Abstract
COVID‐19 was first reported as an unknown group of pneumonia in Wuhan City, Hubei province of China in late December of 2019. The rapid increase in the number of cases diagnosed with COVID‐19 and the lack of experienced radiologists can cause diagnostic errors in the interpretation of the images along with the exceptional workload occurring in this process. Therefore, the urgent development of automated diagnostic systems that can scan radiological images quickly and accurately is important in combating the pandemic. With this motivation, a deep convolutional neural network (CNN)‐based model that can automatically detect patterns related to lesions caused by COVID‐19 from chest computed tomography (CT) images is proposed in this study. In this context, the image ground‐truth regarding the COVID‐19 lesions scanned by the radiologist was evaluated as the main criteria of the segmentation process. A total of 16 040 CT image segments were obtained by applying segmentation to the raw 102 CT images. Then, 10 420 CT image segments related to healthy lung regions were labeled as COVID‐negative, and 5620 CT image segments, in which the findings related to the lesions were detected in various forms, were labeled as COVID‐positive. With the proposed CNN architecture, 93.26% diagnostic accuracy performance was achieved. The sensitivity and specificity performance metrics for the proposed automatic diagnosis model were 93.27% and 93.24%, respectively. Additionally, it has been shown that by scanning the small regions of the lungs, COVID‐19 pneumonia can be localized automatically with high resolution and the lesion densities can be successfully evaluated quantitatively.
Keywords: classification, computer‐aided diagnosis, convolutional neural networks, coronavirus, COVID‐19, deep learning, radiology
1. INTRODUCTION
In late December 2019, a new type of coronavirus mutated from the SARS‐CoV family has first reported an unknown group of pneumonia in Wuhan City, Hubei province of China. 1 , 2 The gene sequence of this new unknown virus, leading to a severe acute respiratory syndrome, was found to be 70% similar to the SARS‐CoV virus and was named SARS‐CoV‐2 by the International Virus Taxonomy Committee. 3 The same symptoms in healthcare professionals interacting with patients revealed that SARS‐CoV‐2 can be infected from one person to another. Within a short period, the virus spread to different parts of China and showed intercontinental spread within a few weeks. This outbreak, which spread rapidly and has a rarity in human history, was declared as a pandemic by the World Health Organization (WHO) on February 11, 2020 and was named the new coronavirus disease 2019 (COVID‐19). As of May 6, 2020, WHO confirmed that there are approximately 3.5 million positive cases worldwide. Unfortunately, 247 503 positive cases resulted in death. 4 The fact that the mortality rate in positive cases worldwide remaining at a high level of mortality as 6.89% reveals the importance of developing speed diagnosis and treatment methods for COVID‐19 in a short time.
COVID‐19 symptoms, which may differ from person to person, generally appear as severe respiratory failure syndromes such as high fever, sore throat, shortness of breath, and dry cough. 5 In the clinical area, reverse transcription‐polymerase chain reaction (RT‐PCR) testing is used as a key indicator for people who are suspected of having these symptoms. In the detection of nucleic acid forms from COVID‐19, test outputs must be confirmed by gene sequencing for blood and respiratory samples. 6 However, the difficulty of the application of the PCR test, limitations arising from production dynamics, and low sensitivity increase the interest in radiological imaging methods as an alternative. Studies have demonstrated that radiology imaging methods have specific features related to the COVID‐19 outbreak. X‐ray and computed tomography (CT) technics are the most effective imaging methods used by physicians. COVID‐19 lesions can be diagnosed by analyzing them with radiography techniques. 7 Additionally, the damage caused by the pandemic in the lung, and the changes in abnormalities with the treatment applied can be followed from CT images and evaluations can be made about the condition of the patient. Therefore, it is of great importance to comprehensively handle radiological imaging methods.
The main finding of COVID‐19 is pneumonia. Bilateral pneumonia accompanies 75% of cases related to the disease. Pneumonia‐induced abnormalities occur in CT images as ground‐glass opacities (GGO), paving‐stone appearance, ground glass, and consolidation, vascular enlargement in the lesion, traction bronchiectasis, and pleural thickening. 8 , 9 The course of the disease can be evaluated by examining the abnormalities caused by COVID‐19 lesions from CT images by radiologists. In particular, GGO is one of the main parameters considered in the detection and evaluation of COVID‐19. Changes occur as the disease increases or decreases in these abnormalities. An increase in the number and size of GGO, an increase in the number of lobes held, paving stone appearances, the development of multifocal consolidations, interlobular septal thickening and pleural effusion can be observed with the disease progression. 10 , 11 In patients with a tendency to heal, naturally decreases occur in GGO numbers and sizes, number of lesions, and lobes held. Due to the extraordinary rate of increase in the number of patients and the lack of specialists in radiology, one‐to‐one manual examination of the changes in these abnormalities can lead to remarkable time consumption and human‐induced errors. Therefore, automatic analysis of COVID‐19 pneumonia and the course of the disease with changes in lesions from CT or X‐ray images by computer‐aided diagnosis (CAD) systems can alleviate the burden of health centers suffering from a lack of radiologists. In this study, the successful performance of deep learning algorithms, which have been one of the artificial intelligence technologies in the diagnosis of various diseases from biomedical images, has been the main source of motivation for the detection and localization of lesions from CT images related to COVID‐19 pneumonia.
Studies related to the automatic diagnosis and evaluation of various diseases with a machine learning approach from biomedical images have been the focus of attention in recent years. Since the deep learning approach can successfully deliver the desired outputs without the need for a separate feature extraction increases its popularity in the field of biomedical image processing day by day. Moreover, feature extraction that forms the backbone of conventional methods and the ability to overcome the time and effort spent in its selection with deep learning makes deep learning algorithms more advantageous than other conventional methods. It is known from the literature that deep learning performs successfully in the screening of diabetic retinopathy from fundoscopic images, 12 metastatic breast cancer cells from pathological images, 13 and brain tumors from magnetic resonance imaging. 14 Automatic detection of lung nodules from X‐ray and CT images for early diagnosis of lung cancer, 15 , 16 detection of interstitial lung disease patterns from high‐resolution CT images, 17 and characterization and detection of different levels of tuberculosis findings are the application areas where the deep learning‐based models achieve superior success in chest radiological images. 18 The performance of deep learning‐based CAD systems is the driving force for researchers to move toward this approach for automatic early detection and evaluation of COVID‐19.
For the COVID‐19 pandemic, which only spreads across all world within weeks, it is essential to develop CAD systems that can assist clinicians in their diagnosis and follow‐up. Inspired by the performance of deep learning in biomedical images in the past, researchers have attempted to demonstrate the functionality of deep learning‐based algorithms in the early diagnosis of COVID‐19. In this context, many successful deep learning‐based CAD models have been introduced to the literature in a short period of time. Ardakani et al 19 proposed 10 different CNN models for the diagnosis of COVID‐19 from CT images. Researchers demonstrated that the deep learning technique can be a supporting tool for physicians. They achieved the highest success for ResNet‐101 and Xception architectures. Thus, they gave the researchers an idea about which architecture can be preferred. Amyar et al 20 tried to perform multitask learning based COVID‐19 lesion detection and segmentation from CT images. They obtained the classification accuracy as 86%. Ucar and Korkmaz 21 implemented a multiclass classification using deep Bayes‐SqueezeNet architecture. They demonstrated that COVID‐19, normal, and non‐COVID‐19 pneumonia can be classified from X‐ray images with their proposed model. Chen et al 22 used data replication for the segmentation of lesion patterns originated by COVID‐19 formed as GGO, consolidation, and pleural thickening. They observed that it effectively changed the classification performance. Zheng et al 23 proposed a unique model for the automatic early diagnosis of COVID‐19 and called it DeCovNET. With their proposed model, they obtained 90.8% classification accuracy from CT images. In the literature, it is seen that CT and X‐ray images have been used in early diagnosis and segmentation studies based on deep learning. Contrary to X‐ray images, CT images were used for the automatic detection and high‐resolution localization of COVID‐19 pneumonia lesions in this study, since CT images provide three‐dimensional visualization of the anatomical structure and have high sensitivity and low inter‐interpretive differences in lesion detection in the lungs. 24
The important subjects that will contribute to the literature in engineering and clinical fields with this study are highlighted below:
A novel deep convolutional neural network (CNN) that can automatically detect patterns of lesions caused by COVID‐19 from chest CT images was presented. In the deep learning‐based automatic diagnosis models recommended in the literature, the lungs are usually handled with full or large‐scale clipped regions. Unlike the literature, this study shows that lesion patterns can be detected for very small lung regions.
Detection of lesions in the lungs by radiologists is a tiring activity that requires a lot of effort. These challenges can be overcome with the proposed high precision deep learning‐based scanning system.
It is demonstrated that by taking into consideration small lung regions, pneumonic infiltration lesions in the CT of patients with COVID‐19 can be localized with high accuracy and advanced resolution than the literature by using the proposed model.
The patient's response to the treatment is evaluated by the attentive examination of the lesion density by radiologists. It is possible to add quantitative values to the qualitative evaluations made by radiologists with the proposed model.
The remainder of this article is structured as follows: In the material and method section, details of CT images collected on COVID‐19 pneumonia, pretreatment steps applied to CT images and proposed CNN model are described. The performance of the deep learning‐based model proposed for automatic diagnosis and localization of lesions is explained in detail in the experimental result section with model performance criteria. In this context, the results of the deep learning‐based state‐of‐the‐art models proposed in the literature are compared with the results obtained in this study and significant differences are interpreted in the discussion section with a different perspective. Finally, the study is concluded with the conclusion section.
2. MATERIALS AND METHODS
2.1. Axial CT image acquisition
In this study, CT images used for training and testing of the proposed model were taken from two different publicly available sources. One of the sources is the platform shared by researchers from different regions of the world. 25 It is an updateable data source where researchers from different regions are constantly adding new images. From this accessible database, 19 axial CT images of 9 patients were obtained. The other source includes the majority of the radiological images we used in the study. This resource contains accessible axial CT images shared by the Italian Society of Medical and Interventional Radiology. 26 110 axial CT images taken from 60 patients diagnosed with COVID‐19 were included in this study and an image data pool containing 129 COVID‐19 pneumonia lesion‐related abnormalities in total was formed. As a result, 102 CT images were approved by the expert radiologist for evaluation. Figure 1 shows axial CT images taken from three patients diagnosed with COVID‐19 randomly selected from the collected data set.
FIGURE 1.
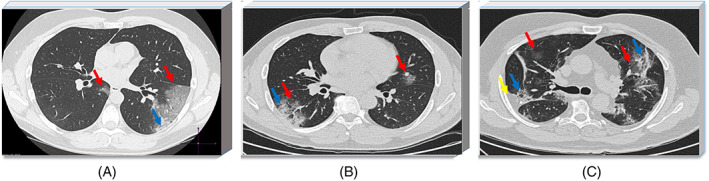
Axial computed tomography (CT) images of patients diagnosed with COVID‐19: A, Ground‐glass opacities (red arrow) and consolidation (blue arrow) in the right lobe, B, ground‐glass and consolidation in the left lobe, ground‐glass opacity in the right lobe, C, pleural effusion in the left lobe (yellow arrow ground‐glass opacities and consolidation appear in both lobes) 26 [Color figure can be viewed at wileyonlinelibrary.com]
2.2. Data preprocessing
An experienced radiologist marked the lesions before the researchers processed the COVID‐19 lesions findings. Thus, the training and testing processes of CNN‐based automatic diagnostic models can be carried out successfully and reliably. In this context, the marking of COVID‐19 lesions from axial CT images collected in this study was performed by a specialist radiologist. Accordingly, abnormalities originated from COVID‐19 pneumonia such as GGO, a paving stone appearance, ground‐glass and consolidation, vascular enlargement in the lesion, traction bronchiectasis, and pleural thickening were carefully marked on the CT images. Figure 2 shows three sample CT images with lesion findings marked by the radiologist.
FIGURE 2.

COVID‐19 lesions manually scanned by the radiologist: A, Ground‐glass opacities and consolidation, B, ground‐glass and consolidation, and C, pleural effusion, ground‐glass opacities, and consolidation in the left lobe [Color figure can be viewed at wileyonlinelibrary.com]
For the automatic detection and localization of abnormalities such as GGO and consolidation or pleural effusion caused by COVID‐19 pneumonia in CT images, data sets of different classes must be created with the proposed deep CNN model. In this context, all CT images obtained in different sizes were resized to 1200 × 800 pixels and standardized for segmentation. The regions scanned for the lesions by the radiologist were labeled as COVID‐positive and other regions without any lung lesion were labeled as COVID‐negative. Given the densities of the shaded regions related to the lesions, it was decided that 28 × 28 size segmentation would be suitable for capturing both COVID‐positive and COVID‐negative characteristic features. The segmentation process provided a detailed analysis of the regions where COVID‐19 lesions spread in CT images. From the CT images scanned by the radiologist, a total of 16 040 CT image segments belonging to the 10 420 COVID‐negative and 5620 COVID‐positive classes with a size of 28 × 28 pixels were obtained. A sufficient number of images were obtained for efficient training of the deep learning‐based model. Furthermore, the analysis made for 28 × 28 pixels patterns allowed the localization process to be performed more specifically. The segmentation process of CT images diagnosed with COVID‐19 is demonstrated in Figure 3.
FIGURE 3.
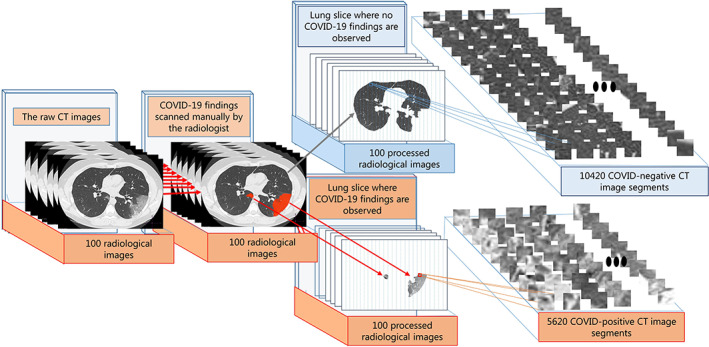
The segmentation process of computed tomography (CT) images diagnosed with COVID‐19 pneumonia: Scanned regions related to lesions on 1200 × 800 CT images (COVID‐positive), obtaining 28 × 28 sub‐CT images for other regions (COVID‐negative) without any findings of lesions [Color figure can be viewed at wileyonlinelibrary.com]
2.3. Deep CNNs
The conventional classifier algorithm based CAD systems consists of hierarchical methodologies, including the detection of patterns related to the problem from any biomedical image, the process of feature extraction, which usually requires intense labor and time, and then the classification of the extracted features. However, in recent studies, it has been accepted that the most distinctive features of patterns can be extracted with multilayered systems. 27 , 28 Deep learning algorithms take part in this structure by being distinguished from conventional classifiers, in terms of both architectural structure and layer types. In this study, the CNN model, which is one of the deep learning techniques and has an effective potential especially in computer vision, was used to detect and localize lesions caused by COVID‐19 from axial CT images. CNN's ability to successfully extract complex image features and perform nearly perfect in the medical area has been our primary source of motivation.
The CNN tries to learn high‐level hidden information in data using hierarchical architectures. 29 By training the convolutional layers quickly, they can successfully adapt to the biomedical image classification process. The training process of the proposed network structure plays a key role in achieving the desired outcomes. The training of the CNN algorithm, which is a powerful tool in image processing, takes two steps. These stages are described as forward and backward oriented processes. In the CNN algorithm, the raw image given to the input of the network is tried to be expressed as weight and bias value in each layer. Then, the gradient values of each parameter are calculated according to the chain rule. The parameters considered as a result of these processes are updated according to their gradient values. The update of each weight and bias value in the CNN model during the training process is given in Equations (1) and (2), respectively.
| (1) |
| (2) |
The decision‐making process for the final output in the CNN algorithm is similar to the artificial neural network. Here, W , B , l, λ , x, n, m, t, and C correspond to weight, bias, number of layers, regulation parameter, learning coefficient, the total number of training samples, momentum coefficient, update step, and cost function, respectively. 30 Although the CNN architecture, which is based on the feed‐forward network structure, has many variations, it generally consists of three basic layers. These are convolutional, pooling, and fully connected layers.
2.3.1. Convolutional layer
It is the main part of the CNN structure, and the algorithm derives its real power from convolution processing. In this layer, it creates new feature maps from input images or previous feature maps. 31 The convolutional layer consists of a series of convolution kernels. The convolution kernel slides over the image to take the weighted sum of the pixels it touches and calculates the outputs that make up the feature map. Each neuron in the feature map establishes a series of weighted connections with neighboring neurons in the previous layer. 32 The mathematical expression of the operations carried out in the convolutional layer is given as follows;
| (3) |
where the input image given to the convolutional layer is the w JxI dimension convolutional kernel, b is bias, and y is the feature map resulting from convolutional. All neurons in a feature map have constrained weights equal. However, different feature maps in the same convolutional layer have different weights. So, many features can be extracted from each region. 33
2.3.2. Pooling layer
The pooling layer generally follows the convolutional layer. 28 , 29 The purpose of this layer is to reduce the spatial size and complexity of feature maps. In this way, it speeds up the training of the network and enables the decision mechanism to work efficiently. In addition, the pooling process can overcome possible overfitting problems by better generalizing the learning phase of the deep model. It is often stated in the literature that max‐pooling layer gives more successful result. 34 Maximum pooling, which captures invariances efficiently, is considered the most commonly used layer in ESA architectures. Therefore, in this study, the max‐pooling operator was chosen to reduce the size and complexity of feature maps extracted from CT image segments in the convolutional layer.
2.3.3. Fully connected layer
This layer generally comes after the last convolutional or pooling layer. The working principle of fully connected layers is like conventional artificial neural networks. 29 , 35 During the learning of the network, approximately 90% of the parameters are located in this layer. In ESA algorithms, feature maps generated after the last pooling or convolution layer are transformed into vectors of the column. Feature vectors obtained by flatten process are given as input to fully connected layer. Thus, decision‐making is achieved.
2.3.4. Activation functions
Rectified linear units (ReLUs) and softmax functions were used in this study for the automatic detection and localization of COVID‐19 lesions. The activation functions in the deep structure keep the outputs in the predetermined limit so that the proposed models offer desired and accurate outputs. In the CNN approach proposed for COVID‐19 automatic detection, ReLU and softmax activation functions are used instead of sigmoid functions. The mathematical expression of the ReLU activation function is given in Equation (4). In the ReLU activation function, if the input value is less than zero, the output is zero, if it is greater than zero, the output is equal to the input. ReLU activation function is very similar to biological nerve cells with this ability.
| (4) |
The softmax function creates a probability distribution function for k outputs. It is preferred in very problematic classification processes. In this study, the softmax function was used to distinguish the findings of COVID‐19 lesions from healthy lung region (Equation (5)).
| (5) |
where k is the total number of classes and j is classes.
2.4. The CNN architecture for diagnosis and localization of COVID‐19 pneumonia
In this study, a six‐layer CNN architecture is proposed for the detection of COVID‐19 pneumonia lesions from CT images of lung region. In architecture, there are two convolutional and two max‐pooling layers as the structure that captures the characteristic features from the images. Feature maps produced by these layers, inspired by the visual cortex, are subjected to flatten. One‐dimensional feature vectors produced by the flatten process are classified through fully connected layers that work like a typical feed‐forward neural network. In the first convolutional layer, 32 convolution kernels of 5 × 5 dimensions and in the second convolutional layer, 64 convolution kernels of 5 × 5 dimensions were used. In the pooling layers following the convolutional layers, the max‐pooling operator was chosen, and its kernel dimension was set as 2 × 2 size. Accordingly, the dimensions of the feature maps made in the convolutional layers were halved and the spatially invariant was increased. The optimum performance in diagnosing lesions related to COVID‐19 was determined by trial and error method. Considering many combinations, the most effective hierarchical architecture decided in detection and localization is given in Table 1.
TABLE 1.
The proposed CNN architecture
| Layers | Kernel size | Numbers of kernels | Number of neurons | Stride |
|---|---|---|---|---|
| 1. Convolution | 5 × 5 | 32 | — | 1 |
| Max‐pooling | 2 × 2 | 32 | — | 2 |
| 2. Convolution | 5 × 5 | 64 | — | 1 |
| Max‐pooling | 2 × 2 | 64 | — | 2 |
| Fully connected | — | — | 2048 | — |
| Fully connected | — | — | 2 | — |
Abbreviation: CNN, convolutional neural network.
In the pre‐processing step, 1200 × 800 pixels axial CT images evaluated by an experienced radiologist were divided into two sections as regions where lesions are observed and not observed. The segregated lung regions were taken into account as sub‐CT image segments in 28 × 28 pixels. As a result of the first convolutional layer from each image segment related to the COVID‐positive and COVID‐negative classes, 24 × 24 dimensions feature maps were produced and then resized to 12 × 12 with the applied maximum pooling layer. In the second convolutional layer, new feature maps in size 8 × 8 were generated from the feature maps of 12 × 12 size and were resized to 4 × 4 in the last pooling layer. Thus, the approximation and detail features of lesion findings from the lung CT image segments were attempted to be captured. Figure 4 shows the graphical representation of the proposed CNN structure.
FIGURE 4.
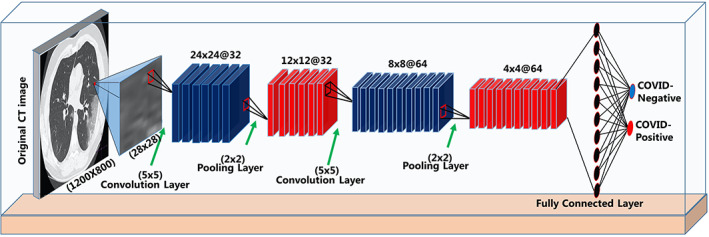
Graphical representation of the proposed convolutional neural network (CNN) model [Color figure can be viewed at wileyonlinelibrary.com]
2.5. Training and testing process of the proposed CNN‐based CAD system
Classification of 16 040 CT image segments obtained during the pre‐processing, 10 420 of which were COVID‐negative and 5620 of which were COVID‐positive, was carried out by considering a certain methodology with the proposed deep learning model. The CT images of the two classes were divided into 80% for the network training and 20% for the network testing. Then, 25% of the images allocated to training were used as a validation data set so that the network does not overfitting and can make a good generalization. To reliably evaluate the ability of the network to automatically detect patterns related to lesions, fivefold cross‐validation was applied. Thus, all images in the COVID‐negative and COVID‐positive clusters were included in both the training and testing processes. Evaluation of the performance of the network by applying the fivefold cross‐validation is given in Equation (6).
| (6) |
In the training of the model proposed for the automatic detection and localization of COVID‐19 lesions, the stochastic gradient descent with momentum (SGDM) optimization method was preferred. As is known from the literature, it can give more successful results than other adaptive optimization methods. 36 In the training of the network, the appropriate initial learning rate was determined as 0.001 to avoid a long period in which the network can get stuck during learning or rapid learning without optimal weight values. Momentum coefficient was taken as 0.9 to reduce possible oscillations and shorten the time to reach the target with the SGDM method. For the proposed model to perform successful learning, mini‐batch size 128 and maximum epoch number 10 are the other parameter values selected.
Successful training of CNN architectures in image processing problems is associated with determining hyperparameters for the network. In this study, parameters such as the number of filters and filter size used in the convolution layer, pooling type, dropout rate, initial learning rate, and the number of neurons in the fully connected layer were determined by trial and error. The evaluation process was made based on combinations of different values of these parameters. The combination that offers the most successful results is considered as optimum hyperparameters.
2.6. Evaluation criteria of the proposed model
The success of the model proposed in this study for the automatic detection and localization of COVID‐19 lesions was evaluated in terms of the accuracy, sensitivity, specificity, precision, and F1‐score criteria. These criteria are often preferred to evaluate the performance of automatic diagnostic systems. Accuracy is the success of the proposed model in the classification of lung CT image segments labeled COVID‐positive and COVID‐negative, sensitivity represents the accuracy of the findings it detects as COVID‐19, and specificity indicates the accuracy of the image segments that it perceives as COVID‐negative. In this study, the accuracy of the data tested was determined using the F1‐score criterion.
3. RESULTS
In this study, all of the methods based on deep learning suggested for the automatic diagnosis and localization of COVID‐19 lesions were implemented in MATLAB (MathWorks, Natick, Massachusetts) programming environment running on Intel Core i7 8700, NVIDIA GeForce ROG‐STRIX 256 bit 8GB GPU and 16 GB RAM memory. Experimental findings on the ability of the proposed model to diagnose lesions and quantitatively interpret their density by showing the lung sections where the lesions spread are discussed under two separate headings.
3.1. Detection of patterns related to COVID‐19 lesions
Regions of the lungs, where no lesion related findings were observed during pre‐processing of the CT images and regions observed as GGO, consolidation and pleural effusion of lesions were separated into distinct segments. The regions related to healthy and lesions were epoched to two‐dimensional 28 × 28 pixels subimage segments, taking into account the lesion pattern volumes manually scanned by the radiologist. Then, 10 420 CT image segments without any findings were labeled as COVID‐negative and 5620 CT image segments where the findings related to the lesions observed in various forms were labeled as COVID‐positive. In this way, by performing binary classification, the diagnostic performance of the proposed model was evaluated.
The unbalanced distribution between data sets to be classified in conventional machine learning or deep learning algorithms negatively affects classification performance. 37 The overfitting problem may occur for the class, which constitutes the majority in the data distribution by the proposed classifier model. This possibility is undesirable and can be observed by inconsistencies in model performance metrics. Taking into consideration the mentioned handicaps, the classification process in this study was carried out by following two different data distribution strategies. Due to the existing unbalanced distribution of the number of image segments between classes, the first strategy aimed to balance the data distribution. In this strategy, 5620 images belonging to the COVID‐positive class and 5620 image segments randomly selected from the COVID‐negative class were used. Thus, an equal number of CT image samples were obtained for the two classes. In the second classification strategy, all image segments of both classes were used, and the classification performance offered by the proposed model against both conditions was compared. Table 2 shows the number of images reserved for training, validation, and testing for both classification strategies.
TABLE 2.
Different distribution strategies for the image segments that form the classes
| Balanced distribution | The all data | |||||
|---|---|---|---|---|---|---|
| Train | Validation | Test | Train | Validation | Test | |
| COVID‐positive | 3372 | 1124 | 1124 | 3372 | 1124 | 1124 |
| COVID‐negative | 3372 | 1124 | 1124 | 6252 | 2084 | 2084 |
| Total | 6744 | 2248 | 2248 | 9624 | 3208 | 3208 |
The CT images data were randomly divided into five equal portions. Four out of five portions of the image segments were used to train CNN while the remaining one‐fifth of the images used to test the performance of the system. This strategy was repeated five times by shifting the test and training dataset. In order to reliably evaluate the ability of the network to automatically detect patterns related to lesions, fivefold cross‐validation was applied. Confusion matrices for the classification performance obtained for the two strategies applied in the image data distribution are demonstrated in Figure 5 for overall and each folding stage.
FIGURE 5.
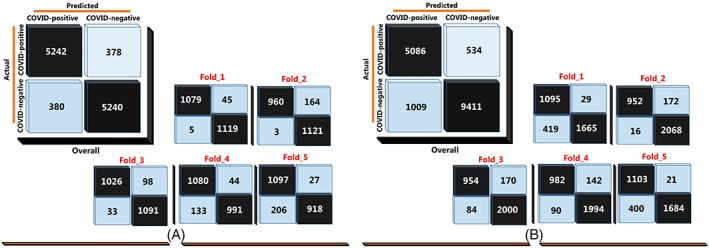
Overall and test confusion matrices for each fold in COVID‐19 detection with the proposed model: A, Balanced distribution, and, B, Unbalanced distribution [Color figure can be viewed at wileyonlinelibrary.com]
The performance of the proposed model was evaluated considering the frequently used model performance criteria. The results obtained for both data distributions are given in Table 3.
TABLE 3.
Model performance values of the proposed model performance related to different data distribution strategies
| Accuracy | Sensitivity | Specificity | Precision | F1‐score | |
|---|---|---|---|---|---|
| Balanced distribution | 0.9326 | 0.9327 | 0.9324 | 0.9387 | 0.9331 |
| The all data | 0.9038 | 0.9050 | 0.9032 | 0.8551 | 0.8714 |
As can be seen in Table 3, the proposed model performed more efficiently in the strategy in which the distribution between classes was balanced. It was able to classify CT image patterns related to COVID‐19 lesions with 93.26% accuracy. In the strategy in which all data were included in the classification process and an unbalanced distribution observed, the proposed model provided a classification accuracy of 90.38%. Given the number and size of images considered, it is clear that the proposed model works quite efficiently. The sensitivity and specificity model performance criteria are indicators to understand that the deep learning‐based model can make a good generalization for any class. The sensitivity and specificity criteria obtained in the balanced distribution of classes were 93.27% and 93.24%, respectively. For the strategy in which all images are included in the process, sensitivity and specificity were observed as 90.50% and 90.32%, respectively. In general, it can be observed that the performance is at a more successful level for a balanced distribution. For the balanced distribution strategy, the relevant ROC curves and area under the ROC curve (AUC) values for each fold process are shown in Figure 6.
FIGURE 6.
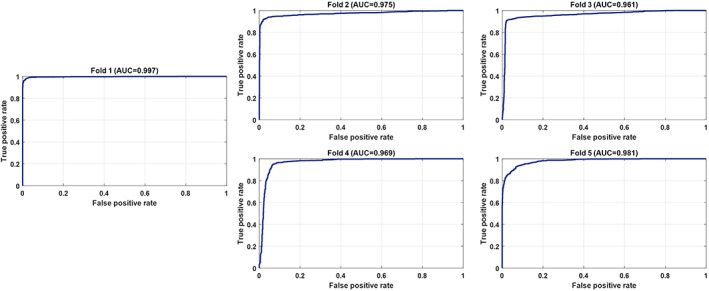
The ROC curves and area under the ROC curve (AUC) values for each fold related to a balanced distribution [Color figure can be viewed at wileyonlinelibrary.com]
As seen in Figure 6, the average AUC value obtained with the fivefold cross‐validation was 97.58%. The learning curves of the proposed model related to the training and validation data sets used for each fold are shown in Figure 7.
FIGURE 7.
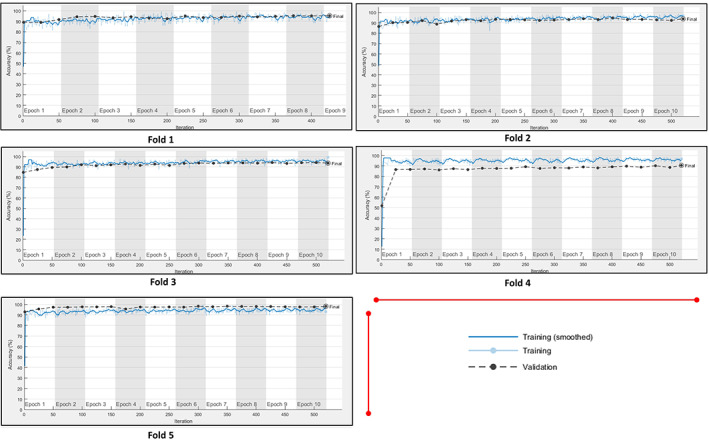
The learning curves of the proposed model related to the training and validation data sets used for each fold [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Automatic localization of COVID‐19 findings
In this section, it is showed that the advanced model can localize abnormalities with a higher resolution than the literature, taking into account approximately 0.63% smaller regions from the lungs and scanning COVID‐19 pneumonia lesions for these regions. For all CT images, considering all lung regions and 28 × 28 pixels images taken into consideration, the image segments used correspond to an average of 0.63% slices of the lungs. The mathematical equation is defined as follows;
| (7) |
where N represents the raw CT images taken from the patient, and convp and convn represent the COVID‐positive and COVID‐negative image segments, respectively.
Abnormalities such as GGO, paving stone, ground glass and consolidation, vascular enlargement in the lesion, traction bronchiectasis, and pleural effusion have been shown to be localized correctly from the CT images. For six samples randomly selected from CT images taken from patients diagnosed with COVID‐19, the localization of lesion findings done by a radiologist manually and automatic localization performed by the proposed model in this study were compared in Figure 8. The localization process was applied to different images that were not previously used in training and testing the network.
FIGURE 8.
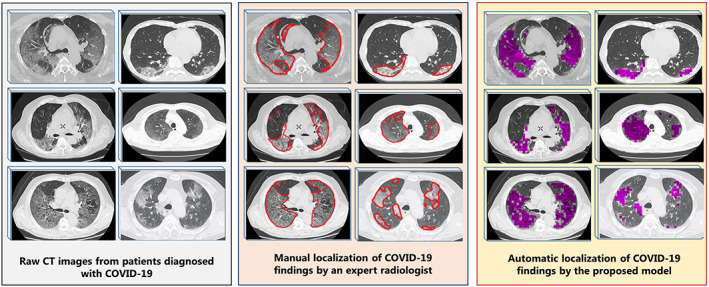
Automatic localization of COVID‐19 pneumonia findings with the model proposed: Raw computed tomography (CT) images without any processing, manual screening of COVID‐19 findings by an expert radiologist (red borders), visualization of the findings by automatically scanning the same images with the proposed model (the findings are represented in purple) [Color figure can be viewed at wileyonlinelibrary.com]
As shown in Figure 8, the localization of the findings made by applying the proposed model overlaps with those made by the radiologist. With the correct automatic localization process used in this study, it was shown that time loss and a huge workload caused by manual examination of radiologists can be overcome with this system. Furthermore, the quantitative evaluation of the density of the lesions in the lung can successfully be carried out by localization. The quantitative evaluation of the density of the lesions was applied according to the formula given below.
| (8) |
where covid(p) and covid(n) represent the reel COVID‐positive and COVID‐negative image segments, respectively. covid(p ′) represents the COIVD positive lesion segments marked by the proposed model. Lesion densities are shown in Figure 9, quantitatively for six randomly selected samples from CT images from patients diagnosed with COVID‐19.
FIGURE 9.
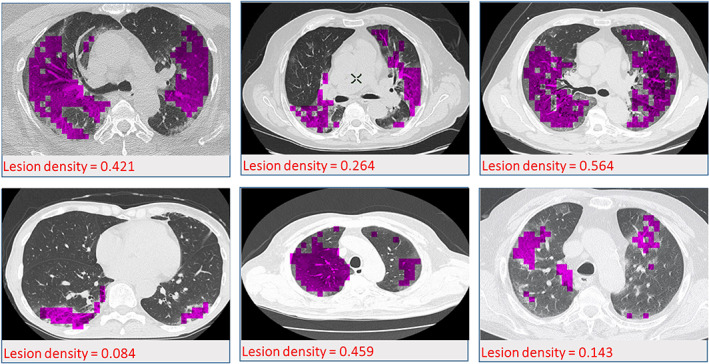
Quantitative display of lesion densities for six randomly selected samples from patients diagnosed with COVID‐19 [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The main tools used in the diagnosis of COVID‐19 are the developed RT‐PCR and radiological images. It is inevitable to include radiological imaging techniques such as CT and X‐ray imaging in the diagnosis process, to make the correct diagnosis to patients who display suspicious symptoms but whose other tests may show false negativity. Although the RT‐PCR test was negative in the first attempts, there are also examples that CT images have positive findings and the diagnosis reached by CT imaging technique appears to be accurate. Therefore, this situation puts an important responsibility to the experienced and competent experts in the field of CT in radiology during the pandemic process. The rapid increase in the number of cases diagnosed with COVID‐19 increases the need for radiologists experienced in the medical area. 23 , 38 Manual examination of the chest radiology images by a relatively small number of radiologists will inevitably lead to human‐induced errors in the interpretation of the CT images, along with the heavy workload. Due to the handicaps mentioned above in the medical area, the development of the CAD system that can automatically diagnose COVID‐19 lesions from lung CT images has been our primary motivation. Based on this motivation, a deep learning model was developed that can automatically detect and localize COVID‐19 findings using CT images. Unlike the literature, it was possible to detect COVID‐19 lesions even in very small regions of the lungs. In this context, 10 420 CT image segments without any lesions were classified as COVID‐negative, and 5620 CT image segments, in which the findings related to the lesions were detected in various forms, were classified as COVID‐positive. In this way, a large and comprehensive data set was considered, unlike small data sets handled in the automatic diagnosis models proposed in the literature. CNN architecture recommended in the literature was used as the classification algorithm. The 93.26% classification accuracy of the model developed shows that the steps followed in automatic detection of lesion findings are correct.
It is known from the literature that the proposed deep learning architectures give successful results on the images obtained by the radiography method in the early and accurate diagnosis of COVID‐19. However, there are some remarkable issues in these studies. The first one that stands out is that the radiological images or image segments of images used are limited. In this context, Hemdan et al 39 proposed a deep learning‐based CAD system that can assist radiologists in the diagnosis of COVID‐19. They tried to classify 25 COVID‐negative and 25 COVID‐positive labeled X‐ray images using seven different deep convolutional network architectures and compare the performances obtained for different architectures. They achieved the highest success as 90%. Sethy and Behera 40 proposed a hybrid model for automatic detection of COVID‐19 from X‐ray images. In their strategy, they first extracted the feature using the ResNet‐50 architecture, one of the deep learning algorithms and tried to classify the extracted feature vectors with a support vector machine, one of the conventional classification algorithms. With the hybrid model they proposed, they achieved a 95.38% success in binary classification. The classification process was carried out by considering 50 radiological images. Ozturk et al 41 proposed the DarkCovidNet‐19 deep architecture with 17 convolutional layers for automatic diagnosis of COVID‐19 cases from X‐ray images. The deep architecture they proposed for 500 images related to any finding, 500 pneumonia (non‐COVID), and 125 COVID‐19 positive image groups showed 87.02% classification success. With our trained model for a total of 16 040 image segments, it can be said that a deep network with better‐generalized weight values was designed for automatic detection of COVID‐19 findings, considering the number of images examined in the mentioned studies.
As in this study, there are many studies where the segmentation approach is applied to detect COVID‐19 lesions automatically from CT images. With the segmentation process, a large number of images can be obtained from CT or X‐ray images, and the findings of the lesions can be scanned in small regions of the lungs. Accordingly, more data can be provided for the training of the developed deep learning architectures. However, the most outstanding deficiency in studies, which this approach applied, is that the segmentation process is done for large lung regions. In this context, Butt et al 42 aimed early imaging of COVID‐19 lesions in CT images with their proposed deep learning‐based model. It was tried to increase the segmentation of CT image data sets in the size of 60 × 60. With the model they recommend based on ResNeT and location attention, they achieved a diagnostic success of 86.7%. Ying et al 38 proposed a model that can detect COVID‐19 findings from CT images with detail relation extraction neural network (DRE‐Net) architecture. Segmentation was performed by dividing CT images into 15 equal parts, and image segments for 777 slices of COVID‐19 and 708 slices with no lesion findings were obtained. They achieved a classification accuracy of 86% by using DRE‐Net. Ardakani et al 19 collected CT images from 108 patients diagnosed with COVID‐19 and 86 different viral pneumonia and achieved a total of 1020 image segments for both classes by epoching in 60 × 60 pixels subimages. They achieved 99.51% accuracy. Considering that image sizes before the segmentation in the mentioned studies were 368 × 368 and 512 × 512 pixels, it can be understood that the CT image segments taken into account in the classification process correspond to the large lung regions. However, CT images of 1200 × 800 pixels size were separated into 28 × 28 pixels size segments in our study. Thus, automatic detection of COVID‐19 lesions was made possible for very small regions of the lungs. With this aspect of our study, scanning at a higher resolution than the literature can be made. Although there are data sets extended by segmentation in current studies, it can be said that they are still far behind the number of image segments achieved in this study.
Since COVID‐19 is yet a new disease, there are classification‐based studies conducted in the literature where data sets for the COVID‐19 class have been enlarged by obtaining artificial images from the original images by several data augmentation methods. However, it is noteworthy that the classification achievements have changed significantly with the increase of COVID‐19 image segments by the addition of artificial images by researchers in these studies. As an example of these studies, Ucar and Korkmaz 21 proposed a deep learning‐based model that can automatically diagnose COVID‐19 from X‐ray images. With their proposed SqueezeNet with Bayes optimization‐based deep architecture, they classified X‐ray images from healthy individuals and X‐ray images where pneumonia (bacterial, non‐COVID‐19) and COVID‐19 findings were observed. Data augmentation was applied to overcome the imbalance between classes due to the small number of COVID‐19 related images. Along with mirroring the original images, artificial images were obtained by techniques such as adding noise, increasing, and decreasing brightness. While they offered a classification accuracy of 76.37% for the model they proposed using original data, they achieved an accuracy of 98.26% with artificial augmented images. COVID‐19 classification success was increased from 70% for original images to 100% with data augmentation. Chen et al 22 suggested a U‐Net based architecture for segmentation of COVID‐19 infections from CT images. To increase the number of patterns, they generated artificial images by rotating the original CT images at different angles. While the success achieved using raw images was 79%, it was increased to 89% by the addition of artificial images. The imbalance in the number of patterns that form the classes will certainly affect the classification performance. However, the application of various data augmentation methods to achieve a balanced distribution has directly affected the performance of the models proposed in these studies. This situation negatively affects the reliability of the performances of the proposed models. In our proposed model, original image segments were used in the unbalanced and balanced distribution of the patterns that form the classes. Classification models made with original images can be said to be more consistent and reliable, with 90.38% and 93.26% classification successes for both unbalanced and balanced distribution, respectively. The automated COVID‐19 diagnostic model proposed in this study and other deep learning‐based state‐of‐the‐art models are compared in Table 4.
TABLE 4.
Comparison of the proposed automatic COVID‐19 diagnostic model with other deep learning‐based state‐of‐the‐art models
| Study | Year | Type of radiological images | Methods | Class labels | Overall accuracy (%) |
|---|---|---|---|---|---|
| Amyar et al 20 | 2020 | Chest CT | Deep learning‐based multitask model |
COVID‐19 Non‐COVID |
86 |
| Ying et al 38 | 2020 | Chest CT | DRE‐Net |
COVID‐19 Pneumonia (bacterial) |
86 |
| Xu et al 42 | 2020 | Chest CT | ResNet with location attention |
COVID‐19 Influenza viral pneumonia Healthy |
86.7 |
| Ozturk et al 41 | 2020 | Chest X‐ray | DarkCovidNet‐19 |
COVID‐19 NO‐finding Pneumonia (non‐COVID) |
87.02 |
| Li and Zhu 43 | 2020 | Chest X‐ray | DenseNet |
Pneumonia Normal COVID‐19 |
88.9 |
| Hemdan et al 39 | 2020 | Chest X‐ray | COVIDX‐Net |
COVID‐positive COVID‐negative |
90 |
| Zheng et al 23 | 2020 | Chest CT | 3D deep CNN |
COVID‐positive (data augmentation) COVID‐negative |
90.8 |
| Wang et al 44 | 2020 | Chest X‐ray | Tailored deep CNN |
Normal Pneumonia COVID‐19 |
92.6 |
| Sethy and Behera 40 | 2020 | Chest X‐ray | ResNet50 + SVM |
COVID‐positive COVID‐negative |
95.38 |
| Ucar et al 21 | 2020 | Chest X‐ray | Squeeze‐Net with Bayes optimization |
Normal Pneumonia (bacterial) COVID‐19 (data augmentation) |
98.26 |
| Proposed model | 2020 | Chest CT | CNN |
COVID‐positive COVID‐negative |
93.26 |
Abbreviations: CNN, convolutional neural network; DRE‐Net, detail relation extraction neural network; SVM, support vector machine.
As seen in Table 4, the model proposed in this study provides more successful results in comparison to the literature in the detection of COVID‐19 lesions from radiological images. In rare studies in the literature where better classification accuracy is obtained in the diagnosis of COVID‐19, handicaps are predominant due to the use of artificial image segments or very few radiological images. Therefore, it is clear that the proposed model will contribute to the literature in the early and accurate diagnosis of COVID‐19.
In patients diagnosed with COVID‐19, lesions occurring in the lungs over time are examined by qualitative evaluations from CT images by radiologists. It has been shown that quantitative evaluation of the intensity of COVID‐19 lesions in the lungs over time can be successfully made with the proposed deep learning‐based model. Thus, qualitative reports prepared by radiologists can be enriched by including quantitative data in the proposed model. The segmentation process is based on a proposed original approach, eliminating the need to use separate algorithms such as U‐net. Also, the location of the detected segments as well as their quantitative measurement makes this approach a powerful method.
5. CONCLUSION
In this study, it has been shown that lesions caused by COVID‐19 pneumonia can be detected from the proposed CNN‐based model CT images. The developed model managed to detect patterns related to COVID‐19 lesions with an accuracy of 93.26%. In addition to this success, the detection of very small regions in the lungs caused by lesions from chest CT images is one of the most important outputs of the proposed model. With the determination of the lesions correctly for very small lung regions, the change of COVID‐19 lesion densities over time in the lungs of infected patients can quantitatively be evaluated. The proposed model can be used as an auxiliary system by clinicians in the diagnosis and evaluation of the disease with high accuracy in health centers, where other molecular diagnostic tests are insufficient during the outbreak.
Polat H, Özerdem MS, Ekici F, Akpolat V. Automatic detection and localization of COVID‐19 pneumonia using axial computed tomography images and deep convolutional neural networks. Int J Imaging Syst Technol. 2021;31:509–524. 10.1002/ima.22558
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in covid‐chestxray‐datase at https://github.com/ieee8023/covid-chestxray-dataset/, reference number 25 and Italian Society of Medical and Interventional Radiology at https://www.sirm.org/en/2020/03/30/covid‐19‐management‐strategy‐in‐radiology/, reference number 26.
REFERENCES
- 1. Toğaçar M, Ergen B, Cömert Z. COVID‐19 detection using deep learning models to exploit social mimic optimization and structured chest X‐ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pereira RM, Bertolini D, Teixeira LO, Silla CN Jr, Costa YMG. Covid‐19 identification in chest X‐ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed. 2020;194:105532. 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. International Committee on Taxonomy of Viruses (ICTV) Website . https://talk.ictvonline.org/. Accessed February 14, 2020.
- 4. World Health Organization (WHO) Website . https://www.who.int/emergencies/diseases/novel-coronavirus-2019/. Accessed May 6, 2020.
- 5. Al‐Balas M, Al‐Balas HI, Al‐Balas H. Surgery during the COVID‐19 pandemic: a comprehensive overview and perioperative care. Am J Surg. 2020;219(6):903‐906. 10.1016/j.amjsurg.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease‐19 (COVID‐19): relationship to duration of infection. Radiology. 2020;295(3):200463. 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID‐19) pneumonia: a multicenter study. AJR Am J Roentgenol. 2020;214(5):1072‐1077. 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 9. Salehi S, Abedi A, Balakrishnan S, Gholamrezanezhad A. Coronavirus disease 2019 (COVID‐19): a systematic review of imaging findings in 919 patients. AJR Am J Roentgenol. 2019;215:1‐7. 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 10. Pan Y, Guan H, Zhou S, et al. Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019‐nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 2020;30(6):3306‐3309. 10.1007/s00330-020-06731-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pan F, Ye T, Sun P, et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID‐19) pneumonia. Radiology. 2020;295(3):200370‐200721. 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Islam MM, Yang HC, Poly TN, Jian WS, Li YC. Deep learning algorithms for detection of diabetic retinopathy in retinal fundus photographs: a systematic review and meta‐analysis. Comput Methods Programs Biomed. 2020;191:105320. 10.1016/j.cmpb.2020.105320. [DOI] [PubMed] [Google Scholar]
- 13. Yang X, Wu L, Zhou K, et al. Deep learning signature based on staging CT for preoperative prediction of sentinel lymph node metastasis in breast cancer. Acad Radiol. 2019;27(9):1226‐1233. 10.1016/j.acra.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 14. Deepak S, Ameer PM. Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med. 2019;111:103345. 10.1016/j.compbiomed.2019.103345. [DOI] [PubMed] [Google Scholar]
- 15. Li X, Shen L, Xie X, et al. Multi‐resolution convolutional networks for chest X‐ray radiograph based lung nodule detection. Artif Intell Med. 2019;103:101744. [DOI] [PubMed] [Google Scholar]
- 16. Shariaty F, Mousavi M. Application of CAD systems for the automatic detection of lung nodules. Inform Med Unlocked. 2019;15:100173. 10.1016/j.imu.2019.100173. [DOI] [Google Scholar]
- 17. Agarwala S, Kale M, Kumar D, et al. Deep learning for screening of interstitial lung disease patterns in high‐resolution CT images. Clin Radiol. 2020;75(6):481.e1‐481.e8. 10.1016/j.crad.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 18. Gao XW, James‐Reynolds C, Currie E. Analysis of tuberculosis severity levels from CT pulmonary images based on enhanced residual deep learning architecture. Neurocomputing. 2020;392:233‐244. 10.1016/j.neucom.2018.12.086. [DOI] [Google Scholar]
- 19. Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID‐19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amyar A, Modzelewski R, Ruan S. Multi‐task deep learning based CT imaging analysis for covid‐19: classification and segmentation. medRxiv preprint. 2020;126:104037. 10.1101/2020.04.16.20064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ucar F, Korkmaz D. COVIDiagnosis‐Net: deep Bayes‐SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID‐19) from X‐ray images. Med Hypotheses. 2020;140:109761. 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen X, Yao L, Zhang Y. Residual attention U‐net for automated multi‐class segmentation of COVID‐19 chest CT images. arXiv Preprint arXiv. 2020. [Google Scholar]
- 23. Zheng C, Deng X, Fu Q, et al. A weakly‐supervised framework for COVID‐19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020;39(8):2615‐2625. 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 24. Lee SM, Seo JB, Yun J, et al. Deep learning applications in chest radiography and computed tomography. J Thorac Imaging. 2019;34(2):75‐85. 10.1097/RTI.0000000000000387. [DOI] [PubMed] [Google Scholar]
- 25. Covid‐chestxray‐dataset . https://github.com/ieee8023/covid-chestxray-dataset/. Accessed April 18, 2020.
- 26. Italian Society of Medical and Interventional Radiology . https://www.sirm.org/en/2020/03/30/covid-19-management-strategy-in-radiology/. Accessed April 18, 2020.
- 27. Raghavendra U, Fujita H, Bhandary SV, Gudigar A, Tan JH, Acharya UR. Deep convolution neural network for accurate diagnosis of glaucoma using digital fundus images. Inform Sci. 2018;441:41‐49. 10.1016/j.ins.2018.01.051. [DOI] [Google Scholar]
- 28. Amorim WP, Tetila EC, Pistori H, Papa JP. Semi‐supervised learning with convolutional neural networks for UAV images automatic recognition. Comput Electron Agric. 2019;164:104932. 10.1016/j.compag.2019.104932. [DOI] [Google Scholar]
- 29. Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2‐13. 10.1016/j.compmedimag.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 30. Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med. 2018;100:270‐278. 10.1016/j.compbiomed.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 31. Li P, Zhao W. Image fire detection algorithms based on convolutional neural networks. Case Stud Therm Eng. 2020;19:100625. 10.1016/j.csite.2020.100625. [DOI] [Google Scholar]
- 32. LeCun Y, Kavukcuoglu K, Farabet C. Convolutional networks and applications in vision. In: ISCAS 2010–IEEE International Symposium on Circuits and Systems. Nano‐Bio Circuit Fabrics and Systems, 2010. 10.1109/ISCAS.2010.5537907 [DOI]
- 33. Rawat W, Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017;29(9):2352‐2449. 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 34. Scherer D, Müller A, Behnke S. Evaluation of pooling operations in convolutional architectures for object recognition. Lect Notes Comp Sci. 2010;6354:92‐101. 10.1007/978-3-642-15825-4_10. [DOI] [Google Scholar]
- 35. Traore BB, Kamsu‐Foguem B, Tangara F. Deep convolution neural network for image recognition. Eco Inform. 2018;48:257‐268. 10.1016/j.ecoinf.2018.10.002. [DOI] [Google Scholar]
- 36. Wilson A, Roelofs R, Stern M, Srebro N, Recht B. The marginal value of adaptive gradient methods in machine learning. NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems, 2017.
- 37. Buda M, Maki A, Mazurowski MA. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 2018;106:249‐259. 10.1016/j.neunet.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 38. Ying S, Zheng S, Li L, et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID‐19) with CT images. MedRxiv. 2020. 10.1101/2020.02.23.20026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hemdan EED, Shouman MA, Karar ME. COVIDX‐net: a framework of deep learning classifiers to diagnose COVID‐19 in X‐ray images. arXiv Preprint arXiv. 2020. [Google Scholar]
- 40. Sethy PK, Behera SK. Detection of Coronavirus Disease (COVID‐19) Based on Deep Features, 2020. 10.20944/preprints202003.0300.v1 [DOI] [Google Scholar]
- 41. Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of covid‐19 cases using deep neural networks with X‐ray images. Comput Biol Med. 2020;121:103792. 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Butt C, Gill J, Chun D, Babu BA. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020;220:1‐7. 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li X, Li C, Zhu D. COVID‐Xpert: an AI powered population screening of COVID‐19 cases using chest radiography images. ArXiv. 2020. [Google Scholar]
- 44. Wang L, Wong A. COVID‐net: a tailored deep convolutional neural network design for detection of COVID‐19 cases from chest radiography images. ArXiv. 2020; arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in covid‐chestxray‐datase at https://github.com/ieee8023/covid-chestxray-dataset/, reference number 25 and Italian Society of Medical and Interventional Radiology at https://www.sirm.org/en/2020/03/30/covid‐19‐management‐strategy‐in‐radiology/, reference number 26.


