Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RNA‐dependent RNA polymerase (RdRp) has been identified to be a mutation hot spot, with the P323L mutation being commonly observed in viral genomes isolated from North America. RdRp forms a complex with nonstructural proteins nsp7 and nsp8 to form the minimal replication/transcription machinery required for genome replication. As mutations in RdRp may affect formation of the RdRp–nsp7–nsp8 supercomplex, we analyzed viral genomes to identify mutations in nsp7 and nsp8 protein sequences. Based on in silico analysis of predicted structures of the supercomplex comprising of native and mutated proteins, we demonstrate that specific mutations in nsp7 and nsp8 proteins may have a role in stabilization of the replication/transcription complex.
Keywords: mutation, nsp7, nsp8, RdRp, SARS‐CoV‐2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a single‐stranded RNA virus responsible for the ongoing coronavirus disease 2019 (COVID‐19) pandemic that originated in China in December 2019. RNA viruses have a high mutation rate, contributing to their evolvability and virulence. 1 SARS‐CoV‐2 is no exception; studies have demonstrated that this virus has a high rate of mutations, 2 with some genomic positions showing a mutation rate as high as approximately 30%. 3
The SARS‐CoV‐2 replication/transcription machinery consists of an RNA‐dependent RNA polymerase (RdRp, also known as nsp12) working in tandem with nsp7 and nsp8 proteins, forming a RdRp–nsp7–nsp8 supercomplex. 4 This supercomplex may represent the minimal machinery required for nucleotide polymerization, as determined for the closely related SARS‐CoV virus, 5 , 6 and is a potential antiviral target. 7
Mutations in the polymerase genes of viruses are reported to enhance their virulence. 8 , 9 , 10 Mutations in the RdRp gene sequence were first reported in virus isolates from Europe in February 2020, and then in virus isolates from North America in March 2020. 2 In particular, a mutation at position 14408, which leads to an amino acid substitution (P323L) in RdRp, was identified as a mutation hotspot, and in silico analysis suggests that this change may affect stability of the tertiary complex of RdRp. 11 It is speculated that this mutation, which falls outside the catalytic site of RdRp, may impact proofreading capability of the replication supercomplex. 2 Though RdRp residues involved in interacting with nsp7 and nsp8 are yet to be identified, mutations in RdRp may also affect its interaction with these associated proteins and contribute to decreased fidelity of genome replication.
As an increased mutation rate may be deleterious to the virus, we hypothesized that there may arise compensatory mutations, 12 either in RdRp or in proteins that it interacts with, to counteract the effect of mutations in RdRp. Toward this end, we analyzed mutations in nsp7 and nsp8 proteins in SARS‐CoV‐2 sequences and attempted to correlate them with mutations in RdRp (Figure S1).
2. METHODS
2.1. Genome sequence analysis
Genome sequences of SARS‐CoV‐2 isolated from Homo sapiens available in the NCBI GenBank database 13 till July 14, 2020 (n = 9692) were analyzed. Nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/) was carried out using nsp7, nsp8, and nsp12 gene sequences from the reference Wuhan‐Hu‐1 genome (GenBank Accession NC_045512.2) as query sequences. Pairwise alignment of protein sequences was carried out using lalign. 14
The χ 2 test was used to assess significance of associations between mutations in nsp7 or nsp8 and RdRp proteins.
2.2. Protein structure modeling and analysis
Protein structures were modeled using SWISS‐MODEL (https://swissmodel.expasy.org/). Sequences of native and mutant RdRp, nsp7, and nsp8 proteins (Table S1) were used to model supercomplex structures using the SARS‐CoV‐2 RdRp–nsp7–nsp8 structure 6yyt.1 as reference. 15 Predicted structures were downloaded as PDB files and analyzed using BioLuminate (Schrödinger, Inc.).
Briefly, PDB structures were imported in BioLuminate and processed using the Protein Preparation Wizard. Protein structures were optimized by assigning bonds, fixing structural defects, removing unwanted parts, assigning protonation and tautomeric states, and refining the structure. Optimized protein structures were analyzed by performing protein interaction analysis, wherein specific interactions like hydrogen bonds, salt bridges, pi stackings, surface complementarity and buried SASA at protein interfaces were analyzed.
3. RESULTS AND DISCUSSION
Twenty‐three distinct mutations in nsp7 protein sequences were identified in a total of 218 virus isolates (Tables S2 and S3), while 34 distinct mutations were identified in nsp8 protein sequences of 130 isolates (Tables S4 and S5). Three residues in nsp7 were mutated to two different amino acids: S26 to F and A, I68 to T and V, and R79 to S and G. Three residues in nsp8 were mutated to two different amino acids: A14 to S and V, A27 to T and V, and I156 to V and L. The most commonly occurring mutations in nsp7 protein were S25L (n = 165) and S26F (n = 28), and the most common mutations in nsp8 were M129I (n = 34) and I156V (n = 32). Interestingly, only one instance of the same viral genome carrying mutations in both nsp7 (S25L) and nsp8 (A21V) was observed (Genbank accession number MT370871.1).
As the nsp7 and nsp8 proteins form a supercomplex with RdRp, mutations in these two proteins may affect complex formation and may have an effect on genome replication. To determine whether any compensatory mutations are present in RdRp, RdRp protein sequences were analyzed in strains that carry nsp7 or nsp8 mutations. It was observed that genomes carrying both frequently occurring mutations in nsp7 (S25L and S26F) also had the P323L mutation in RdRp (Table S2), while genomes carrying both frequently occurring mutations in nsp8 (M129I and I156V) had native RdRp sequences (Table S4). nsp7 S25L and S26F mutations are significantly associated with the P323L mutation in RdRp in the 218 genomes carrying mutations in nsp7 (χ 2 test, p < .001); nsp8 M129I and I156V mutations are significantly associated with native RdRp in the 130 genomes carrying mutations in nsp8 (χ 2 test, p < .001).
The P323L mutation is commonly observed in RdRp sequences and is speculated to have an effect on RdRp function. 2 The effect of mutations in nsp7 and nsp8, if any, is unknown, though it is possible that these mutations may also have an impact on the RdRp–nsp7–nsp8 supercomplex.
To investigate the effect of mutations in nsp7 and nsp8 on the RdRp–nsp7–nsp8 supercomplex, in silico analysis of predicted supercomplex structures was carried out. When serine at position 25 of nsp7 was mutated to leucine, nsp7 S26 was observed to interact with nsp8 D163 in the RdRp–nsp7–nsp8 supercomplex (Figure 1). Due to the nsp7 S25L mutation, surface complementarity increased by 10%, indicating that this mutation may play a beneficial role in supercomplex stability (Table S6). The P323L mutation in RdRp also increased surface complementarity by 10% due to better interaction with N118 of nsp8 (Figure 2A). Similarly, an increase in surface complementarity was observed when serine at position 26 of nsp7 was mutated to phenylalanine (Table S6).
Figure 1.
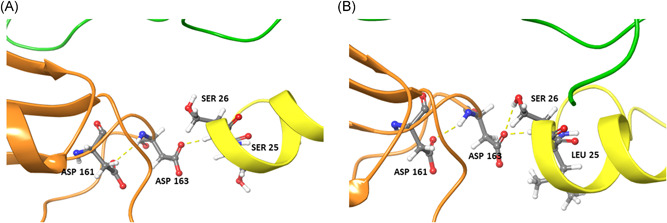
Interactions between residues of (A) native and (B) mutant nsp7 (yellow), nsp8 (orange) and RdRp (green) proteins. The S25L mutation in nsp7 leads to a H‐bond formation between nsp7 S26 and nsp8 D163, which is not observed in the native RdRp–nsp7–nsp8 supercomplex
Figure 2.
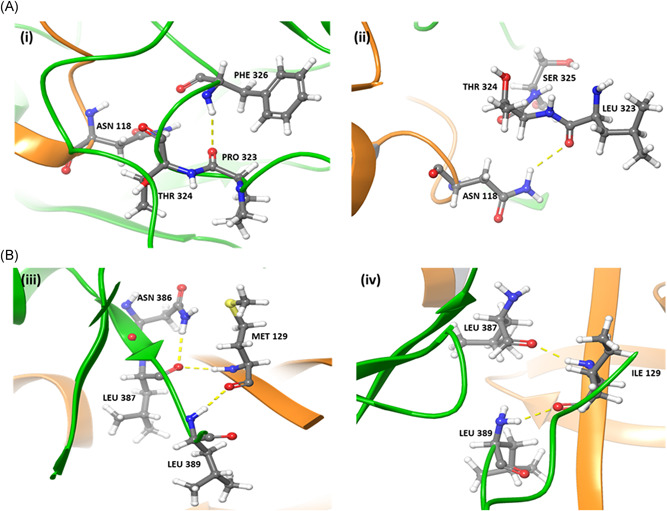
(A) Interactions between residues of (i) native and (ii) mutant RdRp (green) and nsp8 (orange) proteins. The P323L mutation in RdRp leads to a H‐bond formation with nsp8 N118, which is not observed when native RdRp interacts with nsp8. (B) Interactions between residues of (iii) native and (iv) mutant nsp8 (orange) and RdRp (green) proteins. A steric clash is observed between nsp8 M129 and RdRp N386, which is abolished in the nsp8 M129I mutant
As the M129I mutation in nsp8 is associated with native RdRp, the RdRp–nsp7–nsp8 M129I supercomplex was modeled. It was observed that the M129I mutation abolishes the steric clash between M129 of nsp8 and N386 of RdRp (Figure 2B). The M129I mutation therefore appears to stabilize the interaction between nsp8 and RdRp in the supercomplex.
Recent studies have reported correlations between SARS‐CoV‐2 spike protein mutations and disease severity. 16 , 17 No studies have so far documented pathogenicity of SARS‐CoV‐2 strains as a function of mutations in RdRp, nsp7, or nsp8. As the RdRp–nsp7–nsp8 supercomplex plays a crucial role in the life cycle of RNA viruses, 18 mutations in these proteins may impact viral replication and, by extension, virulence.
4. CONCLUSION
Parallel evolution events have been suggested for SARS‐CoV‐2 genomes isolated from different hosts, wherein the same mutation arises in two different hosts. 19 In this report, we show that mutant nsp7 proteins are significantly associated with mutant RdRp. Based on in silico analysis of modeled RdRp–nsp7–nsp8 supercomplex structures, we hypothesize that these are complementary mutations that may help in proper assembly of the replication machinery of SARS‐CoV‐2 and may have a role in maintaining fidelity of genome replication. On the other hand, the most frequently occurring mutation in nsp8, which is significantly associated with native RdRp, leads to increased stability of the supercomplex, indicating that this may a beneficial mutation.
CONFLICT OF INTERESTS
The authors declare they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Shamlan M. S. Reshamwala and Shalini S. Deb conceived the study; Shamlan M. S. Reshamwala, Shalini S. Deb, and Santosh B. Noronha analyzed genome sequence data; Vishakha Likhite and Mariam S. Degani carried out protein structure analysis; Shamlan M. S. Reshamwala, Vishakha Likhite, Mariam S. Degani, Shalini S. Deb, and Santosh B. Noronha wrote the manuscript and approved it for submission.
Supporting information
Supporting information.
Reshamwala SMS, Likhite V, Degani MS, Deb SS, Noronha SB. Mutations in SARS‐CoV‐2 nsp7 and nsp8 proteins and their predicted impact on replication/transcription complex structure. J Med Virol. 2021;93:4616–4619. 10.1002/jmv.26791
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article.
REFERENCES
- 1. Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16:e3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pachetti M, Marini B, Benedetti F, et al. Emerging SARS‐CoV‐2 mutation hot spots include a novel RNA‐dependent‐RNA polymerase variant. J Transl Med. 2020;18:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C, Liu Z, Chen Z, et al. The establishment of reference sequence for SARS‐CoV‐2 and variation analysis. J Med Virol. 2020;92:667‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368:779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirchdoerfer RN, Ward AB. Structure of the SARS‐CoV nsp12 polymerase bound to nsp7 and nsp8 co‐factors. Nat Commun. 2019;10:2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhai Y, Sun F, Li X, et al. Insights into SARS‐CoV transcription and replication from the structure of the nsp7‐nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980‐986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon CJ, Tchesnokov EP, Woolner E, et al. Remdesivir is a direct‐acting antiviral that inhibits RNA‐dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785‐6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dreier C, Resa‐Infante P, Thiele S, et al. Mutations in the H7 HA and PB1 genes of avian influenza a viruses increase viral pathogenicity and contact transmission in guinea pigs. Emerg Microbes Infect. 2019;8:1324‐1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu C, Xu B, Wu Y, et al. A single amino acid at position 431 of the PB2 protein determines the virulence of H1N1 swine influenza viruses in mice. J Virol. 2020;94:e01930‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu W, Zhu Y, Qin K, et al. Mutations in polymerase genes enhanced the virulence of 2009 pandemic H1N1 influenza virus in mice. PLoS One. 2012;7:e33383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chand GB, Banerjee A, Azad GK. Identification of novel mutations in RNA‐dependent RNA polymerases of SARS‐CoV‐2 and their implications on its protein structure. PeerJ. 2020;8:e9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fitzsimmons WJ, Woods RJ, McCrone JT, et al. A speed‐fidelity trade‐off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 2018;16:e2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41:D36‐D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang X, Miller W. A time‐efficient, linear‐space local similarity algorithm. Adv Appl Math. 1991;12:337‐357. [Google Scholar]
- 15. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. Structure of replicating SARS‐CoV‐2 polymerase. Nature. 2020;584:154‐156. [DOI] [PubMed] [Google Scholar]
- 16. Isabel S, Graña‐Miraglia L, Gutierrez JM, et al. Evolutionary and structural analyses of SARS‐CoV‐2 D614G spike protein mutation now documented worldwide. Sci Rep. 2020;10:14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Q, Wu J, Wang H, et al. Structural basis for RNA replication by the SARS‐CoV‐2 polymerase. Cell. 2020;182:417‐428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS‐CoV‐2 genomes. Proc Natl Acad Sci USA. 2020;117:9241‐9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
The data that supports the findings of this study are available in the supplementary material of this article.


