Abstract
Current diagnostic standards involve severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) detection in nasopharyngeal swabs (NPS), but saliva is an attractive and noninvasive option for diagnosis. The objectives were to determine the performance of saliva in comparison with NPS for detecting SARS‐CoV‐2 and to compare the optimized home brew reverse‐transcription polymerase chain reaction (RT‐PCR) with a commercial RT‐PCR. Paired NPS and saliva specimens were prospectively collected and tested by RT‐PCR from patients presenting at an emergency room with signs and symptoms compatible with coronavirus disease‐2019. A total of 348 samples from 174 patients were tested by RT‐PCR assays. Among 174 patients with symptoms, 63 (36%) were SARS‐CoV‐2 positive in NPS using the optimized home‐brew PCR. Of these 63 patients, 61 (98%) were also positive in saliva. An additional positive SARS‐CoV‐2 saliva was detected in a patient with pneumonia. Kappa Cohen's coefficient agreement between NPS and saliva was 0.96 (95% confidence interval [CI], 0.90–0.99). Median Ct values in NPS versus saliva were 18.88 (interquartile range [IQR], 15.60–23.58; range, 11.97–38.10) versus 26.10 (IQR, 22.75–30.06; range, 13.78–39.22), respectively (p < .0001). The optimized home‐brew RT‐PCR demonstrated higher analytical and clinical sensitivity compared with the commercial RT‐PCR assay. A high sensitivity (98%) and agreement (kappa 0.96) in saliva samples compared to NPS was demonstrated when using an optimized home‐brew PCR even when the viral load in saliva was lower than in NPS. This noninvasive sample is easy to collect, requires less consumable and avoids discomfort to patients. Importantly, self‐collection of saliva can diminish exposure to healthcare personnel.
Keywords: COVID 19, nasopharyngeal swab, PCR, saliva, SARS‐CoV‐2
1. INTRODUCTION
The number of patients infected with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) causing coronavirus disease‐2019 (COVID‐19) remains increasing during the ongoing pandemic. A crucial strategy for controlling transmission relies on expanding diagnosis. The current diagnostic standard involves detection of SARS‐CoV‐2 by reverse‐transcription polymerase chain reaction (RT‐PCR) using nasopharyngeal swabs (NPS). However, NPS are associated with patient's discomfort or complications—such as iatrogenic cerebrospinal fluid leak—as well as an increasing healthcare worker's exposure to SARS‐CoV‐2. 1 , 2
Saliva is an attractive and noninvasive option for diagnosing SARS‐CoV‐2 and sampling enables self‐collection without causing discomfort or pain. In addition, self‐collection of saliva can reduce the exposure to healthcare personnel by avoiding cough, sneezing, and/or aerosolization during sampling. Furthermore, saliva testing requires less consumables, offering a significant benefit when there is shortage of supplies. 3
The value of using saliva for SARS‐CoV‐2 detection has been variable between different studies. Such variability may be related to the type of collection, processing, detection techniques, and/or PCR assays. 4
The objectives of this study were to determine the performance of saliva in comparison to NPS for detecting SARS‐CoV‐2 and to compare our optimized home brew RT‐PCR with a commercial RT‐PCR.
2. MATERIALS AND METHODS
Paired NPS and saliva were prospectively collected from patients presenting at the emergency room (ER) in CEMIC University Hospital from August to September 2020. Patients more than or equal to 18 years old with signs or symptoms potentially due to COVID‐19 were invited to participate in the study. This study was approved by the Ethics Committee of CEMIC (Protocol: 1298/20).
Signs or symptoms of COVID‐19 included fever (>37.5°C), throat, abdominal or chest pain, rhinorrhea, cough, dyspnea, myalgias, headache, anosmia, or dysgeusia. All patients were evaluated by a trained ER physician.
NPS were obtained and placed in a sterile tube with 2 ml viral transport media (Minimun Essential Medium; Gibco); l‐Glutamine 200 mM; HEPES 1 N; Bovine Albumin 5%; Sigma; Sodium Bicarbonate 7.5%; Penicillin, Streptomycin, and Amphotericin; pH = 7.2). Patients were also instructed to collect saliva by themselves in a plastic sterile container without any transport media. Both NPS and saliva samples were transported at room temperature to the laboratory within 2 h of sample collection. NPS collected in 2 ml of viral transport medium were vortexed in a biosafety cabinet and an aliquot was used for nucleic acid extraction. Saliva samples were conserved at 4°C until processed in a biosafety cabinet within 48 h from arrival. Viscous saliva samples were mechanically disrupted by adding 500 µl viral transport medium.
Nucleic acid was extracted from 100 µl and eluted in 15 µl using manual columns (Quick‐RNA™ Viral Kit, Zymo Research Corp.) following manufacturer's recommendation. An in‐house one‐step real‐time RT‐PCR assay targeting the E gene of SARS‐CoV‐2 was performed. 5 Optimization of this assay was done to achieve a higher analytical sensitivity. Specifically, magnesium final concentration was increased to 3.8 mM and cycling conditions were modified into three PCR steps. Such steps included an initial transcription stage (10 min at 55°C), followed by 2 min at 95°C, and subsequently by 45 cycles (95°C for 15 s, 55°C for 30 s, and 72°C for 15 s). Quality control amplification was confirmed by testing the human RNAse P gene. This optimized PCR assay was compared to a commercial RT‐PCR that amplifies SARS‐CoV‐2 E gene and S gene and includes an internal amplification control (Real Star® SARS‐CoV‐2 RT‐PCR Kit 1.0.; altona Diagnostics Argentina S.R.L.). Real time assays were performed in a CFX 96 Deep Well™ Real Time System (BioRad).
A positive result was considered when the human RNAse gene or the internal amplification control were positive and the cycle threshold (Ct) value was less than 40. Analytical sensitivity was determined with a quantified SARS‐CoV‐2 positive material (altona Diagnostic, Argentina S.R.L.). Patient demographics were presented using descriptive statistics.
Any patient with at least one positive test for SARS‐CoV‐2 was considered true positive. Sensitivity, agreement, and Cohen's kappa coefficient were calculated. Ct values and matched positive and discrepant samples were compared by a Wilcoxon signed rank sum test (GraphPad Prism 5.00).
3. RESULTS
A total of 174 patients with signs or symptoms consistent with COVID‐19 were enrolled. Clinical and demographic characteristics of these patients are described in Table 1. The median age in the population was 38 years old (interquartile range [IQR], 31–50) and 59.8% were females. The majority of patients (95%) had symptoms of upper tract respiratory infection and 36% had fever. The median time from the onset of symptoms to sample collection was 2 days (IQR, 1–4). Nine patients (5.2%) required hospitalization and one of them was admitted in the intensive care unit. Among SARS‐CoV‐2 RT‐PCR positive patients, the most common symptoms were fever (36%), cough (46%), and odynophagia (49%).
Table 1.
Baseline characteristics in patients presenting to the emergency room with COVID‐19 symptoms
| SARS‐CoV2 positive PCR | SARS‐CoV2 negative PCR | ||||
|---|---|---|---|---|---|
| Characteristics | Total (n = 174) | Total (n = 64)a | Discharged (n = 55) | Admitted (n = 9) | Discharged (n = 110) |
| Gender, n (%) | |||||
| Female | 104 (59.8) | 39 (60.9) | 33 (60.0) | 6 (66.7) | 65 (59.1) |
| Male | 70 (40.2) | 25 (39.1) | 22 (40.0) | 3 (33.3) | 45 (40.9) |
| Age (in years) | |||||
| Median (IQR) | 38 (31–50) | 38 (31–50.5) | 35 (30–46.3) | 55 (41–67) | 38.5 (31–48.5) |
| Mean (range) | 41.1 (17–88) | 41.8 (21–88) | 39.3 (21–78) | 56.2 (35–88) | 40.7 (17–81) |
| Clinical syndrome, n (%) | |||||
| URTI | 165 (94.8) | 55 (85.9) | 55 (100) | 0 | 110 (100) |
| Pneumonia | 9 (5.2) | 9 (14.1) | 0 | 9 (100) | 0 |
Abbreviations: COVID‐19, coronavirus disease 2019; IQR, interquartile range; NPS, nasopharyngeal swabs; PCR, polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; URTI, upper respiratory tract infection.
61 patients were positive in NPS and saliva; one patient was positive only in saliva; two patients were positive only in NPS.
A total of 348 samples (174 NPS and 174 saliva) were tested for SARS‐CoV‐2 by RT‐PCR assays. Of 174 patients with symptoms, 63 (36%) were SARS‐CoV‐2 RT‐PCR positive in NPS by the optimized home‐brew PCR assay. Of these 63 positive patients, 61 (98%) were also positive in saliva and one additional patient was positive only in saliva. The median Ct values in NPS versus saliva were 18.88 (IQR, 15.60–23.58; range, 11.97–38.10) versus 26.10 (IQR, 22.75–30.06; range, 13.78–39.22), respectively (p < .0001; Figure 1). The median Ct value in saliva for RNAse P was 22.1 (IQR, 21.2–23.2; range, 19–28.1). Kappa Cohen's coefficient agreement between NPS and saliva was 0.96 (95% confidence interval [CI], 0.92–0.99). Discordant results between NPS and saliva occurred in 3/174 (1.7%) patients showing Ct values higher than 28. Specifically, E gene Ct values in these three patients were 28.92 versus 41.00, 29.48 versus 41.00, and 44.00 versus 31.80, in NPS versus saliva, respectively. This last patient SARS‐CoV‐2 RT‐PCR positive in saliva but negative in NPS developed pneumonia (CT scan image compatible with COVID‐19) requiring hospitalization. Workup on this patient for additional respiratory pathogens including other respiratory viruses by PCR and bacteria cultures in blood and respiratory samples were negative. The other two patients who were SARS‐CoV‐2 RT‐PCR positive in NPS but negative in saliva had provided a low volume of saliva (<500 µl).
Figure 1.
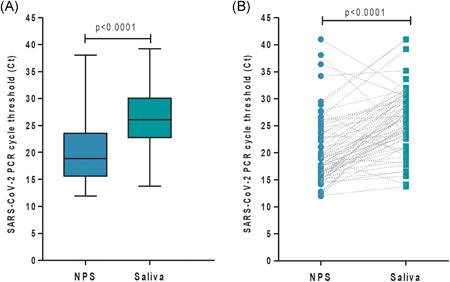
SARS‐CoV‐2 Ct in saliva and nasopharyngeal swabs (NPS). (A) Ct median from positive nasopharyngeal swabs (n = 63) and saliva samples (n = 62) were compared (p < .0001). (B) Patients matched positive and discrepant samples (n = 64) represented by the connecting lines were compared by a Wilcoxon rank sum test (p < .0001). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The comparison of the optimized home brew RT‐PCR and the commercial assay showed different analytical and clinical sensitivities. The limit of detection of the optimized home brew RT‐PCR assay and the commercial assay for SARS‐CoV‐2 was 1 copy/µl and 10 copies/µl, respectively. Of 62 patients with positive saliva by the optimized home brew RT‐PCR, 54 (87%) were positive by the commercial RT‐PCR. Agreement of both RT‐PCR assays was 94.6% (kappa 0.9, 95% CI, 0.83–0.97). Saliva samples that were positive with the optimized home brew RT‐PCR but negative with the commercial kit had Ct values higher than 28 (mean Ct value 31.82; range, 28.93–39.22). Of 63 patients with positive NPS by the optimized home brew RT‐PCR, 60 (95%) were positive by the commercial RT‐PCR. All 3 discordant patients had a Ct value higher than 35.
4. DISCUSSION
Nasopharyngeal swabbing has become a standard diagnostic test for detection of SARS‐CoV‐2. However, NPS requires an invasive sampling and further exposes healthcare workers to the pandemic virus. Saliva is a noninvasive sample which can be easily obtained for viral diagnosis. 4 In this study, a high sensitivity (98%) and agreement (kappa 0.96) in saliva samples compared to NPS was demonstrated when using an optimized home brew PCR even in saliva samples with a lower viral load. In addition, saliva was able to detect SARS‐CoV‐2 in one patient with negative NPS who developed pneumonia requiring hospitalization. High detection rate in saliva has been previously demonstrated in symptomatic and asymptomatic adults suggesting that it can be used as a suitable specimen for detection of SARS‐CoV‐2. 6 , 7 , 8 In fact, such viral detection may be related to the high expression of ACE2 receptors on the salivary glands and tongue. 9
In contrast, some studies have shown a lower sensitivity in saliva compared with NPS. 10 Saliva collection and/or processing as well as sensitivity of the PCR assay may play a role in such lower performances. 10 , 11 In fact, the need of optimizing saliva collection and processing has been previously suggested. 12 In this study, saliva collection without any transport media or any nucleic acid stabilization proved to be adequate to achieve a high detection rate and sensitivity. Furthermore, optimization of the in‐house RT‐PCR assay increased the limit of detection compared with previous reports and to the commercial assay also evaluated in this study. 5 As viral loads in saliva can be lower than in NPS, optimized and sensitive assays are recommended. Discrepant results between both PCR assays were observed in saliva samples when Ct values were higher than 28 and occurred in NPS when Ct values were higher than 35. These results underscore the importance of highly sensitive assays for accurate diagnosis.
One limitation of this study is that patients were relatively young and with mild symptoms. Saliva production may be diminished in older and/or less collaborative patients. Therefore, the performance of this test in other populations (e.g., older and sicker) will be further investigated.
In conclusion, our study demonstrates that testing self‐collected saliva can be as sensitive as the NPS for diagnosing COVID‐19 among ambulatory patients. Saliva collection and processing is important to achieve adequate diagnosis. The use of pure saliva without transport media or stabilizators showed no RNA degradation. This noninvasive sample is easy to collect, requires less consumables, and avoids discomfort to patients. In addition, self‐collection of saliva can diminish exposure to SARS‐CoV‐2 in healthcare personnel. Further studies are needed to evaluate the role of saliva testing in other populations and settings.
CONFLICT OF INTERESTS
Martin Stryjewski is a consultant to Basilea, a speaker for Pfizer and principal investigator in Argentina for NIH grant UM1AI104681. The rest of the authors have no conflict of interest.
AUTHOR CONTRIBUTIONS
Marcela Echavarria: Conceptualization, Funding Acquisition, Investigation, Project Administration, Resources, Visualization, Writing – Original Draft Preparation, Writing – Review & Editing; Noelia Soledad Reyes: Data Curation, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing; Pamela Elizabeth Rodriguez: Data Curation, Formal Analysis, Methodology, Writing – Original Draft Preparation, Writing – Review & Editing; Martin Ypas: Methodology, Resources, Writing – Review & Editing; Carmen Ricarte: Methodology, Writing – Review & Editing; María Pilar Rodriguez: Resources, Writing – Review & Editing; Matias Gastón Perez: Resources, Writing – Review & Editing; Alejandro Seoane: Resources, Writing – Review & Editing; Alfredo Martinez: Resources, Writing – Review & Editing; Cristina Videla: Resources, Writing – Review & Editing; Martin Estanislao Stryjewski: Conceptualization, Formal Analysis, Writing – Original Draft Preparation, Writing – Review & Editing; Guadalupe Carballal: Conceptualization, Writing – Original Draft Preparation, Writing – Review & Editing.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jmv.26839.
ACKNOWLEDGMENTS
We are very grateful to all patients and to Fernando Olivera, Carlos Gonzalez, Luis Beligoy, Priscila Vaudagna, Belén Tebe, Marina Reyes, Rita Andrade, Florencia Morales Corinales, Lucía Llamosas Camba, Lucía Yury, Augusto Silva, Sofía Cohendoz, Sofía Paz, Magdalena Grad, Alejandra Abramovsky, Ailen Martiarena, María Sambuco, Natalia Boccia, Claudia Suarez, Lucca Ahumada, and Andrea Galvis at the Emergency Department for patients' enrollment. We want to thank altona Diagnostics Argentina SRL for providing their kits. This study was partially supported by a grant from Fondo para la Investigación Científica y Tecnológica (FONCYT) [IP‐COVID 19: 0938] awarded to Dr. Marcela Echavarría and an internal fund from CEMIC and Fundación “Norberto Quirno” N° 01/20.
Echavarria M, Reyes NS, Rodriguez PE, et al. Self‐collected saliva for SARS‐CoV‐2 detection: A prospective study in the emergency room. J Med Virol. 2021;93:3268–3272. 10.1002/jmv.26839
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Muniz IAF, Linden LV, Santos ME, et al. SARS‐CoV‐2 and saliva as a diagnostic tool: A real possibility. Pesqui Bras Odontopediatria Clin Integr. 2020;20:1‐7. [Google Scholar]
- 2. Sullivan CB, Schwalje AT, Jensen M, et al. Cerebrospinal fluid leak after nasal swab testing for coronavirus disease 2019. JAMA Otolaryngol Surg. 2020;146:1‐2. [DOI] [PubMed] [Google Scholar]
- 3. Pasomsub E, Watcharananan SP, Boonyawat K, et al. Saliva sample as a non‐invasive specimen for the diagnosis of coronavirus disease 2019: a cross‐sectional study. Clin Microbiol Infect. 2020. 10.1016/j.cmi.2020.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a noninvasive specimen for detection of SARS‐CoV‐2. J Clin Microbiol. 2020;58:1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corman V, Bleicker T, Brünink S, et al. Diagnostic detection of 2019‐nCoV by real‐time RT‐RCR. Charité Berlin. 2020:1‐13. [Google Scholar]
- 6. Rao M, Rashid FA, Sabri FSAH, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS‐CoV‐2 [published online ahead of print August 06, 2020]. Clin Infect Dis. 2020. https://academic.oup.com/cid/advance-article/doi/10.1093/cid/ciaa1156/5882012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wyllie AL, Fournier J, Casanovas‐Massana A, et al. Saliva is more sensitive for SARS‐CoV‐2 detection in COVID‐19 patients than nasopharyngeal swabs [published online ahead of print April 22, 2020]. medRxiv. 2020. 10.1101/2020.04.16.20067835 [DOI] [Google Scholar]
- 8. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS‐CoV‐2 _ Elsevier Enhanced Reader.pdf. J Infect. 2020;81:45‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu H, Zhong L, Deng J, et al. High expression of ACE2 receptor of 2019‐nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. To KKW, Tsang OTY, Yip CCY, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sutjipto S, Lee PH, Tay JY, et al. The effect of sample site, illness duration, and the presence of pneumonia on the detection of SARS‐CoV‐2 by real‐time reverse transcription PCR. Open Forum Infect Dis. 2020;7:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Landry ML, Criscuolo J, Peaper DR. Challenges in use of saliva for detection of SARS CoV‐2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


