SUMMARY:
The past decade has seen impressive advances in the types of neuroimaging information that can be acquired in patients with traumatic brain injury. However, despite this increase in information, understanding of the contribution of this information to prognostic accuracy and treatment pathways for patients is limited. Available techniques often allow us to infer the presence of microscopic changes indicative of alterations in physiology and function in brain tissue. However, because histologic confirmation is typically lacking, conclusions reached by using these techniques remain solely inferential in almost all cases. Hence, a need exists for validation of these techniques by using data from large population samples that are obtained in a uniform manner, analyzed according to well-accepted procedures, and correlated with closely monitored clinical outcomes. At present, many of these approaches remain confined to population-based research rather than diagnosis at an individual level, particularly with regard to traumatic brain injury that is mild or moderate in degree. A need and a priority exist for patient-centered tools that will allow advanced neuroimaging tools to be brought into clinical settings. One barrier to developing these tools is a lack of an age-, sex-, and comorbidities-stratified, sequence-specific, reference imaging data base that could provide a clear understanding of normal variations across populations. Such a data base would provide researchers and clinicians with the information necessary to develop computational tools for the patient-based interpretation of advanced neuroimaging studies in the clinical setting.
The recent “Joint ASNR-ACR HII-ASFNR TBI Workshop: Bringing Advanced Neuroimaging for Traumatic Brain Injury into the Clinic” on May 23, 2014, in Montreal, Quebec, Canada, brought together neuroradiologists, neurologists, psychiatrists, neuropsychologists, neuroimaging scientists, members of the National Institute of Neurologic Disorders and Stroke, industry representatives, and other traumatic brain injury stakeholders to attempt to reach consensus on issues related to and develop consensus recommendations in terms of creating both a well-characterized normative data base of comprehensive imaging and ancillary data to serve as a reference for tools that will allow interpretation of advanced neuroimaging tests at an individual level of a patient with traumatic brain injury. The workshop involved discussions concerning the following: 1) designation of the policies and infrastructure needed for a normative data base, 2) principles for characterizing normal control subjects, and 3) standardizing research neuroimaging protocols for traumatic brain injury. The present article summarizes these recommendations and examines practical steps to achieve them.
Traumatic brain injury (TBI) is an important, common cause of morbidity and mortality, with approximately 275,000 hospitalizations and 52,000 deaths each year.1 Among the leading causes of TBI in the civilian population are falls, motor vehicle collisions, assaults, and sports-related injuries.1,2 In addition, a particularly high incidence of TBI is seen in the military population. Among individuals with long-term sequelae of TBI, symptoms can range from subtle mild cognitive impairment to severe disability. In many cases, the diagnosis is clear when a patient presents with physical findings of intracranial injury, and neuroimaging findings indicate injuries that may require emergent surgical intervention and medical therapy. Examples of such injuries include (but are not limited to) intraparenchymal and extra-axial hematomas, which can result in life-threatening mass effect and brain herniation. Imaging soon after the time of injury can also give information regarding future prognostic parameters, including early mortality and late morbidity.3 Commonly used neuroimaging methods for the clinical evaluation of TBI include head CT and certain brain MR imaging sequences (eg, T1-weighted, T2-weighted, FLAIR, diffusion-weighted, and some type of T2* imaging including gradient-echo imaging or susceptibility-weighted imaging), which can detect acute intracranial sequelae as well as the more chronic effects of TBI, such as encephalomalacia or prior hemorrhage.
Neuroimaging methods beyond those used in conventional imaging (so-called “advanced” techniques) show promise in assessing mild TBI but do not yet play a central standardized role in diagnosis and management of mild TBI because they require further validation. Such techniques include quantitative diffusion imaging methods, such as diffusion tensor imaging (as opposed to the diffusion-weighted imaging listed earlier) and diffusional kurtosis imaging (DKI). These advanced neuroimaging methods allow one to infer tissue microstructural information based on directionality of microscopic water diffusion. DTI is an imaging technique used in a number of brain applications, most notably in fiber tracking for surgical planning. DKI is an extension of DTI and enables the calculation of all the usual DTI metrics. The potential advantage of DKI over DTI is the added metrics quantifying non-Gaussian diffusion, which may supply new information to better characterize brain tissue. This feature may be particularly important in assessment of gray matter, because gray matter water diffusion is nearly isotropic, which limits the value of fractional anisotropy and other metrics of diffusional anisotropy obtainable with DTI. DKI is only now being investigated in application to mild TBI.
Other “advanced” neuroimaging methods also include cerebral perfusion/permeability MR imaging, MR spectroscopy, resting-state functional MR imaging, positron-emission tomography, and magnetoencephalography. Although the aforementioned have yet to be established in clinical practice for TBI diagnosis, many studies have demonstrated the ability of these methods to identify statistically significant effects of mild TBI by using, specifically, group data.4–30 In particular, when comparing groups of individuals having mild TBI with normal control subjects (as opposed to diagnosing mild TBI in an individual), these techniques have shown quantitative abnormalities in the brain related to subacute and long-term traumatic sequelae (eg, cognitive dysfunction), frequently in the absence of intracranial abnormalities detected by nonquantitative visual inspection alone on conventional neuroimaging methods.19,25,31–42 Such intriguing data suggest a potentially important clinical role for advanced imaging methods, especially in the large population of patients with mild TBI who are symptomatic but whose conventional neuroimaging examination shows no visible abnormality. In these cases, advanced neuroimaging techniques may hold the most promise as sensitive diagnostic and prognostic tools for identifying clinically relevant abnormalities that are otherwise imperceptible on conventional imaging. Until such a time, however, these advanced structural and functional neuroimaging methods do not play a role in clinical practice for the diagnosis of mild TBI in individual patients.
The current method for interpreting neuroimaging studies in the patient care setting relies on a qualitative (ie, solely visual) assessment of acquired imaging information by radiologists or subspecialty-trained neuroradiologists with the generation of a narrative report that is placed into the patient's medical record. These assessments are geared toward the detection and characterization of macroscopically visible lesions and often include a summary differential diagnosis provided for any detected abnormalities. However, advanced neuroimaging approaches such as DTI, blood oxygen level–dependent, and magnetoencephalography generate data that are best assessed through quantitative approaches rather than subjective visual observation of altered intensity within a given brain structure or region. Various types of computational tools are available for analysis of these data for population-based research. However, well-characterized methods for quantitative analysis of advanced imaging data in an individual subject or patient do not exist. Thus, the quantitative data obtained from advanced imaging techniques represent a real departure from the imaging information radiologists interpret in standard clinical care. Therefore, a careful approach is needed before bringing these techniques into routine diagnostic use. The lack of well-accepted, uniform, cross-platform, and user-friendly analysis tools and the fact that these imaging techniques so far have shown promise solely for comparing groups of subjects (as opposed to diagnosis of a single individual) are additional barriers for bringing these potentially informative techniques into the clinic.
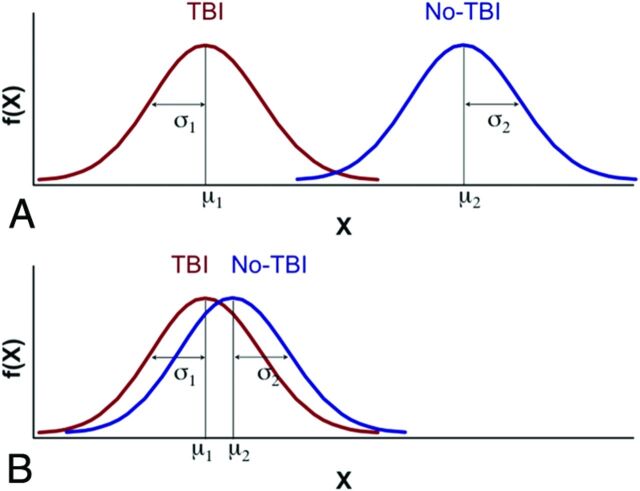
One feature that has hampered even more widespread use of advanced imaging techniques in TBI research has been the lack of a large data base of normal individuals to which patients with mild TBI could be compared. Such a data base would allow determination of the range of normal variation in a large population, which is needed before data in a different (eg, diseased) individual or population can be reliably considered to be normal or abnormal. Examples of other diagnostic tests in current clinical practice serve to emphasize this point. Just as one could not use important laboratory tests, such as measurement of hematocrit values and troponin values, without having first established the variations of normal, so we must first determine the operating parameters of advanced imaging tests before more widespread utilization in a research setting, let alone a clinical setting. Instead, at present, many TBI research studies are based on relatively small groups of normal subjects that are unlikely to represent the range of variability present in the general population at large. Hence, the need exists for construction of large-scale data bases that contain information indicating the standard variation of normal values in an age-stratified fashion and sufficient representations of abnormal to generate a library of key features that possess both diagnostic and prognostic value. Constructing a large comprehensive normative data base has a number of challenges, including the following: 1) defining normality; 2) distinguishing acute findings from chronic abnormalities in patients with pre-existing conditions (Figure); 3) creating image-acquisition protocols that are robust across multiple vendor platforms, methodologies, and institutions; and 4) constructing accessible repositories for data sharing and applying the necessary informatics tools to identify the critical features most sensitive and specific for characterizing the effects of mild TBI.
Figure.
Hypothetic distribution of an advanced neuroimaging-dependent measure for TBI (red) and non-TBI (blue) groups. A, Idealized separation of distributions between the TBI and non-TBI groups due to a very selective “normal” non-TBI control group (ie, supernormal, with no history of TBI, and no neurologic, psychiatric, or other detectable abnormalities), which would be amenable to conventional statistical analyses based on the general linear model. B, Marked overlap of distributions between the TBI and non-TBI groups due to a non-TBI control group comprising subjects with pre-existing abnormalities present in the general population, which would be unlikely to yield a statistically significant differentiation by using the general linear model. Despite relatively marked overlap between distributions, classification approaches may be able to identify features unique to each group and therefore discriminate whether an individual belongs in the TBI or non-TBI group. To pursue implementation of such a binary classification, it will be necessary to characterize the variability associated with neuroimaging methods expected from the general population in the absence of TBI, which can be facilitated by constructing a large comprehensive normative data base.
The purpose of this review is to provide consensus recommendations regarding the creation of such neuroimaging data repositories and associated metadata. This data base would serve as a reference for more uniform interpretation of data across research sites and will, ultimately, aid in clinical translation. A comprehensive, well-characterized, and widely accessible normative neuroimaging data base will provide the best chance of resolving important mild TBI-associated abnormalities that are occult to conventional imaging. This multifaceted approach is expected to play an important role in eventually understanding the importance of findings in individual patients and transforming the care of those who suffer from the effects of mild TBI.
Optimal Data Base Approach
A number of repositories and informatics systems currently exist that provide an excellent opportunity for advancing knowledge about ways to diagnose, classify, and monitor TBI. These repositories and toolsets use different sharing models and have different focuses and strengths. Characteristics of strong repositories include not just ease of input of data and sophisticated, intuitive retrieval engines; the expectations of a durable future data base now include extensibility and sophisticated informatics and data-mining tools to increase both relevance and fidelity. A registry listing such National Institutes of Health (NIH) data bases is available at http://www.nlm.nih.gov/NIHbmic/nih_data_sharing_repositories.html. A list of the processing tools made available by these informatics systems should be added to this registry listing, or it should be linked to other resources such as the Neuroinformatics Tools and Resources (NITRC) (http://www.nitrc.org/) and Neuroimaging Informatics Technology Initiative (http://nifti.nimh.nih.gov/). These include (Table 1), but are not limited to, the following:
The National Database for Autism Research (http://ndar.nih.gov) is an NIH-based research data repository that aims at accelerating progress in autism spectrum disorders research through data sharing, data harmonization, and the reporting of research results. The National Database for Autism Research also aims to provide a platform and portal to other research repositories, allowing for aggregation and secondary analysis of data. Interested researchers must complete dedicated forms and then obtain institutional-level approval to either deposit data or access the data base contents.43 The NIH considers the National Database for Autism Research as the prime example of a disease-specific data base supporting the concept of Research Domain Criteria. The NIH is examining the use of the National Database for Autism Research model for data bases for mental health disorders as well as other neurologic conditions (eg, TBI).
The Federal Interagency TBI Research (FITBIR) Informatics System (https://fitbir.nih.gov/) is the result of a collaboration which began in 2011 between the NIH and the US Department of Defense. It is among the first instances of applying the National Database for Autism Research model to other disease types. Its purpose is to create a national resource for archiving and sharing clinical data from research studies on TBI, along with appropriate control data. The ultimate goals are to support data sharing across the entire TBI research field, facilitate collaboration between laboratories, and promote interconnectivity with other informatics platforms. FITBIR uses the National Institute of Neurologic Disorders and Stroke Common Data Elements (CDEs) (http://www.commondataelements.ninds.nih.gov/tbi.aspx#tab=Data_Standards) for a data dictionary but also permits use of unique data elements, as needed. The FITBIR data-sharing policy allows for controlled access to contributing, as well as noncontributing, scientific investigators. All FITBIR technologic modules are open-source except for the Global Unique Identifier, which is restricted to ensure subject de-identification.
The Longitudinal On-line Research and Imaging System (LORIS) for Neuroinformatics (https://cbrain.mcgill.ca/collaborative-platform/loris) is an open-source informatics system with 3 major components: a data repository for archiving data, tools for data-acquisition management, and a pipeline for analyzing data. It has been in existence for over 10 years and was previously supported by the NIH/National Institute of Mental Health to develop an imaging resource for normal brain development. It is a modular and extensible Web-based data-management system which seeks to collect all aspects of data from a multicenter study. A subject-centric internal organization allows researchers to capture and subsequently extract all information (whether longitudinal or cross-sectional) from any subset of a study cohort. It has error-checking and quality-control procedures, security, data management, data querying, and administrative functions. LORIS aims to provide the following: 1) continuous project coordination and monitoring of data acquisition, 2) data storage/cleaning/querying, and 3) an interface with arbitrary external data processing. LORIS has only been formally available for a few years but is working to become a complete solution and is now being used in numerous international neurodevelopmental and neurodegenerative research projects.43 It now contains data from numerous other research projects, which can be accessed through the CBRAIN portal (http://mcin-cnim.ca/neuroimagingtechnologies/cbrain/).
The Laboratory of Neuroimaging (LONI) Image and Data Archive (http://ida.loni.usc.edu) uses a centralized model for working with “Big Data.” LONI, which was established in 1983, focuses on the construction of brain atlases based on detailed representations of anatomy in a standardized 3D coordinate system. The Image and Data Archive contains neuroimaging datasets (eg, T1-weighted, T2-weighted, fMRI, DTI, and so forth) from tens of thousands of subjects. It also contains demographic, phenomic, and genetic data from most enrolled subjects. It is by far the largest data base of its kind and has been specifically constructed to serve as a repository for large-scale, multisite neuroimaging projects. Such projects include the Alzheimer Disease Neuroimaging Initiative (ADNI), data from the Michael J. Fox Foundation work on Parkinson Disease, the Parkinson's Progressive Measures Initiative, and many other multisite projects. Data in the Image and Data Archive is not “controlled” by LONI per se but is subject to user-specified access policies by which various levels of access are possible and determined by the data collectors. Data are easily searched by using a user-specified data dictionary. Users can create query-specific data collections that can then be easily retrieved. Collections made in the Image and Data Archive can also be accessed directly from within the LONI Pipeline workflow processing environment, which simplifies access and avoids movement of large collections of data. Several articles have been published which discuss the Image and Data Archive and its interactivity with the LONI Pipeline. In addition to providing an advanced statistical analysis method, LONI has developed sophisticated visualizations to enable effective communication of the results. The NIH-funded study of 3000 children and adults with TBI includes LONI investigators and will be able to take advantage of the LONI Pipeline and other tools.
The Alzheimer Disease Neuroimaging Initiative (http://www.adni-info.org/) project is directed by Dr Michael Weiner from University of California, San Francisco, and represents a major success for neuroimaging data basing, sharing, and re-use. ADNI comprises at least 10 different institutions, all performing standardized imaging and acquiring phenotypic and genetic data on patients with Alzheimer disease, subjects with mild cognitive impairment, and age-matched normal control subjects. ADNI 1.0 had more than 900 subjects in its collection. The current ADNI 2.0 seeks to substantially increase that total with an enhanced protocol (including DTI) as well as full-scale genetic sequencing. All ADNI data are stored in the Laboratory of Neuroimaging Image Data Archive. The data have been included in hundreds of peer-reviewed research articles.
The Human Connectome Project is an NIH-funded effort to use advanced DTI and fMRI methods at field strengths of 3T and 7T. There are 2 components: 1) the Washington University–University of Minnesota consortium, and 2) the Massachusetts General Hospital–University of Southern California consortium. The Washington University Consortium focuses on large-scale but commodity-level imaging of functional imaging in twins, whereas the Massachusetts General Hospital–University of Southern California initiative is devoted to advanced technologic development of DTI acquisition. Data from Washington University are stored using a customized version of the Extensible Neuroimaging Archive Toolkit (http://www.xnat.org/), whereas the Massachusetts General Hospital–University of Southern California datasets are stored in the Laboratory of Neuroimaging Image Data Archive.
The American College of Radiology's (ACR's) Translational Research Informatics and Data Management Grid (TRIAD) and Data Archive and Research Toolkit (DART) are an end-to-end informatics framework for research data submission, processing, archiving, and analysis. TRIAD is a standards-based workflow and transfer engine for the incorporation of images and data. It has been in use for clinical trials and related data-acquisition functions (eg, data registries and quality programs such as accreditation) for approximately a decade. Workflow features include automatic de-identification, presubmission automated validation, and distributed workflow for preanalytic review and quality control. DART is the latest version of the data warehouse and archiving functionality provided within the TRIAD informatics environment. The data warehouse has collected DICOM studies and related clinical data for archival and ongoing analysis across a wide array of clinical domains. DART has expanded the ACR archive to include flexible tools for analysis and algorithm development, the ability to support diverse data types (including digital pathology images, biospecimen/biorepository information, and operational research reports), and the ability to ergonomically and rapidly search and query the data. The platform is flexible and designed to provide a low barrier of entry to data input and a high degree of interoperability with other repositories. As a result, DART serves as an independent repository and analytic platform for research but also serves as part of a larger national ecosystem where data can either be accessible via federated means or staged, refined, and submitted for input into more controlled platforms such as FITBIR, where conformance to standards such as TBI CDEs is a prerequisite for inclusion.
The Autism Brain Imaging Data Exchange (ABIDE) (http://fcon_1000.projects.nitrc.org/indi/abide/) is a grassroots initiative that aims to address the complexity of autism by curating previously collected data for the purpose of data sharing in the broader scientific community. One of the subprojects is the “1000 Functional Connectome Project,” which currently has resting-state fMRI and phenotypic data from more than 500 individuals with autism spectrum disorders and equivalent numbers of typically developing children. ABIDE has received data from more than 16 international sites. This is a decentralized process with few restrictions, but it does require curation of the data. The complete collection actually exists in several locations (LONI, LORIS, and NITRC).
Table 1:
Existing imaging repositories
| NDAR | FITBIR | LORIS | LONI IDA | DART | XNAT | INDI/FCP | COINS | BIRN HID | NIDB | |
|---|---|---|---|---|---|---|---|---|---|---|
| Core developers | National Institutes of Health | National Institutes of Health NINDS | Montreal Neurological Institute | University of Southern California | American College of Radiology | Washington University | NITRC | MIND Research Network | National Institutes of Health NIGMS | Olin Neuropsychiatry Research Center |
| Types of data stored | Neuroimaging | Neuroimaging | Neuroimaging | Neuroimaging | Imaging | Neuroimaging | Neuroimaging | Neuroimaging | Neuroimaging | Neuroimaging |
| Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | Phenomic | |
| Genetic | Demographic | Genetic | Genetic | Demographic | Demographic | Demographic | Demographic | Demographic | ||
| Demographic | Other: extensible | Demographic | Demographic | Other: extensible | Other: extensible | Other: extensible | Other: extensible | |||
| Other: extensible | Other: extensible | Other: extensible | Other: extensible | |||||||
| Common data elements | Required use | Required use | Flexible usage | Flexible usage | Flexible usage | Flexible usage | None | None | None | None |
| Workflow | Yes | |||||||||
| Continuous project coordination | No | No | Yes | No | Yes | No | No | No | No | No |
| Monitoring of data acquisition | No | No | Yes | No | Yes | No | No | No | No | Yes |
| Data cleaning | No | No | Yes | Yes | Yes | Yes | No | No | Yes | Yes |
| Data storage | Developer hosting | Developer hosting | Developer hosting or local storage | Developer hosting or local storage | Developer hosting or local storage | Local storage | Developer hosting | Developer hosting | Local storage | Local storage |
| Data query | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
| Data download | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Interface with external data-processing | No | No | Yes | Yes | Yes | Yes | No | No | Yes | No |
| Integrated data-processing tools | No | No | No | Yes | Yes? | Yes | No | No | No | No |
| Disease-specific | Yes: autism | Yes: TBI | No | No | No | No | No | No | No | No |
| Major datasets shared | Data from 77,575 individuals related to autism | In development | ABIDE | ABIDE | Data from 150,000 individuals across a wide array of clinical domains | HCP | ABIDE | ABIDE | ABIDE | ABIDE |
| AddNeuroMed | AIBL | ADHD-200 | MCIC Schizophrenic | Brainscape | ||||||
| CBRAIN | ADNI | CoRR | NKI Rockland | ELUDE | ||||||
| CITA | ADNIDOD | NKI Rockland | fBIRN | |||||||
| GUSTO | HCP | MIRIAD | ||||||||
| IBIS | ICBM | OASIS | ||||||||
| MAVAN | PPMI | |||||||||
| NeuGrid | ||||||||||
| NeuroDevNet | ||||||||||
| Access policies | Government-defined with institutional agreement | Government-defined with institutional agreement | User-specified | User-specified | User-specified | User-specified | Unrestricted | User-specified | User-specified | User-specified |
| Data contribution | Required as a condition of many NIH grants | Required as a condition of many NIH grants | Required for some multisite projects | Required for some multisite projects | Required for some multisite projects | Voluntary | Voluntary | Voluntary | Voluntary | |
| Voluntary | Voluntary | Voluntary | Voluntary |
Note:—NDAR indicates National Database for Autism Research; NIGMS, National Institute of General Medical Sciences; INDI/FCP, International Neuroimaging Data Sharing Initiative/1000 Functional Connectome Project; IDA, Image Data Archive; COINS, Collaborative Informatics Neuroimaging Suite; NIDB, Neuroinformatics Database; HCP, Human Connectome Project; AIBL, Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing; ELUDE, Efficient Longitudinal Upload of Depression in the Elderly; CITA, Centro de Investigación y Terapias Avanzadas; ADNIDOD, Study of Brain Aging in Vietnam War Veterans; GUSTO, Singapore's birth cohort study; MIRIAD, Multisite Imaging Research In the Analysis of Depression; BIRN, Biomedical Informatics Research Network; fBIRN, Functional Bioinformatics Research Network; IBIS, Infant Imaging Brain Study; ICBM, International Consortium for Brain Mapping; OASIS, Open Access Structural Imaging Series; MAVAN, Maternal Adversity, Vulnerability, and Neurodevelopment; PPMI, Parkinson's Progressive Measures Initiative; XNAT, Extensible Neuroimaging Archive Toolkit; NINDS, National Institute of Neurologic Disorders and Stroke; ADHD, attention deficit/hyperactivity disorder; CoRR, Consortium for Reliability and Reproducibility; NKI Rockland, Nathan Kline Institute Rockland; MCIC, MIND Clinical Imaging Consortium.
Because of the excellent existing data bases and informatics systems, there does not appear to be a compelling rationale for creating a new platform at this time. Rather, it seems appropriate that the existing systems should be leveraged and utilized by the TBI community. Harmonization of imaging, phenotypic, and outcome data to enable data sharing and queries across platforms so that they can be federated by using tools such as Mica-Opal (http://wiki.obiba.org/pages/viewpage.action?pageId=22544438) will be an important step toward encouraging the use of such data bases for TBI imaging research purposes. Furthermore, federation of data bases and data pooling will be greatly facilitated through the systematic use of the National Institute of Neurologic Disorders and Stroke Common Data Elements,44–47 which have been developed by more than 200 scientific experts and include more than 1000 CDEs for TBI and another 8000 for other neurologic disorders (http://www.commondataelements.ninds.nih.gov/tbi.aspx#tab=Data_Standards). Importantly, for prospective data collection, consultation of both the Federal Interagency Traumatic Brain Injury Research informatics system and the National Institute of Neurologic Disorders and Stroke Web site is recommended to ensure that data conform to the guidelines and specification of these variables. For data that were collected before the formation of these recommended elements, attempts to recategorize or adjust data to fit the recommended elements are encouraged when possible, by using tools such as those available in ACR DART to map data to the CDEs.
For a uniform approach toward TBI data sharing to succeed, it is important that issues regarding data policies be addressed and synchronization of data sharing policies across systems be performed. While it will often be important to establish embargo periods before data can be shared beyond the principal investigators, those periods should be minimized to facilitate use of data by other investigators. The data sharing policy employed by FITBIR is an example of the complexity that can be found in such policies. On the one hand, FITBIR has a staged an embargo period by which core Common Data Elements are accessible on an ongoing basis as data are collected. However, the FITBIR data-sharing policy also allows the principal investigators to control access to the data needed for the primary analyses as described by them in their original application until completion of the study, when it becomes available 6 months after the grant has ended for investigators who have also contributed data to the FITBIR and 12 months later for other investigators. Thus, substantial delays in data access may be experienced unless collaboration is agreed on by the principal investigators, which could limit research advances across the field of inquiry. Finally, serious consideration should be given to providing limited access to industry partners (possibly in exchange for a fee) for the sole purpose of promotion of development of analysis programs.
The informed consent of the patients whose data are incorporated in these data bases should include the language required to accommodate the above-discussed sharing policies. Specifically, informed consent documents (and Health Insurance Portability and Accountability Act authorization documents) should explicitly include a statement as to whether data will be contributed to an existing (or future) data repository. Template institutional review board applications for retrospective and prospective data collection (including consent forms for prospective data collections) should be prepared and made widely available to the TBI community, especially to junior investigators. These documents should explicitly indicate whether research scans will be reviewed for incidental findings (and by whom) and the method of reporting critical findings to the study's primary investigator or data safety officer/medical monitor.
The development of better interfaces to streamline the process of uploading imaging and other data is recommended. Uploading imaging processes should include automated anonymization to erase personal health information as well as, ideally, a defacing algorithm to remove subject facial features. Adequate quality control should also be implemented, either by the data submitter or by the data base manager. The granularity of this quality control should be defined on the basis of the characteristics, purpose, and complexity of the studies but should, at least, ensure that the appropriate imaging protocols have been used and that the core CDEs have been collected.
Considerations for a Data Base of Normal Control Subjects
A comprehensive understanding of control groups is essential to providing a context against which the presumed effects of mild TBI can be interpreted. Normative standards may need to be considered across the range of ages, genders, and a broad range of otherwise unsuspected individual and related population identifiers. Recommendations regarding collection of certain data elements are provided below along with a brief rationale behind the recommendation. Specific selection of inclusion/exclusion criteria will vary by study and the question being addressed. Furthermore, some studies may benefit from more than 1 comparison group.
Consistent with the CDEs initiative,48,49 the recommendations consist of a 2-tiered system, with core elements that studies should collect under most circumstances (Table 2) and preferred elements for studies that permit more detailed data collection. Core recommendations should be easily obtainable, with minimal cost or effort. Toolboxes such as the NIH toolbox (http://www.nihtoolbox.org/Pages/default.aspx) and Immediate Post-Concussion Assessment and Cognitive Testing (IMPACT) testing50 have been designed and developed to facilitate the collection of neurologic, cognitive, and psychological variables relevant to mild TBI.
Table 2:
Core (required) demographic variables that should be collected for any normative data base
| Recommended Core (Required) Variables | CDE Variable Name | Permissible Values | Notes, Including Those from the CDE Web Site |
|---|---|---|---|
| Age | Age value | 0–120 yr? | For children younger than 1 year born at <36 weeks' gestation, it is recommended to also collect gestational age. Because of potential violation of privacy legislation and specifically HIPAA regulations, the calculated age should be recorded rather than using the actual date of birth. |
| Gender | Gender type | Female, male, unknown, unspecified, not reported | Note that “gender” refers to self-reported gender of the participant. Gender is the socially constructed identity of sex and is equated with phenotypic sex. Gender may differ from the sex of an individual determined genetically. The use of “gender” as opposed to “sex” is to facilitate the ease of self-report data collection. |
| Handedness | Hand preference type | Left hand, right hand, both hands, unknown | Hand which the participant/subject uses predominantly, not necessarily the hand he/she writes with exclusively |
| Race | Race USA category | American Indian or Alaska native, Asian, black or African-American, Native Hawaiian or other Pacific Islander, white, unknown, not reported | The patient's self-declared racial origination, independent of ethnic origination, using OMB-approved categories. We note that these may not be applicable to all non-USA regions. |
| Ethnicity | Ethnicity USA category | Hispanic or Latino, not Hispanic or Latino, unknown, not reported | Category of ethnicity the participant/subject most closely identifies with. We note that these may not be applicable to all non-USA regions. |
| Socioeconomic status | Education level USA-type | Never attended/kindergarten only; 1st grade; 2nd grade; 3rd grade; 4th grade; 5th grade; 6th grade; 7th grade; 8th grade; 9th grade; 10th grade; 11th grade; 12th grade, no diploma; high school graduate; GED or equivalent; some college, no degree; associate degree: occupational/technical/vocational program; associate degree: academic program; bachelor's degree (eg, BA, AB, BS, BBA); master's degree (eg, MA, MS, MEng, MEd, MBA); professional school degree (eg, MD, DDS, DVM, JD); doctoral degree (eg, PhD, EdD); unknown | Highest grade or level of school participant/subject has completed or the highest degree received. Note that for children, these should reflect the highest education level of the primary caregiver. |
| Job classification category | Official/manager, professional, technician, sales worker, administrative support worker, craft worker, operative, laborer/helper, service worker, social worker, unknown, none | Category that classifies work performed by participant or, in the case of children, the work performed by the primary caregiver. | |
| Family income range | $15,000 to $24,999, $25,000 to $34,999, $35,000 to $49,999, $50,000 to $74,999, $75,000 to $99,999, ≥$100,000, refused, unknown, <$15,000; | Range, in US dollars, of the annual pretax, prededuction total income, of the household of which the participant/subject is a member | |
| Academic achievement | Education service type | Special education, regular education, none, unknown, early intervention | Type of educational services received (as a child) |
Note:—GED indicates General Educational Development; HIPPA, Health Insurance Portability and Accountability Act; OBM, Office of Budget and Management.
The following core (ie, required) CDEs for demographic variables should be collected in all studies, based on supporting evidence in the literature that they influence imaging results in 1 or more imaging modalities: age, sex, and handedness. For handedness, the preferred recommendation is the use of a measure designed to measure hand preference, such as the Edinburgh Handedness Inventory.
Medical and psychiatric history screening for inclusion in normative data bases should routinely involve questions related to the following conditions. For adults, core recommendations are listed in Table 2 in addition to other information on medical illnesses, such as hypertension, cardiovascular risk factors, and pain syndromes. For children, it is recommended to record the following information: estimated gestational age at the time of birth, presence or absence of eventful or high-risk pregnancy, maternal substance abuse during pregnancy, type of delivery (eg, vaginal, cesarean delivery, and/or vacuum/forceps birth), hospitalization after birth for more than 3 days, history of developmental disorder, presence or absence of attention deficit/hyperactivity disorder, learning disorder or autistic spectrum disorder, systematic exposure to a contact sport, history of physical trauma, specific questioning of previous concussions, presence of childhood obesity, presence of CNS or non-CNS tumor, presence of neurologic disorders (eg, tics, seizures, febrile seizures), history of exposure to general anesthesia, history of toxic exposure (eg, lead ingestion, smoke inhalation), medication history, history of diabetes, asthma, substance use, and sleep disorder. These criteria are not necessarily exclusion criteria, but collection of these data points is important in understanding the degree to which the data base is homogeneous and for understanding what factors could, in theory, be implicated in affecting test results.
General level of intelligence function or academic achievement is considered an important factor in studies utilizing control participants to ensure general comparability between groups. Although screening with standardized measures may not be practical in all situations, we recommend collection of at least limited information to ensure group comparability, which may also be used as a factor in later analysis. Level of education attainment should be considered a core requirement. Furthermore, collection of standardized measures is highly recommended because it may enhance participant screening and group characterization for general cognitive functioning (or intelligence quotient). The selection of specific measures will depend on the age group under assessment and population being studied. If used, a screening measure such as the Wechsler Abbreviated Scale of Intelligence, Version II, may be used in both children and adults (for general intelligence). The Wechsler Test of Adult Reading or another reading test may also be used in adults. The mean and distribution of the subject group and control group should be comparable. Depending on the study, investigators may wish to consider a lower limit exclusion of 2 SDs below the mean for such tests as the Wechsler Abbreviated Scale of Intelligence, Version II, and the Wechsler Test of Adult Reading for the specific metric under consideration, because this is generally consistent with definitions of “impairment” and would be unusual in a population of typically developing individuals.
Level of psychological functioning (including the presence of psychiatric disorders or substance abuse) may also contribute to brain integrity, and several psychiatric disorders have been shown to be associated with altered brain structure or function.51 Major psychiatric disorders, including major mood, anxiety, and thought disorders, may require exclusion from normative populations. Other disorders may have an increased representation in studies of TBI, including attention deficit/hyperactivity disorder (in children) and posttraumatic stress disorder (in military studies); these may require measurement of severity to use in later analysis and to ensure group comparability. The Child Behavior Checklist screener (behavioral) is recommended for use as a screening tool. Finally, the presence of substance abuse disorders in normal subjects (as well as in study patients) requires consideration because it can produce both behavioral and structural changes.
At present, insufficient evidence exists to determine the effect of the following factors on imaging variables: race, ethnicity, and socioeconomic status (ie, level of education, primary caregiver's level of education, job classification, and family income range; Table 2). Nonetheless, further evaluation of these demographic variables is warranted; thus, continued collection of such data is recommended until evaluation is complete.
Evidence and guidelines regarding appropriate comparison groups for military and sports-related concussion are lacking, and a great need exists for further research in this area. Our knowledge regarding the effect of numerous variables, the size of the effects, and which control group or groups are most ideal is still incomplete. Therefore, studies involving combat-related TBI in either active duty service members or veterans or studies involving sports-related concussion may require additional special consideration for control participants, and proposed guidelines may require some flexibility based on the study question.
Imaging Protocols
The choice of advanced imaging protocols is very important both when considering a method of obtaining meaningful data and developing a means of data sharing across sites. A wide variety of users exists for such data, including individuals interested in defining characteristics that distinguish the mild TBI population from normal subjects, investigators concerned with assessment of treatment effect, and researchers involved with designating the means by which imaging techniques can, after sufficient validation among a wide group of investigators, eventually be used for diagnosis. These various forms of use require that technologic features of advanced imaging protocols have, on the one hand, qualities such as excellent test performance characteristics (eg, reproducibility and accuracy) and, on the other hand, features that allow them to be used on multiple types of scanners and in various scanning environments.
We now provide recommendations in terms of imaging protocols for TBI research. Please note that these recommendations are intended for TBI research and are not meant to provide guidelines for clinical use in TBI. The recommendations have been designed as a core imaging protocol that will allow imaging of a sufficient number of normal control subjects for a normative data base. However, a wider, more inclusive population, including patients with TBI, could be generated by using documented variants of this core imaging protocol and tailored to fit more closely the research question posed by each study. For an initial, normative data base, homogeneity of the data is essential, and acquisition of the imaging data on 3T MR imaging scanners is strongly recommended.
One condition for meaningful use of a data base is a high level of quality control in selection of data before deposition in the data base. For a normal subject data base, such quality control would consist of the following: 1) determination of whether adherence to the recommended imaging protocol has been performed, 2) recording of specific details of advanced imaging sequences (eg, features such as gradient table, number of diffusion directions, number of channels in the head coil, and voxel dimensions), and 3) evaluation of the quality of images obtained in both conventional and advanced imaging sequences. Because these factors are critical to success, we recommend that a skilled neuroradiologist currently certified by the American Board of Radiology and having a Certificate of Added Qualification in Neuroradiology review all imaging studies before their entry into the normative data base. Ideally, this neuroradiologist should have worked for several years at a center treating a high volume of patients with TBI and also have an understanding of the image features needed for high-quality DTI and resting-state fMRI data. Besides a review before the data uploading, the imaging data should also be reviewed by a team of qualified referees after submission to the data base.
The recommended imaging protocol for the normative data base should include routine (conventional) MR imaging pulse sequences and advanced MR imaging pulse sequences. The recommended routine, conventional MR imaging pulse sequences include the following:
High-resolution 3D T1 (eg, 3D MPRAGE or echo-spoiled gradient echo)
High-resolution thin-section (3D) T2
High-resolution 3D T2 FLAIR
3D T2* (eg, SWI).
Recommended advanced MR imaging pulse sequences include diffusion tensor imaging and resting-state functional MR imaging. Details for each are listed below.
For DTI (Institute of Medicine's Forum on Neuroscience and Nervous System Disorders, Health Arm of the National Academy of Science; Developing Standards for Diffusion Tensor Imaging and Diffusion Spectrum Imaging through Public-Private Partnerships, 2013), we recommend a 2-tiered protocol set that allows flexibility in imaging, depending on the specific DTI information sought (Table 3). A so-called “Core” protocol is specified that consists of a basic, “bare-bones” DTI acquisition that should be readily available on virtually all scanners and will be maximally inclusive of TBI investigators across sites. A “Preferred” protocol is then specified with 2 variations to allow for time trade-offs in DTI imaging, which often mean balancing the number of diffusion gradients, the voxel size, number of sections, and number of acquisitions to complete imaging in a reasonable time. Specifically, the 2 variations of “Preferred” protocol allow investigators to use either a protocol with a very high number of diffusion gradient directions (at least 30, ideally 64 directions recommended) or one that provides a means to measure non-Gaussian diffusion (ie, diffusional kurtosis imaging), which requires at least 3 b-values (DKI protocols are available at http://www.musc.edu/cbi/dki/). Use of DKI may become more widespread once multiband echo-planar and compressed sensing techniques become more readily available. The DTI (or DKI) protocol used should be recorded when resultant imaging data are expected to be included in a data base. For all DTI protocols, we recommend a head coil with at least 8 channels, a scanner gradient strength of at least 40–50 mT/m, a slew rate of at least 150–200 T/m/s, and voxel dimensions as close as possible to being isotropic (eg, ∼2- to 3-mm thickness in the z-axis). Source images should be inspected for quality assurance before fitting the tensor model. Recommendations for postprocessing are beyond the scope of this roadmap.
Table 3:
“Core” and “preferred” imaging protocols for diffusion-weighted imaging
| Parameter | “Core” | “Preferred” No. 1 (High Angular Resolution) | “Preferred” No. 2 (More b-Values) |
|---|---|---|---|
| Orientation | Axial | Axial | Axial |
| Coil | Any | Phased array ≥8 channels | Phased array ≥8 channels |
| Readout | EPI | EPI | EPI |
| TR (ms) | ∼9000 | ∼9000 | ∼9000 |
| TE (ms) | Min | Min (<100) | Min (<100) |
| FOV (mm2) | 256 × 256 or 350 (66% phase) | 256 × 256 or 350 (66% phase) | 256 × 256 or 350 (66% phase) |
| Matrix size | 128 × 128 | 128 × 128 | 128 × 128 |
| Sections/thickness (mm) | Any/≤3 | 59/2.7 | 59/2.7 |
| Section gap (mm) | 0 | 0 | 0 |
| Voxel size (mm) | ≤3 in all dimensions | Isotropic 2.73 | Isotropic 2.73 |
| Directions | ≥12 | 64 | ≥12 |
| Dual-echo | Any | No | No |
| Fat-suppression | Any | Yes | Yes |
| Phase-encode direction | A to P | A to P | A to P |
| BW (Hz/pixel) | 1346 | 1346 | 1346 |
| Parallel imaging factor | Any | 2 | 2 |
| b-value (s/mm2) | 2 (0, ∼1000) | 2 (0, 1300) | ≥3 (eg, 0, ∼1000, ∼2000) |
| No. b=0 images | 1 | 1 per 8 directions if allowed | 1 per 8 directions if allowed |
Note:—Min indicates minimum; A to P, anterior to posterior; BW, bandwidth.
For resting-state functional MR imaging, standard gradient echo echo-planar imaging–based blood oxygen level–dependent fMRI is recommended by using a dedicated head coil having at least 8 channels to allow for parallel imaging. Whole-brain coverage (including the cerebellum) should be standard, and the concomitant acquisition of a high-resolution T1-weighted gradient-echo sequence for anatomic coregistration is needed. An isotropic voxel resolution of 3 mm is satisfactory with current standard scanning tools to attempt to simultaneously maximize SNR and resolution. Generally, an axial acquisition should be performed parallel to the anterior/posterior commissure line. Acquisition should be obtained with the subject awake and in a restful state, wearing some method of head-movement constraint. Both eyes-open and eyes-closed imaging techniques have been successfully performed; evidence suggests that the eyes-open imaging method more readily ensures that subjects remain awake.
Mandatory steps for preprocessing of fMRI data include the following: quality assessment for motion, spatial Gaussian smoothing, temporal bandpass filtering, and a regression model to remove physiologic nuisance covariates and allow coregistration of anatomic and functional data and realignment of the functional data. Spatial transformation to a common brain space will often be necessary to accomplish group analyses. A variety of reasonable approaches are available, including the Functional MR Imaging of the Brain Nonlinear Image Registration Tool (http://www.fmrib.ox.ac.uk//fsl), Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra (http://nipy.sourceforge.net/nipype/users/examples/fmri_spm_dartel.html), and Advanced Normalization Tools (http://stnava.github.io/ANTs/). Individuals interested in pursuing resting-state fMRI in this population are referred to some commonly used and readily available software packages (including but not limited to the Functional MR Imaging of the Brain Software Library, http://www.fmrib.ox.ac.uk/fsl; Analysis of Functional Neuroimages, http://afni.nimh.nih.gov/afni/; and Statistical Parametric Mapping, http://www.fil.ion.ucl.ac.uk/spm/) for preprocessing of data. A few tools that can be used for connectivity analyses include fMRI of the Brain Software Library (http://www.fmrib.ox.ac.uk/fsl), Conn (http://www.nitrc.org/projects/conn/), and Data Processing Assistant for Resting-State fMRI (http://www.nitrc.org/projects/conn/). The Human Connectome project has a suite of resting-state functional MR imaging–related sources available for use (http://www.humanconnectome.org/about/project/resting-fmri.html).
Phantom calibration of the MR imaging scanners should be performed on at least a weekly basis. A variety of standardized phantom and measurement protocols is now available. These include the ADNI or ACR phantom for conventional, structural MR imaging sequences (http://www.acr.org/∼/media/ACR/Documents/Accreditation/MRI/LargePhantomGuidance.pdf),the National Institute of Standards and Technology (NIST) diffusion phantom for DWI data, and the Biomedical Informatics Research Network phantom for functional MR imaging data (http://www.birncommunity.org/tools-catalog/function-birn-stability-phantom-qa-procedures/). The results of phantom scanning can be compared and contrasted across sites to ensure that multisite consortia remain in agreement in terms of basic scanner and image quality control metrics.
Conclusions
The “Joint ASNR-ACR HII-ASFNR TBI Workshop: Bringing Advanced Neuroimaging for TBI into the Clinic,” on May 23, 2014, in Montreal, Quebec, Canada, identified a number of excellent already-existing data bases and informatics systems, which obviates the need to create a new platform at this time. Rather, to facilitate the creation of a normative data base, we make recommendations to harmonize collection of imaging, phenotypic, and outcomes data. Such a process would enable data sharing and queries across platforms to allow the data bases to be federated. Federation of data bases and data pooling will be greatly facilitated through the systematic use of the National Institute of Neurologic Disorders and Stroke Common Data Elements and through harmonization of data-sharing policies. Better interfaces to streamline the process of uploading imaging and other data should be developed.
In terms of the demographic variables to be collected for the normative data base, a 2-tier system is recommended, with core elements that studies should collect under most circumstances and preferred guidelines for more detailed data collection for studies that permit this. These core recommendations are generally easily obtainable, with minimal cost or effort. Studies involving combat-related TBI in either active duty service members or veterans or from studies involving sports-related concussion may require additional special consideration for control participants, and proposed guidelines may require some flexibility based on the study question being asked.
The recommended conventional imaging sequences include high-resolution 3D T1 (3D MPRAGE), high-resolution thin-section T2, high-resolution 3D T2 FLAIR, and 3D T2* (for instance SWI); and the recommended advanced imaging sequences include high-quality DTI (or DKI) and resting-state fMRI (ie, in conformance with the image acquisition parameters outlined earlier). Image acquisition on 3T scanners is recommended, along with (at least) monthly calibration of the MR imaging scanners by using a phantom. A skilled neuroradiologist should review all the imaging data (including review for incidental findings) before deposition of the data in the normative data base, and the imaging data should also be reviewed once within the data base by a team of qualified referees.
We did not specifically discuss imaging-processing tools and pipelines or ways to share them across institutions and platforms. We intend for these issues to be the topic of a future workshop.
Acknowledgments
We wish to acknowledge the generous support of the American Society of Neuroradiology, the Head Injury Institute of the American College of Radiology, the American Society of Functional Neuroradiology, GE Healthcare and Philips Healthcare, whose contributions made possible the “Joint ASNR-ACR HII-ASFNR TBI Workshop: Bringing Advanced Neuroimaging for TBI into the Clinic,” held on Friday May 23, 2014, in Montreal, Quebec, Canada, as a parallel session during the 52nd Annual Meeting of the ASNR and The Foundation of the ASNR Symposium 2014.
ABBREVIATIONS:
- ABIDE
Autism Brain Imaging Data Exchange
- ACR
American College of Radiology
- ADNI
Alzheimer Disease Neuroimaging Initiative
- AFSNR
American Society of Functional Neuroradiology
- ASNR
American Society of Neuroradiology
- CDE
Common Data Element
- DART
Data Archive and Research Toolkit
- DKI
diffusional kurtosis imaging
- FITBIR
Federal Interagency TBI Research
- LONI
Laboratory of Neuroimaging
- LORIS
Longitudinal On-line Research and Imaging System
- NIH
National Institutes of Health
- NITRC
Neuroinformatics Tools and Resources
- TBI
traumatic brain injury
- TRIAD
Translational Research Informatics and Data Management Grid
Appendix
American College of Radiology Head Injury Institute Contributors: Trevor Andrews, PhD, Philips Healthcare and University of Vermont, College of Medicine, Burlington, Vermont; Sylvain Bouix, PhD, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts; Michael Brant-Zawadzki, MD, FACR, Hoag Hospital, Newport Beach, California; Asim Choudhri, MD, University of Tennessee Health Science Center, Memphis, Tennessee; Cameron Craddock, PhD, Center for the Developing Brain, Child Mind Institute, New York, New York; Alan Evans, PhD, McConnell Brain Imaging Centre, Montreal Neurological Institute, McGill University, Montreal, Quebec, Canada; Damien Galanaud, MD, PhD, Hôpital de la Pitié Salpêtrière, Paris, France; Alisa Gean, MD, University of California, San Francisco, and San Francisco General Hospital, San Francisco, California; Jam Ghajar, MD, PhD, FACS, Stanford University, Stanford, California; Rajiv Gupta, MD, PhD, Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts; E. Michael Harned, MD, Indiana University School of Medicine, Indianapolis, Indiana; Joseph A. Helpern, PhD, Medical University of South Carolina, Charleston, South Carolina; Nadja Kadom, MD, Boston Medical Center and Boston University, Boston, Massachusetts; Inga Koerte, MD, Ludwig-Maximilian-University, Munich, Germany, and Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts; Roland Lee, MD, FACR, University of California, San Diego, San Diego, California; Joseph Maldjian, MD, PhD, Wake Forest School of Medicine, Winston-Salem, North Carolina; Srini Mukundan, MD, PhD, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts; Sumit N. Niogi, MD, PhD, NY Presbyterian Hospital–Weill Cornell Medical College, New York, New York; Ofer Pasternak, PhD, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts; Cyrus Raji, MD, PhD, University of California, Los Angeles, Medical Center, Los Angeles, California; Dan Rettmann, PhD, GE Healthcare, Milwaukee, Wisconsin; Michael I. Rothman, MD, Bethlehem, Pennsylvania; Martha Shenton, PhD, Brigham and Women's Hospital and Harvard Medical School, Boston, Massachusetts; Tamar D. Shirman, PhD, Philips Healthcare, Best, the Netherlands; Jeannette Söderberg, PhD, International Neuroinformatics Coordinating Facility, Karolinska Institutet, Stockholm, Sweden; Kenneth A. Spero, MD, Northwest Community Hospital, Arlington Heights, Illinois; Jack Darrell Van Horn, MEng, PhD, The Institute for Neuroimaging and Informatics and Laboratory of Neuro Imaging, Keck School of Medicine of University of Southern California, Los Angeles, California; Michael D. Wirt, MD, PhD, COL, MC, FS Commander, USAISROTSG Radiology Consultant JBSA, Fort Sam Houston, Houston, Texas; David W. Wright, MD, FACEP, Emory University School of Medicine, Atlanta, Georgia; Lyubomir Zagorchev, PhD, Philips Healthcare, Best, the Netherlands.
Footnotes
Disclosures: Max Wintermark—RELATED: Grant: ASNR,* ASFNR,* ACR,* Comments: paid to ASNR, not my institution; UNRELATED: Grants/Grants Pending: GE Healthcare,* Philips Healthcare.* Christopher G. Filippi—UNRELATED: Consultancy: Regeneron Pharmaceuticals, Syntactx, Comments: For both of these companies, I interpret MR images that are part of research trials; Grants/Grants Pending: NIH, Comments: Coinvestigator on NIH grant, 1R01CA161404-01A; Other: Co-Principal Investigator on Coulter Grant (software development) and coinvestigator on Irving Foundation Grant (new MR imaging sequence development) and mentor to Radiological Society of North American grant recipients (currently a medical student and a resident). Randy Horton—RELATED: Consulting Fee or Honorarium: American College of Radiology, Comments: I am a paid consultant to the ACR; Support for Travel to Meetings for the Study or Other Purposes: American College of Radiology, Comments: I am a paid consultant to the ACR, and they paid for my travel to the meeting where the information in this publication was first developed; Payment for Writing or Reviewing the Manuscript: American College of Radiology, Comments: The ACR compensated me for my review time on this manuscript. Yvonne W. Lui—UNRELATED: Grants/Grants Pending: NIH R01.* Meng Law—RELATED: Grant: Toshiba America Medical Systems*; Consulting Fee or Honorarium: Bracco Diagnostics.* Gordon Sze—UNRELATED: Expert Testimony: a very few legal firms. James Provenzale—RELATED: Support for Travel to Meetings for the Study or Other Purposes: ACR, Comments: support for travel to the ACR/ASNR workshop; UNRELATED: Grants/Grants Pending: US Department of Veterans Affairs.* Aaron Field—UNRELATED: Consultancy: Asterias Biotherapeutics (paid consultant); Grants/Grants Pending: NIH,* National MS Society,* Comments: Research grant support. Elisabeth Wilde—RELATED: Support for Travel to Meetings for the Study or Other Purposes: ACR, Comments: ASNR meeting for workshop related to work associated with this manuscript; UNRELATED: Grants/Grants Pending: NIH,* Department of Defense,* Veterans Administration,* Comments: Grants not associated with this work. Pratik Mukherjee—UNRELATED: Board Membership: GE Healthcare (Medical Advisory Board), RGN Neuroscience (Scientific Advisory Board); Grants/Grants Pending: GE Healthcare (research grant and industry contract)*; Stock/Stock Options: RGN Neuroscience (stock options). *Money paid to the institution.
REFERENCES
- 1. Faul M, Xu L, Wald M, et al. Traumatic brain injury in the United States: emergency department visits, hospitalizations and deaths 2002–2006. Centers for Disease Control and Prevention, National Center for Injury Prevention and Control 2010. http://www.cdc.gov/traumaticbraininjury/pdf/blue_book.pdf. Accessed January 17, 2015.
- 2. Gilchrist J, Thomas KE, Xu LK, et al. Nonfatal traumatic brain injuries related to sports and recreation activities among persons aged ≤19 years—United States, 2001–2009. MMWR Morbid Mortal Wkly Rep 2011;60:1337–42 [PubMed] [Google Scholar]
- 3. Maas AI, Hukkelhoven CW, Marshall LF, et al. Prediction of outcome in traumatic brain injury with computed tomographic characteristics: a comparison between the computed tomographic classification and combinations of computed tomographic predictors. Neurosurgery 2005;57:1173–82; discussion 1173–82 [DOI] [PubMed] [Google Scholar]
- 4. Wilde EA, Ramos MA, Yallampalli R, et al. Diffusion tensor imaging of the cingulum bundle in children after traumatic brain injury. Dev Neuropsychol 2010;35:333–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arfanakis K, Haughton VM, Carew JD, et al. Diffusion tensor MR imaging in diffuse axonal injury. AJNR Am J Neuroradiol 2002;23:794–802 [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar R, Gupta RK, Husain M, et al. Comparative evaluation of corpus callosum DTI metrics in acute mild and moderate traumatic brain injury: its correlation with neuropsychometric tests. Brain Inj 2009;23:675–85 [DOI] [PubMed] [Google Scholar]
- 7. Miles L, Grossman RI, Johnson G, et al. Short-term DTI predictors of cognitive dysfunction in mild traumatic brain injury. Brain Inj 2008;22:115–22 [DOI] [PubMed] [Google Scholar]
- 8. Newcombe VF, Williams GB, Nortje J, et al. Concordant biology underlies discordant imaging findings: diffusivity behaves differently in grey and white matter post acute neurotrauma. Acta Neurochir Suppl 2008;102:247–51 [DOI] [PubMed] [Google Scholar]
- 9. Newcombe VFJ, Williams GB, Nortje J, et al. Analysis of acute traumatic axonal injury using diffusion tensor imaging. Br J Neurosurg 2007;21:340–48 [DOI] [PubMed] [Google Scholar]
- 10. Wozniak JR, Krach L, Ward E, et al. Neurocognitive and neuroimaging correlates of pediatric traumatic brain injury: a diffusion tensor imaging (DTI) study. Arch Clin Neuropsychol 2007;22:555–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McAllister TW, Ford JC, Ji S, et al. Maximum principal strain and strain rate associated with concussion diagnosis correlates with changes in corpus callosum white matter indices. Ann Biomed Eng 2012;40:127–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chokshi FH, Munera F, Rivas LA, et al. 64-MDCT angiography of blunt vascular injuries of the neck. AJR Am J Roentgenol 2011;196:W309–15 [DOI] [PubMed] [Google Scholar]
- 13. Bazarian JJ, Zhong J, Blyth B, et al. Diffusion tensor imaging detects clinically important axonal damage after mild traumatic brain injury: a pilot study. J Neurotrauma 2007;24:1447–59 [DOI] [PubMed] [Google Scholar]
- 14. Bazarian JJ, Zhu T, Blyth B, et al. Subject-specific changes in brain white matter on diffusion tensor imaging after sports-related concussion. Magn Reson Imaging 2012;30:171–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lewine JD, Davis JT, Sloan JH, et al. Neuromagnetic assessment of pathophysiologic brain activity induced by minor head trauma. AJNR Am J Neuroradiol 1999;20:857–66 [PMC free article] [PubMed] [Google Scholar]
- 16. Huang MX, Theilmann RJ, Robb A, et al. Integrated imaging approach with MEG and DTI to detect mild traumatic brain injury in military and civilian patients. J Neurotrauma 2009;26:1213–26 [DOI] [PubMed] [Google Scholar]
- 17. Huang MX, Nichols S, Robb A, et al. An automatic MEG low-frequency source imaging approach for detecting injuries in mild and moderate TBI patients with blast and non-blast causes. Neuroimage 2012;61:1067–82 [DOI] [PubMed] [Google Scholar]
- 18. Tarapore PE, Findlay AM, Lahue SC, et al. Resting state magnetoencephalography functional connectivity in traumatic brain injury. J Neurosurg 2013;118:1306–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo Q, Xu D, Roskos T, et al. Complexity analysis of resting state magnetoencephalography activity in traumatic brain injury patients. J Neurotrauma 2013;30:1702–09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Batoulis H, Recks MS, Holland FO, et al. Blockade of tumour necrosis factor-alpha in experimental autoimmune encephalomyelitis reveals differential effects on the antigen-specific immune response and central nervous system histopathology. Clin Exp Immunol 2014;175:41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stamatakis EA, Wilson JT, Hadley DM, et al. SPECT imaging in head injury interpreted with statistical parametric mapping. J Nucl Med 2002;43:476–83 [PubMed] [Google Scholar]
- 22. Lorberboym M, Lampl Y, Gerzon I, et al. Brain SPECT evaluation of amnestic ED patients after mild head trauma. Am J Emerge Med 2002;20:310–13 [DOI] [PubMed] [Google Scholar]
- 23. Lewine JD, Davis JT, Bigler ED, et al. Objective documentation of traumatic brain injury subsequent to mild head trauma: multimodal brain imaging with MEG, SPECT, and MRI. J Head Trauma Rehabil 2007;22:141–55 [DOI] [PubMed] [Google Scholar]
- 24. Chen SHA, Kareken DA, Fastenau PS, et al. A study of persistent post-concussion symptoms in mild head trauma using positron emission tomography. J Neurol Neurosurg Psychiatry 2003;74:326–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peskind ER, Petrie EC, Cross DJ, et al. Cerebrocerebellar hypometabolism associated with repetitive blast exposure mild traumatic brain injury in 12 Iraq War veterans with persistent post-concussive symptoms. Neuroimage 2011;54(suppl 1):S76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang MX, Nichols S, Baker DG, et al. Single-subject-based whole-brain MEG slow-wave imaging approach for detecting abnormality in patients with mild traumatic brain injury. Neuroimage Clin 2014;5:109–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alexander AL, Lee JE, Lazar M, et al. Diffusion tensor imaging of the brain. Neurotherapeutics 2007;4:316–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stokum JA, Sours C, Zhuo J, et al. A longitudinal evaluation of diffusion kurtosis imaging in patients with mild traumatic brain injury. Brain Inj 2015;29:47–57 [DOI] [PubMed] [Google Scholar]
- 29. Grossman EJ, Ge Y, Jensen JH, et al. Thalamus and cognitive impairment in mild traumatic brain injury: a diffusional kurtosis imaging study. J Neurotrauma 2012;29:2318–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benson RR, Meda SA, Vasudevan S, et al. Global white matter analysis of diffusion tensor images is predictive of injury severity in traumatic brain injury. J Neurotrauma 2007;24:446–59 [DOI] [PubMed] [Google Scholar]
- 31. Kraus MF, Susmaras T, Caughlin BP, et al. White matter integrity and cognition in chronic traumatic brain injury: a diffusion tensor imaging study. Brain 2007;130:2508–19 [DOI] [PubMed] [Google Scholar]
- 32. Warner MA, de la Plata CM, Spence J, et al. Assessing spatial relationships between axonal integrity, regional brain volumes, and neuropsychological outcomes after traumatic axonal injury. J Neurotrauma 2010;27:2121–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharp DJ, Ham TE. Investigating white matter injury after mild traumatic brain injury. Curr Opin Neurol 2011;24:558–63 [DOI] [PubMed] [Google Scholar]
- 34. Jorge RE, Acion L, White T, et al. White matter abnormalities in veterans with mild traumatic brain injury. Am J Psychiatry 2012;169:1284–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adamson C, Yuan WH, Babcock L, et al. Diffusion tensor imaging detects white matter abnormalities and associated cognitive deficits in chronic adolescent TBI. Brain Inj 2013;27:454–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baek SO, Kim OL, Kim SH, et al. Relation between cingulum injury and cognition in chronic patients with traumatic brain injury; diffusion tensor tractography study. Neurorehabilitation 2013;33:465–71 [DOI] [PubMed] [Google Scholar]
- 37. Palacios EM, Sala-Llonch R, Junque C, et al. Resting-state functional magnetic resonance imaging activity and connectivity and cognitive outcome in traumatic brain injury. JAMA Neurol 2013;70:845–51 [DOI] [PubMed] [Google Scholar]
- 38. Venkatesan UM, Dennis NA, Hillary FG. Chronology and chronicity of altered resting-state functional connectivity after traumatic brain injury. J Neurotrauma 2015;32:252–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maller JJ, Thomson RH, Pannek K, et al. The (Eigen)value of diffusion tensor imaging to investigate depression after traumatic brain injury. Hum Brain Mapp 2014;35:227–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schönberger M, Ponsford J, Reutens D, et al. The relationship between mood disorders and MRI findings following traumatic brain injury. Brain Inj 2011;25:543–50 [DOI] [PubMed] [Google Scholar]
- 41. Pitkänen A, Kemppainen S, Ndode-Ekane XE, et al. Posttraumatic epilepsy: disease or comorbidity? Epilepsy Behav 2014;38C:19–24 [DOI] [PubMed] [Google Scholar]
- 42. Gross H, Kling A, Henry G, et al. Local cerebral glucose metabolism in patients with long-term behavioral and cognitive deficits following mild traumatic brain injury. J Neuropsychiatry Clin Neurosci 1996;8:324–34 [DOI] [PubMed] [Google Scholar]
- 43. Das S, Zijdenbos AP, Harlap J, et al. LORIS: a web-based data management system for multi-center studies. Front Neuroinform 2011;5:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yue JK, Vassar MJ, Lingsma HF, et al. Transforming research and clinical knowledge in traumatic brain injury pilot: multicenter implementation of the common data elements for traumatic brain injury. J Neurotrauma 2013;30:1831–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hicks R, Giacino J, Harrison-Felix C, et al. Progress in developing common data elements for traumatic brain injury research: version two—the end of the beginning. J Neurotrauma 2013;30:1852–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berger RP, Beers SR, Papa L, et al. Common data elements for pediatric traumatic brain injury: recommendations from the biospecimens and biomarkers workgroup. J Neurotrauma 2012;29:672–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adelson PD, Pineda J, Bell MJ, et al. Common data elements for pediatric traumatic brain injury: recommendations from the working group on demographics and clinical assessment. J Neurotrauma 2012;29:639–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilde EA, Whiteneck GG, Bogner J, et al. Recommendations for the use of common outcome measures in traumatic brain injury research. Archi Physi Med Rehabil 2010;91:1650–60.e17 [DOI] [PubMed] [Google Scholar]
- 49. McCauley SR, Wilde EA, Anderson VA, et al. Recommendations for the use of common outcome measures in pediatric traumatic brain injury research. J Neurotrauma 2012;29:678–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Maas AI, Steyerberg EW, Marmarou A, et al. IMPACT recommendations for improving the design and analysis of clinical trials in moderate to severe traumatic brain injury. Neurotherapeutics 2010;7:127–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patel R, Spreng RN, Shin LM, et al. Neurocircuitry models of posttraumatic stress disorder and beyond: a meta-analysis of functional neuroimaging studies. Neurosci Biobehav Rev 2012;36:2130–42 [DOI] [PubMed] [Google Scholar]