To the Editor,
The recent SARs‐CoV‐2 created focal news when the paradoxical spread was reported in the Wuhan province of China, further jeopardizing the existence of the human race on the planet earth. Due to its rapid expansion throughout the world, the COVID‐19 outbreak was declared as a global pandemic by the World Health Organization (WHO) on March 20, 2020. The updates of November 13, 2020, have been reported 53,429,139 infections and 1,304,470 deaths. Fever, shortness of breath, coughing, myalgia, dyspnea, and radiological indications of ground‐glass lung opacity compatible with atypical pneumonia are the signs exhibited by most patients with COVID‐19. However, some patients have also been reported to have asymptomatic or mildly symptomatic (Chen et al., 2020; Huang et al., 2020; Lu et al., 2020). The whole proteome of the SARS‐CoV‐2 is encoded by ~30 kb a genome. The whole‐genome encodes three major components, including non‐structural (NS), structural, and accessory proteins (Durojaiye, Clarke, Stamatiades, & Wang, 2020). The genes found on the 3′‐terminus encode the four structural proteins constituting the main envelope of the virus and eight accessory proteins. Among the structural proteins small envelope protein (E), spike surface glycoprotein (S), nucleocapsid protein (N), and the membrane protein (M) while the eight accessory proteins codes for 3a, 3b, p6, 7a, 7b, 8b, 9b and ORF14 (Wu et al., 2020). In contrast, the two overlapping genes, ORFIa and ORF1ab, encode the non‐structural proteins (polypeptides pp1a and pp1ab) and form a replication/transcription complex (RTC). These pp1a and pp1ab polypeptides are translated and then proteolytically cleaved by two main viral proteases, (papain‐like protease) PLpro and (3‐chymotrypsin‐like protease) 3CLpro or main proteases (Mpro) (Perlman & Netland, 2009). PLpro is accountable for the cleavage of non‐structural proteins (nsp 1–3). In contrast, the 3CLpro (Figure 1A) cleaves the polyprotein at 11 discrete sites downstream of nsp4 to yield different non‐structural proteins that play a crucial role in the viral life cycle. Previous studies have indicated that 3CLpro (Mpro) plays an important role in cell proliferation and maturation. So, the inhibition of this target would significantly contribute to controlling the COVID‐19 (Needle, Lountos, & Waugh, 2015). Because of these multi‐faceted aspects, 3CLpro has been deemed as a promising drug development target for anti‐coronaviruses (Hatada et al., 2020).
FIGURE 1.
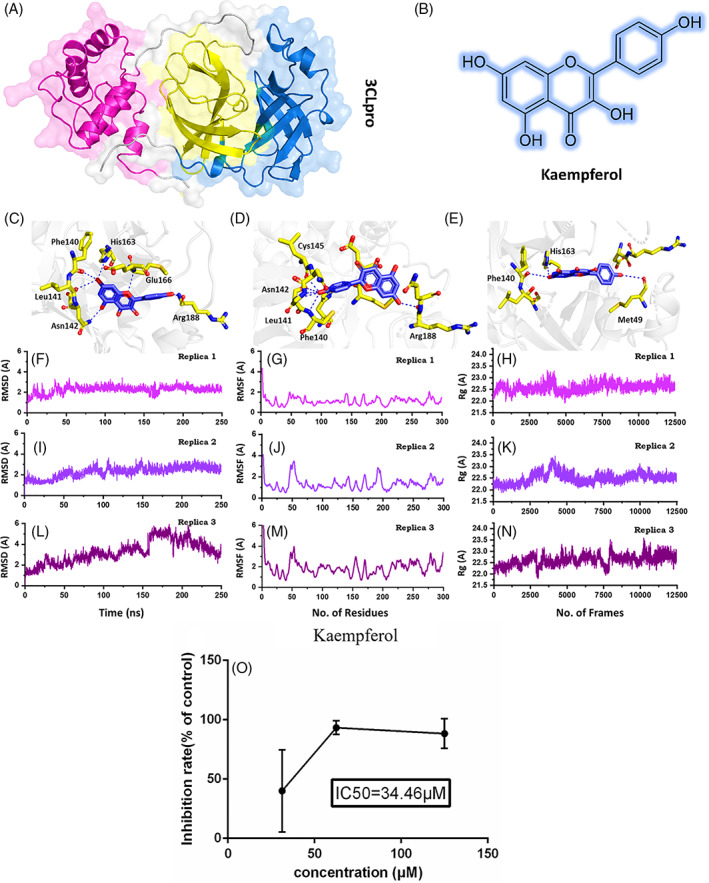
(A) Shows the structure of 3CLpro and its different domains. (B) Shows the 2D structure of kaempferol. (C–E) Show the binding mode of kaempferol each conformation. (F–H) Show the RMSd of the three best conformations. (I–K) Show the RMSf of the three best conformations. (L–N) Show the Rg of the three best conformations. (O) The inhibitory effects of kaempferol on SARS‐CoV‐2 in Vero E6 cells [Colour figure can be viewed at wileyonlinelibrary.com]
In this study, we have used both in silico and in vitro approaches to confirm the activity of an active compound Kaempferol (Figure 1B), which was reported in our previous study to potentially interact with the SARs‐CoV‐2 main protease 3CLpro (Khan et al., 2020). The compound found through molecular search from the Traditional Chinese Medicine (TCM) was re‐docked here against the active site of the main protease. AutoDock Vina (Trott & Olson, 2010) with exhaustiveness set as 64 was used to dock kaempferol against the main protease (3CLpro). The docking predictions revealed the docking scores for the 10 conformations. Among the 10 conformations, the docking score for the first three conformations was −6.4 kcal/mol. Previously, these residues His41, Met49, Tyr54, Phe140, Leu141, Asn142, Cys145, His163, Met165, Glu166, Leu167, Phe185, Asn187, Arg188, and Gln192 comprised the active site. Therefore, the first three conformations were analyzed for potential interactions with these residues. Conformation 1 (Figure 1C) formed six hydrogen bonds with the active site residues, including Phe140, Leu141, Asn142, His163, Glu166, and Arg188. A pie‐sulfur interaction was formed by Cys145, while the Met165 formed a pie‐alkyl interaction. The second conformation (Figure 1D) with the docking score (−6.4 kcal/mol) also formed six hydrogen bonds with the key active site residues. Among the key interactions, five were formed with Phe140, Leu141, Asn142, and Cys145, while one hydrogen bond was formed with Arg188. The conformation 3 formed only three hydrogen bonds with the key residues. As shown in (Figure 1E) Met49, Phe140, and His163 are involved in the interaction with Kaempferol. Hence, the docking predictions significantly confirm that kaempferol potentially interacts with the same active site residue even in different conformations, thus verify its activity against the 3CLpro.
To further validate the potential of kaempferol as an active drug, a biophysical investigation was performed. Using Amber20 (Salomon‐Ferrer, Case, & Walker, 2013), a 250 ns simulation for each conformation was performed to reveal the dynamic behavior of Kaempferol‐3CLpro complexes. The parameters were set as used in the previous study (Khan et al., 2020). To demonstrate the structural stability of the complexes, root mean square deviation (RMSD) of each complex was calculated. As given in Figure 1F–H, all the complexes reached the stability at 1.8 Å and reached the equilibrium state at 15 ns. For conformation 1 the average RMSD was reported to be 2.0 Å. The system remained stable during the 250 ns simulation time. No significant convergence was reported. The stability graph of the conformation 2 followed the same pattern as conformation 1 except the average RMSD remained relatively higher as 2.2 Å. The system faced a little convergence between 90 and 110 ns; however, the system remained stable during the simulation time.
On the other hand, conformation 3 was relatively unstable, and the RMSD converged after 150 ns but then decreased afterward. The average RMSD for this conformation was reported to be 2.8 Å. Their results significantly justify that the kaempferol is stable in the 3CLpro cavity and acts as a stronger inhibitor of the 3CLpro. The residual flexibility (RMSF) given in Figure 1I–K show similar behavior of residual flexibility. Differences in the residual flexibility justify the differential binding of each conformation and its impact on the protein's internal dynamics. To further confirm the compactness of the kaempferol‐3CLpro complexes, radius of gyration (Rg) was calculated. Conformation 1 (Figure 1L) and 2 (Figure 1M) possess a similar pattern of compactness during the 250 ns time. An average Rg for these two complexes was reported to be 22.0 Å. On the other hand, the compactness of the conformation 3 (Figure 1N) remained higher relatively and converged during the simulation time. The Rg value for conformation 3 increased after 150 ns. Conclusively, these results suggest that conformation 1 and 2 remained more stable and compact than conformation 3 relatively. The binding conformation 1 and 2 are the best representation of the kaempferol binding.
Furthermore, the total binding free energy was calculated by using the MM‐GBSA module (Genheden & Ryde, 2015). Using the free energy approach vdW, electrostatic, SASA, and the total binding free energy was calculated. The vdW for each conformation was reported to be −33.113, −33.808, −23.505 kcal/mol, while the electrostatic contribution for each was reported to be −9.359, −10.192, −4.9419 kcal/mol, respectively. The total binding energy for conformation 1 was reported to be −25.67 kcal/mol. For conformation 2, the total binding energy was −26.81 kcal/mol, while for the third conformation, the binding energy −17.21 kcal/mol was reported. Hence, these results significantly verify that kaempferol possesses strong anti‐coronavirus activity and thus tested against SARs‐CoV‐2 experimentally. The free energy results are given in Table 1.
TABLE 1.
Molecular docking, free energy, and experimental analysis results, including drug concentration and inhibition rate in percentage are tabulated
| Complex | Docking score | VdW | Electrostatic | SASA | ΔG |
|---|---|---|---|---|---|
| Conformation 1 | −6.4 kcal/mol | −33.113 | −9.359 | −3.646 | −25.67 kcal/mol |
| Conformation 2 | −6.4 kcal/mol | −33.808 | −10.192 | −3.564 | −26.81 kcal/mol |
| Conformation 3 | −6.4 kcal/mol | −23.505 | −4.9419 | −2.793 | −17.21 kcal/mol |
| Virus | Kaempferol (μM) | Inhibition rate (%) | |||
| SARS‐CoV‐2 | 125.00 | 88.33 ± 12.58 | |||
| 62.50 | 93.33 ± 5.77 | ||||
| 31.25 | 40.00 ± 34.64 | ||||
For the experimental testing of kaempferol, the drug molecule was obtained from Bide Pharmatech Limited located in Shanghai, China. From ATCC, Vero E6 cells or the African green monkey kidney epithelial cells were purchased. The cell was cultured in Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. SARS‐CoV‐2 (Genebank accession no. MT123290.1) was clinical isolates from Guangzhou Customs Technical Center BSL‐3 Laboratory (Research Institute of Highly Pathogenic Microorganisms, State Key Laboratory of Respiratory Diseases). Reed Muench method (Haggett & Gunawardena, 1964) (TCID50 [50% tissue culture infective dose] = 10−6/100 μL) was utilized to determine the 50% tissue culture infective dose. Stocks of viruses were gathered and deposited at −80°C. In a biosafety level‐3 (BLS‐3) laboratory, all the infection experiments were conducted.
To investigate the antiviral effects of kaempferol against SARS‐CoV‐2, the CPE inhibition assay under the nontoxic concentration of kaempferol was employed. The related protocol was employed, as reported in the previous study (Ma et al., 2020). Drug concentrations 125, 62.5, and 31.25 μM for kaempferol were tested. Each test was done at least in triplicate and repeated two times. Following 72 hr of incubation, the infected cells showed 100% cytopathic effect (CPE) under the light microscope. The percentage of CPE in kaempferol‐treated cells was recorded. The range of concentrations and the number of standards for each tested preparation were performed according to guidelines for an accurate IC50 (the 50% inhibition concentration) estimation (Sebaugh, 2011). Cellular toxicity or cytotoxicity of kaempferol was appraised in Vero E6 cells using a non‐radioactive cell proliferation assay (MTT) (Sebaugh, 2011). To investigate the inhibitory activity of kaempferol against the SARS‐CoV‐2, CPE inhibition assay was performed (Figure 1O). It has been reported that SARS‐CoV‐2 causes a severe CPE in infected Vero E6 cells, including cell detachment, rounding, and death. A decrease in SARS‐CoV‐2‐induced CPE after the incubation of 72 hr revealed the antiviral efficacy of kaempferol and remdesivir. It was found that kaempferol (62.50–125.00 μg/ml) significantly abridged the CPE instigated by an infection in Vero E6 cells (Table 1). These findings have shown that kaempferol has been able to protect cells from virus‐induced cell death and could be a promising SARS‐CoV‐2 antiviral drug.
Our compounds kaempferol shares structural similarity with Scutellarein, Dihydromyricetin, Quercetagetin, Myricetin, 5,6‐Dihydroxyflavone, 6,7‐Dihydroxyflavone, Chrysin, Herbacetin, and Baicalein which are flavonoids and have been reported to have strong inhibitory activity against the 3CLpro from SARS‐COV‐2 (Dai et al., 2020). Similarly, Park et al. (2017) reported 4‐hydroxyisolonchocarpin as an active compound against the SARS‐CoV also possess a similar scaffold as kaempferol with the IC50 value 202.7 μM. Furthermore, Ampelopsin, a Flavanonole, with similar structural arrangement reported to inhibit the 3CLpro. The IC50 value for Ampelopsin was reported to be 364 μM (Nguyen et al., 2012). Kaempferol has also been previously reported to inhibit the SARS 3CLpro (IC50 = 116.3 μM and MERS 3CLpro (IC50 = 35.3 μM) (Jo, Kim, Shin, & Kim, 2020; Schwarz et al., 2014). Hence, this comparative analysis shows that kaempferol possesses stronger inhibitory effects than the related group of compounds, which are previously reported to inhibit the coronavirus.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
The computations were partially performed at the Pengcheng Lab and the Center for High‐Performance Computing, Shanghai Jiao Tong University. We acknowledge their help. Dong‐Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant Nos. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), the Natural Science Foundation of Henan Province (162300410060), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2017ZD14).
Funding information National Natural Science Foundation of China, Grant/Award Number: 32070662; Shanghai Jiao Tong University, Grant/Award Number: YG2017ZD14; Natural Science Foundation of Henan Province, Grant/Award Number: 162300410060; Science and Technology Commission of Shanghai Municipality, Grant/Award Number: 19430750600; Ministry of Science and Technology of China, Grant/Award Number: 2016YFA0501703
Contributor Information
Wang Heng, Email: wang_heng126@126.com.
Dong‐Qing Wei, Email: dqwei@sjtu.edu.cn.
REFERENCES
- Chen, N. , Zhou, M. , Dong, X. , Qu, J. , Gong, F. , Han, Y. , … Wei, Y. (2020). Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. The Lancet, 395(10223), 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, W. , Zhang, B. , Jiang, X.‐M. , Su, H. , Li, J. , Zhao, Y. , … Liu, F. (2020). Structure‐based design, synthesis and biological evaluation of peptidomimetic aldehydes as a novel series of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durojaiye, A. B. , Clarke, J.‐R. D. , Stamatiades, G. A. , & Wang, C. (2020). Repurposing cefuroxime for treatment of COVID‐19: A scoping review of in silico studies. Journal of Biomolecular Structure and Dynamics, 38, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S. , & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand‐binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggett, P. , & Gunawardena, K. (1964). Determination of population thresholds for settlement functions by the Reed‐Muench method. The Professional Geographer, 16(4), 6–9. [Google Scholar]
- Hatada, R. , Okuwaki, K. , Mochizuki, Y. , Handa, Y. , Fukuzawa, K. , Komeiji, Y. , … Tanaka, S. (2020). Fragment molecular orbital based interaction analyses on COVID‐19 main protease‐inhibitor N3 complex (PDB ID: 6LU7). Journal of Chemical Information and Modeling, 60, 3593–3602. [DOI] [PubMed] [Google Scholar]
- Huang, C. , Wang, Y. , Li, X. , Ren, L. , Zhao, J. , Hu, Y. , … Gu, X. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet, 395(10223), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S. , Kim, S. , Shin, D. H. , & Kim, M.‐S. (2020). Inhibition of SARS‐CoV 3CL protease by flavonoids. Journal of Enzyme Inhibition and Medicinal Chemistry, 35(1), 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. , Ali, S. S. , Khan, M. T. , Saleem, S. , Ali, A. , Suleman, M. , … Wei, D.‐Q. (2020). Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS‐CoV‐2 main protease (3CLpro). Journal of Biomolecular Structure and Dynamics, 38, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Zhu, N. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Q. , Pan, W. , Li, R. , Liu, B. , Li, C. , Xie, Y. , … Huang, J. (2020). Liu Shen capsule shows antiviral and anti‐inflammatory abilities against novel coronavirus SARS‐CoV‐2 via suppression of NF‐κB signaling pathway. Pharmacological Research, 158, 104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needle, D. , Lountos, G. T. , & Waugh, D. S. (2015). Structures of the Middle East respiratory syndrome coronavirus 3C‐like protease reveal insights into substrate specificity. Acta Crystallographica Section D: Biological Crystallography, 71(5), 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T. T. H. , Woo, H.‐J. , Kang, H.‐K. , Kim, Y.‐M. , Kim, D.‐W. , Ahn, S.‐A. , … Kim, D. (2012). Flavonoid‐mediated inhibition of SARS coronavirus 3C‐like protease expressed in Pichia pastoris . Biotechnology Letters, 34(5), 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.‐Y. , Yuk, H. J. , Ryu, H. W. , Lim, S. H. , Kim, K. S. , Park, K. H. , … Lee, W. S. (2017). Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. Journal of Enzyme Inhibition and Medicinal Chemistry, 32(1), 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S. , & Netland, J. (2009). Coronaviruses post‐SARS: Update on replication and pathogenesis. Nature Reviews Microbiology, 7(6), 439–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon‐Ferrer, R. , Case, D. A. , & Walker, R. C. (2013). An overview of the Amber biomolecular simulation package. Wiley Interdisciplinary Reviews: Computational Molecular Science, 3(2), 198–210. [Google Scholar]
- Schwarz, S. , Sauter, D. , Wang, K. , Zhang, R. , Sun, B. , Karioti, A. , … Schwarz, W. (2014). Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Medica, 80(2–3), 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaugh, J. (2011). Guidelines for accurate EC50/IC50 estimation. Pharmaceutical Statistics, 10(2), 128–134. [DOI] [PubMed] [Google Scholar]
- Trott, O. , & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , … Pei, Y.‐Y. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]


