Abstract
Coronavirus disease 2019 (COVID‐19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). In China, Yinqiao powder is widely used to prevent and treat COVID‐19 patients with Weifen syndrome. In this study, the screening and verification of active ingredients, target selection and DisGeNET scoring, drug–ingredient–gene network construction, protein–protein interaction network construction, molecular docking and surface plasmon resonance (SPR) analysis, gene ontology (GO) functional analysis, gene tissue analysis, and kyoto encyclopedia of genes and genomes (KEGG) pathway analysis were used to explore the active ingredients, targets, and potential mechanisms of Yinqiao powder in the treatment of COVID‐19. We also predicted the therapeutic effect of Yinqiao powder using TCM anti‐COVID‐19 (TCMATCOV). Yinqiao powder has a certain therapeutic effect on COVID‐19, with an intervention score of 20.16. Hesperetin, eriodictyol, luteolin, quercetin, and naringenin were the potentially effective active ingredients against COVID‐19. The hub‐proteins were interleukin‐6 (IL‐6), mitogen‐activated protein kinase 3 (MAPK3), tumor necrosis factor (TNF), and tumor protein P53 (TP53). The potential mechanisms of Yinqiao powder in the treatment of COVID‐19 are the TNF signaling pathway, T‐cell receptor signaling pathway, Toll‐like receptor signaling pathway, and MAPK signaling pathway. This study provides a new perspective for discovering potential drugs and mechanisms of COVID‐19.
Keywords: COVID‐19, molecular docking, network pharmacology, signaling pathway, Yinqiao powder
1. INTRODUCTION
In December 2019, several patients with severe acute respiratory syndrome, which is now called COVID‐19 by the World Health Organization, were found in Wuhan, China (Sun, Lu, Xu, Sun, & Pan, 2020). The disease is highly transmissible, pathogenic, and recurrent, and large outbreaks have occurred in many countries and regions around the world (Arshad, Baloch, Ahmed, Arshad, & Iqbal, 2020). On December 18, 2020, China reported a total of more than 95,491 cases, and the total number of cases worldwide has exceeded 50 million, affecting more than 200 countries and regions. Whole‐genome sequencing and phylogenetic analysis indicate that COVID‐19 is caused by SARS‐CoV‐2, which is related to the phylogeny of the SARS bat virus, suggesting that bats may be the main host. The main source and the intermediate source of the transfer to humans are unclear, but the rapid transfer from person to person has been widely confirmed (Shereen, Khan, Kazmi, Bashir, & Siddique, 2020). To make matters worse, specific antiviral drugs or vaccines for COVID‐19 are still undergoing clinical trials.
After more than 2,000 years of development, traditional Chinese medicine (TCM) has formed a comprehensive and unique system from disease diagnosis to prognosis, which plays an important role in the prevention and treatment of human infectious diseases (Sun, Sun, Yan, Li, & Xin, 2020). In addition to strict prevention and control measures, many provinces in China have issued TCM prevention and treatment plans for COVID‐19 in response to the outbreak of COVID‐19, which has achieved remarkable results. Big data mining analysis found that Yinqiao powder was the basic formulation for the Weifen syndrome of COVID‐19 (Fan et al., 2020). Pharmacological studies have also shown that Yinqiao powder has an antitussive and expectorant effect, improves lung function, alleviates acute lung injury, alleviates pulmonary fibrosis, enhances the antiviral immune response, and alleviates the adverse effects of modern drugs (Rothan & Byrareddy, 2020). In fact, the active ingredients of Yinqiao powder are complicated. Some active ingredients found in Yinqiao powder by high‐performance liquid chromatography analysis may have anti‐COVID‐19 effects, such as rutin and hesperidin (Shu et al., 2012). Rehman, AlAjmi, and Hussain (2020) and Liang et al. (2020) found that the binding affinity of rutin for the main protease (3CLpro) of SARS‐CoV‐2 was much higher than that of chloroquine, hydroxychloroquine, and remdesivir, and it may inhibit COVID‐19 by downregulating IL‐6. Balmeh et al. found that hesperidin has an inhibitory effect on human angiotensin‐converting enzyme 2 (ACE2), TMPRSS2, GRP78, and AT1R receptors and may have the ability to treat COVID‐19 infection (Balmeh, Mahmoudi, Mohammadi, & Karabedianhajiabadi, 2020). Although some biologically active substances and their molecular mechanisms for the treatment of COVID‐19 have been under investigation, there is a lack of in‐depth exploration of Yinqiao powder in the treatment of COVID‐19.
Network pharmacology is an emerging discipline. Through the “disease–gene–protein–drug” interaction, the effect of drugs on disease can be systematically and comprehensively observed, thereby revealing the complex relationship between Chinese medicine and disease (Lin et al., 2019). This research method shows integrity and systematic characteristics, which are consistent with the overall theory of Chinese medicine (Li & Zhang, 2013). In previous studies, network pharmacology has been successfully used to reveal the potential active ingredients, targets, and mechanisms of Chinese medicine in disease treatment, such as the Maxing Shigan Decoction (Luo et al., 2020) and the Huashi Baidu Formula for COVID‐19 (Xun, Jialei, Shaoju, & Bin, 2020).
In this study, we used absorption, distribution, metabolism, and excretion (ADME) information to screen the potential active ingredients and targets of Yinqiao powder for COVID‐19 in the TCMSP database, verified the active ingredients with high‐resolution liquid chromatography‐mass spectrometry (LC‐MS), and predicted the efficacy of Yinqiao powder using TCMATCOV. The next step was the use of the disease‐association score of the DisGeNET database to screen the co‐genes of COVID‐19 and Yinqiao powder. Then, we used the STRING database to perform protein–protein interaction (PPI) and topological analyses, performed molecular docking of the identified hub‐proteins with their corresponding active ingredients to distinguish the binding activity and site, and used surface plasmon resonance (SPR) analysis to evaluate the affinity. Finally, we performed GO functional, tissue location, and KEGG analyses to identify the mechanism of Yinqiao powder in the treatment of COVID‐19, which is shown in Figure 1. We expect that the results will enhance our understanding of the effective, potential active ingredients of Yinqiao powder for COVID‐19 and reveal the biological basis of pharmaceutically acceptable targets, thereby promoting the development of effective COVID‐19 therapeutic drugs.
FIGURE 1.
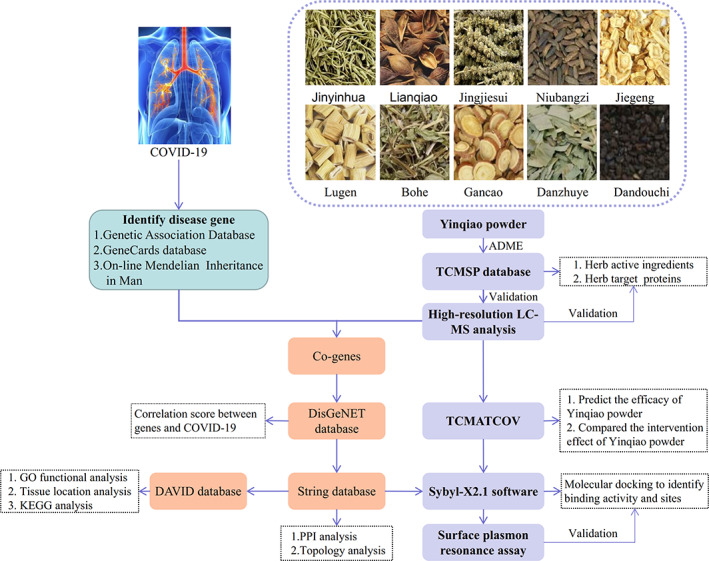
Flow chart of this study [Colour figure can be viewed at wileyonlinelibrary.com]
2. MATERIALS AND METHODS
2.1. Acquisition of active ingredients
The TCMSP database (http://lsp.nwu.edu.cn/, Version 2.3) covers 499 kinds of Chinese medicines registered in the Chinese Pharmacopoeia and provides ADME information on the active ingredients of common Chinese medicines (Ru et al., 2014). Totally, 176 ingredients of Yinqiao powder were obtained from the TCMSP database, and the potential active ingredients were further screened according to ADME. Oral bioavailability (OB) refers to the relative amount of the drug taken from the liver to the blood circulation after oral drug absorption through the gastrointestinal tract. It is an objective indicator for evaluating drug absorption and affects the effectiveness of clinical drug trials (Xu et al., 2012). The TCMSP database is based on a dataset composed of 805 different drugs or drug‐like molecules. The internal model of the OBioavail 1.1 algorithm was used to calculate the OB value, and ingredients with OB ≥ 70% were screened as potential candidate compounds (Sietsema, 1989). In particular, if an active ingredient was present in two or more different drugs for Yinqiao powder, the active ingredient only needed to have an oral availability of OB ≥ 35%. Drug‐likeness (DL) reflects the pharmacokinetic characteristics of compounds in humans. The TCMSP database uses the Tanimoto parameters to calculate the DL value, which helps to screen out the highly effective active drug ingredients and improve the hit rate of candidate drug molecules (Yongye & Medina‐Franco, 2013). We selected DL ≥ 0.18 to screen potential active ingredients and downloaded their structure in the mol2 format.
2.2. High‐resolution LC‐MS analysis
To verify the active ingredients screened above, LC‐MS technology was used to analyze the Yinqiao powder. The decocting program of Yinqiao powder adopts the best strategy screened by the previous orthogonal experiment (Shu et al., 2012). In short, according to the records of Systematized Identification of Warm (Pathogen) Diseases, we chose Forsythiae fructus (Lianqiao, 30 g), Lonicera japonica (Jinyinhua, 30 g), Platycodonis radix (Jiegeng, 18 g), Lophatheri herba (Danzhuye, 12 g), Glycyrrhizae radix et rhizoma (Gancao, 15 g), Sojae semen praeparatum (Dandouchi, 15 g), Great Burdock Achene (Niubangzi, 18 g), and reed rhizome (Lugen, 30 g, purchased from the First Affiliated Hospital of Guangzhou University of Chinese Medicine), then added 12× water, soaked for 30 min, decocted for 10 min, added Schizonepetae herba (Jingjiesui, 12 g) and Menthae haplocalycis herba (Bohe, 18 g), and decocted for 5 min. The mixture was filtered, and 200 μl of the filtrate was mixed with 1 ml of 80% methanol liquid (methanol:water = 8:2); this mixture was vortexed to mix evenly and centrifuged for 10 min at 4°C at 20,000 rpm. Extracts were analyzed by high‐resolution LC‐MS (Q‐Exactive, Thermo Scientific™ Orbitrap Fusion™). The specific protocol was similar to that in previous studies with slight modifications (Fagbohun et al., 2020). Briefly, the chromatographic column was an RP C18 column of size 150 mm × 2.1 mm, 1.8 μm. The mobile phase consisted of solvent A composed of 0.1% (v/v) formic acid in water and B composed of 0.1% (v/v) formic acid in acetonitrile. The separation started with 98% of eluent A, dropping linearly to 5% within 20 min. The retention time of the drug was 5 min, and after 1 min, it increased again to 98% within 4 min. The total run time was 30 min, the flow rate was 0.3 ml/min, the injection volume was 5 μl, and the column temperature was maintained at 35°C. The LC‐MS detection and analysis were conducted in full mass spectrometry‐selected ion monitoring mode followed by data‐dependent MS2 (dd‐MS2), which was equipped with positive and negative polarity switching scanning from m/z 150.0 to 2,000. The capillary temperature was 300°C, the sheath flow was 40 arb, the spray voltage was 3,800 V, and the auxiliary temperature was 350°C. The overall mass resolution for MS was set to 70,000 and the mass resolution for dd‐MS2 was set to 17,500. The detailed analysis method is shown in Supplementary Material 1. The data were matched with the mzCloud (https://www.mzcloud.org/), mzVault (https://mytracefinder.com/tag/mzvault/), and MassList (www.maldi-msi.org/mass) databases.
2.3. Acquisition of protein targets of active ingredients
The TCMSP database contains information on 3,311 protein targets, and the relationship between these protein targets and drugs is obtained using the SysDT prediction algorithm (Zhang et al., 2019). A total of 190 protein targets of the verified potentially active ingredients was obtained and downloaded from the TCMSP database. At the same time, published articles were searched to identify the potential protein targets of the active ingredients.
The UniProt database contains three subdatabases: Swiss‐Port, PRI‐PSD, and TrEMBL, and is the most comprehensive protein database that contains information (Bateman et al., 2019). The UniProt database (http://www.uniprot.org/uniprot/, updated on June 22, 2017) was used to retrieve the protein ID and gene name of the potential active ingredient and was limited to Homo sapiens, thereby obtaining the gene of the potential active ingredient. Then, the structure of the corresponding protein was downloaded from the PDB database (http://www.rcsb.org/) in the PDB format.
2.4. Screening of COVID‐19‐related gene targets
The Genetic Association Database (GAD, https://geneticassociationdb.nih.gov/) collects, standardizes, and archives research data on human genetic associations, making it easy for scientific access (Becker, Barnes, Bright, & Wang, 2004). The GeneCards database (http://www.genecards.org/) integrates gene‐centric data from approximately 150 data sources, including genome, transcriptome, proteome, genetics, as well as clinical and functional information (Rebhan, Chalifa‐Caspi, Prilusky, & Lancet, 1997). Online Mendelian Inheritance in Man (OMIM, http://www.ncbi.nlm.nih.gov/omim) is a comprehensive database of human genes and genetic traits. This database focuses on the relationship between phenotype and genotype and contains information about all Mendelian inherited diseases and more than 12,000 human genes (Schorderet, 1991). We used the keywords “coronavirus” and “SARS‐coronavirus” in the GAD, GeneCards, and OMIM databases to search for the COVID‐19‐related genes, and then removed duplicate genes and false positive genes. The gene targets of the disease were matched with the gene targets of Yinqiao powder to obtain co‐genes, which were used as the potential gene targets of the active ingredient of Yinqiao powder to treat COVID‐19.
2.5. Correlation score between gene targets and COVID‐19
The DisGeNET database (http://www.disgenet.org/web/DisGeNET/menu, version 5.0) is a detection platform that can be used to study the molecular basis of human disease and its complications, analyze the characteristics of disease genes, and assess the correlation between genes and disease (Pinero et al., 2020). We obtained 5,742 associations between genes and diseases in the DisGeNET database, and screened genes associated with viral diseases in a disease class.
2.6. Construction and analysis of the drug–ingredient–gene target network
The potential gene targets of Yinqiao powder's active ingredients for treating COVID‐19 and the active ingredients of Yinqiao powder were imported into Cytoscape software (version 3.4.0) to construct the Yinqiao powder's active ingredients and co‐gene target network. Topological analysis was used to analyze the connection between the active ingredients and the targets.
2.7. Classic anti‐COVID‐19 prescription validation
The TCMATCOV (http://tcmatcov.bbtcml.com, version 1.0, co‐invented by the Institute of Chinese Materia Medica China Academy of Chinese Medical Sciences and Beijing Proteome Research Center) platform can predict the efficacy of TCM against COVID‐19 (Guo et al., 2020). It uses the quantitative evaluation algorithm of multitarget drugs for the disturbance of disease networks to predict the potential drug efficacy of the target drugs. Based on the TCMATCOV platform, Qingfei Paidu decoction was used as a positive control and Banxia Baizhu Tianma decoction was used as a negative control to predict the interference score of Yinqiao powder on COVID‐19. Qingfei Paidu decoction has been found to have a therapeutic effect on COVID‐19 through in silico and experimental studies (Yang et al., 2020). Banxia Baizhu Tianma decoction has been used to treat vertigo (Guo, Su, Wang, Luo, & Lai, 2017).
2.8. Construction and analysis of the PPI network
The STRING database (https://string-db.org/, version 10.5) is a database containing a large number of protein interaction relationships, involving a total of 9,643,763 proteins and 1,380,838,440 interactions, which are detected by experiments or predicted by bioinformatics methods (Szklarczyk et al., 2017). We imported the potential gene targets of Yinqiao powder into the STRING database, restricted the species to Homo sapiens, and obtained the protein interaction relationship for further study. The medium confidence was set to 0.4, and the results were saved in the TSV format. Node 1, node 2, and the combined score information were imported into Cytoscape software to perform the interactive network. We also used the Generate style from the statistics tool in Cytoscape to set the node size.
2.9. Molecular docking
The Sybyl software (Tripos, version X2.1) was used for molecular docking. This module has the characteristics of high program running speed, accuracy, and reliability. Surflex‐Dock scores are expressed in –log10 (K d) units to represent binding affinities (Jain, 1996). Surflex‐Dock contains the following information: First, the total score of Surflex‐Dock is expressed by –log (K d). Second, “crash” refers to the degree to which a ligand improperly penetrates into a protein and the degree of interpenetration between ligand atoms separated by a rotatable bond. It is advantageous for the crash score to be close to 0. Third, “polar scoring” refers to the contribution of polar interaction to the total score, and it can be used to exclude docking results without hydrogen bonding.
3CLpro is an established drug target for the design of inhibitors to stop viral replication (Kneller et al., 2020). ACE2 is the binding protein for SARS‐CoV‐2 to invade the human body (He, Tao, Yan, Huang, & Xiao, 2020). Therefore, the top four proteins recognized by the PPI network, ACE2 proteins (https://covid-19.uniprot.org/), and 3CLpro (https://www.rcsb.org/structure/6M2Q) were used for molecular docking with Sybyl software to identify the binding ability of the active ingredients and targets and screen potential targets and efficient active ingredients. At the same time, we used the commonly used clinical antiviral drug interferon alpha (IFN‐α) (https://pubchem.ncbi.nlm.nih.gov/compound/71306834) as a positive control drug. Molecular docking was performed according to the Sybyl docking manual (https://v.youku.com/v_show/id_XNDU0Mjg0NTYw.html?refer=seo_operation.liuxiao.liux_00003308_3000_YvmIba_19042900). First, a docking file was created. Second, docking ligands, including removed water molecules, added polar hydrogen atoms, and extracted original ligands, were used to identify the binding sites. Third, molecular docking was performed, and the results were saved. Fourth, the docking score was analyzed to evaluate the binding activity. Finally, Schrodinger Suites (version 2019‐1, Materials) was used to draw the 3D map of the corresponding molecular docking.
2.10. SPR assay
To verify the binding ability of ACE2 to the most effective active ingredients screened by molecular docking, SPR was performed with a NeoSPR‐M100 surface plasmon resonance instrument (Hangzhou Neoline Technology Co. Ltd., China). The ACE2 protein (DD04531k1g0wj, Cusabio Biotech Co. Ltd.) was fixed on a carboxyl sensor chip (NS‐SCHC, Hangzhou Neoline Technology Co. Ltd., China) with an amide bond, and the active ingredients at 2.73, 10.93, 21.85, 61.19, 87.4, and 139.86 μM were injected sequentially into the chamber. The concentration of ACE2 was 25 μg/ml, the pH was 4.0, the volume was 100 μl, the flow rate was 20 μl/min, and the amount was 292 PU. The flow rate of the active ingredients was 20 μl/min, the binding time and dissociation time were both 150 s, and the chip was regenerated with 40 mM NaOH. The data were retrieved and analyzed using the TraceDrawer software, assuming a 1:1‐binding model.
2.11. GO functional analysis and tissue location analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/, version 6.8) provides systematic and comprehensive biological function annotation information, including biological process (BP), cellular component (CC), and molecular function (MF), for genes or proteins and can find the most significantly enriched biological annotations (Dennis et al., 2003). The potential genes for the treatment of COVID‐19 with the active ingredients of Yinqiao powder were imported into the DAVID database, limiting the species to Homo sapiens, GO functional analysis and tissue location analysis were performed, and the results were saved. Finally, GO terms with a p‐value ≤.05 and FDR ≤ .05 were selected for further analysis. The top biological processes and tissue positioning were screened, and GraphPad Prism 5.0 software was used to draw biological processes and tissue location maps.
2.12. KEGG analysis and signal pathway integration
The UniProt ID of Yinqiao powder was imported into the KEGG database (http://www.kegg.jp/), and the species was limited to Homo sapiens to obtain the signaling pathways of Yinqiao powder in the treatment of COVID‐19. The KEGG‐enriched advanced bubble chart was designed using Omicshare Tools (https://www.omicshare.com/) and integrated common signal paths.
3. RESULTS
3.1. Screening of active ingredients in Yinqiao powder
A total of 222 active ingredients of Yinqiao powder were obtained from the TCMSP database. According to the OB and DL of Yinqiao powder, 30 active ingredients were selected, including hesperetin, licopyranocoumarin, luteolin, quercetin, β‐sitosterol, β‐carotene, and naringenin. Details are shown in Table S1. However, only hesperetin, eriodictyol, luteolin, quercetin, and naringenin were verified by mass spectrometry, as shown in Table 1, Figure S1, and Supplementary Material 2.
TABLE 1.
Qualitative analysis of chemical components in Yinqiao powder
| No. | Molecule ID | Name | Formula | Structure | Molecular weight | RT (min) | Area | Annotation source: mzCloud search | mzCloud best match (%) | Annotation source: mzVault search | Annotation source: MassList match |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MOL002341 | Hesperetin | C16 H14 O6 |
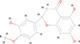
|
302.07898 | 14.464 | 666,388.8127 | Full match | 88.5 | Full match | Full match |
| 2 | MOL005190 | Eriodictyol | C15 H12 O6 |
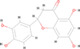
|
288.06324 | 13.345 | 770,446.0006 | Full match | 76 | Full match | Full match |
| 3 | MOL000006 | Luteolin | C15 H10 O6 |
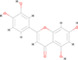
|
286.04784 | 15.247 | 1,440,073.476 | Full match | 78.9 | Partial match | Full match |
| 4 | MOL000098 | Quercetin | C15 H10 O7 |
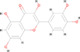
|
302.04253 | 12.725 | 300,998.8408 | Full match | 86.2 | Full match | Full match |
| 5 | MOL004328 | Naringenin | C15 H12 O5 |
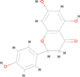
|
272.06841 | 14.208 | 714,046.1477 | Full match | 90.3 | Full match | Full match |
Abbreviations: area, compound peak area; mzCloud best match, MzCloud database matching score (the higher the value, the higher the credibility of the result identified); RT, chromatographic retention time.
3.2. Target prediction
A total of 190 gene targets were identified by the above five active ingredients of Yinqiao powder in the UniProt database. By comparing 349 gene targets related to COVID‐19 in the Genetic Association Database, GeneCards database, and OMIM database, 43 co‐gene targets, such as PTGS1, PTGS2, and DPP4, that may be related to Yinqiao powder for the treatment of COVID‐19 were selected. Details are shown in Table S2.
3.3. DisGeNET score between co‐genes and COVID‐19
In the DisGeNET database, 209 associations were obtained. After deleting duplicate and false results, 33 genes were associated with viral diseases or coronavirus infections, which are shown in Table S3. There were eight genes with DisGeNET scores ≥ 0.1 (TNF, TP53, IL6, IL10, STAT1, IFNG, CXCL10, and BCL2), and two genes were related to COVID‐19 (MCL1 and ACE2).
3.4. Construction of drug–ingredient–gene target network
Information on the active ingredients and gene targets of Yinqiao powder was imported into Cytoscape software to construct a drug–ingredient–gene target network, as shown in Figure 2a. There were 6 drug nodes, 5 active ingredient nodes, 43 gene nodes, and 89 kinds of nodes that were connected. The triangle nodes represent the potential anti‐COVID‐19 drugs of Yinqiao powder, which include Jinyinhua, Lianqiao, Jingjiesui, Jiegeng, Bohe, and Gancao. The square‐shaped nodes represent the active ingredients of Yinqiao powder, which include hesperetin, eriodictyol, luteolin, quercetin, and naringenin and are considered to have potential anti‐COVID‐19 effects. The elliptical nodes represent the genes, and the edges represent the correlation between drugs, active ingredients, and gene targets. The results showed the multicomponent, multitarget characteristics of Yinqiao powder against COVID‐19.
FIGURE 2.
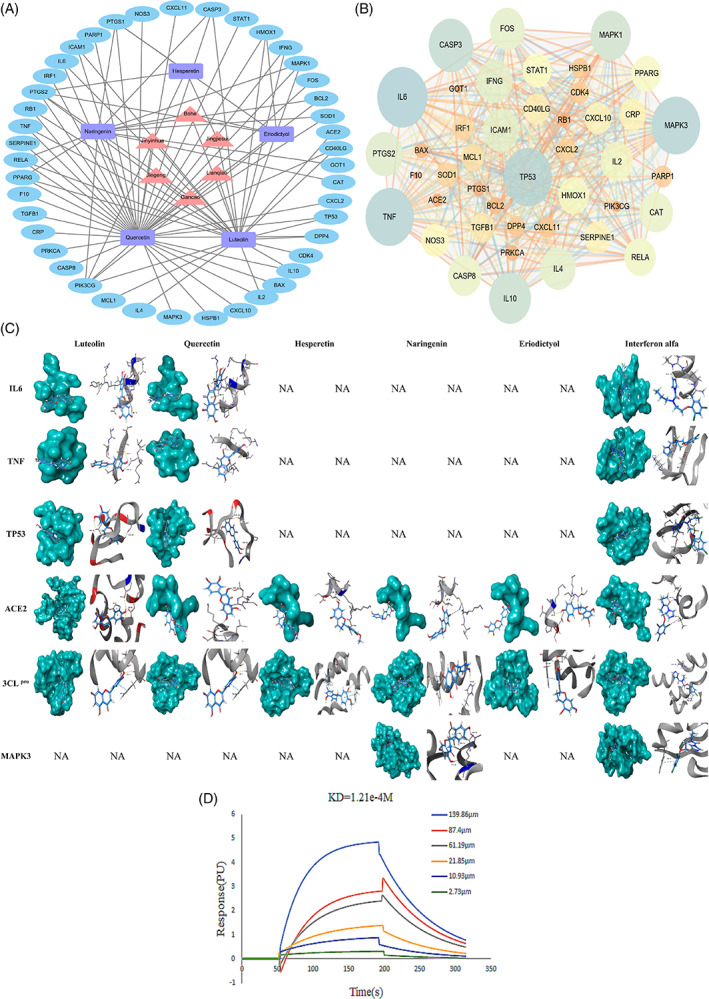
Drug–ingredient–gene target network, protein interaction network, and molecular docking of Yinqiao powder. (a) Drug–ingredient–gene target network of Yinqiao powder. The triangle (▲) is the drug of Yinqiao powder, the rectangle (■) is the main active ingredients of Yinqiao powder, the oval nodes (●) is the potential targets for treating COVID‐19 of Yinqiao powder. (b) Protein interaction network of Yinqiao powder. The size of the node represents the value of the degree (low values to small sizes). (c) Molecular docking. (d) Surface plasmon resonance (SPR) analysis of luteolin and ACE2 [Colour figure can be viewed at wileyonlinelibrary.com]
3.5. Prediction of Yinqiao powder for the treatment of COVID‐19 by TCMATCOV
Using TCMATCOV, we compared the intervention effect of Yinqiao powder with Qingfei Paidu decoction and Banxia Baizhu Tianma decoction on COVID‐19. The platform uses network topology analysis to evaluate drug efficacy, including the average connectivity, average shortest path, connectivity centrality, and tightness centrality. Negative average connectivity, connectivity centrality, and tightness centrality represent the destructiveness of the drug for the disease target network. A positive value of the average shortest path indicates the destructiveness of the drug to the disease target network. The larger the value of these indicators, the stronger the destructive effect of the target drug on the disease target network. Based on the above four values, the total interference score was calculated and the disturbance effect of the drug on the disease target network was evaluated at the overall level. The higher the total interference score, the higher the degree of damage to the network stability of the drug. The results suggested that Yinqiao powder and Qingfei Paidu decoction had very close intervention effects on COVID‐19, with an intervention score of 20.16 versus 23.28, which was significantly higher than that of Banxia Baizhu Tianma decoction (14.52) (Table S4). In addition, from the four dimensions of average connectivity, shortest path, connectivity centrality, and tightness centrality, it can be seen that Yinqiao powder has a significant intervention effect on COVID‐19 and that the effect is similar to the effect of Qingfei Paidu decoction, suggesting that Yinqiao powder could be used to treat COVID‐19.
3.6. Construction and analysis of the PPI network
The PPI network is shown in Figure 2b. The nodes represent proteins, and the lines represent the associations between proteins. A total of 43 nodes and 440 edges were involved. The average node degree was 20.47. Topological analysis found that four proteins ranked in the top four in degree, betweenness centrality, and closeness centrality, namely IL6, mitogen‐activated protein kinase 3 (MAPK3), tumor necrosis factor (TNF), and tumor protein P53 (TP53) (Table S5).
3.7. Molecular docking
The molecular docking results in comparison to those of IFN‐α, which is currently a common treatment for COVID‐19, are highlighted in Table S6 and Figure 2c. It is generally believed that when the docking score is above 1.0, the molecule has a certain binding activity between the ingredients and the protein targets. A docking score >3.0 indicates that the ingredients have good binding activity to the protein targets, while a docking score >5.0 indicates strong binding activity (Lin et al., 2019). The crash scores were close to 0, indicating that the docking results were reliable. The polar score and total score were similar, indicating that the total score basically reflects the docking of the ingredients and protein targets. Luteolin and ACE2 obtained the highest docking total score, mainly by hydrogen bonds or π–π bonds, which was higher than highest docking total score for the binding of IFN‐α and ACE2. SPR assays indicated that luteolin is bound to ACE2 with a dissociation constant (K d) of 121 μM (Figure 2d). At the same time, 10 molecular docking results with a docking total score of 3.0–5.0 accounted for 58.82%, including luteolin and IL6, TP53, 3CLpro; quercetin and IL6, TP53, 3CLpro; hesperetin and 3CLpro; naringenin and MAPK3, 3CLpro; eriodictyol and 3CLpro. These five active ingredients have good binding ability with 3CLpro, which means that they may be highly effective active ingredients for Yinqiao powder to treat COVID‐19.
3.8. GO enrichment analysis
GO enrichment analysis refers to a directed acyclic graph composed of the number of proteins or genes at a certain functional level, including BP, MF, and CC. There were 455 BPs involved with the genes. The top five BPs (Figure 3a) were the regulation of apoptosis (25 genes/58.14%), regulation of programmed cell death (25 genes/58.14%), regulation of cell death (25 genes/58.14%), regulation of cell proliferation (25 genes/58.14%), and response to organic substances (23 genes/53.49%). There were 37 CCs involved with the genes, and the top 2 CCs (Figure 3b) were extracellular regions (19 genes/44.19%) and membrane‐enclosed lumens (18 genes/41.86%). There were 53 MFs involved with the genes. The top three MFs (Figure 3c) were identity protein binding (11 genes/25.58%), protein dimerization activity (10 genes/23.26%), and cytokine activity (10 genes/23.26%). Tissue analysis (Figure 3f) showed that genes were mainly highly expressed in nine areas, of which the top four areas were placenta (15 genes/34.88%), lung (10 genes/23.26%), liver (8 genes/18.60%), blood (8 genes/18.60%).
FIGURE 3.
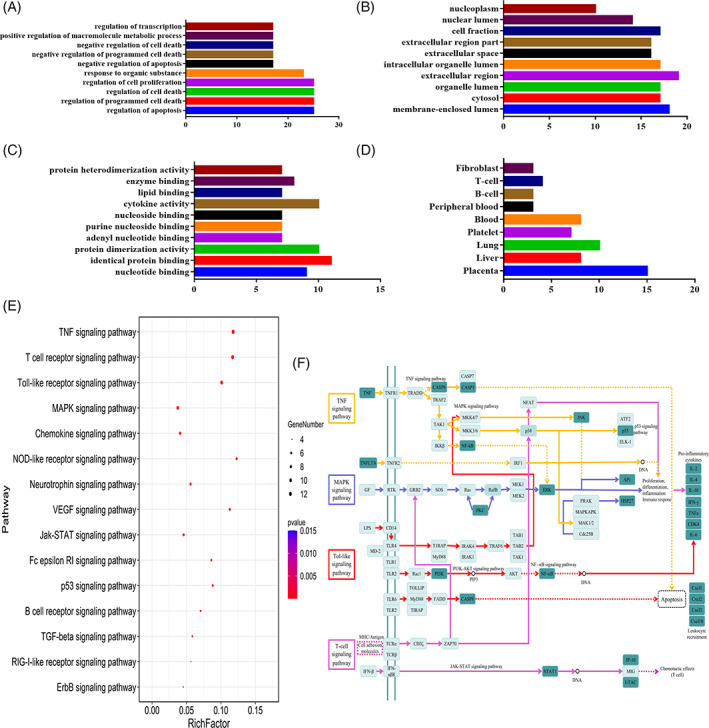
Gene ontology (GO) enrichment and KEGG pathway analyses of potential targets for treating COVID‐19 from main active ingredients of Yinqiao powder: (a) biological process (BP); (b) cellular component (CC); (c) molecular function (MF); (d) gene location; (e) enriched KEGG pathways of potential targets for treating COVID‐19 from main active ingredients of Yinqiao powder; (f) the potential anti‐COVID‐19 pathway of the main active ingredients of Yinqiao powder (↑‐arrows indicate the promotion effect, ⊤‐arrow indicates the inhibition effect). The target is marked in light color, and the potential target of Yinqiao powder for COVID‐19 is marked in dark color [Colour figure can be viewed at wileyonlinelibrary.com]
3.9. KEGG pathway analysis
KEGG analysis results showed that the gene targets of Yinqiao powder for COVID‐19 involve a total of 57 signaling pathways. The top 15 important signaling pathways are shown in Figure 3e. The top four signaling pathways closely related to COVID‐19 were the TNF signaling pathway (13 genes/30.23%), the T‐cell receptor signaling pathway (12 genes/27.91%), the Toll‐like receptor signaling pathway (11 genes/25.58%), and the MAPK signaling pathway (10 genes/23.26%). The main signaling pathway of Yinqiao powder in the treatment of COVID‐19 is shown in Figure 3f, which involves a total of 24 gene targets, accounting for 55.81% of the co‐genes. Some gene targets, such as p38, JNK, and MKK3, play a role in multiple pathways, suggesting that they may be the key genes involved in COVID‐19.
4. DISCUSSION
Yinqiao powder comes from the Systematized Identification of Warm by Wu Jutong in the Qing Dynasty. Using TCMATCOV, we also found that Yinqiao powder has a certain therapeutic effect on COVID‐19, with an intervention score of 20.16. Drug–ingredient–gene target network analysis revealed that Jinyinhua, Lianqiao, Jingjiesui, Jiegeng, Bohe, and Gancao have potential anti‐COVID‐19 effects. Some of these drugs have been confirmed to have obvious advantages in improving the clinical symptoms of COVID‐19 patients. For example, Zhang, Lei, Xu, Wei, & Hu (2020) found that antiviral drugs combined with Jinyinhua oral solution have a significant benefit in improving fever, fatigue, and cough symptoms and reducing lung injury in patients with COVID‐19. A retrospective study of historical classics and Chinese programs to control COVID‐19 found that Jinyinhua, Lianqiao, and Gancao were commonly used to control COVID‐19 (Luo et al., 2020). Zhang, Liang, Kong, and Xiao (2019) found that moxifloxacin combined with Jiegeng was significantly better than moxifloxacin alone in improving cough symptoms in patients with pneumonia and could reduce the WBC and CRP levels in patients.
In our study, 30 ingredients of Yinqiao powder were screened from the TCMSP database based on the OB and DL, and five ingredients, namely hesperetin, eriodictyol, luteolin, quercetin, and naringenin, were verified by LC‐MS analysis. All of them were flavonoids and were associated with potential anti‐COVID‐19 genes. A previous study confirmed that flavonoids have antiinflammatory, antioxidant, antiproliferative, antithrombotic, cardioprotective, and neuroprotective effects (Gujar & Wairkar, 2020). In addition, some of these ingredients have been proven to have anti‐COVID‐19 effects. For example, hesperetin was found to have high affinity for the spike protein and the helicase and protease sites on the ACE2 receptor (Ngwa et al., 2020). Luteolin was found in a variety of anti‐COVID‐19 Chinese medicine prescriptions, such as Maxing Shigan decoction (Wang et al., 2020) and Tanreqing injection (Kong et al., 2020). Kong et al. (2019) found that luteolin inhibits inflammation by inhibiting the expression of cyclic adenosine monophosphate‐phosphodiesterases (PDEs) or PDE4 activity and intracellular cell adhesion molecule (ICAM)‐1 and soluble ICAM‐1 in serum in pulmonary microvascular endothelial cells. Quercetin was also found in a variety of anti‐COVID‐19 Chinese medicine prescriptions, such as Xiaochaihu decoction (Sun, Zhang, Liu, & Sun, 2020) and Huoxiang Zhengqi oral liquid (Deng et al., 2020). Wang, Leng, Guo, and Jin (2019) confirmed that quercetin could not only reduce the counts of lymphocytes, monocytes, and neutrophils in mice with pneumonia but also downregulate the levels of serum TNF‐α, IL‐6, and IL‐1β to inhibit the inflammatory response, which may be related to the inhibition of the IKK/nuclear factor‐κB (NF‐κB)/IκB signaling pathway. A previous study also found that naringenin could reduce lung injury by inhibiting the expression of inflammatory cytokines, including IL‐6, IL‐1β, TNF‐α, and TGF‐β, mediated by autophagy and pulmonary fibrosis (Lin, Tan, Kan, Xiao, & Jiang, 2018). Therefore, flavonoids may be the main active ingredient in treatments used for COVID‐19.
In our study, PPI analysis found that there were four anti‐COVID‐19 hub‐proteins, namely IL6, MAPK3, TNF, and TP53, some of which have been confirmed by clinical trials. Clinical studies have found that the accelerated development of the disease in patients with COVID‐19 was closely related to the high inflammation state (Saleh, Peyssonnaux, Singh, & Edeas, 2020). Han et al. (2020) also found that the levels of serum cytokines in patients with COVID‐19, including TNF‐α, IFN‐γ, IL‐2, IL‐4, IL‐6, IL‐10, and CRP, were higher than those in healthy people. Huang found that neutrophil counts, proinflammatory cytokines (TNF‐α, IL‐6, IL‐1β, and IL‐2R), and coagulation dysfunction biomarkers (D‐dimer, PT, and Fbg) were closely related to the adverse clinical outcomes of COVID‐19 patients through logistic regression analysis (Huang et al., 2020). Xiong et al. (2020) performed transcriptome sequencing of RNA isolated from bronchoalveolar lavage fluid and peripheral blood mononuclear cell specimens of COVID‐19 patients and found that SARS‐CoV‐2 infection may cause lymphocyte apoptosis and that TP53 was an important gene in regulating apoptosis. Di Paola et al. (2009) found that the MAPK3 protein is involved in the inflammatory response of lung injury, and the inhibition of MAPK3/MAPK1 can reduce the level of proinflammatory cytokines, such as TNF‐α and IL‐1β, in lung injury. Therefore, the antagonistic effect of Yinqiao powder on COVID‐19 may be closely related to the regulation of these inflammation‐related proteins.
To further screen the potential active ingredients, we used molecular docking to identify the binding activity of the active ingredients and anti‐COVID‐19 protein. We found that luteolin and ACE2 have the best binding ability, mainly through hydrogen bonds or π–π bonds, which means that luteolin has a certain role in preventing COVID‐19 infection. Jimilihan et al. (2020) also confirmed that luteolin has good binding activity with ACE2 and that hydrogen bonding plays a key role in the recognition and stability of the active ingredients and proteins. SPR analysis showed that luteolin combined with ACE2 had a K d of 121 μM, and the corresponding –log10 (K d) value was 3.917, which was lower than the total score of 5.63. This indicated that the molecular docking results obtained by Surflex‐Dock may overestimate the actual affinity between the ingredients and the targets. This phenomenon has also been reported in previous studies (Jain, 1996). However, this affinity was still far lower than that of COVID‐19 and ACE2 (K d 121 μM vs. 15 nM) (Wrapp et al., 2020). In fact, the role of Yinqiao powder in preventing and treating COVID‐19 not only depends on the competitive combination of luteolin and ACE2 but also may be related to the joint control of the inflammatory response and antiviruses with multiple ingredients and targets. Interestingly, we also discovered that luteolin and IL6, TP53, and 3CLpro; quercetin and IL‐6; TP53 and 3CLpro; hesperetin and 3CL pro; naringenin and MAPK3 and 3CLpro; and eriodictyol and 3CLpro have good affinity, which means that they may be the effective active ingredients of Yinqiao powder to treat COVID‐19. These results have been partially confirmed by previous studies. For example, Palombo et al. (2019) confirmed that luteolin could inhibit STAT3 activity in mice to counteract the proliferative effect of the IL‐22/IL‐6 signaling pathway and could be used as a drug candidate for the treatment of inflammation and proliferative diseases. Lin et al. (2020) confirmed that quercetin could inhibit the expression of inflammatory factors induced by LPS through mouse experiments, such as cyclooxygenase‐2 (PTGS2) and IL6. Glinsky (2020) found that quercetin, luteolin, and eriodictyol were structurally similar and could be developed as inhibitors for SARS‐CoV‐2 infection through drug docking screening and gene expression profile analysis. However, the direct inhibitory effects of hesperetin, eriodictyol, luteolin, quercetin, and naringenin on SARS‐CoV‐2 have not been verified in clinical trials or animal experiments.
GO analysis revealed that the main BP involved in the anti‐COVID‐19 gene was the regulation of apoptosis, the main CC was the extracellular region, and the main MF was protein binding. Some of these results have been verified by previous studies. For example, Saleh et al. (2020) believe that the highly inflammatory state of COVID‐19 patients and triggered oxidative stress may lead to mitochondrial dysfunction, eventually leading to platelet damage and apoptosis. Couture et al. (2020) found that luteolin could inhibit membrane synthesis and cell proliferation to activate apoptosis, which may be related to the increased expression of related genes such as Fas, Cdkn1a, Atp7b, and TP53, and the increased accumulation of cleaved caspase 3 and PARP. Wrapp et al. (2020) believe that the key to COVID‐19 infection lies in the binding between the SARS‐CoV‐2 spike protein and ACE2 protein, and understanding the structure and affinity of these two proteins will help in the development of antiviral drugs. However, the role of the extracellular region in COVID‐19 is still unclear. In addition, it is interesting to find that the anti‐COVID‐19 genes were mainly expressed in the lung and liver; this finding has also been confirmed in previous studies. For example, Zhang observed that COVID‐19 patients have obvious abnormalities in coagulation, which may be caused by liver damage and inflammatory storms (Zhang et al., 2020). In addition, inflammatory storms can cause lung damage (Han et al., 2020).
KEGG analysis revealed that the four important signaling pathways closely related to COVID‐19 were the TNF signaling pathway, the T‐cell receptor signaling pathway, the Toll‐like receptor signaling pathway, and the MAPK signaling pathway. These signaling pathways are related to the inflammatory response, and some have been found to be involved in the pathogenesis of COVID‐19. For example, Karki et al. found that some proinflammatory factors in the TNF signaling pathway are also involved in the immune response process of COVID‐19 patients, and TNF‐α and IFN‐γ have a synergistic effect in inducing inflammation, tissue damage, and death in COVID‐19 patients (Karki et al., 2020). DiNicolantonio and McCarty (2020) found that the Toll‐like receptor signaling pathway was involved in thrombosis in patients with COVID‐19. In addition, Yang et al. (2020) found that the thrombin and Toll‐like receptor signaling pathways may be important antiinflammatory pathways of Maxing Shigan decoction for the treatment of COVID‐19 through transcriptomics analysis. The role of the T‐cell receptor signaling pathway in COVID‐19 is still unclear, but Sallenave and Guillot (2020) found that JAK1‐STAT5 could increase CD8+ T cells and decrease lymphocytes in critical patients with COVID‐19 by inhibiting the expression of IL‐2/IL‐2R. Grimes and Grimes (2020) believe that the p38 MAPK pathway plays a key role in the release of proinflammatory cytokines (such as IL‐6), pro‐vasoconstriction, and prothrombotic activity, and is related to acute lung injury and myocardial dysfunction. Although these signaling pathways were related to the occurrence and development of COVID‐19, there is currently no experiment to further verify the specific role of Yinqiao powder against COVID‐19 in these signaling pathways.
5. CONCLUSION
In this study, we found that hesperetin, eriodictyol, luteolin, quercetin, and naringenin were potential effective active ingredients against COVID‐19 The antagonistic effect of Yingqiao powder on the inflammatory storm caused by COVID‐19 may be related to the regulation of IL‐6, MAPK3, TNF, and TP53 targets. The specific pathways were the TNF signaling pathway, the T‐cell receptor signaling pathway, the Toll‐like receptor signaling pathway, and the MAPK signaling pathway. Our study provides a new perspective for discovering potential drugs and mechanisms of COVID‐19.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
Haixiong Lin and Xiaotong Wang conceived and designed the study. Minyi Liu and Xiaopeng Ye revised the protocol. Haixiong Lin and Xiaotong Wang extracted the data. Junjie Feng, Zhen Shen, Huijun Yang, Minling Huang, and Zige Li checked the data. Minling Huang and Junyan Gao performed statistical analysis. Haixiong Lin and Xiaotong Wang wrote the manuscript. Haixiong Lin and Xiaotong Wang interpreted the results. Haixiong Lin, Xiaotong Wang, and Xiaopeng Ye reviewed and advice. All authors contributed constructive comments on the paper.
Supporting information
Figure S1 Chromatographic retention time and mass spectrogram of Yinqiao powder ingredients
Supplementary Material 1 High‐resolution liquid chromatography‐mass spectrometry (LC‐MS) analysis of Yinqiao powder
Supplementary Material 2 Total ion chromatography of Yinqiao powder
Table S1 Main active ingredients in Yinqiao powder
Table S2 Information of potential gene targets for treating COVID‐19 from Yinqiao powder
Table S3 Genes associated with viruses or COVID‐19 and their DisGeNET scores
Table S4 The intervention effect of Yinqiao powder, Qingfei Paidu decoction, and Banxia Baizhu Tianma decoction on COVID‐19
Table S5 Topological analysis of protein and protein interaction network
Table S6 Molecular docking of Yinqiao powder ingredients with hub‐proteins
ACKNOWLEDGMENTS
This work was supported, in part, by International Program for Postgraduates, Guangzhou University of Chinese Medicine (GZYXB[2019]114), the Excellent Doctoral Dissertation Incubation Grant of Guangzhou University of Chinese Medicine (GZYXB2020‐18), and the Excellent Doctoral Dissertation Incubation Grant of First Clinical School of Guangzhou University of Chinese Medicine (YB201902).
Lin H, Wang X, Liu M, et al. Exploring the treatment of COVID‐19 with Yinqiao powder based on network pharmacology. Phytotherapy Research. 2021;35:2651–2664. 10.1002/ptr.7012
Haixiong Lin and Xiaotong Wang contributed equally to this work.
Funding information Excellent Doctoral Dissertation Incubation Grant of First Clinical School of Guangzhou University of Chinese Medicine, Grant/Award Number: YB201902; Excellent Doctoral Dissertation Incubation Grant of Guangzhou University of Chinese Medicine, Grant/Award Number: GZYXB2020‐18; International Program for Postgraduates, Guangzhou University of Chinese Medicine, Grant/Award Number: GZYXB[2019]114
Contributor Information
Xiaotong Wang, Email: wxtong642@foxmail.com.
Xiaopeng Ye, Email: 865172054@qq.com.
DATA AVAILABILITY STATEMENT
The data and materials generated or analyzed during this study are available from the corresponding author on reasonable request.
REFERENCES
- Arshad, A. S. , Baloch, M. , Ahmed, N. , Arshad, A. A. , & Iqbal, A. (2020). The outbreak of coronavirus disease 2019 (COVID‐19)—An emerging global health threat. Journal of Infection and Public Health, 13(4), 644–646. 10.1016/j.jiph.2020.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmeh, N. , Mahmoudi, S. , Mohammadi, N. , & Karabedianhajiabadi, A. (2020). Predicted therapeutic targets for COVID‐19 disease by inhibiting SARS‐CoV‐2 and its related receptors. Informatics in Medicine Unlocked, 20, 100407. 10.1016/j.imu.2020.100407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. , Martin, M. , Orchard, S. , Magrane, M. , Alpi, E. , Bely, B. , … Bolleman, J. (2019). UniProt: A worldwide hub of protein knowledge. Nucleic Acids Research, 47(D1), D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, K. G. , Barnes, K. C. , Bright, T. J. , & Wang, S. A. (2004). The genetic association database. Nature Genetics, 36(5), 431–432. 10.1038/ng0504-431 [DOI] [PubMed] [Google Scholar]
- Couture, R. , Mora, N. , Al Bittar, S. , Najih, M. , Touaibia, M. , & Martin, L. J. (2020). Luteolin modulates gene expression related to steroidogenesis, apoptosis, and stress response in rat LC540 tumor Leydig cells. Cell Biology and Toxicology, 36(1), 31–49. 10.1007/s10565-019-09481-9 [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Liu, B. , He, Z. , Liu, T. , Zheng, R. , Yang, A. , … Xu, Y. (2020). Study on active compounds from Huoxiang Zhengqi oral liquid for prevention of coronavirus disease 2019 (COVID‐19) based on network pharmacology and molecular docking. Chinese Traditional and Herbal Drugs, 51(5), 1113–1122. 10.7501/j.issn.0253-2670.2020.05.004 [DOI] [Google Scholar]
- Dennis, G. J. , Sherman, B. T. , Hosack, D. A. , Yang, J. , Gao, W. , Lane, H. C. , & Lempicki, R. A. (2003). DAVID: Database for annotation, visualization, and integrated discovery. Genome Biology, 4(5), P3. https://link.springer.com/article/10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- Di Paola, R. , Crisafulli, C. , Mazzon, E. , Genovese, T. , Paterniti, I. , Bramanti, P. , & Cuzzocrea, S. (2009). Effect of PD98059, a selective MAPK3/MAPK1 inhibitor, on acute lung injury in mice. International Journal of Immunopathology and Pharmacology, 22(4), 937–950. 10.1177/039463200902200409 [DOI] [PubMed] [Google Scholar]
- DiNicolantonio, J. J. , & McCarty, M. (2020). Thrombotic complications of COVID‐19 may reflect an upregulation of endothelial tissue factor expression that is contingent on activation of endosomal NADPH oxidase. Open Heart, 7(1), e001337. 10.1136/openhrt-2020-001337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbohun, O. F. , Babalola, O. O. , Agboola, F. K. , Joseph, J. S. , Malindisa, S. , & Msagati, T. A. M. (2020). Evaluation of phytochemicals, antioxidants, trace elements in Kigelia africana fruit extracts and chemical profiling analysis using UHPLC‐qTOF‐MS2 spectrometry. Biological Trace Element Research, 195(2), 679–695. 10.1007/s12011-019-01869-2 [DOI] [PubMed] [Google Scholar]
- Fan, T. , Chen, Y. , Bai, Y. , Ma, F. , Wang, H. , Yang, Y. , … Lin, Y. (2020). Analysis of medication characteristics of traditional Chinese medicine in treating coronavirus disease‐19 based on data mining. Journal of Zhejiang University (Medical Sciences), 49(1), 1–18. 10.3785/j.issn.1008-9292.2020.03.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky, G. V. (2020). Tripartite combination of candidate pandemic mitigation agents: Vitamin D, quercetin, and estradiol manifest properties of medicinal agents for targeted mitigation of the COVID‐19 pandemic defined by genomics‐guided tracing of SARS‐CoV‐2 targets in human cells. Biomedicine, 8(5), 129. 10.3390/biomedicines8050129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes, J. M. , & Grimes, K. V. (2020). p38 MAPK inhibition: A promising therapeutic approach for COVID‐19. Journal of Molecular and Cellular Cardiology, 144, 63–65. 10.1016/j.yjmcc.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar, K. , & Wairkar, S. (2020). Nanocrystal technology for improving therapeutic efficacy of flavonoids. Phytomedicine, 71, 153240. 10.1016/j.phymed.2020.153240 [DOI] [PubMed] [Google Scholar]
- Guo, F. , Zhang, Y. , Tang, S. , Tang, X. , Xu, H. , Liu, Z. , … Yang, H. (2020). TCMATCOV—A bioinformatics platform to predict efficacy of TCM against COVID‐19. Chinese Journal of Traditional Chinese Medicine, 45(10), 2257–2264. 10.19540/j.cnki.cjcmm.20200312.401 [DOI] [PubMed] [Google Scholar]
- Guo, Z. , Su, Z. , Wang, Z. , Luo, X. , & Lai, R. (2017). The effect of chinese herbal medicine Banxia Baizhu Tianma decoction for the treatment of vertebrobasilar insufficiency vertigo: A systematic review and meta‐analysis of randomized controlled trials. Complementary Therapies in Medicine, 31, 27–38. 10.1016/j.ctim.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Han, H. , Ma, Q. , Li, C. , Liu, R. , Zhao, L. , Wang, W. , … Xia, Y. (2020). Profiling serum cytokines in COVID‐19 patients reveals IL‐6 and IL‐10 are disease severity predictors. Emerging Microbes & Infections, 9(1), 1123–1130. 10.1080/22221751.2020.1770129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, J. , Tao, H. , Yan, Y. , Huang, S. , & Xiao, Y. (2020). Molecular mechanism of evolution and human infection with SARS‐CoV‐2. Viruses‐Basel, 12(4284), 1–19. 10.3390/v12040428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Zhang, M. , Chen, C. , Zhang, H. , Wei, Y. , Tian, J. , … Dai, H. (2020). Clinical characteristics of COVID‐19 in patients with pre‐existing ILD: A retrospective study in a single center in Wuhan, China. Journal of Medical Virology, 92(11), 2742–2750. 10.1002/jmv.26174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, A. N. (1996). Scoring noncovalent protein‐ligand interactions: A continuous differentiable function tuned to compute binding affinities. Journal of Computer‐Aided Molecular Design, 10(5), 427–440. 10.1007/BF00124474 [DOI] [PubMed] [Google Scholar]
- Jimilihan, S. , Maimaitiming, N. , Ainiwaer, W. , Maierdan, Y. , Muhadaisi, N. , Nulibiya, M. , & Zhou, W. (2020). Study on the active components in the adjuvant treatment of novel coronavirus pneumonia (COVID‐19) with Jinhua Qinggan granules based on network pharmacology and molecular docking. Journal of Chinese Medicinal Materials, 43(5), 1275–1283. http://kns.cnki.net/kcms/detail/44.1286.R.20200323.1926.002.html [Google Scholar]
- Karki, R. , Sharma, B. R. , Tuladhar, S. , Williams, E. P. , Zalduondo, L. , Samir, P. , … Kanneganti, T. (2020). COVID‐19 cytokines and the hyperactive immune response: Synergism of TNF‐alpha and IFN‐gamma in triggering inflammation, tissue damage, and death. BioRxiv: The Preprint Server for Biology, 361048. 10.1101/2020.10.29.361048 [DOI] [Google Scholar]
- Kneller, D. W. , Galanie, S. , Phillips, G. , O'Neill, H. M. , Coates, L. , & Kovalevsky, A. (2020). Malleability of the SARS‐CoV‐2 3CL Mpro active‐site cavity facilitates binding of clinical antivirals. Structure, 28, 1–8. 10.1016/j.str.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. , Huo, G. , Liu, S. , Li, F. , Chen, W. , & Jiang, D. (2019). Luteolin suppresses inflammation through inhibiting cAMP‐phosphodiesterases activity and expression of adhesion molecules in microvascular endothelial cells. Inflammopharmacology, 27(4), 773–780. 10.1007/s10787-018-0537-2 [DOI] [PubMed] [Google Scholar]
- Kong, Y. , Wu, H. , Chen, Y. , Lai, S. , Yang, Z. , & Chen, J. (2020). Mechanism of Tanreqing injection on treatment of coronavirus disease 2019 based on network pharmacology and molecular docking. Chinese Traditional and Herbal Drugs, 51(7), 1785–1794. 10.7501/j.issn.0253-2670.2020.07.012 [DOI] [Google Scholar]
- Li, S. , & Zhang, B. (2013). Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chinese Journal of Natural Medicines, 11(2), 110–120. 10.3724/SP.J.1009.2013.00110 [DOI] [PubMed] [Google Scholar]
- Liang, C. , Niu, W. , Wu, F. , Cao, W. , Wu, Z. , Chao, Y. C. , & Peng, F. (2020). Network pharmacology for the identification of phytochemicals in traditional Chinese medicine for COVID‐19 that may regulate interleukin‐6. Bioscience Reports, BSR20202583. 10.1042/BSR20202583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Wang, X. , Wang, L. , Dong, H. , Huang, P. , Cai, Q. , … Jiang, Z. (2019). Identified the synergistic mechanism of Drynariae Rhizoma for treating fracture based on network pharmacology. Evidence‐Based Complementary and Alternative Medicine, 2019, 7342635. 10.1155/2019/7342635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. , Peng, Q. , Zhang, J. , Li, X. , Huang, J. , Duan, S. , & Zhang, W. (2020). Quercetin prevents lipopolysaccharide‐induced experimental preterm labor in mice and increases offspring survival rate. Reproductive Sciences, 27(4), 1047–1057. 10.1007/s43032-019-00034-3 [DOI] [PubMed] [Google Scholar]
- Lin, Y. , Tan, D. , Kan, Q. , Xiao, Z. , & Jiang, Z. (2018). The protective effect of Naringenin on airway remodeling after mycoplasma pneumoniae infection by inhibiting autophagy‐mediated lung inflammation and fibrosis. Mediators of Inflammation, 2018, 1–10. 10.1155/2018/8753894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, F. , Chen, J. , Yuan, J. , Han, Z. , Liu, D. , Zhao, Y. , & Wang, H. (2020). Exploring the mechanism of Maxing Shigan decoction in treating new coronavirus pneumonia based on network pharmacology. Shaanxi Journal of Traditional Chinese Medicine, 41(5), 555–559. 10.3969/j.issn.1000-7369.2020.05.001 [DOI] [Google Scholar]
- Luo, H. , Tang, Q. , Shang, Y. , Liang, S. , Yang, M. , Robinson, N. , & Liu, J. (2020). Can Chinese medicine be used for prevention of corona virus disease 2019 (COVID‐19)? A review of historical classics, research evidence and current prevention programs. Chinese Journal of Integrative Medicine, 26(4), 243–250. 10.1007/s11655-020-3192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwa, W. , Kumar, R. , Thompson, D. , Lyerly, W. , Moore, R. , Reid, T. , … Toyang, N. (2020). Potential of flavonoid‐inspired phytomedicines against COVID‐19. Molecules, 25(11), 2707. 10.3390/molecules25112707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo, R. , Caporali, S. , Falconi, M. , Iacovelli, F. , Della Rocca, B. M. , Lo Surdo, A. , … Terrinoni, A. (2019). Luteolin‐7‐O‐β‐D‐glucoside inhibits cellular energy production interacting with HEK2 in keratinocytes. International Journal of Molecular Sciences, 20(11), 2689. 10.3390/ijms20112689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinero, J. , Ramirez‐Anguita, J. M. , Sauch‐Pitarch, J. , Ronzano, F. , Centeno, E. , Sanz, F. , & Furlong, L. I. (2020). The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Research, 48(D1), D845–D855. 10.1093/nar/gkz1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan, M. , Chalifa‐Caspi, V. , Prilusky, J. , & Lancet, D. (1997). GeneCards: Integrating information about genes, proteins and diseases. Trends in Genetics, 13(4), 163. 10.1016/s0168-9525(97)01103-7 [DOI] [PubMed] [Google Scholar]
- Rehman, M. T. , AlAjmi, M. F. , & Hussain, A. (2020). Natural compounds as inhibitors of SARS‐CoV‐2 main protease (3CLpro): A molecular docking and simulation approach to combat COVID‐19. Current Pharmaceutical Design, 26(1), 1–13. 10.2174/1381612826999201116195851 [DOI] [PubMed] [Google Scholar]
- Rothan, H. A. , & Byrareddy, S. N. (2020). The epidemiology and pathogenesis of coronavirus disease (COVID‐19) outbreak. Journal of Autoimmunity, 109, 102433. 10.1016/j.jaut.2020.102433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru, J. , Li, P. , Wang, J. , Zhou, W. , Li, B. , Huang, C. , … Yang, L. (2014). TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics, 6, 13. 10.1186/1758-2946-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, J. , Peyssonnaux, C. , Singh, K. K. , & Edeas, M. (2020). Mitochondria and microbiota dysfunction in COVID‐19 pathogenesis. Mitochondrion, 54, 1–7. 10.1016/j.mito.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallenave, J. , & Guillot, L. (2020). Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS‐CoV‐2 in Covid‐19: Key therapeutic targets? Frontiers in Immunology, 11, 1229. 10.3389/fimmu.2020.01229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet, D. F. (1991). Using OMIM (on‐line Mendelian inheritance in man) as an expert system in medical genetics. American Journal of Medical Genetics, 39(3), 278–284. 10.1002/ajmg.1320390307 [DOI] [PubMed] [Google Scholar]
- Shereen, M. A. , Khan, S. , Kazmi, A. , Bashir, N. , & Siddique, R. (2020). COVID‐19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. 10.1016/j.jare.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, Y. , Qin, K. , Cai, H. , Liu, X. , Yin, F. , Zhang, J. , & Cai, B. (2012). Study on the optimal traditional decoction process for Yinqiao San by orthogonal experiments. China Journal of Traditional Chinese Medicine and Pharmacy, 27(10), 2651–2655. https://www.cnki.com.cn/Article/CJFDTOTAL-BXYY201812080.htm [Google Scholar]
- Sietsema, W. K. (1989). The absolute oral bioavailability of selected drugs. International Journal of Clinical Pharmacology, Therapy, and Toxicology, 27(4), 179–211. [PubMed] [Google Scholar]
- Sun, J. H. , Sun, F. , Yan, B. , Li, J. Y. , & Xin, L. (2020). Data mining and systematic pharmacology to reveal the mechanisms of traditional Chinese medicine in mycoplasma pneumoniae pneumonia treatment. Biomedicine & Pharmacotherapy, 125, 109900. 10.1016/j.biopha.2020.109900 [DOI] [PubMed] [Google Scholar]
- Sun, K. , Zhang, X. , Liu, J. , & Sun, R. (2020). Network pharmacological analysis and mechanism prediction of Xiaochaihu decoction in treatment of COVID‐19 with syndrome of pathogenic heat lingering in lung and obstructive cardinalate. Chinese Traditional and Herbal Drugs, 51(7), 1750–1760. 10.7501/j.issn.0253-2670.2020.07.009 [DOI] [Google Scholar]
- Sun, P. , Lu, X. , Xu, C. , Sun, W. , & Pan, B. (2020). Understanding of COVID‐19 based on current evidence. Journal of Medical Virology, 92(6), 548–551. 10.1002/jmv.25722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D. , Morris, J. H. , Cook, H. , Kuhn, M. , Wyder, S. , Simonovic, M. , … von Mering, C. (2017). The STRING database in 2017: Quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Research, 45(D1), D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. , Leng, C. , Guo, K. , & Jin, S. (2019). Effect of quercetin on mice Staphylococcus aureus induced pneumonia and its mechanism of IKK/NF‐κB/IκB signaling pathway. Pharmacology and Clinics of Chinese Materia Medica, 35(4), 53–57. 10.13412/j.cnki.zyyl.2019.04.010 [DOI] [Google Scholar]
- Wang, Z. , Sun, Y. , Qu, R. , Liu, B. , Fan, Z. , Tian, J. , & Lu, T. (2020). Network pharmacological study on mechanism of Maxing Shigan decoction in treatment of coronavirus disease 2019 (COVID‐19). Chinese Traditional and Herbal Drugs, 51(8), 1996–2003. 10.7501/j.issn.0253-2670.2020.08.003 [DOI] [Google Scholar]
- Wrapp, D. , Wang, N. , Corbett, K. S. , Goldsmith, J. A. , Hsieh, C. , Abiona, O. , … McLellan, J. S. (2020). Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. , Liu, Y. , Cao, L. , Wang, D. , Guo, M. , Jiang, A. , … Chen, Y. (2020). Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID‐19 patients. Emerging Microbes & Infections, 9(1), 761–770. 10.1080/22221751.2020.1747363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. , Zhang, W. , Huang, C. , Li, Y. , Yu, H. , Wang, Y. , … Ling, Y. (2012). A novel chemometric method for the prediction of human oral bioavailability. International Journal of Molecular Sciences, 13(6), 6964–6982. 10.3390/ijms13066964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun, S. , Jialei, T. , Shaoju, X. , & Bin, Y. (2020). The molecular mechanism of treating COVID‐19 with Huashi Baidu formula based on network pharmacology. Journal of Chinese Medicinal Materials, 43(8), 2050–2055. http://kns.cnki.net/kcms/detail/44.1286.R.20200430.1759.006.html [Google Scholar]
- Yang, R. , Liu, H. , Bai, C. , Wang, Y. , Zhang, X. , Guo, R. , … Wang, Y. (2020). Chemical composition and pharmacological mechanism of Qingfei Paidu decoction and Ma Xing Shi Gan decoction against coronavirus disease 2019 (COVID‐19): In silico and experimental study. Pharmacological Research, 157, 104820. 10.1016/j.phrs.2020.104820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongye, A. B. , & Medina‐Franco, J. L. (2013). Systematic characterization of structure‐activity relationships and ADMET compliance: A case study. Drug Discovery Today, 18(15–16), 732–739. 10.1016/j.drudis.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Xue, K. , Gao, Y. , Huai, Y. , Wang, W. , Miao, Z. , … Qian, A. (2019). Systems pharmacology dissection of action mechanisms of Dipsaci Radix for osteoporosis. Life Sciences, 235, 116820. 10.1016/j.lfs.2019.116820 [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , He, L. , Chen, H. , Lu, S. , Xiong, Y. , Liu, J. , … Liu, L. (2020). Manifestations of blood coagulation and its relation to clinical outcomes in severe COVID‐19 patients: Retrospective analysis. International Journal of Laboratory Hematology, 42(6), 766–772. 10.1111/ijlh.13273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Lei, L. , Xu, Y. , Wei, D. , & Hu, F. (2020). Clinical efficacy of Jinyinhua oral liquid in the treatment of 80 patients with coronavirus disease 2019. China Pharmaceuticals, 29(9), 23–26. 10.3969/j.issn.1006-4931.2020.09.006 [DOI] [Google Scholar]
- Zhang, Z. , Liang, D. , Kong, X. , & Xiao, M. (2019). Effects of Jiegeng on inflammatory markers in patients with community acquired pneumonia. Clinical Journal of Chinese Medicine, 11(16), 77–79. 10.3969/j.issn.1674-7860.2019.16.029 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Chromatographic retention time and mass spectrogram of Yinqiao powder ingredients
Supplementary Material 1 High‐resolution liquid chromatography‐mass spectrometry (LC‐MS) analysis of Yinqiao powder
Supplementary Material 2 Total ion chromatography of Yinqiao powder
Table S1 Main active ingredients in Yinqiao powder
Table S2 Information of potential gene targets for treating COVID‐19 from Yinqiao powder
Table S3 Genes associated with viruses or COVID‐19 and their DisGeNET scores
Table S4 The intervention effect of Yinqiao powder, Qingfei Paidu decoction, and Banxia Baizhu Tianma decoction on COVID‐19
Table S5 Topological analysis of protein and protein interaction network
Table S6 Molecular docking of Yinqiao powder ingredients with hub‐proteins
Data Availability Statement
The data and materials generated or analyzed during this study are available from the corresponding author on reasonable request.


