Abstract
Over the last six decades production of laboratory rodents have been refined with the aim of eliminating all pathogens, which could influence research results. This has, however, also created rodents with little diversity in their microbiota. Until 10 years ago the impact of the microbiota on the outcome of rodent studies was ignored, but today it is clear that the phenotype of rodent models differs essentially in relation to the environment of origin, i.e. different breeders or different rooms. In this review, we outline the mechanisms behind gut bacterial impact on rodent models of immune mediated diseases, and how differences in environment of origin leads to phenotypic model differences within research areas such as infectious diseases and vaccine development, the metabolic syndrome, gut immunity and inflammation, autoimmunity and allergy. Finally, we sum up some tools to handle this impact to increase reproducibility and translatability of rodent models.
Introduction
In the late fifties of the twentieth century, a new era of specific pathogen free (SPF) rodent breeding was initiated by the use of caesarian section and barrier protection (Foster 1958), at first attempting to eliminate strong pathogens such as Citrobacter rodentium (by then C. freundii type 4280) (Barthold et al. 1976) and zoonoses, such as Streptobacillus moniliformis (Kaspareit-Rittinghausen et al. 1990). In the coming decades, and especially enforced by the first set of common European standards for health monitoring issued by the Federation of European Laboratory Animal Associations (Kraft et al. 1994) in the mid-nineties, the process of pathogen eradication was speeded up, and now also included microorganisms with a more discrete impact on the research models, such as various Helicobacter spp. (Fox et al. 1994). In parallel to this development, the rearing of germ-free animals, so-called gnotobiotechnology, was developed from the thirties (Glimstedt 1936; Reyniers et al. 1946), which made it clear that bacteria played an essential role in many characteristics of the mammal host (Falk et al. 1998; Gustafsson and Norin 1977; O'Mahony et al. 2009). To minimize the phenomenon that germ-free rodents seem to be predisposed to opportunistic infections, when removed from their isolators and introduced into barrier rooms, the so-called Schaedler Flora was developed and later modified as the Altered Schaedler Flora (ASF) (Dewhirst et al. 1999; Schaedler et al. 1965a). Today, the starting point for most commercial breeding colonies of rodents will be rederivation by caesarian section or embryo transfer and subsequent association with the eight ASF bacteria, and such animals can be bred for generations still harbouring these bacteria (Alexander et al. 2006; Schaedler et al. 1965b; Stehr et al. 2009). Most likely, the remaining members of the microbiome of a barrier bred colony are transferred from either humans currently emitting several microbe-carrying particles (Whyte et al. 1983), or from the diet (Zhang et al. 2010), and the microbiome of laboratory rodents is, therefore, essentially different from that of feral or conventionally bred rodents (Itoh et al. 1983; Wilson et al. 2006). Moreover, establishment of the gut microbiota among genetically similar mice from different commercial vendors was already decades ago reported to be different (Hirayama et al. 1990a; O'Rourke et al. 1988). Even different colonies of the same strain within one vendor may differ (Hufeldt et al. 2010), and the same colony may differ over time (Fahey et al. 2017; O'Rourke et al. 1988). This can be problematic, as the microbiome with its more than 1 million genes is responsible for much of the phenotypic expression of an animal model (Bleich and Hansen 2012; Ellekilde et al. 2014; Hansen et al. 2014a; Holmes and Nicholson 2005), which also includes an essential impact on the immune system (Fig. 1). This includes stimulation of the innate immune system, when microbial molecules known as microbial-associated molecular patterns (MAMP), without any forms of previous memory-inducing stimulation, react with pattern-recognition receptors (PRR) in the host (Tlaskalova-Hogenova et al. 2004). It also includes stimulation of the adaptive immune system, as one single bacterial molecule may express several epitopes, which after presentation by antigen presenting cells, will turn naïve T cells, into specific CD4+ T cells, including T helper cells of type 1, type 2, type 3 and type 17 as well as regulatory T cells, or CD8+ cytotoxic T cells (D'Elios et al. 2011; Korn et al. 2009; Mosmann and Coffman 1989; Ohkura et al. 2013). In mice, the microbiota establishes itself around weaning, and this establishment to a certain extent guides the future immune responses of the host (Hansen et al. 2012b), as early life microbial encounters induce gut oral tolerance, which may counteract inflammatory or autoimmune disease later in life (Castro-Sanchez and Martin-Villa 2013; Gratz et al. 2013; Hsieh et al. 2012). Variation in microbiota stimulation at the breeding facility may therefore be a likely cause of phenotypic variation later in life, because many of the models are driven by specific T cell types (Bleich and Hansen 2012). Current microbial stimulation of the gut lamina propria dendritic cells induces secretion of IL-12 from these, which will favour T helper cell type 1 differentiation on the cost of T helper cell type 2 differentiation (Hsieh et al. 1993) (Fig. 1). Therefore, animals with a low diversity or even being germ-free will everything else equal be dominated by T helper cells type 2 in their immune responses (Mazmanian and Kasper 2006).
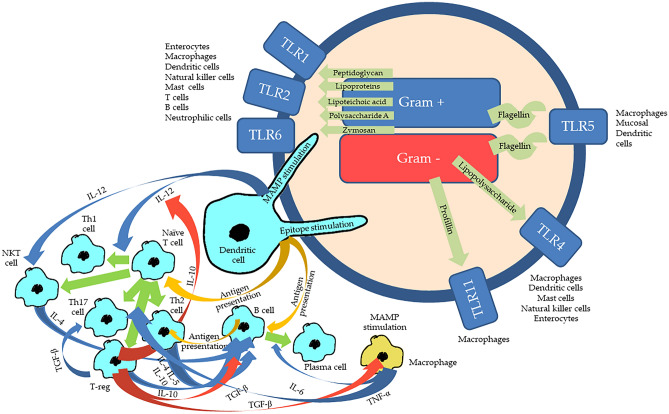
Fig. 1.
Immune activation by bacteria in the gut. In the gut the immune cells and the enterocytes possess different types of Toll-like receptors (TLR), which can be stimulated by different types of microbial associated molecular patterns (MAMP), which dependent on cell type and TLR will cause different forms of innate immune activation and cytokine production. MAMPS are such as peptidoglycan, lipoproteins, lipoteichoic acid, polysaccharide A and zymosan from Gram positive bacteria, lipopolysaccharides and profillin from Gram negative bacteria, and flagellin from bacterial flagellas. Epitopes, which are different parts of bacterial molecules with an adaptive immune stimulatory potential, will be presented by antigen presenting cells (yellow arrows), such as the dendritic cells, to T and B cells. The cytokine production initiated by the innate MAMP-TLR stimulation will upscale (blue arrows) or downscale (red arrows) the differentiation (green arrows) of T and B cells into effector cells. Stimulation by IL-12 secreted by the dendritic cells as a response to MAMP stimulation will facilitate the differentiation of naïve T cells, into T helper cells type 1 (Th1) on the cost of T helper cells type 2 (Th2), while this process is downscaled by IL-10 from the regulatory T cells (T-reg). IL-12 will also stimulate the natural killer T (NKT) cells. The B cells will present the antigen for the Th2s, which will respond back with IL-4 and IL-5 to turn the B cell into an antigen producing plasma cell. This process is upscaled by IL-6 caused by the MAMP-TLR stimulation of the macrophages, and B cells will be stimulated by IL-10 from T-regs and Th2, while TGF-β from the T-regs will downscale activities of both B cells and macrophages. TGF-β will upscale the activity of Th17 cells. The activated macrophages will produce TNF-α, which will stimulate a number of different T cells. There are far more cytokines and cell types than this, there are other pattern recognition receptors than the TLRs, and in relation to gut virus infections cytotoxic T cells will also be relevant
The microbiota in addition to bacteria also includes protozoa and fungi, and the bacteria also have their own viruses, the so-called phages, which also can be shown to have an impact on animal model expression (Rasmussen et al. 2019, 2020). Therefore, the microbiota of genetically closely related strains from different vendors may be essentially different (Hirayama et al. 1990b; Hufeldt et al. 2010), and, therefore, in some (Rasmussen et al. 2019), but not in all cases (Siersbæk et al. 2020), be responsible for the lack or reproducibility of rodent studies. Recently, the SPF principles for rodent breeding have also been debated, as pathogens are important for the basic activation and the function of the immune system (Tao and Reese 2017). Therefore, it has been argued that especially within immunology one of the reasons for failures of translating results from the preclinical to the clinical phase of research is due to the naïve pathogen status of the animal models (Masopust et al. 2017). In relation to the abundances of cytotoxic T cells and T helper cells, a laboratory mouse unlike pet shop and wild mice more resemble the immune system of a newborn than that of an adult human being (Beura et al. 2016).
Therefore, the aim of this review is to elucidate how the commensal as well as the pathogenic microbiota impact the expression, translatability and reproducibility of rodent models on immune mediated diseases. In this review, we discuss major findings to date and challenges faced when using very clean SPF mice compared to the use of conventional, pet shop or feral mice for immunological research.
Microbiota impact on reproducibility of various immunological mouse models
Infectious diseases and vaccine development
It is quite clear from the literature that the microbiota has a profound impact on disease severity and expression in infection models. For instance, it can be shown by antibiotic treatment that the microbiota enhances the severity of some gastrointestinal infections, such as human poliovirus (Kuss et al. 2011), murine norovirus (Jones et al. 2014) and mouse mammary tumour virus MMTV (Kane et al. 2011), while it alleviates severity for some respiratory infections, such as human influenza virus (Ichinohe et al. 2011; Steed et al. 2017; Wu et al. 2013), Streptococcus pneumonia (Brown et al. 2017), Klebsiella pneumonia (Brown et al. 2017) and Mycobacterium tuberculosis (Dumas et al. 2018; Khan et al. 2016). If mice infected with C. rodentium harbour a complex microbiota, they will at some time point of infection express some virulence genes (Kamada et al. 2012). This will relocate the agent to the gut lumen to be outcompeted by the commensal microbiota (Kamada et al. 2012). In contrast, germ-free mice are unable to eradicate C. rodentium (Kamada et al. 2012). A very nice example of the impact a vendor can have, was published by a group that noted that the source of C57BL/6 mice influenced the severity of an infection model of malaria (Villarino et al. 2016). The degree of parasitemia, weight loss and mortality were greater in mice from Envigo (Harlan) and Charles River compared to the more resistant mice from Jackson and Taconic, which harboured substantially different microbial communities in their gastrointestinal tract (Villarino et al. 2016). The causal role of the gut microbiota was nicely demonstrated by cecal transplant to germ-free mice (Villarino et al. 2016). Even mice from the same vendor, changed susceptibility to both malaria and Salmonella infection over time due to a change in the gut microbiota (Mandal et al. 2020), demonstrating the importance of not only considering the origin of the mice, but also the timing when comparing studies of the same character. In particular, vendor differences in Escherichia coli cause variation in host response to Salmonella infection by competing with Salmonella for resources (Velazquez et al. 2019). Moreover, transferring the clearly differing natural gut microbiome from a population of wild mice, which genetically were closely related to laboratory mice, made the recipients exhibit reduced inflammation and increased survival following an otherwise lethal influenza virus infection (Rosshart et al. 2017). In another study with ‘dirty mice’, in which laboratory mice caught pathogens when co-housed with pathogen infected pet shop mice those laboratory mice, which survived the high mortality that it induced, experienced lower severity of disease, when infected with Plasmodium berghei and Listeria monocytogenes (Beura et al. 2016). It has for the same reason been debated, whether it would be a good idea to test vaccines both in pathogen free and pathogen infected mice to improve validity of infection models (Masopust et al. 2017). Antibody responses induced by influenza vaccination in germ-free or antibiotic-treated TLR5 deficient mice were impaired, but restored by oral reconstitution with a flagellated, but not aflagellated, strain of Eschericia coli (Oh Jason et al. 2014). Antibiotic exposure in infant but not adult mice impaired antibody responses to vaccination against Meningococcus B (MenB) and C (MenC), Pneumoccus (PCV13), Mycobacteria (BCG) as well as against the polyvalent 6-in-1 Hexa vaccine, while restoring the commensal microbiota rescued the impaired antibody responses (Lynn et al. 2018). So, as microbiota differs substantially between different colonies of the same strain of mice (Ericsson et al. 2015; Hufeldt et al. 2010; Rasmussen et al. 2019; Siersbæk et al. 2020), there are good reasons to assume that there is an impact of the microbiota on the disease expression in infection models, and that this also has an impact on vaccine efficiency.
Metabolic syndrome
Inflammation is a key element of the metabolic syndrome, as much of the peripheral insulin resistance is caused by cytokine interaction with the peripheral cell insulin receptors (Boucher et al. 2014). The most commonly applied animal model for this is the diet induced obese C57BL/6 mouse (Varga et al. 2010). This model can also be done on leptin or leptin receptor deficient mice (Varga et al. 2010). Much focus has been on microbiota impact on these models, since Turnbaugh et al. successfully transferred the obese phenotype to lean germ-free mice through a microbiota transplantation 15 years ago (Turnbaugh et al. 2006). The phenotype in that study was only characterized in relation to obesity and lipid parameters (Turnbaugh et al. 2006), but later Vrieze et al. showed that similar microbiota transplantation from lean to obese humans for a short period improved insulin resistance (Vrieze et al. 2012). In mice high levels of IL-10, IL-12, TNF-α as well as regulatory T cells correlate to impaired glucose tolerance (Ellekilde et al. 2014), as it does in humans (Claesson et al. 2012). One explanation may be that lipopolysaccharides (LPS) from the Gram negative bacteria of the gut or the diet enters the blood stream, known as metabolic endotoxinaemia, and by TLR4 stimulation induces cytokine secretion which interacts with the insulin receptor (Cani et al. 2007; Lindenberg et al. 2019). The gut microbiota of both C57BL/6N (Rasmussen et al. 2019) and C57BL/6J (Siersbæk et al. 2020) differ substantially between mice from different vendors. Differences in the microbiota, i.e. both bacteria and phages, of C57BL/6N mice could explain much of the phenotypic difference between mice from different vendors (Rasmussen et al. 2019). Even differences in microbiota between different facilities from the same vendor can affect the metabolic phenotype of high-fat diet induced obese CD-1 mice (Unger et al. 2020). The development of obesity and expression of the IFN-γ gene in the liver in these mice was reduced by phage transplantation in C57BL/6N mice (Rasmussen et al. 2020). In contrast, the microbiota seemed to have little impact on the phenotypic differences observed in C57BL/6J mice from different vendors (Siersbæk et al. 2020). Variation in important measures of barrier function, metabolic endotoxemia, and low-grade inflammation related to high-fat feeding is also strongly affected by the microbial differences observed between conventional and SPF housing. Several of the obesity-related metabolic dysfunctions disappear in too clean environments (Müller et al. 2016), including the inflammatory response of the liver, which is pivotal to develop a proper animal model for non-alcoholic fatty liver disease (Chen et al. 2020b). There does not seem to be any studies on the development of the cardiovascular part of the metabolic syndrome in mice in relation to how this differs between different environments, such as commercial vendors, but the involvement of the microbiota seems to become increasingly well documented. Hypertension seems to be influenced by the gut microbiota, as fecal microbiota transplantation to germ-free mice increases blood pressure (Li et al. 2017). As antibiotics and germ-free status influence the host differently, there are conflicting results between studies of germ-free and antibiotic treated mice (Bayer et al. 2019). The development of atherosclerosis is not altered in germ-free LDL receptor deficient mice (Kiouptsi et al. 2020), but microbiota depletion with antibiotics in mice has been shown to suppress the development of atherosclerosis in mice (Chen et al. 2020a; Man et al. 2020; Rune et al. 2016). This is not similarly evident in rats (Rune et al. 2018). Dietary lipid intake, gut microbiota and development of atherosclerosis is linked in mice, which seems to be mediated through the production of inducing metabolites, such as trimethylamine N-oxide (TMAO) (Wang et al. 2011). TMAO promotes the release of pro-inflammatory cytokines, such as IL-1β, IL-6 and IL-18 (Seldin et al. 2016; Yue et al. 2017), which may be mediated by stimulation of TLR5 by flagellin, which promotes hepatic production of ApoE (Yiu et al. 2020).
Gut immunity and inflammation
The development of inflammatory bowel disease (IBD), i.e. T helper cell type 2 dominated ulcerative colitis and the T helper cell type 1 dominated Crohn’s disease, is obviously under strong impact of the microbiota (Nell et al. 2010), as the gut microbiota is a key element in the disease etiology (Nell et al. 2010). The ability of mouse models to develop IBD is, therefore, under strong influence of the presence or absence of specific bacteria. From the first attempts to turn conventional IBD prone mice into SPF mice, it has been realized that germ-free status eliminates or dramatically alleviates symptoms of IBD in mice, such as in senescence accelerated P1/Yit mice (Matsumoto et al. 1998), as well as in mice deficient of IL-10 (Sellon et al. 1998), IL-2 (Contractor et al. 1998), T-cell receptor-α (Dianda et al. 1997) and intestine specific transforming growth factor-β signalling (Hahm et al. 2001). IL-10 deficient mice spontaneously develop chronic colitis, when maintained in conventional facilities, but they fail to express a similar phenotype under SPF conditions. The presence of Helicobacter hepaticus is, however, sufficient for the SPF IL-10 deficient mice to develop a more severe intestinal inflammatory response (Kullberg et al. 1998). Another important example is Faecalibacterium prausnitzii (Carlsson et al. 2013), which also in humans is an important anti-inflammatory agent with a potential to alleviate severity of IBD (Lopez-Siles et al. 2018). Yet another very important example is segmented filamentous bacteria (SFB or Candidatus Savagella), which strongly influences IgA production, T-cell response, and intestinal T helper cell type 17 induction (Ericsson et al. 2014; Gaboriau-Routhiau et al. 2009), as well as the functional specializations of antigen presenting cells able to induce regulatory T cells or T helper cell type 17 (Denning et al. 2011). The first descriptions of the T helper cell type 17 were published, when a research group discovered that C57BL/6 mice obtained from several university and commercial SPF sources had relatively high proportions of T helper cells type 17 (Ivanov et al. 2008). In contrast, C57BL/6 mice obtained from the Jackson Laboratory consistently had only very few, and, therefore, the adaptive transfer model of Crohn’s disease could not be induced in Jackson mice (Ivanov et al. 2008). Another important factor is the properties of the intestinal mucus layer in preventing intestinal bacteria from penetrating and getting in contact with the intestinal epithelial lining. The mucus phenotype was found to differ between different rooms of the same facility which would be expected to have an essential impact on IBD models (Jakobsson et al. 2015). Wild mice have mucus impenetrable to bacteria and are also resistant to induction of IBD and colitis-associated tumorigenesis (Rosshart et al. 2017). So, it is beyond any discussion that the gut microbiota is of crucial importance for the reproducibility of IBD studies between mice from different commercial vendors, housing conditions and in comparison to wild or dirty mice. This would also be expectable in other animal models of intestinal inflammation. As an example, mice expressing the human DQ8 gene, which confers moderate susceptibility to gluten-induced immunopathology, i.e. celiac disease, develop increased response to dietary gluten in conventional conditions compared to SPF conditions with a more restricted microbial composition (Galipeau et al. 2015). The attenuated response in the SPF mice was reversed by introducing a Proteobacterium from a patient with celiac disease (Galipeau et al. 2015).
Autoimmunity
Environmental factors are at play in defining susceptibility to type 1 diabetes in both humans and animal models. Disease incidence in non-obese diabetic (NOD) mice and Bio-Breeding (BB) rats, which spontaneously develop hyperglycemia, vary substantially depending on barrier status in different facilities (Klöting et al. 1984; Pozzilli et al. 1993; van der Werf et al. 2007). One study transplanted microbiota from a low-incidence NOD colony to a high-incidence colony but failed to transfer protection due to a few bacterial taxa not successfully transferred, including Akkermansia muciniphila (Hanninen et al. 2018). However, upon weekly gavage of the high incidence colony with A. muciniphila, the authors found a delay in diabetes onset along with an improvement in gut barrier function and regulatory immunity, demonstrating the significance of harboring this specific taxon in NOD mouse facilities (Hanninen et al. 2018). A. muciniphila was first associated with a reduced incidence of type 1 diabetes in NOD mice that were treated with vancomycin, which eliminates most taxa and, thereby, favours growth of A. muciniphila (Hansen et al. 2012a). Interestingly, another study with vancomycin treated NOD mice shortly after reported a similar increase in the abundance of A. muciniphila, but the diabetes incidence of this population was higher compared to untreated mice (Brown et al. 2016). The contradicting results may likely be explained by the remaining microbiota composition, as the mice in the first study from Taconic were not colonized by SFB unlike the Jackson mice in the study by Brown et al. The presence of SFB was already a decade ago shown to be associated with the diabetes incidence of a NOD colony (Barfod et al. 2015), and antibiotic treatment of the NOD mice colonies at Jackson eliminated SFB from their colony which caused a noteworthy increase in the diabetes incidence in their otherwise low incidence colony (Fahey et al. 2017). Importantly, A. muciniphila and SFB have no effect on diabetes incidence in mono-colonized NOD mice (Hansen et al. 2016; Yurkovetskiy et al. 2013), and thus appear to exert their protective properties through interaction with other commensals. The gut microbiota is also a strong mediator of dietary interventions, and it is not surprising that the result of a dietary change depends on the host’s microbial composition. A gluten-free diet, for example, had an alleviating effect in NOD mice, in which A. muciniphila was propagated by the diet (Hansen et al. 2014b; Marietta et al. 2013), but failed to reduce diabetes incidence in NOD mice, in which A. muciniphila, for unknown reason, was reduced (Liu et al. 2016). Treatment with the antibiotic fusidic acid reduces the incidence of type 1 diabetes in BB rats (Kaas et al. 2001) probably by preventing the impact of IL-1 and IL-6 on the islets (Bendtzen et al. 1992).
Susceptibility and severity of arthritis in a variety of rodent strains also depends on the gut microbiota environment, which often exhibit reduced bacterial diversity in laboratory animal facilities. In the collagen induced mouse model for rheumatoid arthritis (RA), susceptibility is highest in mice enriched with Lactobacillus, and transplantation of feces from mice with high susceptibility was shown to increase the frequency of arthritis induction compared to recipients of feces from arthritis-resistant mice (Liu et al. 2016). Also for the SKG mouse model, disease severity depends on the microbial environment; arthritis develops spontaneously in conventional facilities, while it must be induced in SPF facilities (Sakaguchi et al. 2003). This phenotype seems to be mainly depending on a single species Prevotella copri, which is sufficient to induce arthritis, when mono-colonized in germ-free SKG mice by induction of intestinal Th17 cells (Maeda et al. 2016). The other well-known Th17-inducing bacteria, SFB, also drives autoimmune arthritis, as shown in SFB mono-colonized K/BxN mice (Wu et al. 2010). At first, the authors noticed low auto-antibody titers and attenuated arthritis development in K/BxN mice from Jackson, which typically lack this bacterium and a proper Th17 response (Wu et al. 2010). Therefore, they tested the importance of SFB by transferring germ-free K/BxN mice to a SFB-negative SPF facility, while treating some of them with oral gavage with feces from SFB-monocolonized mice (Wu et al. 2010). The SFB-treated mice showed accelerated arthritis development and appearance of gut Th17 cells, which migrated systemically to trigger the autoimmune response (Wu et al. 2010).
Allergy
Atopic individuals with a predisposition to developing allergic asthma, food allergies, anaphylaxis and childhood eczema, i.e. atopic dermatitis, are becoming increasingly prevalent, and in industrialized countries they represent 20–25% of the population (Al-Herz 2018). There is evidence that the skin and gut microbiomes contribute to allergic reactions of the skin, which is usually the first allergic response in IgE sensitized children, which is then followed by allergic rhinitis and asthma later in life (Bantz et al. 2014). Overgrowth of skin colonizing Staphylococcus aureus is well known to precede onset of disease in atopic dermatitis patients (Meylan et al. 2017), and its abundance correlates with severity of dermal symptoms (Tauber et al. 2016). Perhaps for the same reason, it is not strange that in SPF facilities, in which S. aureus is often notoriously eliminated, certain spontaneous animal models of atopic dermatitis, such as the NC/Nga and DS-Nh mice, fail to display skin lesions and IgE levels (Hikita et al. 2002; Suto et al. 1999). This is comparable to human atopic dermatitis, and to mice raised in conventional facilities (Hikita et al. 2002; Suto et al. 1999). In contrast, a diverse gut microbiota early in life has been shown to prevent high IgE levels and risk of allergy later in life (Cahenzli et al. 2013). However, besides the presence in conventional mice of fur mites and S. aureus, both strongly involved in the induction of dermal symptoms, mechanistic pathways underlying gut microbial influences on IgE levels and development of allergies are likely more complex and species specific. As an example, intestinal Prevotella spp. were found to be strongly associated with low IgE levels and a low-responding phenotype in mice with oxazolone-induced dermatitis, and prebiotic treatment of the mice propagated Prevotella spp. and reduced the number of high-responders (Laigaard et al. 2020). In addition, the high-and low-responding phenotypes of mice with oxazolone-induced dermatitis were transferable with the gut microbiome to germ-free mice, strongly indicating the importance of its composition (Zachariassen et al. 2017). Of note, wild mice share the majority of their skin microbial taxa with laboratory mice which therefore would not be expected to influence the outcome on atopic dermatitis models drastically (Belheouane et al. 2020). This is however in contrast to the more diverse gut microbiota evident in wild life mice (Ericsson et al. 2017), which unlike the skin microbiota may play an essential role comparing wild mice with laboratory mice.
Not only the gut and skin microbiota, but also the respiratory microbiome vary with vendor, and it is strongly correlated with the local immune response in the lung (Dickson et al. 2018). As such, a large variation exists in airway responsiveness among mice from different vendors used as asthma models (Chang et al. 2012). Also, the gut microbiota contributes to development of pulmonary inflammation. Germ-free mice have more NKT cells, both systemically and in the lung, compared to SPF mice, and they are more prone to allergic asthma (Hansen et al. 2012b; Olszak et al. 2012). Abundance of invariant natural killer T (iNKT) cells was especially dependent on stimulation with a glycosphingolipid from Bacteroides fragilis in the very early life (An et al. 2014), which pinpoints that in the early time window before the animals are shipped to the research facilities, the gut microbiota compositions associated with vendor’s facilities are important. Specifically, vendor differences strongly affect iNKT cell phenotypes and functions, and, for example, the percentage of iNKT cells in the spleen is significantly lower in Taconic compared Jackson mice (Wingender et al. 2012). It is, however, important to note that the vendor effect is model specific, as for example an LPS induced lung inflammation model was unaffected by vendor despite clear differences in gut microbiota and pulmonary TNF-α levels (Wolff et al. 2020).
Strategies for handling impact on the mouse models of immune diseases
Having obtained a basic understanding on how essential the microbiota is for immune-mediated disease models, the next steps will be to handle this in biomedical research.
First step will be simply to be aware that rodents from different environments, such as vendors and rooms, have different microbiota, and that this most likely will lead to differences in phenotypes. Based upon the huge amount of literature available scientists should carefully consider in the study planning, which animals of which origin would be most feasible for a specific study. Also, they should carefully report the exact origin of the animals applied in manuscripts, as guided by the ARRIVE guidelines (Kilkenny et al. 2010). This also includes precision in the description of the diet supplied, as diet is a very strong microbiota modifier (Moreno-Indias et al. 2020). Also, the vendors must soon respond to this new situation and be willing to supply information of the status of their animals; at least for a short list of bacteria generally relevant for many research areas (Hansen et al. 2019). With a rapid development and reduction in costs of microbiota sequencing these methods have become generally available, and much more cost efficient than the bacterial pathogen cultivation used by the vendors ever since the introduction of the SPF concept. In the longer run, routine sequencing will also lead to experience in accounting for the microbiota impact on animal models. Sequencing data will allow incorporation of the microbiota composition as a factor in the statistical analyses. Thereby, we will get even more information on specific bacterial species relevant for disease in the animals, which is also highly relevant for developing microbiota-mediated therapeutics or personalized medicine for human patients. A special discussion topic will be, whether we should start testing vaccines and immune interventions on ‘dirty’ animals, i.e. animals infected by the well-known pathogens. This has been excellently reviewed by Masopust et al. who argues that this should be seen as a supplement rather than an alternative to the use of SPF mice (Masopust et al. 2017). It will be a major challenge if we were to re-introduce eradicated pathogens in breeding colonies, and this does not seem very likely to happen. Also, it should be considered that captivity bred mice, such as a pet shop mice, although infected with all the well-known pathogens do not have an immune system comparable to a wild mouse, which for instance has a much lower abundance of CD8+ cells; probably due to a lower viral exposure (Beura et al. 2016).
Next step could be to develop rodents tailor-made with microbiota specific for certain research purposes. This may not necessarily be attractive as a general trend, as the diversity among rodents used would, if combined with a thorough microbiota characterization as described above, give detailed information on microbiota impact on disease development. On the other side, it does not make much sense to use animals, which cannot develop specific disease phenotypes or respond to certain interventions, because they lack specific bacteria. A simple approach is, therefore, to ensure that at least these are harboured by the animal. This could for example be SFB’s in mice used for IBD research (Ivanov et al. 2009), A. muciniphila in mice used for type 1 diabetes research (Hanninen et al. 2018; Hansen et al. 2012a), or Bifidobacterium spp. in mice used to test oligosaccharides (Hansen et al. 2013). This is not necessarily a simple task, and Hanninen et al. had to pass over germ-free status, mono-association and then fecal matter transplants to associate NOD mice with A. muciniphila (Hanninen et al. 2018). In this package of tools also belongs the opportunity to modify the microbiota into a desirable state through the diet, e.g. by using diets with a more humanized profile to optimize human microbiota transplantation studies (Moreno-Indias et al. 2020). The more advanced step will be to give the animals specific complex microbiotas for specific studies. In the present world, this can be difficult as too little is known about what would be the perfect phenotype for a specific disease phenotype in rodents, and whether this would be relevant for the human patients. However, it might within certain research types, such as atopic dermatitis, for which it is known that there is a strong correlation between microbiota and phenotype (Lundberg et al. 2012), be possible to conserve and transplant high responder microbiotas (Zachariassen et al. 2017), as these can be shown to respond to interventions (Debes et al. 2020). The transfer is not necessarily an easy task, as the colonization rate from mice to mice is approximately only 60—80%, and the transplanted mice seem to have a Firmicutes overgrowth (Lundberg et al. 2017). On the other hand, over generations the microbiota seems to be quite stable independent of housing systems (Lundberg et al. 2020). Microbiota transfer can be used to give mice a human microbiota, although the colonization rate will be even lower and key bacteria, such as F. prausnitzii and Bifidobacterium spp., fail to colonize (Lundberg et al. 2020). Another attractive approach could be to transfer a wild mouse microbiota to laboratory mice (Rosshart et al. 2017); a technique that has been further improved by the creation of so-called ‘wildlings’, i.e. C57BL/6 mice born by wild mouse mothers after embryo transfer (Rosshart et al. 2019). One problem is that not only will the commensal microbiota be transplanted, but the pathogens will come as well.
Conclusion
The microbiota has a substantial impact on immune models in rodents and may be the cause of essential phenotypic variation due to differences in the originating environment of the animals. In the handling of the reproducibility crisis, this is a factor not to be ignored.
Author contributions
The authors contributed equally to the literature search and writing of this work, and they both accepted the final manuscript.
Funding
Funding was not obtained for this work.
Compliance with ethical standards
Conflict of interest
Potential conflicts of interests for the authors can be found at https://ivh.ku.dk/english/employees/?pure=en/persons/107126 and https://ivh.ku.dk/english/employees/?pure=en/persons/306048.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Axel Kornerup Hansen, Email: akh@sund.ku.dk.
Camilla Hartmann Friis Hansen, Email: camfriis@sund.ku.dk.
References
- Al-Herz W. A systematic review of the prevalence of atopic diseases in children on the Arabian Peninsula. Med Princ Pract. 2018;27:436–442. doi: 10.1159/000493267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AD, Orcutt RP, Henry JC, Baker J, Bissahoyo AC, Threadgill DW. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–1104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantz SK, Zhu Z, Zheng T. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. J Clin Cell Immunol. 2014;5:202. doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfod KK, Vrankx K, Mirsepasi-Lauridsen HC, Hansen JS, Hougaard KS, Larsen ST, Ouwenhand AC, Krogfelt KA. The murine lung microbiome changes during lung inflammation and intranasal vancomycin treatment. Open Microbiol J. 2015;9:167–179. doi: 10.2174/1874285801509010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, Jonas AM. The etiology of transmissible murine colonic hyperplasia. Lab Anim Sci. 1976;26:889–894. [PubMed] [Google Scholar]
- Bayer F, Ascher S, Pontarollo G, Reinhardt C. Antibiotic treatment protocols and germ-free mouse models in vascular research. Front Immunol. 2019 doi: 10.3389/fimmu.2019.02174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belheouane M, Vallier M, Čepić A, Chung CJ, Ibrahim S, Baines JF. Assessing similarities and disparities in the skin microbiota between wild and laboratory populations of house mice. ISME J. 2020;14:2367–2380. doi: 10.1038/s41396-020-0690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtzen K, Diamant M, Horn T, Pedersen C, Buschard K. Effect of fusidic acid on interleukin-1 (Il-1)-induced and Il-6-induced pancreatic beta-cell functions in rats. J Endocrinol. 1992;132:345–352. doi: 10.1677/joe.0.1320345. [DOI] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, Sekaly RP, Jenkins MK, Vezys V, Haining WN, Jameson SC, Masopust D. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532:512–516. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich A, Hansen AK. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp Immunol Microbiol Infect Dis. 2012;35:81–92. doi: 10.1016/j.cimid.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014 doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Godovannyi A, Ma C, Zhang Y, Ahmadi-Vand Z, Dai C, Gorzelak MA, Chan Y, Chan JM, Lochner A, Dutz JP, Vallance BA, Gibson DL. Prolonged antibiotic treatment induces a diabetogenic intestinal microbiome that accelerates diabetes in NOD mice. ISME J. 2016;10:321–332. doi: 10.1038/ismej.2015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. 2017;8:1512. doi: 10.1038/s41467-017-01803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmee E, Cousin B, Sulpice T, Chamontin B, Ferrieres J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Carlsson AH, Yakymenko O, Olivier I, Hakansson F, Postma E, Keita AV, Soderholm JD. Faecalibacterium prausnitzii supernatant improves intestinal barrier function in mice DSS colitis. Scand J Gastroenterol. 2013;48:1136–1144. doi: 10.3109/00365521.2013.828773. [DOI] [PubMed] [Google Scholar]
- Castro-Sanchez P, Martin-Villa JM. Gut immune system and oral tolerance. Br J Nutr. 2013;109(Suppl 2):S3–11. doi: 10.1017/S0007114512005223. [DOI] [PubMed] [Google Scholar]
- Chang HY, Mitzner W, Watson J. Variation in airway responsiveness of male C57BL/6 mice from 5 vendors. J Am Assoc Lab Anim Sci. 2012;51:401–406. [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Black AS, Sobel AL, Zhao YN, Mukherjee P, Molparia B, Moore NE, Muench GRA, Wu JJ, Chen WX, Pinto AFM, Maryanoff BE, Saghatelian A, Soroosh P, Torkamani A, Leman LJ, Ghadiri MR. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol. 2020;38:1288–1297. doi: 10.1038/s41587-020-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Wang YC, Chiu CC, Lee YP, Hung SW, Huang CC, Chiu CF, Chen TH, Huang WC, Chuang HL. Housing condition-associated changes in gut microbiota further affect the host response to diet-induced nonalcoholic fatty liver. J Nutr Biochem. 2020;79:108362. doi: 10.1016/j.jnutbio.2020.108362. [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Contractor NV, Bassiri H, Reya T, Park AY, Baumgart DC, Wasik MA, Emerson SG, Carding SR. Lymphoid hyperplasia, autoimmunity, and compromised intestinal intraepithelial lymphocyte development in colitis-free gnotobiotic IL-2-deficient mice. J Immunol. 1998;160:385–394. doi: 10.4049/jimmunol.160.1.385. [DOI] [PubMed] [Google Scholar]
- D'Elios MM, Benagiano M, Della Bella C, Amedei A. T-cell response to bacterial agents. J Infect Dev Ctries. 2011;5:640–645. doi: 10.3855/jidc.2019. [DOI] [PubMed] [Google Scholar]
- Debes KP, Evdina NA, Laigaard A, Larsen JM, Zachariassen LF, Hansen CH, Hansen AK. Betamethasone treatment for atopic dermatitis in gut microbiota transplanted mice. Comp Med. 2020;70:6–15. doi: 10.30802/AALAS-CM-18-000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733–747. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. Phylogeny of the defined murine microbiota: altered schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/AEM.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol. 1997;150:91–97. [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am J Respir Crit Care Med. 2018;198:497–508. doi: 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A, Corral D, Colom A, Levillain F, Peixoto A, Hudrisier D, Poquet Y, Neyrolles O. The host microbiota contributes to early protection against lung colonization by Mycobacterium tuberculosis. Front Immunol. 2018;9:2656. doi: 10.3389/fimmu.2018.02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellekilde M, Krych L, Hansen CH, Hufeldt MR, Dahl K, Hansen LH, Sorensen SJ, Vogensen FK, Nielsen DS, Hansen AK. Characterization of the gut microbiota in leptin deficient obese mice—correlation to inflammatory and diabetic parameters. Res Vet Sci. 2014;96:241–250. doi: 10.1016/j.rvsc.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med. 2014;64:90–98. [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS ONE. 2015;10:19. doi: 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Montonye DR, Smith CR, Franklin CL. Modeling a superorganism—considerations regarding the use of "dirty" mice in biomedical research yale. J Biol Med. 2017;90:361–371. [PMC free article] [PubMed] [Google Scholar]
- Fahey JR, Lyons BL, Olekszak HL, Mourino AJ, Ratiu JJ, Racine JJ, Chapman HD, Serreze DV, Baker DL, Hendrix NK. Antibiotic-associated manipulation of the gut microbiota and phenotypic restoration in NOD mice. Comp Med. 2017;67:335–343. [PMC free article] [PubMed] [Google Scholar]
- Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157–1170. doi: 10.1128/MMBR.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster HL. Large scale production of rats free of commonly occurring pathogens and parasites. Proc Anim Care Panel. 1958;8:92–100. [Google Scholar]
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ, Gorelick PL, Ward JM. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol. 1994;32:1238–1245. doi: 10.1128/jcm.32.5.1238-1245.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Galipeau HJ, McCarville JL, Huebener S, Litwin O, Meisel M, Jabri B, Sanz Y, Murray JA, Jordana M, Alaedini A, Chirdo FG, Verdu EF. Intestinal microbiota modulates gluten-induced immunopathology in humanized mice. Am J Pathol. 2015;185:2969–2982. doi: 10.1016/j.ajpath.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimstedt G. Metabolism of bacteria free animals. I. General methods. Skand Arch Physiol. 1936;73:48–62. doi: 10.1111/j.1748-1716.1936.tb01451.x. [DOI] [Google Scholar]
- Gratz IK, Rosenblum MD, Abbas AK. The life of regulatory T cells. Ann N Y Acad Sci. 2013;1283:8–12. doi: 10.1111/nyas.12011. [DOI] [PubMed] [Google Scholar]
- Gustafsson BE, Norin KE. Development of germfree animal characteristics in conventional rats in antibiotics. Acta Pathol Microbiol Scand. 1977;85B:1–8. doi: 10.1111/j.1699-0463.1977.tb01667.x. [DOI] [PubMed] [Google Scholar]
- Hahm KB, Im YH, Parks TW, Park SH, Markowitz S, Jung HY, Green J, Kim SJ. Loss of transforming growth factor beta signalling in the intestine contributes to tissue injury in inflammatory bowel disease. Gut. 2001;49:190–198. doi: 10.1136/gut.49.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanninen A, Toivonen R, Poysti S, Belzer C, Plovier H, Ouwerkerk JP, Emani R, Cani PD, De Vos WM. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- Hansen CHF, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, Hansen A. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- Hansen CHF, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, Tlaskalova-Hogenova H, Hansen AK. Patterns of early gut colonization shape future immune responses of the host. PLoS ONE. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CHF, Frøkiær H, Christensen AG, Bergström A, Licht TR, Hansen AK, Metzdorff SB. Dietary xylooligosaccharide downregulates IFN-γ and the low-grade inflammatory cytokine IL-1β systemically in mice. J Nutr. 2013;143:533–540. doi: 10.3945/jn.112.172361. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Hansen CHF, Krych L, Nielsen DS. Impact of the gut microbiota on rodent models of human disease. World J Gastroenterol: WJG. 2014;20:17727. doi: 10.3748/wjg.v20.i47.17727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CH, Krych L, Buschard K, Metzdorff SB, Nellemann C, Hansen LH, Nielsen DS, Frokiaer H, Skov S, Hansen AK. A maternal gluten-free diet reduces inflammation and diabetes incidence in the offspring of NOD mice. Diabetes. 2014;63:2821–2832. doi: 10.2337/db13-1612. [DOI] [PubMed] [Google Scholar]
- Hansen CH, Yurkovetskiy LA, Chervonsky AV. Cutting edge: commensal microbiota has disparate effects on manifestations of polyglandular autoimmune inflammation. J Immunol. 2016;197:701–705. doi: 10.4049/jimmunol.1502465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Nielsen DS, Krych L, Hansen CHF. Bacterial species to be considered in quality assurance of mice and rats. Lab Anim. 2019;53:281–291. doi: 10.1177/0023677219834324. [DOI] [PubMed] [Google Scholar]
- Hikita I, Yoshioka T, Mizoguchi T, Tsukahara K, Tsuru K, Nagai H, Hirasawa T, Tsuruta Y, Suzuki R, Ichihashi M, Horikawa T. Characterization of dermatitis arising spontaneously in DS-Nh mice maintained under conventional conditions: another possible model for atopic dermatitis. J Dermatol Sci. 2002;30:142–153. doi: 10.1016/S0923-1811(02)00070-1. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Endo K, Kawamura S, Mitsuoka T. Comparison of the intestinal bacteria in specific pathogen free mice from different breeders. Jikken Dobutsu. 1990;39:263–267. doi: 10.1538/expanim1978.39.2_263. [DOI] [PubMed] [Google Scholar]
- Hirayama K, Endo K, Kawamura S, Mitsuoka T. Comparison of the intestinal bacteria in specific pathogen free mice from different breeders. Jikken dobutsu Exp Anim. 1990;39:263–267. doi: 10.1538/expanim1978.39.2_263. [DOI] [PubMed] [Google Scholar]
- Holmes E, Nicholson J. Variation in gut microbiota strongly influences individual rodent phenotypes. Toxicol Sci. 2005;87:1–2. doi: 10.1093/toxsci/kfi259. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med. 2010;60:336–342. [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Mitsuoka T, Sudo K, Suzuki K. Comparison of fecal flora of mice based upon different strains and different housing conditions. Z Versuchstierkd. 1983;25:135–146. [PubMed] [Google Scholar]
- Ivanov II, Frutos RD, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson ME. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164–177. doi: 10.15252/embr.201439263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science (New York, N.Y.) 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–1329. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science (New York, N.Y.) 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspareit-Rittinghausen J, Wullenweber M, Deerberg F, Farouq M. Pathological changes in Streptobacillus moniliformis infection of C57bl/6J mice. Berl Munch Tierarztl Wochenschr. 1990;103:84–87. [PubMed] [Google Scholar]
- Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol. 2016;7:529. doi: 10.3389/fimmu.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiouptsi K, Pontarollo G, Todorov H, Braun J, Jackel S, Koeck T, Bayer F, Karwot C, Karpi A, Gerber S, Jansen Y, Wild P, Ruf W, Daiber A, van der Vorst E, Weber C, Doring Y, Reinhardt C. Germ-free housing conditions do not affect aortic root and aortic arch lesion size of late atherosclerotic low-density lipoprotein receptor-deficient mice. Gut Microbes. 2020;11:1809–1823. doi: 10.1080/19490976.2020.1767463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöting I, Stark O, Brdicka R. Incidence of the insulin-dependent diabetes mellitus in BB rats: their genetic heterogeneity and susceptibility to infection. Folia Biol (Praha) 1984;30:33–42. [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kraft V, Blanchet H, Boot R, Hansen AK, Hem A, van Herck H, Kunstyr I, Milite G, Needham JR, Nicklas W, Perrot A, Rehbinder C, Richard Y, De-Vroey G, Deeny A. Recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit breeding colonies. Lab Anim. 1994;28:1–12. doi: 10.1258/002367794781065933. [DOI] [PubMed] [Google Scholar]
- Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun. 1998;66:5157–5166. doi: 10.1128/IAI.66.11.5157-5166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science (New York, N.Y.) 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas A, Hagerman I, Buschard K. Fusidic acid reduces incidence of diabetes in BB rats. Diabetologia. 2001;44:379. [Google Scholar]
- Laigaard A, Krych L, Zachariassen LF, Ellegaard-Jensen L, Nielsen DS, Hansen AK, Hansen CHF. Dietary prebiotics promote intestinal Prevotella in association with a low-responding phenotype in a murine oxazolone-induced model of atopic dermatitis. Sci Rep. 2020;10:21204. doi: 10.1038/s41598-020-78404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B, Zhang W, Weldon R, Auguste K, Yang L, Liu X, Chen L, Yang X, Zhu B, Cai J. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. doi: 10.1186/s40168-016-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg FCB, Ellekilde M, Thorn AC, Kihl P, Larsen CS, Hansen CHF, Metzdorff SB, Aalbaek B, Hansen AK. Dietary LPS traces influences disease expression of the diet-induced obese mouse. Res Vet Sci. 2019;123:195–203. doi: 10.1016/j.rvsc.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, Zou Q, Zhong B, Wu L, Wei H, Fang Y. Role of the gut microbiome in modulating arthritis progression in mice. Sci Rep. 2016;6:30594. doi: 10.1038/srep30594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Enrich-Capó N, Aldeguer X, Sabat-Mir M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Alterations in the abundance and co-occurrence of Akkermansia muciniphila and Faecalibacterium prausnitzii in the colonic mucosa of inflammatory bowel disease subjects. Front Cell Infect Microbiol. 2018;8:281. doi: 10.3389/fcimb.2018.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg R, Clausen SK, Pang W, Nielsen DS, Möller K, Josefsen K, Hansen AK. Gastrointestinal microbiota and local inflammation during oxazolone-induced dermatitis in BALB/cA mice. Comp Med. 2012;62:371–380. [PMC free article] [PubMed] [Google Scholar]
- Lundberg R, Bahl MI, Licht TR, Toft MF, Hansen AK. Microbiota composition of simultaneously colonized mice housed under either a gnotobiotic isolator or individually ventilated cage regime. Sci Rep. 2017;7:42245. doi: 10.1038/srep42245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg R, Toft MF, Metzdorff SB, Hansen CH, Licht TR, Bahl MI, Hansen AK. Human microbiota-transplanted C57BL/6 mice and offspring display reduced establishment of key bacteria and reduced immune stimulation compared to mouse microbiota-transplantation. Sci Rep. 2020;10:1–16. doi: 10.1038/s41598-020-64703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, Young GP, Marshall HS, Wesselingh SL, Rogers GB, Lynn DJ. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23:653–660.e655. doi: 10.1016/j.chom.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, Hirota K, Matsushita M, Furuta Y, Narazaki M, Sakaguchi N, Kayama H, Nakamura S, Iida T, Saeki Y, Kumanogoh A, Sakaguchi S, Takeda K. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- Man AWC, Li H, Xia N. Resveratrol and the interaction between gut microbiota and arterial remodelling. Nutrients. 2020;12:119. doi: 10.3390/nu12010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal RK, Denny JE, Waide ML, Li Q, Bhutiani N, Anderson CD, Baby BV, Jala VR, Egilmez NK, Schmidt NW. Temporospatial shifts within commercial laboratory mouse gut microbiota impact experimental reproducibility. BMC Biol. 2020;18:83. doi: 10.1186/s12915-020-00810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH, Murray JA, White BA, Kudva YC, Rajagopalan G. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS ONE. 2013;8:e78687. doi: 10.1371/journal.pone.0078687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Sivula CP, Jameson SC. Of mice, dirty mice, and men: using mice to understand human immunology. J Immunol. 2017;199:383–388. doi: 10.4049/jimmunol.1700453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Okabe Y, Setoyama H, Takayama K, Ohtsuka J, Funahashi H, Imaoka A, Okada Y, Umesaki Y. Inflammatory bowel disease-like enteritis and caecitis in a senescence accelerated mouse P1/Yit strain. Gut. 1998;43:71–78. doi: 10.1136/gut.43.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Kasper DL. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat Rev Immunol. 2006;6:849–858. doi: 10.1038/nri1956. [DOI] [PubMed] [Google Scholar]
- Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, Vial Y, Prod'hom G, Greub G, Kypriotou M, Christen-Zaech S. Skin colonization by Staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol. 2017;137:2497–2504. doi: 10.1016/j.jid.2017.07.834. [DOI] [PubMed] [Google Scholar]
- Moreno-Indias I, Lundberg R, Krych L, Metzdorff SB, Kot W, Sørensen DB, Nielsen DS, Hansen CHF, Hansen AK. A Humanized diet profile may facilitate colonization and immune stimulation in human microbiota-colonized mice. Front Microbiol. 2020;11:1336. doi: 10.3389/fmicb.2020.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Müller VM, Zietek T, Rohm F, Fiamoncini J, Lagkouvardos I, Haller D, Clavel T, Daniel H. Gut barrier impairment by high-fat diet in mice depends on housing conditions. Mol Nutr Food Res. 2016;60:897–908. doi: 10.1002/mnfr.201500775. [DOI] [PubMed] [Google Scholar]
- Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- O'Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EMM, Cryan JF, Dinan TG. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- O'Rourke J, Lee A, McNeill J. Differences in the gastrointestinal microbiota of specific pathogen free mice: an often unknown variable in biomedical research. Lab Anim. 1988;22:297–303. doi: 10.1258/002367788780746142. [DOI] [PubMed] [Google Scholar]
- Oh Jason Z, Ravindran R, Chassaing B, Carvalho Frederic A, Maddur Mohan S, Bower M, Hakimpour P, Gill Kiran P, Nakaya Helder I, Yarovinsky F, Sartor RB, Gewirtz Andrew T, Pulendran B. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Kitagawa Y, Sakaguchi S. Development and maintenance of regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world–recent facts and figures. Immunol Today. 1993;14:193–196. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- Rasmussen TS, de Vries L, Kot W, Hansen LH, Castro-Mejia JL, Vogensen FK, Hansen AK, Nielsen DS. Mouse vendor influence on the bacterial and viral gut composition exceeds the effect of diet. Viruses. 2019;11:588160. doi: 10.3390/v11050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TS, Mentzel CMJ, Kot W, Castro-Mejía JL, Zuffa S, Swann JR, Hansen LH, Vogensen FK, Hansen AK, Nielsen DS. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut. 2020;69:2122–2130. doi: 10.1136/gutjnl-2019-320005. [DOI] [PubMed] [Google Scholar]
- Reyniers JA, Trexler PC, Ervin RF (1946) Rearing germ-free albino rats. Lobund Reports (1):1–84 [PubMed]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171(1015–1028):e1013. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, Leonardi I, Collins N, Blatter JA, Han SJ, Tamoutounour S, Potapova S, St Claire MBF, Yuan WX, Sen SK, Dreier MS, Hild B, Hafner M, Wang D, Iliev ID, Belkaid Y, Trinchieri G, Rehermann B. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019 doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rune I, Rolin B, Larsen C, Nielsen DS, Kanter JE, Bornfeldt KE, Lykkesfeldt J, Buschard K, Kirk RK, Christoffersen B. Modulating the gut microbiota improves glucose tolerance, lipoprotein profile and atherosclerotic plaque development in ApoE-deficient mice. PLoS ONE. 2016;11:e0146439. doi: 10.1371/journal.pone.0146439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rune I, Rolin B, Lykkesfeldt J, Nielsen DS, Krych L, Kanter JE, Bornfeldt KE, Kihl P, Buschard K, Josefsen K, Fels JJ, Mortensen A, Christoffersen B, Kirk RK, Hansen AK. Long-term Western diet fed apolipoprotein E-deficient rats exhibit only modest early atherosclerotic characteristics. Sci Rep. 2018;8:5416. doi: 10.1038/s41598-018-23835-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, Yamazaki S, Sakihama T, Matsutani T, Negishi I, Nakatsuru S, Sakaguchi S. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003;426:454–460. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- Schaedler RW, Dubos R, Costello R. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med. 1965;122:59–66. doi: 10.1084/jem.122.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubs R, Costello R. Association of germfree mice with bacteria isolated from normal mice. J Exp Med. 1965;122:77–82. doi: 10.1084/jem.122.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MM, Meng Y, Qi H, Zhu W, Wang Z, Hazen SL, Lusis AJ, Shih DM. Trimethylamine N‐oxide promotes vascular inflammation through signaling of mitogen‐activated protein kinase and nuclear factor‐κB. J Am Heart Assoc. 2016;5:e002767. doi: 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/IAI.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk MS, Ditzel N, Hejbøl EK, Præstholm SM, Markussen LK, Avolio F, Li L, Lehtonen L, Hansen AK, Schrøder HD. C57BL/6J substrain differences in response to high-fat diet intervention. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-70765-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, Stappenbeck TS. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science (New York, N.Y.) 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr M, Greweling MC, Tischer S, Singh M, Blocker H, Monner DA, Muller W. Charles river altered schaedler flora (CRASF (R)) remained stable for four years in a mouse colony housed in individually ventilated cages. Lab Anim. 2009;43:362–370. doi: 10.1258/la.2009.0080075. [DOI] [PubMed] [Google Scholar]
- Suto H, Matsuda H, Mitsuishi K, Hira K, Uchida T, Unno T, Ogawa H, Ra C. NC/Nga mice: a mouse model for atopic dermatitis. Int Arch Allergy Immunol. 1999;120(Suppl 1):70–75. doi: 10.1159/000053599. [DOI] [PubMed] [Google Scholar]
- Tao L, Reese TA. Making mouse models that reflect human immune responses. Trends Immunol. 2017;38:181–193. doi: 10.1016/j.it.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, Viodé C, Schmitt AM, Serre G, Simon M, Paul CF. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol. 2016;137:1272–1274.e1273. doi: 10.1016/j.jaci.2015.07.052. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Hudcovic T, Tuckova L, Cukrowska B, Lodinova-Zadnikova R, Kozakova H, Rossmann P, Bartova J, Sokol D, Funda DP, Borovska D, Rehakova Z, Sinkora J, Hofman J, Drastich P, Kokesova A. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Unger AL, Eckstrom K, Jetton TL, Kraft J. Facility-dependent metabolic phenotype and gut bacterial composition in CD-1 mice from a single vendor: a brief report. PLoS ONE. 2020;15:e0238893. doi: 10.1371/journal.pone.0238893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf N, Kroese FG, Rozing J, Hillebrands JL. Viral infections as potential triggers of type 1 diabetes. Diabetes Metab Res Rev. 2007;23:169–183. doi: 10.1002/dmrr.695. [DOI] [PubMed] [Google Scholar]
- Varga O, Harangi M, Olsson I, Hansen AK. Contribution of animal models to the understanding of the metabolic syndrome: a systematic overview. Obes Rev. 2010;11:792–807. doi: 10.1111/j.1467-789X.2009.00667.x. [DOI] [PubMed] [Google Scholar]
- Velazquez EM, Nguyen H, Heasley KT, Saechao CH, Gil LM, Rogers AWL, Miller BM, Rolston MR, Lopez CA, Litvak Y, Liou MJ, Faber F, Bronner DN, Tiffany CR, Byndloss MX, Byndloss AJ, Bäumler AJ. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat Microbiol. 2019;4:1057–1064. doi: 10.1038/s41564-019-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino NF, LeCleir GR, Denny JE, Dearth SP, Harding CL, Sloan SS, Gribble JL, Campagna SR, Wilhelm SW, Schmidt NW. Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci. 2016;113:2235–2240. doi: 10.1073/pnas.1504887113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–916. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W, Lidwell OM, Lowbury EJ, Blowers R. Suggested bacteriological standards for air in ultraclean operating rooms. J Hosp Infect. 1983;4:133–139. doi: 10.1016/0195-6701(83)90042-7. [DOI] [PubMed] [Google Scholar]
- Wilson KH, Brown RS, Andersen GL, Tsang J, Sartor B. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe. 2006;12:249–253. doi: 10.1016/j.anaerobe.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Wingender G, Stepniak D, Krebs P, Lin L, McBride S, Wei B, Braun J, Mazmanian SK, Kronenberg M. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–428. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff NS, Jacobs MC, Haak BW, Roelofs JJTH, de Vos AF, Hugenholtz F, Wiersinga WJ. Vendor effects on murine gut microbiota and its influence on lipopolysaccharide-induced lung inflammation and Gram-negative pneumonia. Intensive Care Med Exp. 2020;8:47. doi: 10.1186/s40635-020-00336-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Jiang Z-Y, Sun Y-F, Yu B, Chen J, Dai C-Q, Wu X-L, Tang X-L, Chen X-YJCM. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza a virus infection. Curr Microbiol. 2013;67:414–422. doi: 10.1007/s00284-013-0380-z. [DOI] [PubMed] [Google Scholar]
- Yiu JHC, Chan KS, Cheung J, Li J, Liu Y, Wang Y, Fung WWL, Cai JL, Cheung SWM, Dorweiler B, Wan EYF, Tso P, Xu AM, Woo CW. Gut microbiota-associated activation of TLR5 induces apolipoprotein A1 production in the liver. Circ Res. 2020;127:1236–1252. doi: 10.1161/CIRCRESAHA.120.317362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, Yang X, Li J, Chen X, Zhao X, Chen Y, Wen Y. Trimethylamine N-oxide prime NLRP3 inflammasome via inhibiting ATG16L1-induced autophagy in colonic epithelial cells. Biochem Biophys Res Commun. 2017;490:541–551. doi: 10.1016/j.bbrc.2017.06.075. [DOI] [PubMed] [Google Scholar]
- Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariassen LF, Krych L, Engkilde K, Nielsen DS, Kot W, Hansen CHF, Hansen AK. Sensitivity to oxazolone induced dermatitis is transferable with gut microbiota in mice. Sci Rep. 2017;7:44385. doi: 10.1038/srep44385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY, Mao YJ, Zhang XJ, Pang XY, Wei CC, Zhao GP, Chen Y, Zhao LP. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]