Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection has proven to be extremely contagious and has spread rapidly all over the world. A key aspect in limiting the virus diffusion is to ensure early and accurate diagnosis. Serological assays could be an alternative in increasing testing capabilities, particularly when used as part of an algorithmic approach combined with molecular analysis. The aim of this study was to evaluate the diagnostic accuracy of a second generation chemiluminescent automated immunoassay able to detect anti‐SARS‐CoV‐2 immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies. Data are carried out on healthy subjects and other infectious diseases pre‐pandemic sera, as controls, and on two different coronavirus disease 2019 hospitalized patient groups (early and late infection time). Data obtained have been analyzed in terms of precision, linearity, sensitivity and specificity. Specificities are: 100% for anti‐SARS‐CoV‐2 IgG and 98% for anti‐SARS‐CoV‐2 IgM, in all patient groups. Sensitivities are: 97%, 100%, and 98% for anti‐SARS‐CoV‐2 IgG and 87%, 83%, and 86% for anti‐SARS‐CoV‐2 IgM in the early infection, in the late infection and in the total patient group, respectively. The Mindray anti‐SARS‐CoV‐2 IgG and IgM assays demonstrated higher sensitivity and specificity, indicating that IgG and IgM simultaneous detection is useful even in the early phases of infection.
Keywords: biochemical analysis, coronavirus, SARS coronavirus
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection is extremely contagious and is still spreading around the world with minor local differences. Coronavirus disease (COVID‐19) is mild in most cases; in some people, usually elderly and with co‐morbidities, it may progress to pneumonia, acute respiratory distress syndrome and multi organ dysfunction.
As the global impact of this new epidemic is yet uncertain, 1 , 2 a crucial aspect in containing the virus spread is based on ensuring an early and accurate diagnosis and adequate quarantine for infected subjects.
Nucleic acid tests to detect SARS‐CoV‐2 RNA genome have been quickly developed and are now widely employed to diagnose COVID‐19. 3 However, the real‐time polymerase chain reaction (PCR) often fails as a result of suboptimal collection procedures, a low patient viral load due to early stage of the disease or suppressed by host immunity, or if the samples are analyzed at an advanced stage of infection. 4 Unfortunately, a false negative diagnosis allows patients to spread the infection, hindering efforts to contain the virus and producing serious consequences. 5
In such conditions, additional screening methods able to detect the infection despite low viral titers, can be extremely helpful to guarantee a prompt diagnosis of all infected patients.
Specific serological assays are particularly required to understand the epidemiology of SARS‐CoV‐2, but often still need to be accurately validated. Even though, the development of specific immunoglobulin M (IgM) and immunoglobulin A antibodies after symptom onset may occur as quickly as genetic viral material can be detected in respiratory samples, their production time usually ranges from less than 5 days to more than 10 days, thus limiting the serologic tests suitability for diagnosis of acute infections. 6 Nevertheless COVID‐19 patient diagnostic sensitivity may significantly increase when combining serological tests with molecular tests. 7
In particular, serological assessment is able to support a number of highly relevant clinical and epidemiological applications in detecting SARS‐CoV‐2 infection: they can be used to determine the infection prevalence, useful to accurately define the laboratory test positive predictive values; to recognize individuals with a strong antibody response, who might be convalescent plasma donors; to analyze the immune response in a dynamic qualitative and quantitative manner; to evaluate vaccine trials results and the therapeutic antibodies development in the near future. Moreover, validated serological assays are critical for tracking patient contact, identifying viral reservoir hosts and for epidemiological studies, which are primarily needed to help uncover disease burden, especially the rate of asymptomatic individuals, and to obtain better estimates of morbidity and mortality. 8 Furthermore, epidemiological studies can help reveal the extent of virus spread in specific settings, such as households and communities, which could assist in guiding control measures.
Based on these considerations, the aim of this study was to evaluate the diagnostic accuracy of a second generation chemiluminescent automated immunoassay able to detect anti‐SARS‐CoV‐2 immunoglobulin G (IgG) and IgM antibodies, carried out on healthy subjects and other infectious diseases pre‐pandemic sera, as controls, and on two different COVID‐19 patient groups (early infection time and late infection time), hospitalized at “Tor Vergata” University COVID‐Hospital of Rome.
2. MATERIALS AND METHODS
2.1. Patients and serum specimens
Serum samples from “Tor Vergata” University COVID‐Hospital of Rome hospitalized patients were collected from March 16 to April 28, 2020 and were divided as follows: 42 positive SARS‐CoV‐2 patients (mean age 67.8 years ± 16.6 years; 24 males and 18 females), collected on Days 1 to 9 from first positive nasopharyngeal swab (early infection time group); 43 positive SARS‐CoV‐2 patients (mean age 70.0 years ± 13.6 years; 24 males and 19 females), collected on Days 19–41 from first positive nasopharyngeal swab (late infection time group) and 50 negative SARS‐CoV‐2 subjects, including 40 Tor Vergata Hospital physicians and healthcare workers screened for internal surveillance with negative nasopharyngeal swab collected from April 2 to 7, 2020 (control group; mean age 42.5 years ± 10.8 years; 23 males and 17 females), and 10 pre‐pandemic patients collected in 2018 (pre‐pandemic control group; mean age 55.3 years ± 15.6 years; 8 males and 2 females) positive to hepatitis B virus (HBV) or hepatitis C virus (HCV).
2.2. Chemiluminescence immunoassay
The second generation CL‐series SARS‐CoV‐2 IgG and IgM assays are a two‐step chemiluminescent immunoassays for detection of IgG and IgM SARS‐CoV‐2 antibodies in human serum or plasma, performed on the fully automated Mindray CL 1200i analytical system (Shenzhen Mindray Bio‐Medical Electronics Co.; distributed in Italy by Medical Systems S.P.A.). Respect to first generation, kits have been modified by manufacturer to improve anti‐SARS‐CoV‐2 IgG linearity and to reduce anti‐SARS‐CoV‐2 IgG/IgM analytical interference.
Samples react with paramagnetic micro‐particles coated with SARS‐CoV‐2 specific antigens (recombinant N‐Protein and Spike (S) Protein, as declared by the manufacturer). In the second step, diluent solution and alkaline phosphatase‐labeled anti‐human IgG or IgM monoclonal antibodies are added to the reaction, to form sandwich with micro‐particles captured anti‐SARS‐CoV‐2 antibodies. Afterwards, micro‐particles are magnetically captured while other unbound substances are removed by washing in the dispersion dish. Finally, a substrate solution is added and the resulting chemiluminescent reaction is measured as relative light units (RLUs) by a photo‐multiplier. The amount of SARS‐CoV‐2 IgG/IgM antibodies present in the sample is proportional to the RLUs generated during the reaction. The SARS‐CoV‐2 IgG/IgM antibodies results can be determined via a calibration curve, established on two level product calibrators.
First results are generated after 25 min (throughput 180 tests/h). Cut‐off values are: IgG positive greater than 10.00 U/ml and IgM positive greater than 1.00 cut off index (COI), according to the manufacturer's instructions. This test is CE approved.
2.3. Precision evaluation
Precision estimations were calculated by means of quintuplicate measurements on two serum pools (low and high anti‐SARS‐CoV‐2 IgM and IgG concentration), performed each day for a total of five days, following the Clinical and Laboratory Standards Institute (CLSI) EP15‐A3 protocol. 9 The precision results obtained were compared to those claimed by the manufacturer.
2.4. Linearity assessment
Linearity was assessed by serial dilution of two samples with high anti‐SARS‐CoV‐2 IgM and anti‐SARS‐CoV‐2 IgG concentrations, as specified in the CLSI EP06 A: 2003 guideline (paragraph 4.3.1). A low anti‐SARS‐CoV‐2 IgM serum pool value sample (0.11 COI) was used to dilute an anti‐SARS‐CoV‐2 IgM sample with a value of 6.63 COI; in the same way, a low anti‐SARS‐CoV‐2 IgG serum pool value sample (0.60 U/ml) was used to dilute an anti‐SARS‐CoV‐2 IgG sample with a value of 170.18 U/ml. All measurements were performed in triplicate.
2.5. Statistical analysis
Specificity and sensitivity were calculated by ROC curves. Coefficient of variation (CV) percentages are calculated as the standard deviation divided by the mean value. All data were analyzed using Med Calc Ver.18.2.18 (MedCalc Software Ltd).
2.6. Ethical statement
The study was performed according to “Tor Vergata” University COVID‐Hospital of Rome, local ethical approval (protocol no. R.S.44.20). Informed consent was obtained from all subject enrolled in the study. The study was in accordance with the Helsinki Declaration, as revised in 2013.
3. RESULTS
3.1. Precision test
The CV precision data are shown in Table 1 in comparison with those declared by the company, according to the procedure suggested in CLSI protocol. Results for low and high CV % values were satisfactory and with better outcomes than those obtained by the manufacturer (IgG low: 3.84% vs. 4.26%; IgG high: 3.12% vs. 3.85%; IgM high: 3.02% vs. 3.18%), except for anti‐SARS‐CoV‐2 IgM low precision value, resulted moderately higher than those declared (2.71% vs. 2.62%).
Table 1.
Coefficients of variation (CV %) precision data
| TEST | CTRL | Mindray 1200i | |
|---|---|---|---|
| Precision | |||
| Declared CV% | Experimental CV% | ||
| IgG | Low | 4.26 | 3.84 |
| IgG | High | 3.85 | 3.12 |
| IgM | Low | 2.62 | 2.71 |
| IgM | High | 3.18 | 3.02 |
Note: Experimental values are compared with those declared by the manufacturer.
Abbreviations: IgG, immunoglobulin G; IgM, immunoglobulin M.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.2. Linearity assessment
Linearity measurements performed on Mindray Cl 1200i instrument for anti‐SARS‐CoV‐2 IgG and IgM assays are shown in Figure 1. The dilution performed were obtained by mixing high anti‐SARS‐CoV‐2 IgG or IgM concentration sera with low anti‐SARS‐CoV‐2 IgG or IgM concentration serum pools, as described in materials and methods. All serum sample pools tested do not deviate from linearity. Indeed, for anti‐SARS‐CoV‐2 IgG an excellent linear correlation coefficient (R = 0.999, p < .0001) has been found, as well as for anti‐SARS‐CoV‐2 IgM (R = 0.987, p < .0001). The slightly lower value obtained in the IgM linear correlation coefficient, due to the curve that deviates from linearity at high IgM levels, does not affect the determination because is far from the cut‐off value.
Figure 1.
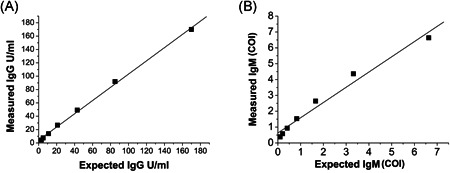
Linearity results. (A) Anti‐severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) immunoglobulin G (IgG) high‐level sample (170.18 U/ml) serially diluted with a low‐level pool (0.60 U/ml). (B) Anti‐SARS‐CoV‐2 immunoglobulin M (IgM) high‐level sample (6.63 cut off index [COI]) serially diluted with a low‐level pool (0.11 COI)
3.3. Specificity and sensitivity
Specificities and sensitivities were calculated using receiver operating characteristic (ROC) curves. Results reporting the analytical parameters for each test (area under curve [AUC], sensitivity and specificity) are shown in Figure 2 and summarized in Table 2.
Figure 2.
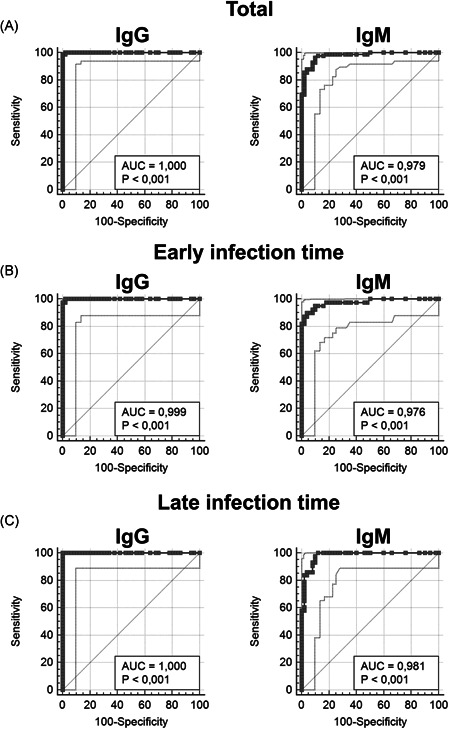
Anti‐SARS‐CoV‐2 IgG and IgM ROC curves results. Total patient group data are shown in Panel A (area under curve [AUC] 1.0 and 0.979, respectively); early infection time group data are shown in Panel B (AUC 0.999 and 0.976, respectively); late infection time group data are shown in Panel C (AUC 1 and 0.981, respectively). IgG, immunoglobulin G; IgM, immunoglobulin M; ROC, receiver operating characteristic; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Table 2.
Area under curve (AUC), sensitivity and specificity for total patient group, early infection time group, and late infection time group
| Control negative group, N = 50 | Total (1–41 days) N = 85 | Total (1–41 days) N = 85 | Early infection time (1–9 days) N = 42 | Early infection time (1–9 days) N = 42 | Late infection time (19–41 days) N = 43 | Late infection time (19–41 days) N = 43 |
|---|---|---|---|---|---|---|
| TEST | IgG | IgM | IgG | IgM | IgG | IgM |
| Sensitivity (%) | 99 | 86 | 97 | 87 | 100 | 83 |
| Specificity (%) | 100 | 98 | 100 | 98 | 100 | 98 |
| Kit cut‐off | >10 U/ml | >1 COI | >10 U/ml | >1 COI | >10 U/ml | >1 COI |
| Area under the ROC curve (AUC); 95% confidence interval | 1; 0,972 to 1,000 | 0.979; 0,937 to 0,996 | 0,999; 0,958 to 1,000 | 0,976; 0,919 to 0,997 | 1; 0,961 to 1,000 | 0,981; 0,929 to 0,998 |
| Laboratory cut‐off | >10 U/ml | >0.46 COI | >5.9 U/ml | >0.79 COI | >10 U/ml | >0.44 COI |
| Sensitivity (%) | 99 | 96 | 100 | 90 | 100 | 100 |
| Specificity (%) | 100 | 90 | 98 | 96 | 100 | 88 |
Abbreviations: COI, cut off index; ROC, receiver operating characteristic.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The ROC curves display optimal AUC values in the total patient group: 1 and 0.979 for anti‐SARS‐CoV‐2 IgG and IgM, respectively. Excellent results were obtained in the anti‐SARS‐CoV‐2 IgG early and late infection time groups, with AUC values of 0.999 and 1, respectively. Finally, anti‐SARS‐CoV‐2 IgM detection shows an AUC values of 0.976 in the early infection time group and 0.981 in the late infection time group.
Specificities are: 100% for anti‐SARS‐CoV‐2 IgG and 98% for anti‐SARS‐CoV‐2 IgM, both in the early infection time group and in the late time infection group, as well as in the total patient group. Sensitivities are: 97%, 100%, and 98% for anti‐SARS‐CoV‐2 IgG in the early infection time group, in the late infection time group and in the total patient group, respectively; 87%, 83%, and 86% for anti‐SARS‐CoV‐2 IgM in the early infection time group, in the late infection time group and in the total patient group, respectively.
We also reported in Table 2 the correlation between the manufacturer's cut‐off and a recalculated best fit cut‐off that emerged from our data analysis (laboratory cut‐off), showing better sensitivities especially for anti‐SARS‐CoV‐2 IgM in all groups (86% vs. 96% in total patient group, 87% vs. 90% in the early infection time group, 83% vs. 100% in the late infection time group).
3.4. Cross‐reactivity test
No cross‐reactivity with antibodies from other pre‐pandemic infectious disease sera (such as HBV and HCV) has been found. Cross‐reactivity studies were carried out on HBV and HCV sera as pre‐pandemic sera from other respiratory virus infections were not available.
4. DISCUSSION
Serological assays represents an alternative in increasing COVID‐19 testing capabilities, particularly when used as part of an algorithmic approach combined with molecular testing. 10 , 11 In fact, specific antibodies detection (IgM and IgG to SARS‐CoV‐2 spike protein) is useful in infected but asymptomatic subjects, to confirm SARS‐CoV‐2 infection in PCR‐negative COVID‐19 patients and in COVID‐19 patients examined many weeks after the disease onset or in those with a low viral load. 12 , 13 In this regard, the Infectious Diseases Society of America recommends that patients with clinical symptoms consistent with COVID‐19 but negative for SARS‐CoV‐2 molecular tests, may be diagnosed by serological examination. 14
In addition, anti‐SARS‐CoV‐2 serology is also a valuable tool for assessing the adapted immunity status of patients, thus providing not only an important complement to RNA testing for specific pathogenic diagnoses. 15 , 16
We have carried out a study to investigate the second generation Mindray CL‐series anti‐SARS‐CoV‐2 IgG and IgM chemiluminescent immunoassay performances. Considering that kits have been modified for better linearity and less interference, data obtained have been analyzed in terms of precision, linearity, sensitivity and specificity.
Our results show that Mindray CL 1200i is a reliable automated instrument with optimal analytical characteristics; the second generation CL‐series SARS‐CoV‐2 IgG and IgM kits display no‐cross reactivity with pre‐pandemic infectious sera as well as significant improvements, related to an advancement in linearity for the anti‐SARS‐CoV‐2 IgG and to a reduction of the analytical interference in anti‐SARS‐CoV‐2 IgG and anti‐SARS‐CoV‐2 IgM, compared to our previous data with first generation testing. 13 Indeed, the prior excellent anti‐SARS‐CoV‐2 IgG specificity of 100% is confirmed and there is an overall increase in the anti‐SARS‐CoV‐2 IgM specificity from 94% to 98%, as well as in anti‐SARS‐CoV‐2 IgM and IgG sensitivities (84% vs. 86% and 94% vs. 98%, respectively). Unfortunately, cross‐reactivity studies are limited due to the small number of samples and to the lack of pre‐pandemic sera from patients with respiratory tract virus infections, such as other human coronavirus.
The use of serological assays in daily practice, compared to the results of our study, confirm their good specificity. Moreover, it should be considered that in general serological tests are cheaper and require a shorter turnaround time which allows a higher productivity compared to molecular testing. However, negative results should always be prudently interpreted based on the clinical context, the patient's medical history according to suspected or documented SARS‐CoV‐2 infection, and also in relation to the diagnostic kits technical characteristics. 17
Furthermore, the second generation Mindray anti‐SARS‐CoV‐2 IgG and IgM assays demonstrated higher sensitivity and specificity both in the early infection time group (97% and 100% for IgG, 87% and 98% for IgM; respectively) and in the late infection time group (100% and 100% for IgG, 83% and 98% for IgM; respectively), compared to other similar studies on serological tests. 15 , 18 , 19 The increase in IgM sensitivity for the early infection time group suggests that their detection is useful even in the first phases of infection and that IgG and IgM simultaneous detection could help to identify more infected people. In conclusion, our data corroborate the strategy that the application of well‐validated serological tests should be strongly recommended in the clinical management and in the public health practice, to improve the control of COVID‐19.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Massimo Pieri and Marzia Nuccetelli conceived of the study and participated in its design, coordination and draft the manuscript. Massimo Pieri, Marzia Nuccetelli, Eleonora Nicolai, and Serena Sarubbi performed the experiment and statistical analysis. Serena Sarubbi and Sandro Grelli participated critical revision of the manuscript for important intellectual content and supervised this study. All authors read and approved the final manuscript.
ETHICS STATEMENT
This study and all the relevant experiments were approved by local Research Ethics Committee (R.S.44.20), and performed in accordance with the Declaration of Helsinki.
ACKNOWLEDGMENTS
The authors would like to thank all the Clinical Biochemistry Laboratory staff of Tor Vergata Hospital, for their support; Mindray (Shenzhen Mindray Bio‐Medical Electronics Co., Shenzen, China; distributed in Italy by Medical Systems S.P.A., Genova, Italy) for kindly having provided kits and instrument for this study.
Pieri M, Nuccetelli M, Nicolai E, Sarubbi S, Grelli S, Bernardini S. Clinical validation of a second generation anti‐SARS‐CoV‐2 IgG and IgM automated chemiluminescent immunoassay. J Med Virol. 2021;93:2523‐2528. 10.1002/jmv.26809
Massimo Pieri and Marzia Nuccetelli contributed equally to this work.
REFERENCES
- 1. Singhal T. A review of coronavirus disease‐2019 (COVID‐19). Indian J Pediatr. 2020;87(4):281‐286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan‐Yeung M, Xu RH. SARS: epidemiology. Respirology. 2003;8(Suppl):S9‐S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019‐nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66(4):549‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. West C, Montori V, Sampathkumar P. COVID‐19 testing: the threat of false‐negative results. Mayo Clin Proc. 2020;95:1127‐1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970‐971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019‐nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75:1564‐1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu H, Stratton CW, Tang YW. An evolving approach to the laboratory assessment of COVID‐19. J Med Virol. 2020;92(10):1812‐1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. EP5‐A3 CLSI. User evaluation of precision of quantitative measurement procedure; approved guideline. Clinical and Laboratory Standards Institute (CLSI). 3rd ed. CLSI EP5‐A3, Wayne; 2014. [Google Scholar]
- 10. Babiker A, Myers CW, Hill CE, Guarner J. SARS‐CoV‐2 testing. Am J Clin Pathol. 2020;153:706‐708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nuccetelli M, Pieri M, Gisone F, Bernardini S. Combined anti‐SARS‐CoV‐2 IgA, IgG, and IgM detection as a better strategy to prevent second infection spreading waves. Immunol Invest. 2020:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pieri M, Ciotti M, Carlozzi N, et al. SARS‐CoV‐2 infection serology validation of different methods: usefulness of IgA in the early phase of infection. Clin Chim Acta. 2020;511:28‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nuccetelli M, Pieri M, Grelli S, et al. SARS‐CoV‐2 infection serology: a useful tool to overcome lockdown? Cell Death Discov. 2020;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang MS, Hock KG, Logsdon NM, et al. Clinical performance of two SARS‐CoV‐2 serologic assays. Clin Chem. 2020;66(8):1055‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lou B, Li T, Zheng S. Serology characteristics of SARS‐CoV‐2 infection since the exposure and post symptoms onset. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 16. Lee CY, Lin RTP, Renia L, Ng LFP. Serological approaches for COVID‐19: epidemiologic perspective on surveillance and control. Front Immunol. 2020;11:879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brochot E, Demey B, Handala L, Francois C, Duverlie G, Castelain S. Comparison of different serological assays for SARS‐CoV‐2 in real life. J Clin Virol. 2020;130:104569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schnurra C, Reiners N, Biemann R, Kaiser T, Trawinski H, Jassoy C. Comparison of the diagnostic sensitivity of SARS‐CoV‐2 nucleoprotein and glycoprotein‐based antibody tests. J Clin Virol. 2020;129:104544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti‐SARS‐CoV‐2 antibodies. J Clin Virol. 2020;128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]


