessed August 18, 2020). [Google Scholar]
- Amini Y., Amel Jamehdar S., Sadri K., Zare S., Musavi D., Tafaghodi M. (2017). Different methods to determine the encapsulation efficiency of protein in PLGA nanoparticles. Biomed. Mater. Eng. 28 613–620. 10.3233/BME-171705 [DOI] [PubMed] [Google Scholar]
- An D., Schneller J. L., Frassetto A., Liang S., Zhu X., Park J.-S., et al. (2017). Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 21 3548–3558. 10.1016/j.celrep.2017.11.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreozzi P., Diamanti E., Py-Daniel K. R., Cáceres-Vélez P. R., Martinelli C., Politakos N., et al. (2017). Exploring the pH sensitivity of Poly(allylamine) phosphate supramolecular nanocarriers for intracellular siRNA delivery. ACS Appl. Mater. Interf. 9 38242–38254. 10.1021/acsami.7b11132 [DOI] [PubMed] [Google Scholar]
- Anraku I., Harvey T. J., Linedale R., Gardner J., Harrich D., Suhrbier A., et al. (2002). Kunjin virus replicon vaccine vectors induce protective CD8+ T-cell immunity. J. Virol. 76 3791–3799. 10.1128/JVI.76.8.3791-3799.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosova Z., Mackova M., Kral V., Macek T. (2009). Therapeutic application of peptides and proteins: parenteral forever? Trends Biotechnol. 27 628–635. 10.1016/j.tibtech.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Arance Fernandez A. M., Baurain J.-F., Vulsteke C., Rutten A., Soria A., Carrasco J., et al. (2019). A phase I study (E011-MEL) of a TriMix-based mRNA immunotherapy (ECI-006) in resected melanoma patients: analysis of safety and immunogenicity. J. Clin. Oncol. 37 2641–2641. 10.1200/JCO.2019.37.15_suppl.2641 [DOI] [Google Scholar]
- Baker B. F., Lot S. S., Condon T. P., Cheng-Flournoy S., Lesnik E. A., Sasmor H. M., et al. (1997). 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 272 11994–12000. 10.1074/jbc.272.18.11994 [DOI] [PubMed] [Google Scholar]
- Balwani M., Sardh E., Ventura P., Peiró P. A., Rees D. C., Stölzel U., et al. (2020). Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Engl. J. Med. 382 2289–2301. 10.1056/NEJMoa1913147 [DOI] [PubMed] [Google Scholar]
- Barenholz (Chezy) Y. (2012). Doxil® — The first FDA-approved nano-drug: lessons learned. J. Controll. Release 160 117–134. 10.1016/j.jconrel.2012.03.020 [DOI] [PubMed] [Google Scholar]
- Bauer M., Strom M., Hammond D. S., Shigdar S. (2019). Anything you can do, i can do better: can aptamers replace antibodies in clinical diagnostic applications? Molecules 24:4377. 10.3390/molecules24234377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. F., Swayze E. E. (2010). RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50 259–293. 10.1146/annurev.pharmtox.010909.105654 [DOI] [PubMed] [Google Scholar]
- Berezhnoy A., Stewart C. A., Mcnamara J. O., II, Thiel W., Giangrande P., Trinchieri G., et al. (2012). isolation and optimization of murine IL-10 receptor blocking oligonucleotide aptamers using high-throughput sequencing. Mol. Ther. 20 1242–1250. 10.1038/mt.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A., Tan A. T. (2020). Challenges of CAR- and TCR-T cell–based therapy for chronic infections. J. Exp. Med. 217:e20191663. 10.1084/jem.20191663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioNTech (2021). Pipeline – BioNTech BioNTech. Available online at: https://www.biontech.de/science/pipeline (accessed January 27, 2021). [Google Scholar]
- Bladen C. L., Salgado D., Monges S., Foncuberta M. E., Kekou K., Kosma K., et al. (2015). The TREAT-NMD DMD global database: analysis of more than 7,000 duchenne muscular dystrophy mutations. Hum. Mutat. 36 395–402. 10.1002/humu.22758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boada C., Zinger A., Tsao C., Zhao P., Martinez J. O., Hartman K., et al. (2020). Rapamycin-loaded biomimetic nanoparticles reverse vascular inflammation. Circ. Res. 126 25–37. 10.1161/CIRCRESAHA.119.315185 [DOI] [PubMed] [Google Scholar]
- Bryant L. M., Christopher D. M., Giles A. R., Hinderer C., Rodriguez J. L., Smith J. B., et al. (2013). Lessons learned from the clinical development and market authorization of glybera. Hum. Gene Ther. Clin. Dev. 24 55–64. 10.1089/humc.2013.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Businesswire (2019). InteRNA Presents Expanded Preclinical Proof-of-concept Data on Lead Oncology Development Candidate INT-1B3 at the 14th Annual OTS Meeting (2018). Available online at: https://www.businesswire.com/news/home/20181001005257/en/InteRNA-Presents-Expanded-Preclinical-Proof-of-concept-Data-Lead (accessed December 26, 2019). [Google Scholar]
- Cao J., An D., Galduroz M., Zhuo J., Liang S., Eybye M., et al. (2019). mRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol. Ther. 27 1242–1251. 10.1016/j.ymthe.2019.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L., Clarke J. C., Yen C., Gregoire F., Albery T., Billger M., et al. (2018). Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 9 330–346. 10.1016/j.omtm.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M. C., Phillips E. J. (2021). Maintaining safety with SARS-CoV-2 vaccines. N. Engl. J. Med. 384 643–649. 10.1056/NEJMra2035343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov (2019). Efficacy, Safety, and Tolerability of Remlarsen (MRG-201) Following Intradermal Injection in Subjects With a History of Keloids - Full Text View - ClinicalTrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03601052 (accessed December 26, 2019). [Google Scholar]
- ClinicalTrials.gov (2020a). An Efficacy Study of Adjuvant Treatment With the Personalized Cancer Vaccine mRNA-4157 and Pembrolizumab in Patients With High-Risk Melanoma (KEYNOTE-942) - Full Text View - ClinicalTrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03897881 (accessed July 10, 2020). [Google Scholar]
- ClinicalTrials.gov (2020d). Long-Term, Prospective, Non-interventional, Safety Extension of a Phase III, Double-blind, Randomized, Placebo-controlled, Multicenter Study to Assess the Safety and Efficacy of VM202 in Subjects With Painful Diabetic Peripheral Neuropathy. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT04055090 (accessed August 25, 2020). [Google Scholar]
- ClinicalTrials.gov (2020c). A Non-Randomized, Open Label, Controlled, Dose-Escalation, Phase I Clinical Trial to Evaluate the Safety, Reactogenicity and Immunogenicity of One or Two Administrations of Candidate Rabies mRNA Vaccine CV7202 in Healthy Adult Subjects. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03713086 (accessed August 4, 2020). [Google Scholar]
- ClinicalTrials.gov (2020e). A Phase 1, Open-Label, Multicenter Study to Assess the Safety and Tolerability of mRNA-5671/V941 as a Monotherapy and in Combination With Pembrolizumab in Participants With KRAS Mutant Advanced or Metastatic Non-Small Cell Lung Cancer, Colorectal Cancer or Pancreatic Adenocarcinoma. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03948763 (accessed August 4, 2020). [Google Scholar]
- ClinicalTrials.gov (2020b). A Phase 1 Randomized, Double Blinded, Placebo Controlled, Ascending Dose Study to Assess the Safety, Tolerability, and Pharmacokinetics of Single Doses of ARCT-810 in Healthy Adult Subjects. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT04416126 (accessed August 4, 2020). [Google Scholar]
- ClinicalTrials.gov (2020f). Safety, Tolerability, and Immunogenicity of Zika Vaccine mRNA-1893 in Healthy Flavivirus Seropositive and Seronegative Adults - Full Text View - ClinicalTrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT04064905 (accessed August 5, 2020). [Google Scholar]
- Condit R. C., Williamson A.-L., Sheets R., Seligman S. J., Monath T. P., Excler J.-L., et al. (2016). Unique safety issues associated with virus-vectored vaccines: Potential for and theoretical consequences of recombination with wild type virus strains. Vaccine 34 6610–6616. 10.1016/j.vaccine.2016.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B., Isaacs C., Cheng L., Asrani K. H., Subramanian R. R. (2018). SERPINA1 mRNA as a treatment for Alpha-1 antitrypsin deficiency. J. Nucleic Acids 2018:e8247935. 10.1155/2018/8247935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conry R. M., LoBuglio A. F., Wright M., Sumerel L., Pike M. J., Johanning F., et al. (1995). Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 55 1397–1400. [PubMed] [Google Scholar]
- Conry R. M., Westbrook B., McKee S., Norwood T. G. (2018). Talimogene laherparepvec: First in class oncolytic virotherapy. Hum. Vaccines Immunother. 14 839–846. 10.1080/21645515.2017.1412896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K. S., Flynn B., Foulds K. E., Francica J. R., Boyoglu-Barnum S., Werner A. P., et al. (2020). Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 383 1544–1555. 10.1056/NEJMoa2024671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo C., Cromer W. E., Molinaro R., Toledano Furman N. E., Hartman K. A., De Rosa E., et al. (2017). Engineered biomimetic nanovesicles show intrinsic anti-inflammatory properties for the treatment of inflammatory bowel diseases. Nanoscale 9 14581–14591. 10.1039/c7nr04734g [DOI] [PubMed] [Google Scholar]
- CovidVax (2020). COVID-19 Vaccine: DNA by Osaka University, AnGes, Takara Bio, Cytiva, Brickell Biotech. Available online at: https://covidvax.org/covid19-vaccine/OsakaUniAnGes/DNA-plasmid-Osaka-University-AnGes-Takara-Bio-Cytiva-Brickell-Biotech (accessed February 22, 2021). [Google Scholar]
- CRISPR (2019c). Pipeline. Available online at: http://www.crisprtx.com/programs/pipeline (accessed December 27, 2019). [Google Scholar]
- CRISPR (2019a). Hemoglobinopathies. Available online at: http://www.crisprtx.com/programs/hemoglobinopathies (accessed December 27, 2019). [Google Scholar]
- CRISPR (2019b). Immuno-Oncology. Available online at: http://www.crisprtx.com/programs/immuno-oncology (accessed December 27, 2019). [Google Scholar]
- CureVac (2020). Pipeline – CureVac. Available online at: https://www.curevac.com/en/pipeline/ (accessed September 1, 2020). [Google Scholar]
- Daley J. (2019). Gene therapy arrives. Nature 576 S12–S13. 10.1038/d41586-019-03716-9 [DOI] [Google Scholar]
- Damase T. R., Miura T. A., Parent C. E., Allen P. B. (2018). Application of the Open qPCR instrument for the in vitro selection of DNA aptamers against Epidermal Growth Factor Receptor and Drosophila C virus. ACS Comb. Sci. 20 45–54. 10.1021/acscombsci.7b00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Chalbatani G. M., Mahmoodzadeh H., Karimloo R., Rezaiean O., Moradzadeh A., et al. (2017). Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. IJBS 13 48–57. [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker B., Claerhout S., Carrasco J., Bar I., Corthals J., Wilgenhof S., et al. (2020). TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: link between T-cell activation and clinical responses in advanced melanoma. J. Immunother. Cancer 8:e000329. 10.1136/jitc-2019-000329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N. M., McKay R., Condon T. P., Bennett C. F. (1994). Inhibition of protein kinase C-alpha expression in human A549 cells by antisense oligonucleotides inhibits induction of intercellular adhesion molecule 1 (ICAM-1) mRNA by phorbol esters. J. Biol. Chem. 269 16416–16424. [PubMed] [Google Scholar]
- Desai P. R., Marepally S., Patel A. R., Voshavar C., Chaudhuri A., Singh M., et al. (2013). Topical delivery of anti-TNFα siRNA and capsaicin via novel lipid-polymer hybrid nanoparticles efficiently inhibits skin inflammation in vivo. J. Control. Release 170 51–63. 10.1016/j.jconrel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devulapally R., Foygel K., Sekar T. V., Willmann J. K., Paulmurugan R. (2016). Gemcitabine and antisense-microRNA Co-encapsulated PLGA–PEG polymer nanoparticles for hepatocellular carcinoma therapy. ACS Appl. Mater. Interf. 8 33412–33422. 10.1021/acsami.6b08153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias N., Stein C. A. (2002). Antisense oligonucleotides: basic concepts and mechanisms. Mol. Cancer Ther. 1 347–355. [PubMed] [Google Scholar]
- Dolgin E. (2021). How COVID unlocked the power of RNA vaccines. Nature 589 189–191. 10.1038/d41586-021-00019-w [DOI] [PubMed] [Google Scholar]
- Dollins C. M., Nair S., Boczkowski D., Lee J., Layzer J. M., Gilboa E., et al. (2008). Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 15 675–682. 10.1016/j.chembiol.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editas (2020). Research and Pipeline Ed. Med. Available online at: https://www.editasmedicine.com/gene-editing-pipeline/ (accessed February 25, 2020). [Google Scholar]
- Ellington A. D., Szostak J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346 818–822. 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- Ezkurdia I., Juan D., Rodriguez J. M., Frankish A., Diekhans M., Harrow J., et al. (2014). Multiple evidence strands suggest that there may be as few as 19 000 human protein-coding genes. Hum. Mol. Genet. 23 5866–5878. 10.1093/hmg/ddu309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fala L. (2018). Yescarta (Axicabtagene Ciloleucel) Second CAR T-Cell Therapy Approved for Patients with Certain Types of Large B-Cell Lymphoma. Available online at: https://www.jons-online.com/special-issues-and-supplements/2018/2018-third-annual-oncology-guide-to-new-fda-approvals/1829-yescarta-axicabtagene-ciloleucel-second-car-t-cell-therapy-approved-for-patients-with-certain-types-of-large-b-cell-lymphoma (accessed August 18, 2020). [Google Scholar]
- FDA (2019). FY 2019 Report from the Director. FDA. Available online at: https://www.fda.gov/vaccines-blood-biologics/fy-2019-report-director (accessed September 8, 2020). [Google Scholar]
- FDA (2020). FDA approval brings first gene therapy to the United States. FDA. Available online at: https://www.fda.gov/news-events/press-announcements/fda-approval-brings-first-gene-therapy-united-states (accessed August 18, 2020). [Google Scholar]
- FDA (2021a). Moderna COVID-19 Vaccine. FDA. Available online at: https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine (accessed February 1, 2021). [Google Scholar]
- FDA (2021b). Pfizer-BioNTech COVID-19 Vaccine. FDA. Available online at: https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/pfizer-biontech-covid-19-vaccine (accessed February 1, 2021). [Google Scholar]
- Feldman R. A., Fuhr R., Smolenov I., Ribeiro A. M., Panther L., Watson M., et al. (2019). mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine 37 3326–3334. 10.1016/j.vaccine.2019.04.074 [DOI] [PubMed] [Google Scholar]
- FierceBiotech (2020). Moderna Raises $500M to Move mRNA Drugs Deeper into Human Tests | FierceBiotech. Available online at: https://www.fiercebiotech.com/biotech/moderna-raises-500m-to-move-mrna-drugs-deeper-into-human-tests (accessed August 6, 2020). [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Frank D. E., Schnell F. J., Akana C., El-Husayni S. H., Desjardins C. A., Morgan J., et al. (2020). Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology 94 e2270–e2282. 10.1212/WNL.0000000000009233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. (1986). Origin of life: the RNA world. Nature 319:618. 10.1038/319618a0 [DOI] [Google Scholar]
- Giles R. V., Spiller D. G., Tidd D. M. (1995). Detection of ribonuclease H-Generated mRNA fragments in human leukemia cells following reversible membrane permeabilization in the presence of antisense oligodeoxynucleotides. Antisense Res. Dev. 5 23–31. 10.1089/ard.1995.5.23 [DOI] [PubMed] [Google Scholar]
- Givlaari (2020). Now Approved: GIVLAARITM (givosiran) GIVLAARI. Available online at: https://www.givlaari.com (accessed January 3, 2020). [Google Scholar]
- Hadinoto K., Sundaresan A., Cheow W. S. (2013). Lipid–polymer hybrid nanoparticles as a new generation therapeutic delivery platform: a review. Eur. J. Pharm. Biopharm. 85 427–443. 10.1016/j.ejpb.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Hanna J., Hossain G. S., Kocerha J. (2019). The potential for microRNA therapeutics and clinical research. Front. Genet. 10:478. 10.3389/fgene.2019.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey T. J., Anraku I., Linedale R., Harrich D., Mackenzie J., Suhrbier A., et al. (2003). Kunjin virus replicon vectors for human immunodeficiency virus vaccine development. J. Virol. 77 7796–7803. 10.1128/JVI.77.14.7796-7803.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y., Hara E., Shingu Y., Minamiguchi D., Nakamura A., Arai S., et al. (2015). siRNA delivery into tumor cells by cationic cholesterol derivative-based nanoparticles and liposomes. Biol. Pharm. Bull. 38 30–38. 10.1248/bpb.b14-00526 [DOI] [PubMed] [Google Scholar]
- Hopkins A. L., Groom C. R. (2002). The druggable genome. Nat. Rev. Drug Discov. 1 727–730. 10.1038/nrd892 [DOI] [PubMed] [Google Scholar]
- Hua Y., Vickers T. A., Baker B. F., Bennett C. F., Krainer A. R. (2007). Enhancement of SMN2 Exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 5:e73. 10.1371/journal.pbio.0050073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C.-F., Xu X., Li L., Ma Y., Jin Q., Viley A., et al. (2018). Development of anti-human mesothelin-targeted chimeric antigen receptor messenger RNA-transfected peripheral blood lymphocytes for ovarian cancer therapy. Hum. Gene Ther. 29 614–625. 10.1089/hum.2017.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. L., Chen R., Trobaugh D. W., Diamond M. S., Weaver S. C., Klimstra W. B., et al. (2015). The 5′ and 3′ ends of alphavirus RNAs – Non-coding is not non-functional. Virus Res. 206 99–107. 10.1016/j.virusres.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- INOVIO (2020). INOVIO Announces Positive Interim Phase 1 Data For INO-4800 Vaccine for COVID-19. Available online at: http://ir.inovio.com/news-releases/news-releases-details/2020/INOVIO-Announces-Positive-Interim-Phase-1-Data-For-INO-4800-Vaccine-for-COVID-19/default.aspx (accessed September 8, 2020). [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., et al. (2020). An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 383 1920–1931. 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger B. W., Dowd K. A., Chen R. E., Desai P., Foreman B., Burgomaster K. E., et al. (2019). Protective efficacy of nucleic acid vaccines against transmission of zika virus during pregnancy in mice. J. Infect. Dis. 220 1577–1588. 10.1093/infdis/jiz338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Doudna J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46 505–529. 10.1146/annurev-biophys-062215-010822 [DOI] [PubMed] [Google Scholar]
- Jirikowski G. F., Sanna P. P., Maciejewski-Lenoir D., Bloom F. E. (1992). Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science 255 996–998. 10.1126/science.1546298 [DOI] [PubMed] [Google Scholar]
- John S., Yuzhakov O., Woods A., Deterling J., Hassett K., Shaw C. A., et al. (2018). Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine 36 1689–1699. 10.1016/j.vaccine.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Kam N. W. S., Liu Z., Dai H. (2005). Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J. Am. Chem. Soc. 127 12492–12493. 10.1021/ja053962k [DOI] [PubMed] [Google Scholar]
- Karikó K. (2019). In vitro-Transcribed mRNA therapeutics: out of the shadows and into the spotlight. Mol. Ther. 27 691–692. 10.1016/j.ymthe.2019.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Welsh F. A., Ludwig J., Kato H., Akira S., et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 16 1833–1840. 10.1038/mt.2008.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlgren M., Simoff I., Keiser M., Oswald S., Artursson P. (2018). CRISPR-Cas9: a new addition to the drug metabolism and disposition tool box. Drug Metab. Dispos. 46 1776–1786. 10.1124/dmd.118.082842 [DOI] [PubMed] [Google Scholar]
- Kaur H., Bruno J. G., Kumar A., Sharma T. K. (2018). Aptamers in the therapeutics and diagnostics pipelines. Theranostics 8 4016–4032. 10.7150/thno.25958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler J. A., Smith A. G., Cha B.-S., Choi S. H., Wymer J., Shaibani A., et al. (2015). Double-blind, placebo-controlled study of HGF gene therapy in diabetic neuropathy. Ann. Clin. Transl. Neurol. 2 465–478. 10.1002/acn3.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hu C., Moufawad, El Achkar C., Black L. E., Douville J., et al. (2019). Patient-customized oligonucleotide therapy for a rare genetic disease. N. Engl. J. Med. 381 1644–1652. 10.1056/NEJMoa1813279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose N., Fox J. M., Sapparapu G., Bombardi R., Tennekoon R. N., Silva A. D., et al. (2019). A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 4:eaaw6647. 10.1126/sciimmunol.aaw6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski P. S., Rudra A., Miao L., Anderson D. G. (2019). Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. J. Am. Soc. Gene Ther. 27 710–728. 10.1016/j.ymthe.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz L. M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K. C., et al. (2016). Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534 396–401. 10.1038/nature18300 [DOI] [PubMed] [Google Scholar]
- Kristen A. V., Ajroud-Driss S., Conceição I., Gorevic P., Kyriakides T., Obici L. (2018). Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener. Dis. Manag. 9 5–23. 10.2217/nmt-2018-0033 [DOI] [PubMed] [Google Scholar]
- Kumari A., Yadav S. K., Yadav S. C. (2010). Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf. B Biointerf. 75 1–18. 10.1016/j.colsurfb.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Lam J. K. W., Chow M. Y. T., Zhang Y., Leung S. W. S. (2015). siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids 4:e252. 10.1038/mtna.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laouini A., Jaafar-Maalej C., Limayem-Blouza I., Sfar S., Charcosset C., Fessi H. (2012). Preparation, characterization and applications of liposomes: state of the art. J. Colloid Sci. Biotechnol. 1 147–168. 10.1166/jcsb.2012.1020 [DOI] [Google Scholar]
- Larrouy B., Blonski C., Boiziau C., Stuer M., Moreau S., Shire D., et al. (1992). RNase H-mediated inhibition of translation by antisense oligodeoxyribo-nucleotides: use of backbone modification to improve specificity. Gene 121 189–194. 10.1016/0378-1119(92)90121-5 [DOI] [PubMed] [Google Scholar]
- Le T. T., Andreadakis Z., Kumar A., Román R. G., Tollefsen S., Saville M., et al. (2020). The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 19 305–306. 10.1038/d41573-020-00073-5 [DOI] [PubMed] [Google Scholar]
- Ledwith B. J., Manam S., Troilo P. J., Barnum A. B., Pauley C. J., Ii T. G. G., et al. (2000). Plasmid DNA vaccines: investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 43 258–272. 10.1159/000053993 [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Kang S., Ahn M., Jang H., Min D.-H. (2018). Development of dual-pore coexisting branched silica nanoparticles for efficient gene–chemo cancer therapy. Small 14:1702564. 10.1002/smll.201702564 [DOI] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Li J.-M., Zhao M.-X., Su H., Wang Y.-Y., Tan C.-P., Ji L.-N., et al. (2011). Multifunctional quantum-dot-based siRNA delivery for HPV18 E6 gene silence and intracellular imaging. Biomaterials 32 7978–7987. 10.1016/j.biomaterials.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Li W., Kerwin J. L., Schiel J., Formolo T., Davis D., Mahan A., et al. (2015). “Structural elucidation of post-translational modifications in monoclonal antibodies,” in ACS Symposium Series, eds Schiel J. E., Davis D. L., Borisov O. V. (Washington, DC: American Chemical Society; ), 119–183. 10.1021/bk-2015-1201.ch003 [DOI] [Google Scholar]
- Liang X., Shen W., Sun H., Migawa M. T., Vickers T. A., Crooke S. T. (2016). Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34 875–880. 10.1038/nbt.3589 [DOI] [PubMed] [Google Scholar]
- Lim K. R. Q., Maruyama R., Yokota T. (2017). Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des. Devel. Ther. 11 533–545. 10.2147/DDDT.S97635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoff A. M., Mehran R., Povsic T. J., Zelenkofske S. L., Huang Z., Armstrong P. W., et al. (2016). Effect of the REG1 anticoagulation system versus bivalirudin on outcomes after percutaneous coronary intervention (REGULATE-PCI): a randomised clinical trial. Lancet 387 349–356. 10.1016/S0140-6736(15)00515-2 [DOI] [PubMed] [Google Scholar]
- Liu M. A. (2019). A comparison of plasmid DNA and mRNA as vaccine technologies. Vaccines 7:37. 10.3390/vaccines7020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S., Sun T., Li H., Hu Z. (2018). Mechanisms of microRNA-mediated gene regulation in unicellular model alga Chlamydomonas reinhardtii. Biotechnol. Biofuels 11:244. 10.1186/s13068-018-1249-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano T., Soldevilla M. M., Casares N., Villanueva H., Bendandi M., Lasarte J. J., et al. (2016). Targeting inhibition of Foxp3 by a CD28 2′-Fluro oligonucleotide aptamer conjugated to P60-peptide enhances active cancer immunotherapy. Biomaterials 91 73–80. 10.1016/j.biomaterials.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Lundstrom K. (2018a). Latest development on RNA-based drugs and vaccines. Future Sci. OA 4:FSO300. 10.4155/fsoa-2017-0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. (2018b). Self-replicating RNA viruses for RNA therapeutics. Molecules 23:3310. 10.3390/molecules23123310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie N., Saville M., Hatchett R., Halton J. (2020). Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382 1969–1973. 10.1056/NEJMp2005630 [DOI] [PubMed] [Google Scholar]
- Maeder M. L., Stefanidakis M., Wilson C. J., Baral R., Barrera L. A., Bounoutas G. S., et al. (2019). Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 25 229–233. 10.1038/s41591-018-0327-9 [DOI] [PubMed] [Google Scholar]
- Maguire A. M., Russell S., Wellman J. A., Chung D. C., Yu Z.-F., Tillman A., et al. (2019). Efficacy, safety, and durability of voretigene neparvovec-rzyl in RPE65 mutation–associated inherited retinal dystrophy. Ophthalmology 126 1273–1285. 10.1016/j.ophtha.2019.06.017 [DOI] [PubMed] [Google Scholar]
- Mahajan R. (2019). Onasemnogene abeparvovec for spinal muscular atrophy: the costlier drug ever. Int. J. Appl. Basic Med. Res. 9 127–128. 10.4103/ijabmr.IJABMR_190_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamcarz E., Zhou S., Lockey T., Abdelsamed H., Cross S. J., Kang G., et al. (2019). Lentiviral gene therapy combined with low-dose busulfan in infants with SCID-X1. N. Engl. J. Med. 380 1525–1534. 10.1056/NEJMoa1815408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. O., Molinaro R., Hartman K. A., Boada C., Sukhovershin R., De Rosa E., et al. (2018). Biomimetic nanoparticles with enhanced affinity towards activated endothelium as versatile tools for theranostic drug delivery. Theranostics 8 1131–1145. 10.7150/thno.22078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.-G., et al. (1993). Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 23 1719–1722. 10.1002/eji.1830230749 [DOI] [PubMed] [Google Scholar]
- Mathew V., Wang A. K. (2019). Inotersen: new promise for the treatment of hereditary transthyretin amyloidosis. Drug Des. Devel. Ther. 13 1515–1525. 10.2147/DDDT.S162913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough K. C., Bassi I., Milona P., Suter R., Thomann-Harwood L., Englezou P., et al. (2014). Self-replicating replicon-RNA delivery to dendritic cells by chitosan-nanoparticles for translation in vitro and in vivo. Mol. Ther. Nucleic Acids 3:e173. 10.1038/mtna.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay P. F., Hu K., Blakney A. K., Samnuan K., Brown J. C., Penn R., et al. (2020). Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 11:3523. 10.1038/s41467-020-17409-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara J. O., Andrechek E. R., Wang Y., Viles K. D., Rempel R. E., Gilboa E., et al. (2006). Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 24 1005–1015. 10.1038/nbt1223 [DOI] [PubMed] [Google Scholar]
- McNamara J. O., Kolonias D., Pastor F., Mittler R. S., Chen L., Giangrande P. H., et al. (2008). Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J. Clin. Invest. 118 376–386. 10.1172/JCI33365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meka A. K., Niu Y., Karmakar S., Hartono S. B., Zhang J., Lin C. X. C., et al. (2016). Facile synthesis of large-pore bicontinuous cubic mesoporous silica nanoparticles for intracellular gene delivery. ChemNanoMat 2 220–225. 10.1002/cnma.201600021 [DOI] [Google Scholar]
- Mendell J. R., Sahenk Z., Lehman K., Nease C., Lowes L. P., Miller N. F., et al. (2020). Assessment of systemic delivery of rAAVrh74.MHCK7.micro-dystrophin in children with duchenne muscular dystrophy. JAMA Neurol. 77 1122–1131. 10.1001/jamaneurol.2020.1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkeninstitute (2020). Milken Institute’s COVID-19 Treatment and Vaccine Tracker tracks the Development of Treatments and Vaccines for COVID-19 at covid-19tracker.milkeninstitute.org #COVID19 #coronavirus #COVID19treatment #COVID19vaccine @MilkenInstitute @FirstPersonSF. Available online at: https://covid-19tracker.milkeninstitute.org/ (accessed September 8, 2020). [Google Scholar]
- Minshull J., Hunt T. (1986). The use of single-stranded DNA and RNase H to promote quantitative ‘hybrid arrest of translation’ of mRNA/DNA hybrids in reticulocyte lysate cell-free translations. Nucleic Acids Res. 14 6433–6451. 10.1093/nar/14.16.6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- miRagen (2019). Pipeline – miRagen MiRagen Ther. Inc. Available online at: http://www.miragen.com/pipeline/ (accessed December 26, 2019). [Google Scholar]
- Moderna (2020). mRNA-2416 | Moderna, Inc. Available online at: https://www.modernatx.com/pipeline/mrna-2416 (accessed August 5, 2020). [Google Scholar]
- Modernatx (2021a). Moderna’s mRNA Clinical Trials: CMV, MMA, Zika, Several Types of Cancer and Other Diseases. Available online at: https://www.modernatx.com/pipeline/modernas-mrna-clinical-trials-cmv-mma-zika-several-types-cancer-and-other-diseases (accessed January 27, 2021). [Google Scholar]
- Modernatx. (2021b). Moderna’s Work on a COVID-19 Vaccine Candidate | Moderna, Inc. Available online at: https://www.modernatx.com/modernas-work-potential-vaccine-against-covid-19 (accessed February 1, 2021). [Google Scholar]
- ModernaTX Inc (2019a). A Phase 1, Randomized, Observer-Blind, Placebo-Controlled, Dose-Ranging Study to Evaluate the Safety, Reactogenicity, and Immunogenicity of Cytomegalovirus Vaccines mRNA-1647 and mRNA-1443 When Administered to Healthy Adults. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03382405 (accessed July 9, 2020). [Google Scholar]
- ModernaTX Inc. (2019b). A Phase 1, Randomized, Placebo-Controlled, Dose Ranging Study to Evaluate the Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of mRNA-1944, Encoding for an Anti-Chikungunya Virus Monoclonal Antibody, in Healthy Adults. clinicaltrials.gov. Available online at: https://clinicaltrials.gov/ct2/show/NCT03829384 (accessed August 4, 2020). [Google Scholar]
- Molina M., Asadian-Birjand M., Balach J., Bergueiro J., Miceli E., Calderón M. (2015). Stimuli-responsive nanogel composites and their application in nanomedicine. Chem. Soc. Rev. 44 6161–6186. 10.1039/C5CS00199D [DOI] [PubMed] [Google Scholar]
- Molinaro R., Corbo C., Martinez J. O., Taraballi F., Evangelopoulos M., Minardi S., et al. (2016). Biomimetic proteolipid vesicles for targeting inflamed tissues. Nat. Mater. 15 1037–1046. 10.1038/nmat4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro R., Pastò A., Corbo C., Taraballi F., Giordano F., Martinez J. O., et al. (2019). Macrophage-derived nanovesicles exert intrinsic anti-inflammatory properties and prolong survival in sepsis through a direct interaction with macrophages. Nanoscale 11 13576–13586. 10.1039/C9NR04253A [DOI] [PubMed] [Google Scholar]
- Möller K., Müller K., Engelke H., Bräuchle C., Wagner E., Bein T. (2016). Highly efficient siRNA delivery from core–shell mesoporous silica nanoparticles with multifunctional polymer caps. Nanoscale 8 4007–4019. 10.1039/C5NR06246B [DOI] [PubMed] [Google Scholar]
- Moyo N., Vogel A. B., Buus S., Erbar S., Wee E. G., Sahin U., et al. (2018). Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol. Ther. Methods Clin. Dev. 12 32–46. 10.1016/j.omtm.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. (2020). 2019 FDA drug approvals. Nat. Rev. Drug Discov. 19 79–84. 10.1038/d41573-020-00001-7 [DOI] [PubMed] [Google Scholar]
- Mulligan M. J., Lyke K. E., Kitchin N., Absalon J., Gurtman A., Lockhart S. P., et al. (2020). Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. medRxiv [Preprint]. 10.1101/2020.06.30.20142570 [DOI] [Google Scholar]
- Naso M. F., Tomkowicz B., Perry W. L., Strohl W. R. (2017). Adeno-associated virus (AAV) as a vector for gene therapy. Biodrugs 31 317–334. 10.1007/s40259-017-0234-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute (2011). Natl. Cancer Inst. Available online at: https://www.cancer.gov/publications/dictionaries/cancer-drug (accessed July 10, 2020). [Google Scholar]
- Nature Biotechnology (2020). Second RNAi drug approved. Nat. Biotechnol. 38:385. 10.1038/s41587-020-0494-3 [DOI] [PubMed] [Google Scholar]
- Neil E. E., Bisaccia E. K. (2019). Nusinersen: a novel antisense oligonucleotide for the treatment of spinal muscular atrophy. J. Pediatr. Pharmacol. Ther. JPPT 24 194–203. 10.5863/1551-6776-24.3.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng E. W. M., Shima D. T., Calias P., Cunningham E. T., Guyer D. R., Adamis A. P. (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5 123–132. 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- Nimjee S. M., White R. R., Becker R. C., Sullenger B. A. (2017). Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 57 61–79. 10.1146/annurev-pharmtox-010716-104558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel 2019 Coronavirus Genome (2020). Virological. Available online at: https://virological.org/t/novel-2019-coronavirus-genome/319 (accessed September 9, 2020). [Google Scholar]
- NOXXON (2020b). NOX-E36. Available online at: https://www.noxxon.com/index.php?option=com_content&view=article&id=20&Itemid=477 (accessed February 21, 2020). [Google Scholar]
- NOXXON (2020a). NOX-A12. Available online at: https://www.noxxon.com/index.php?option=com_content&view=article&id=21&Itemid=478 (accessed February 21, 2020). [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9:402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag E. (2018). Update of The P-BCMA-101-001 Phase 1 Clinical Trial: A Novel Stem Cell Memory CAR-T for Relapsed/Refractory Multiple Myeloma. Available online at: https://poseida.com/publications/ (accessed February 1, 2021). [Google Scholar]
- Paik J., Duggan S. (2019). Volanesorsen: first global approval. Drugs 79 1349–1354. 10.1007/s40265-019-01168-z [DOI] [PubMed] [Google Scholar]
- Pardi N., Hogan M. J., Pelc R. S., Muramatsu H., Andersen H., DeMaso C. R., et al. (2017). Zika virus protection by a single low dose nucleoside modified mRNA vaccination. Nature 543 248–251. 10.1038/nature21428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J. L., Vallet-Regí M. (2020). Mesoporous silica nanoparticles for co-delivery of drugs and nucleic acids in oncology: a review. Pharmaceutics 12:526. 10.3390/pharmaceutics12060526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch B., Schnee M., Vogel A. B., Lange E., Hoffmann B., Voss D., et al. (2012). Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 30 1210–1216. 10.1038/nbt.2436 [DOI] [PubMed] [Google Scholar]
- Regulus (2019). Regulus Therapeutics - Leading the Way in the Discovery and Development of microRNA Therapeutics Regulus Ther. Available online at: http://regulusrx.com (accessed December 26, 2019). [Google Scholar]
- Reid G., Kao S. C., Pavlakis N., Brahmbhatt H., MacDiarmid J., Clarke S., et al. (2016). Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 8 1079–1085. 10.2217/epi-2016-0035 [DOI] [PubMed] [Google Scholar]
- Rigo F., Hua Y., Krainer A. R., Bennett C. F. (2012). Antisense-based therapy for the treatment of spinal muscular atrophy. J. Cell Biol. 199 21–25. 10.1083/jcb.201207087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckman J., Green L. S., Beeson J., Waugh S., Gillette W. L., Henninger D. D., et al. (1998). 2′-Fluoropyrimidine RNA-based Aptamers to the 165-Amino Acid Form of Vascular Endothelial Growth Factor (VEGF165) INHIBITION OF RECEPTOR BINDING AND VEGF-INDUCED VASCULAR PERMEABILITY THROUGH INTERACTIONS REQUIRING THE EXON 7-ENCODED DOMAIN. J. Biol. Chem. 273 20556–20567. 10.1074/jbc.273.32.20556 [DOI] [PubMed] [Google Scholar]
- Russo E. (2003). Special report: the birth of biotechnology. Nature 421 456–457. 10.1038/nj6921-456a [DOI] [PubMed] [Google Scholar]
- Sahin U., Karikó K., Türeci Ö. (2014). mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 13 759–780. 10.1038/nrd4278 [DOI] [PubMed] [Google Scholar]
- Santulli-Marotto S., Nair S. K., Rusconi C., Sullenger B., Gilboa E. (2003). Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Res. 63 7483–7489. [PubMed] [Google Scholar]
- Saraswat P., Soni R. R., Bhandari A., Nagori B. P. (2009). DNA as therapeutics; an update. Indian J. Pharm. Sci. 71 488–498. 10.4103/0250-474X.58169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeer M., Buchholz T., Schleef M. (2017). Plasmid DNA manufacturing for indirect and direct clinical applications. Hum. Gene Ther. 28 856–861. 10.1089/hum.2017.159 [DOI] [PubMed] [Google Scholar]
- Schnee M., Vogel A. B., Voss D., Petsch B., Baumhof P., Kramps T., et al. (2016). An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl. Trop. Dis. 10:e0004746. 10.1371/journal.pntd.0004746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrand B., Berezhnoy A., Brenneman R., Williams A., Levay A., Kong L.-Y., et al. (2014). Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer Immunol. Res. 2 867–877. 10.1158/2326-6066.CIR-14-0007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrand B., Verma B., Levay A., Patel S., Castro I., Benaduce A. P., et al. (2017). Radiation-induced enhancement of antitumor T-cell immunity by VEGF-targeted 4-1BB costimulation. Cancer Res. 77 1310–1321. 10.1158/0008-5472.CAN-16-2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler-Lenz M., Enzmann H., Vamvakas S. (2020). Regulators’ advice can make a difference: european medicines agency approval of zynteglo for beta thalassemia. Clin. Pharmacol. Ther. 107 492–494. 10.1002/cpt.1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian M., Papachristofilou A., Weiss C., Früh M., Cathomas R., Hilbe W., et al. (2014). Phase Ib study evaluating a self-adjuvanted mRNA cancer vaccine (RNActive®) combined with local radiation as consolidation and maintenance treatment for patients with stage IV non-small cell lung cancer. BMC Cancer 14:748. 10.1186/1471-2407-14-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercombe L., Veerati T., Moheimani F., Wu S. Y., Sood A. K., Hua S. (2015). Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 6:286. 10.3389/fphar.2015.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Corey D. R. (2018). Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 46 1584–1600. 10.1093/nar/gkx1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Xiao Z., Votruba A. R., Vilos C., Farokhzad O. C. (2011). Differentially charged hollow Core/Shell Lipid–polymer–lipid hybrid nanoparticles for small interfering RNA delivery. Angew. Chem. Int. Ed. 50 7027–7031. 10.1002/anie.201101554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Park S.-J., Yim Y., Kim J., Choi C., Won C., et al. (2018). Recent advances in RNA therapeutics and RNA delivery systems based on nanoparticles. Adv. Ther. 1:1800065. 10.1002/adtp.201800065 [DOI] [Google Scholar]
- Singh H., Huls H., Cooper L. J. (2014). A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol. Rev. 257 181–190. 10.1111/imr.12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South E., Cox E., Meader N., Woolacott N., Griffin S. (2018). Strimvelis® for treating severe combined immunodeficiency caused by adenosine deaminase deficiency: an evidence review group perspective of a NICE highly specialised technology evaluation. PharmacoEconomics Open 3 151–161. 10.1007/s41669-018-0102-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SparkCures (2020). Descartes-08 | SparkCures Myeloma Clin. Trials SparkCures. Available online at: https://sparkcures.com/treatment/321/descartes-08 (accessed August 5, 2020). [Google Scholar]
- St Jude Children’s Research Hospital (2020). St. Jude gene therapy cures babies with ‘bubble boy’ disease. Available online at: https://www.stjude.org/media-resources/news-releases/2019-medicine-science-news/st-jude-gene-therapy-cures-babies-with-bubble-boy-disease.html (accessed August 18, 2020). [Google Scholar]
- Stephenson M. L., Zamecnik P. C. (1978). Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. U.S.A. 75 285–288. 10.1073/pnas.75.1.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjude (2020). Gene Therapy at St. Jude - St. Jude Children’s Research Hospital. Available online at: https://www.stjude.org/research/initiatives/gene-therapy.html (accessed August 5, 2020). [Google Scholar]
- Sushnitha M., Evangelopoulos M., Tasciotti E., Taraballi F. (2020). Cell membrane-based biomimetic nanoparticles and the immune system: immunomodulatory interactions to therapeutic applications. Front. Bioeng. Biotechnol. 8:627. 10.3389/fbioe.2020.00627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar R. S., Matta H., Chaudhary P. M. (2003). Use of adeno-associated viral vector for delivery of small interfering RNA. Oncogene 22 5712–5715. 10.1038/sj.onc.1206733 [DOI] [PubMed] [Google Scholar]
- Torchilin V. P. (2005). Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 4 145–160. 10.1038/nrd1632 [DOI] [PubMed] [Google Scholar]
- TranslateBio (2019). mRNA Therapeutics and Vaccines | Translate Bio | Pipeline Transl. Bio. Available online at: https://translate.bio/pipeline/ (accessed December 30, 2019). [Google Scholar]
- Tratschin J. D., West M. H., Sandbank T., Carter B. J. (1984). A human parvovirus, adeno-associated virus, as a eucaryotic vector: transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol. Cell. Biol. 4 2072–2081. 10.1128/MCB.4.10.2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249 505–510. 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- Tugues S., Burkhard S. H., Ohs I., Vrohlings M., Nussbaum K., vom Berg J., et al. (2015). New insights into IL-12-mediated tumor suppression. Cell Death Differ. 22 237–246. 10.1038/cdd.2014.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer J. B., Donnelly J. J., Parker S. E., Rhodes G. H., Felgner P. L., Dwarki V. J., et al. (1993). Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259 1745–1749. 10.1126/science.8456302 [DOI] [PubMed] [Google Scholar]
- Ulmer J. B., Geall A. J. (2016). Recent innovations in mRNA vaccines. Curr. Opin. Immunol. 41 18–22. 10.1016/j.coi.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Ura T., Okuda K., Shimada M. (2014). Developments in viral vector-based vaccines. Vaccines 2 624–641. 10.3390/vaccines2030624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ree M. H., van der Meer A. J., van Nuenen A. C., de Bruijne J., Ottosen S., Janssen H. L., et al. (2016). Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment. Pharmacol. Ther. 43 102–113. 10.1111/apt.13432 [DOI] [PubMed] [Google Scholar]
- van Dülmen M., Rentmeister A. (2020). mRNA therapies: new hope in the fight against melanoma: published as part of the Biochemistry series “Biochemistry to Bedside.”. Biochemistry 59 1650–1655. 10.1021/acs.biochem.0c00181 [DOI] [PubMed] [Google Scholar]
- Vickers T. A., Wyatt J. R., Burckin T., Bennett C. F., Freier S. M. (2001). Fully modified 2′ MOE oligonucleotides redirect polyadenylation. Nucleic Acids Res. 29 1293–1299. 10.1093/nar/29.6.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viridian (2021). Pipeline – Viridian Viridian Ther. Inc. Available online at: https://www.viridiantherapeutics.com/pipeline/ (accessed February 1, 2021). [Google Scholar]
- Vogel A. B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K. C., et al. (2018). Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 26 446–455. 10.1016/j.ymthe.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F. R., Sarver N. (1995). Nucleic acid vaccines. Clin. Microbiol. Rev. 8 406–410. 10.1128/CMR.8.3.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E. E., Frenck R., Falsey A. R., Kitchin N., Absalon J., Gurtman A., et al. (2020a). RNA-based COVID-19 vaccine BNT162b2 selected for a pivotal efficacy study. medRxiv [Preprint]. 10.1101/2020.08.17.20176651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E. E., Frenck R. W., Falsey A. R., Kitchin N., Absalon J., Gurtman A., et al. (2020b). Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 383 2439–2450. 10.1056/NEJMoa2027906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li X., Li T., Zhang S., Wang L., Wu X., et al. (2020). The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 1–7. 10.1007/s10096-020-03899-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisenand J. M., Azizian K. T., Henderson J. M., Shore S., Shin D., Lebedev A., et al. (2017). Considerations for the Design and cGMP Manufacturing of mRNA Therapeutics. San Diego, CA: TriLink BioTechnologies. [Google Scholar]
- Wittrup A., Lieberman J. (2015). Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16 543–552. 10.1038/nrg3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J. A., Malone R. W., Williams P., Chong W., Acsadi G., Jani A., et al. (1990). Direct gene transfer into mouse muscle in vivo. Science 247 1465–1468. 10.1126/science.1690918 [DOI] [PubMed] [Google Scholar]
- Wu H., Lima W. F., Zhang H., Fan A., Sun H., Crooke S. T. (2004). Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 279 17181–17189. 10.1074/jbc.M311683200 [DOI] [PubMed] [Google Scholar]
- Wurster C. D., Ludolph A. C. (2018). Nusinersen for spinal muscular atrophy. Ther. Adv. Neurol. Disord. 11:1756285618754459. 10.1177/1756285618754459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyanejad S., Gunst T., de, Schultz I., Boer H., den, et al. (2018). Abstract 4405: Pharmacologic profile of INT-1B3: a novel synthetic microRNA 193a-3p mimic for therapeutic intervention in oncology. Cancer Res. 78 4405–4405. 10.1158/1538-7445.AM2018-4405 [DOI] [Google Scholar]
- Yang H.-W., Huang C.-Y., Lin C.-W., Liu H.-L., Huang C.-W., Liao S.-S., et al. (2014). Gadolinium-functionalized nanographene oxide for combined drug and microRNA delivery and magnetic resonance imaging. Biomaterials 35 6534–6542. 10.1016/j.biomaterials.2014.04.057 [DOI] [PubMed] [Google Scholar]
- Zhan T., Rindtorff N., Betge J., Ebert M. P., Boutros M. (2019). CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 55 106–119. 10.1016/j.semcancer.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Zhang L., Chan J. M., Gu F. X., Rhee J.-W., Wang A. Z., Radovic-Moreno A. F., et al. (2008). Self-assembled lipid–polymer hybrid nanoparticles: a robust drug delivery platform. ACS Nano 2 1696–1702. 10.1021/nn800275r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-W., Li L., Li D., Liu J., Li X., Li W., et al. (2018). The first approved gene therapy product for cancer Ad-p53 (Gendicine): 12 years in the clinic. Hum. Gene Ther. 29 160–179. 10.1089/hum.2017.218 [DOI] [PubMed] [Google Scholar]
- Zhang X., Goel V., Robbie G. J. (2020). Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J. Clin. Pharmacol. 60 573–585. 10.1002/jcph.1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Li M., Zhang Z., Gong T., Sun X. (2016). Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 23 2596–2607. 10.3109/10717544.2015.1038856 [DOI] [PubMed] [Google Scholar]
- Zheng D., Giljohann D. A., Chen D. L., Massich M. D., Wang X.-Q., Iordanov H., et al. (2012). Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proc. Natl. Acad. Sci. U.S.A. 109 11975–11980. 10.1073/pnas.1118425109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P.-P., Kros J. M., Li J. (2018). Approved CAR T cell therapies: ice bucket challenges on glaring safety risks and long-term impacts. Drug Discov. Today 23 1175–1182. 10.1016/j.drudis.2018.02.012 [DOI] [PubMed] [Google Scholar]
- Zhu X., Yin L., Theisen M., Zhuo J., Siddiqui S., Levy B., et al. (2019). Systemic mRNA therapy for the treatment of fabry disease: preclinical studies in wild-type mice, fabry mouse model, and wild-type non-human primates. Am. J. Hum. Genet. 104 625–637. 10.1016/j.ajhg.2019.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T. S., Karsten V., Chan A., Chiesa J., Boyce M., Bettencourt B. R., et al. (2017). Clinical Proof of concept for a novel hepatocyte-targeting GalNAc-siRNA conjugate. Mol. Ther. 25 71–78. 10.1016/j.ymthe.2016.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinger A., Brozovich A., Pasto A., Sushnitha M., Martinez J. O., Evangelopoulos M., et al. (2020). Bioinspired extracellular vesicles: lessons learned from nature for biomedicine and bioengineering. Nanomaterials 10:2172. 10.3390/nano10112172 [DOI] [PMC free article] [PubMed] [Google Scholar]