Abstract
Viral infections remain a major cause of economic loss with an unmet need for novel therapeutic agents. Ivermectin is a putative antiviral compound; the proposed mechanism is the inhibition of nuclear translocation of viral proteins, facilitated by mammalian host importins, a necessary process for propagation of infections. We systematically reviewed the evidence for the applicability of ivermectin against viral infections including SARS‐CoV‐2 regarding efficacy, mechanisms and selective toxicity. The SARS‐CoV‐2 genome was mined to determine potential nuclear location signals for ivermectin and meta‐analyses for in vivo studies included all comparators over time, dose range and viral replication in multiple organs. Ivermectin inhibited the replication of many viruses including those in Flaviviridae, Circoviridae and Coronaviridae families in vitro. Real and mock nuclear location signals were identified in SARS‐CoV‐2, a potential target for ivermectin and predicting a sequestration bait for importin β, stopping infected cells from reaching a virus‐resistant state. While pharmacokinetic evaluations indicate that ivermectin could be toxic if applied based on in vitro studies, inhibition of viral replication in vivo was shown for Porcine circovirus in piglets and Suid herpesvirus in mice. Overall standardized mean differences and 95% confidence intervals for ivermectin versus controls were −4.43 (−5.81, −3.04), p < 0.00001. Based on current results, the potential for repurposing ivermectin as an antiviral agent is promising. However, further work is needed to reconcile in vitro studies with clinical efficacy. Developing ivermectin as an additional antiviral agent should be pursued with an emphasis on pre‐clinical trials in validated models of infection.
Keywords: antiviral, nuclear location signals, ivermectin, SARS‐CoV‐2
Abbreviations
- HIV‐1
Human immunodeficiency virus‐1
- IRF
interferon regulatory factor
- NLSs
nuclear location signals
- NPC
nuclear pore complex
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta‐Analyses
1. INTRODUCTION
Ivermectin (Figure 1a ) is an essential drug with clinical approval for treating different types of parasitic infections in humans and animals. More recently however, several studies have documented antiviral effects of ivermectin and the potential to repurpose it as a therapeutic agent for viral infections [1, 2, 3]. Most scientific investigations in this area have been done in vitro, by infecting mammalian cells and, using this approach, efficacy has been reported against many viruses with most notable effects on enveloped, positive‐sense, single‐stranded flaviviruses including Dengue, West Nile, Yellow fever and Zika [4, 5, 6, 7, 8]. A plausible and well‐characterized antiviral mechanism of ivermectin has been proposed to be the inhibition of nuclear translocation of viral proteins, facilitated by mammalian host importin also known as karyopherin α/β‐1 heterodimerization [2]. Based on this mechanism, ivermectin binds to the importin alpha (armadillo repeat) domain causing thermal stability and a conformational change in alpha‐helicity that prevents binding to importin beta‐1 [5, 9]. This is a eukaryotic cell‐dependent process that may limit infection and replication, or enhance host antiviral responses depending on the specific functions of target cargo proteins [10]. Detailed illustrations of this mode of inhibiting viral replication by ivermectin have been shown for Human immunodeficiency virus‐1 (HIV‐1) via the integrase enzyme, Dengue virus via nonstructural protein 5 (a polymerase for viral RNA synthesis and regulator for immune signalling), Suid herpesvirus via DNA polymerase UL42 and, Yellow fever virus, Dengue virus and West Nile virus via nonstructural protein 3 (a DNA helicase enzyme) [4, 5, 11]. For a detailed recent review of the evidence for ivermectin blocking importin α, see Jans and Wagstaff [2]. Despite these detailed molecular characterizations for some viruses, it is not known whether similar or other structurally divergent nuclear location signals and corresponding target cargo proteins are present and likely to be a target in all other viruses against which ivermectin may be effective. Furthermore, the potential for the in vitro antiviral effects of ivermectin to translate into clinically relevant applications against infections in mammals is yet to be determined. Recent reviews of ivermectin as an antiviral [2, 3] highlight the need to better understand the pharmacological considerations. Therefore, in this study, we sought to undertake a systematic review of all published work on antiviral effects of ivermectin and our objective was to present an integrative, critical appraisal of the qualitative and quantitative antiviral properties of ivermectin for putative applications in agriculture and medicine with examination of SARS‐CoV‐2.
Figure 1.
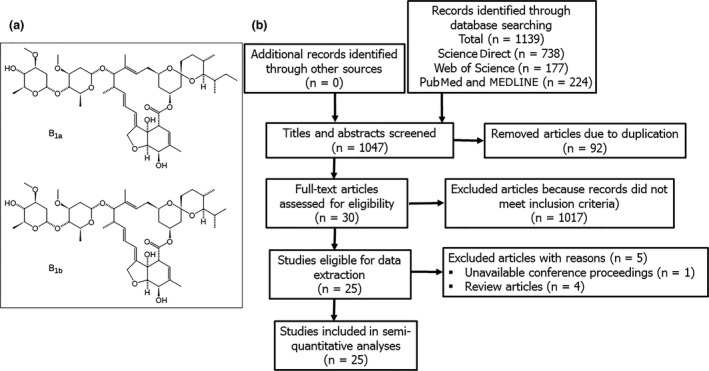
The chemical structure of ivermectin represented by two constituent 22,23‐dihydroavermectin B1a and 22,23‐dihydroavermectin B1b enantiomers (panel a) and, a flow diagram of Preferred Reporting Items of Systematic Review and Meta‐analyses‐PRISMA (panel b).
2. MATERIALS AND METHODS
2.1. Literature search strategy, study inclusion and exclusion criteria
This systematic review was done according to the 2009 Preferred Reporting Items for Systematic Reviews and Meta‐Analyses (PRISMA) statement [12]. The specific aim was to determine whether ivermectin exerted antiviral properties including the prevention of infection, viral replication after infection and infection‐induced mortalities. Major databases including MEDLINE and PubMed, ScienceDirect and Web of Science were searched from dates of inception to August 2020. Our strategy was to capture and analyse all published work on the experimental or clinical use of ivermectin against viruses. The specific search terms were (“Ivermectin”) AND (Virus OR Viral Infection) in medical subject headings as well as keyword searches. Only publications in English were considered; titles and abstracts were screened to generate a reference list. Included studies were also examined for additional references that fit the inclusion criteria. All controlled, primary studies examining antiviral effects of ivermectin were collated irrespective of type or strain of virus, in vitro culture system or animal model of infection used and, the dose and route of administration of ivermectin.
2.2. Data extraction and quality assessment of included studies
Research articles that qualified for in‐depth analysis and data extraction were assessed and qualified by both authors (RTK and LO). All data were extracted from text, tables and figures in the published work except for one study where some of the raw data were acquired directly from the authors [13]. The extracted information included type and strain of the virus, type of cells for in vitro cultures, number and age of animals, conditions of infection and the observed qualitative as well as quantitative effects of ivermectin. Qualitative evaluation of individual studies testing the antiviral effects of ivermectin in mammals was done by a criterion based on SYRCLE’s risk of bias for animal studies [14]. These evaluations include the sex and age of animals used, sample size evaluations and justification, randomization in generating experimental groups and assigning treatments, blinding in assessing experimental outcomes, compliance with relevant welfare regulations and ethics and, the citation of any conflicts of interest. At the time of writing, only one published, peer‐reviewed paper [28] on the in vitro effect of ivermectin against SARS‐CoV‐2 exists, so this review contains in vitro and in vivo studies and no peer‐reviewed human clinical studies (as a subset of in vivo studies) exist to enter our screen.
2.3. Identifying nuclear location signals in viral genomes as a target for ivermectin
As well as the need for many viruses’ genomes to access the nucleus, many viral proteins need to enter the nucleus. Access of proteins into the nucleus is through the ‘lock’ of the nuclear pore complex (NPC) which is ‘unlocked’ by a protein ‘key’; a run of basic amino acids (aa) called the nuclear location signals (NLSs). These NLSs are most often stretches of sequences of the basic amino acids lysine (Lys) and arginine (Arg) [15], and can be preceded by helix‐breaking neutral amino acids, proline (Pro), glutamine (Gln) or glycine (Gly) and less commonly with the negatively charged aspartic acid (Asp) or glutamic acid (Glu). The NLS can be monopartite (Table 1) (e.g. SV‐40 T‐antigen) as hexapeptides with at least 4 basic and no acidic nor bulky amino acids [16] and proceeded by a helix‐breaking residue (Pro, Gln or Gly) [17]. The NLS can also be bipartite with two groups of basic amino acids separated by at least 9 aa (e.g. DNA helicase Q1) or non‐classical (e.g. Pro‐Tyr). These stretches of basic amino acids then bind directly to β importin or α–β heterodimer complexed importins for transport of the protein through the nuclear pore complex into the nucleus.
Table 1.
Definitions of nuclear location signals used herein.
| Classic monopartite | 6 amino acids of which 4 are basic, no acidic or bulky amino acid, preceded by a helix‐breaking proline (Pro), glutamine (Gln) or glycine (Gly); sometimes the negatively charged aspartic acid (Asp) or glutamic acid (Glu), for example Pro‐Lys‐Arg‐Lys‐Lys‐Val‐Arg |
| Chelsky sequence | 4 amino acids, 3 of which are basic, starting dibasic, for example Lys‐Lys/Arg‐x‐Lys/Arg |
| Classic bipartite | 2 basic amino acids separated by at least 9 amino acids from a cluster of at least 3 basic amino acids, for example Arg‐Lys‐15aa‐‐Lys‐Arg‐Gln‐Lys |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The NCBI Entrez virus type genomes were taken as the default sequences. A search for areas in the open reading frames of the viruses for stretches of basic amino acids (Table 1) was conducted manually similar to Zhou et al. [18] and motifs were compared to those listed previously.[15] The substitutions of Arg for Lys and vice versa were taken as interchangeable with no loss of functionality. The ‘&’ was used for any bulky, hydrophobic aa like Ala, Met, Val, Lue, Phe, Tyr, Ile and Trp. ‘X’ was used for any amino acid.
2.4. Data analysis
Data extracted from in vitro cell culture studies were summarized and presented as qualitative descriptions in the results section below. Quantitatively, the selectivity index of ivermectin was evaluated by determining the ratio of the concentration of ivermectin that inhibited viral activity by 50% (EC50) to the concentration that caused cytotoxicity in 50% of utilized mammalian cells (CC50). Studies on antiviral effects of ivermectin in multicellular organisms were stratified into two rational groups including arthropods and mammalian hosts. Extracted data were pooled into meta‐analyses to determine the magnitude of the overall effect of ivermectin on viral infections, replication and viral infection‐induced mortalities using the RevMan 5.3 software. As there were marked differences in the utilized infection models, type of viruses considered, ivermectin doses, routes of administration and duration of treatment, data analysis was based on the random effects model in RevMan 5.3. Data were presented as standardized mean differences with 95% confidence intervals, and a P‐value <0.05 was considered significant. The sensitivity and effect of each study on the overall standardized mean differences was determined by a commonly used leave‐one‐out approach in meta‐analyses. The degree of heterogeneity in the extracted data was evaluated from I2 values with values >50% considered significant.
3. RESULTS
3.1. Qualitative and quantitative antiviral effects of ivermectin based on in vitro studies
A total of 1139 studies were identified from database searches and 92 of these were duplicate records that were removed from further analyses (Figure 1b ). Titles and abstracts of the remaining 1047 studies were screened against the established inclusion criteria, and an additional 1017 articles were removed. Thirty studies met the inclusion criteria but data for one abstract were inaccessible; reported as a conference proceeding, and four studies were reviews presenting no primary data. Accordingly, a total of twenty‐five studies were subjected to qualitative and quantitative analyses (Tables 2 and 3). In vitro studies in cell cultures show that ivermectin exerted a time and concentration‐dependent inhibition of infection and replication, and plaque formation against many viruses representing several families including: Arteriviridae, Circoviridae, Coronaviridae, Flaviviridae, Herpesviridae, Paramyxoviridae, Polyomaviridae, Retroviridade and Togaviridae. However, two studies demonstrated that at concentrations as high as 15–25 µM, ivermectin inhibited replication but did not specifically inhibit cellular attachment and entry following infection with Betaarterivirus in PAM‐pCD163 macrophages [19], and Bovine herpesvirus 1 in MDBK cells [20]. Ivermectin had no effect on infection and replication of Venezuelan equine encephalitis virus in U87MG and Vero cells, and Equine herpesvirus 1 in primary neuronal cells (Table 2). Due to marked variations in the procedural approaches and systems used for in vitro studies reported herein, we sought to evaluate the relative potency and safety margin of ivermectin as an antiviral agent. On aggregate, ivermectin had a wide in vitro safety margin for many viral species including Chikungunya, Dengue, Zika, Yellow fever, Suid herpesvirus 1 and the Kunjin strain of West Nile virus (Figure 2 ). By contrast, EC50 values against polyomavirus, Betaarterivirus, Bovine herpes virus 1, Newcastle disease virus and the NY99 strain of West Nile virus fell within the cytotoxic range for mammalian cells (Table 2 and Figure 2 ). Yellow fever virus had the lowest EC50 value (0.5–5 nM) but relatively high EC50 values (0.4–25 µM) were seen for other viruses studied. Parallel CC50 values of ivermectin in utilized mammalian cells were 5.8 ± 1.1 µM for Vero cells, 8.4 ± 0.8 µM for Huh cells and 13.6 ± 8.3 µM for BHK cells (Figure 2 ).
Table 2.
In vitro evaluation of the antiviral activity of ivermectin in cell cultures.
| Target virus | Cell culture system and conditions | Quantitative and qualitative effects | References |
|---|---|---|---|
|
West Nile virus (Kunjin; MRM61C strain), Zika virus (Asian/Cook Islands/2014 strain), Dengue virus‐2 (New Guinea C; M29095 strain). |
Vero cells, MOI of 1 for 2 hr, followed for 22 hr. Ivermectin at (0–10 µM). |
Concentration‐dependent inhibition of plague formation. EC50 West Nile virus = 0.8 µM EC50 Zika virus = 1.1 µM EC50 Dengue virus 0.4 µM Concentration‐dependent inhibition of replication. EC50 West Nile virus = 0.8 µM EC50 Zika virus = 1.9 µM EC50 Dengue virus 0.6 µM. (n = 2) |
[8] |
|
Dengue virus‐1 (EU081230), Dengue virus‐2 (EU081177), Dengue virus‐2 (mouse adapted S221) |
Huh‐7 cells, MOI of 0.3 for 1 hr, followed for 48 hr. Ivermectin at (0–10 µM). |
Concentration‐dependent inhibition of replication and plague formation. EC50 Dengue virus‐1 (EU081230) = 3.7 µM EC50 Dengue virus‐2 (EU081177) = 2.6 µM EC50 Dengue virus‐2 (S221) = 2.9 µM. (n = 2) |
[7] |
|
Dengue virus‐1 (EU081230), Dengue virus‐2 (EU081177), Dengue virus‐3 (EU081190), Dengue virus‐4 (GQ398256) |
BHK‐21 cells, MOI of 0.3, followed for 48 hr and, Huh‐7 cells, MOI of 0.3 followed for 48 hr. Ivermectin at (0–45 µM). |
Concentration‐dependent inhibition of replication. Values in BHK‐21 cells were: EC50 Dengue virus‐1 (EU081230) = 2.32 µM EC50 Dengue virus‐2 (EU081177) = 2.08 µM EC50 Dengue virus‐3 (EU081190) = 1.66 µM EC50 Dengue virus‐4 (GQ398256) = 1.90 µM Values in Huh‐7 cells were: EC50 Dengue virus‐1 (EU081230) = 2.97 µM EC50 Dengue virus‐2 (EU081177) = 1.74 µM (n = 2) |
[6] |
|
Wild type Chikungunya virus (LR2006 OPY1), Yellow fever virus (17D strain), Wild type Semliki Forest virus, Wild type Sindbis virus |
BHK‐21 cells, MOI of 0.01, followed for 16 hr and, Huh‐7.5 cells, MOI of 0.1 followed for 16 hr. Ivermectin at (0–300 µM). |
Time‐ and concentration‐dependent inhibition of replication. EC50 was evaluated for Chikungunya virus were: EC50 in BHK‐21 cells = 0.6 µM (n = 3) EC50 in Huh 7.5 cells = 1.9 µM. (n = 3) Ivermectin (3 µM) reduced viral titres in BHK‐21 cells by: 2.5 log values for Sindbis virus 3.0 log values for Semliki Forest virus 3.0 log values for Chikungunya virus 4.0 log values for Yellow fever virus |
[51] |
|
Yellow fever virus (17D strain), Dengue virus‐2 (New Guinea C strain) |
Vero‐B cells, MOI of 0.4 for Dengue virus and 1.0 for Yellow fever virus. Followed for 168 hr. Ivermectin at (0–105 µM) |
Ivermectin inhibited virus‐induced cytopathic effect. EC50 Yellow Fever virus (17D) = 0.005 µM EC50 Dengue virus‐2 (New Guinea) >1.0 µM |
[52] |
|
Yellow fever virus (17D strain), Dengue virus‐2 (New Guinea C strain), West Nile virus (NY99; NC‐009942) |
Vero‐B cells for Dengue and Yellow Fever virus, MOI of 0.1 for 2 hr, followed for 96 hr. Vero‐E6 cells for West Nile virus, MOI of 0.1–1 for 2 hr, followed for 72 hr. Ivermectin at (0–5 µM). |
Time‐ and concentration‐dependent inhibition of viral plication. EC50 Yellow Fever virus (17D) = 0.0005 µM EC50 Dengue virus‐2 (New Guinea) = 0.7 µM EC50 West Nile virus (NC‐009942) = 4.0 µM Ivermectin (0.01 µM) caused complete protection against Yellow Fever virus in the plague formation assay. |
[4] |
|
Dengue virus‐2 (New Guinea C strain), Pseudotyped NL4‐3.Luc.R‐E‐ HIV |
Vero cells, MOI of 4 for Dengue virus, for 2 hr, followed for 72 hr and, Hela cells, 200 ng (capsid protein‐equivalent), for 2 hr, followed for 72 hr. Ivermectin (25 & 50 µM). |
Concentration‐dependent inhibition of replication. Ivermectin at 25 and 50 µM inhibited Dengue virus production by 73% and 100% respectively. Ivermectin at 25 and 50 µM inhibited HIV virus production by 36% and 57% respectively. (n = 4) |
[5] |
| Zika virus (PRVABC59 strain) | Vero cells, MOI of 0.1 for 1 hr, followed for 96 hr. Ivermectin at 10 µM. |
Ivermectin (10 µM) reduced viral titres by 1.5 log values. |
[53] |
|
Zika virus (Cambodia, FSS13025) |
C6/36 Aedes albopictus clone, MOI of 0.5, for 1 hr, followed for 5 days. Ivermectin tested at 2 and 10 µM. |
Concentration‐dependent inhibition of replication. Ivermectin at 2 and 10 µM inhibited virus production by 34% and 62% respectively. EC50 >2 µM. (n = 3) |
[23] |
| Porcine circovirus‐2 (SH1 strain) | PK‐15 cells, MOI of 10 for 1 hr, followed for 48 hr. Ivermectin at (57 and 114 µM). | Time‐ and concentration‐dependent inhibition of replication. Decreased by 59% at 24 hr and 72% at 48 hr for 57 µM of ivermectin, and by 81% at 24 hr and 84% at 48 hr for 114 µM of ivermectin. (n = 3) | [25] |
|
Hendra virus (Australia/Horse/1994) |
Vero cells, MOI of 0.05 for 2 hr, followed for 24 hr. Ivermectin at (0–10 µM). | Concentration‐dependent inhibition of replication. Viral titre decreased by 5 log values at 10 µM of ivermectin; EC50 = 2.0 µM. (n = 3) | [54] |
|
Betaarterivirus (Porcine Reproductive and Respiratory Syndrome virus) (VR‐2332) |
PAM‐pCD163 macrophages, MOI of 1 for 1 hr, followed for 48 hr. Ivermectin at (0–15 µM). |
Time‐ and concentration‐dependent inhibition of viral replication. EC50 = 6.7 µM. At 15 µM of ivermectin, both infection and replication were reduced by 95%. No effect on adsorption and cell entry (n = 3) |
[19] |
| Wild type BK Polyomavirus (BKPyV) | RPTE cells, MOI of 104 genomes/cell for 1 hr, followed for 24 hr. Ivermectin tested at 10 µM. | Ivermectin at 10 µM inhibited viral replication by 50%. (n = 3) | [55] |
| Venezuelan Equine Encephalitis virus (VEEV TC83) |
U87MG cells, MOI of 0.1–1, followed for 24 hr. Vero cells, MOI of 0.1–1, followed for 24 hr. Ivermectin tested at 1 µM. |
Ivermectin at 1 µM inhibited viral replication by 30% at 16 and 24 hr post‐infection in U87MG cells. No inhibitory effect was observed in Vero cells. Viral titres in plague assays were not affected at 24 hr post‐infection in both Vero and U87MG cells. | [9] |
| Venezuelan Equine Encephalitis virus (VEEV TC83) | Vero cells, MOI of 0.1, followed for 16 hr. Ivermectin tested at 1 µM. | No significant effect on viral titres in plague assays at 16 hr post‐infection in Vero cells. (n = 3). | [56] |
| Bovine herpesvirus‐1 (IBRV HB06 strain) | MDBK cells, MOI of 0.1–1 for 1 hr, followed for 48 hr. Ivermectin tested at (0–25 µM). |
Dose‐dependent inhibition of viral replication. Viral titre decreased by 4 log values and, ~50% inhibition of virion production with 25 µM of ivermectin. No effect on viral attachment and cell entry. (n = 3). |
[20] |
| Equine herpesvirus‐1 (Jan‐E strain) and (Rac‐H strain) | Primary murine neuronal cells, MOI of 0.3 for 1 hr, followed for 24 hr. Ivermectin tested at (0–75 µM). | No effect on the replication of EHV‐1 (Rac‐H strain). EC50 for EHV‐1 (Jan‐E strain) was >100 µM. | [57] |
| Suid herpesvirus 1 | BHK‐21 cells, MOI of 0.01 for 1 hr, followed for 16–72 hr. Ivermectin at (0–2.5 µM). |
Time‐ and concentration‐dependent inhibition of plague formation. Viral titres reduced by 23%, 68% and 70% for ivermectin concentrations of 0.5, 1.0 and 1.5 µM at 72 hr. No effect on adsorption and cell entry (n = 3) |
[11] |
|
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‐CoV‐2 (Aus/VIC01/2020) |
Vero/hSLAM cells, MOI of 0.1 for 2 hr, followed for 72 hr. Ivermectin tested at (0–10 µM). |
Time‐ and concentration‐dependent inhibition of viral replication. Cell associated virus E‐gene EC50 = 2.8 µM Cell associated virus RdRp‐gene EC50 = 2.5 µM (n = 3). |
[28] |
|
Newcastle disease virus (LaSota vaccine strain) |
9‐day chick embryos, 50% egg infective dose, followed for 6 hours. Ivermectin tested at (0–229 µM) |
Dose‐dependent reduction of viral replication. Log2 reduction EC50 = (71 ± 33) µM (n = 5). |
[58] |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 3.
Evaluation of the antiviral activity of ivermectin in vivo in multicellular organisms.
| Target virus and invertebrate hosts | Infection conditions | Quantitative and qualitative effects | References |
|---|---|---|---|
| Zika virus (Cambodia, FSS13025) | Aedes aegypti fed infected blood MOI = 0.5. Viral replication assessed in midgut 7 days post‐infection. Ivermectin at 10 nM. | Ivermectin had significant mosquitocidal effect but no effect on infection and replication of Zika virus in Aedes aegypti 7 days after infection. Control: 104(103–105) PFU/midgut, Ivermectin: 104 (102–105) PFU/midgut (n = 15) | [23] |
| West Nile virus (Colorado strain) | Wild Culex tarsalis trapped at pilot field trials sites with ivermectin‐treated bird feed (200 mg/kg diet) or control in an endemic area. | Ivermectin had no effect on bird health but was significantly mosquitocidal. Ivermectin had no effect on the number of Culex tarsalis pools and the average infection rates (MLE). Control: 14 (4–24), Ivermectin: 5 (0–9) (n = 136–1316) | [24] |
| Dengue virus‐2 | Aedes albopictus (3–5 days) post‐larvae were fed with Dengue virus contaminated human blood for 4 days and then ivermectin in blood for 6 days at (0–64 ng/mL). |
At (16, 32 and 64) ng/mL, ivermectin significantly reduced viral replication in Aedes albopictus. Control: 85 (81–90)%, Ivermectin 64 µg/mL: 43 (40–45)%. Ivermectin significantly inhibited viral replication. At 64 ng/mL, viral copies/mL reduced by 100 (84–100)% (n = 61–65) |
[21] |
|
Bluetongue virus (BTV‐17) Epizootic haemorrhagic disease virus (EHDV‐2) |
Female Culicoides sonorensis were fed blood from ivermectin‐treated animals, mixed with viruses 7log10 TCID50 EHDV‐2 and 200 µg/kg ivermectin in elk blood. 7log10 TCID50 BTV‐17 and 400 µg/kg ivermectin in sheep blood. |
Ivermectin significantly reduced infection and dissemination of BTV‐17. Control: 60%, Ivermectin 400 µg/kg: 18%. Ivermectin had no effect on infection and dissemination of EHDV‐2. Control: 40%, Ivermectin 200 µg/kg: 38% (n 100). | [13] |
| Gill parvovirus of crayfish and parvo‐like virus of crayfish |
Fresh water crayfish with pre‐existing gill parvovirus were given ivermectin (3, 6 and 7 µg/kg, i.m). Ivermectin (7 µg/kg, i.m) was also given before, same day and after experimental infection with parvo‐like virus. |
Ivermectin decreased the number of hypertrophied nuclei (68%) in gills of crayfish with pre‐existing parvo‐like virus. Control: 1591 ± 392.33 Ivermectin 3 µg/kg: 1039.85 ± 383.96 Ivermectin 7 µg/kg: 671 ± 379.07 (n = 20). Ivermectin modestly extended longevity of crayfish after experimental infection with parvo‐like virus. |
[22] |
| Target virus and vertebrates hosts | |||
| Porcine circovirus‐2 (SH1 strain) | Piglets (30 days old) were infected (5 × 104 TCID50 i.m). Ivermectin (0.2 mg/kg i.m) at days 2, 4 and 6 post‐infection. Monitored 21 days | Ivermectin decreased replication; viral copies/titres in serum, brain, heart, kidneys, liver, lungs spleen and lymph nodes. Day 21 serum viral copies. Control: 5.1 (4.9–5.5) log10, Ivermectin: 2.4 (2.0–2.9) log10 (n = 3) | [25] |
| Zika virus (Senegal strain) | Five‐week‐old Ifnar1−/− mice were infected with 103 PFU in the footpad. Ivermectin (4 mg/kg, i.p), 2 days pre‐infection and then days 1, 2 and 4 post‐infection. |
Ivermectin did not prevent infection or inhibit mortalities. Control: 100% mortality, Ivermectin =100% mortality (n = 7–8) |
[26] |
| Suid herpesvirus 1 | Female BALB/c mice (6–8 weeks old) were infected (~106 TCID50/mL). Ivermectin (0.2 mg/kg) at infection or at 12 hr post‐infection; 10 days monitoring. |
Ivermectin inhibited replication; decreased viral DNA copies/titres in the brain and kidneys. Reduced mortality at day 7. Control: 100%, Ivermectin (at 12 hr) = 50% Ivermectin (at 0 hr) = 40% (n = 10) |
[11] |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 2.
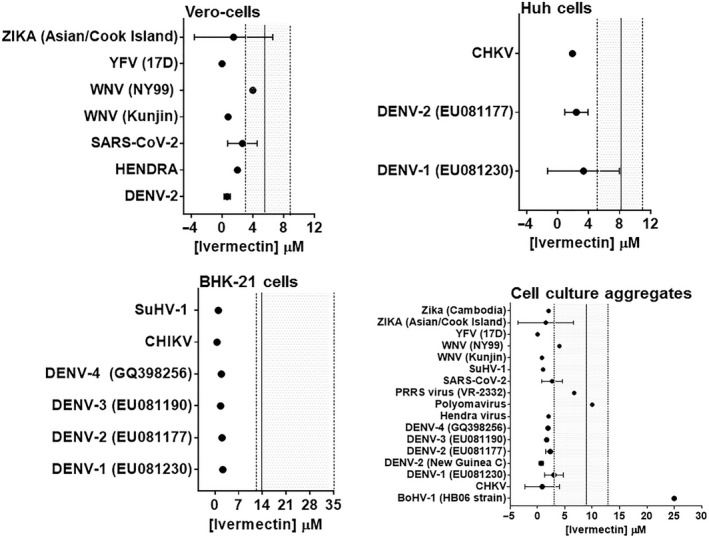
Plots of the quantitative evaluation of the selectivity index of ivermectin against viral infections in different mammalian cell lines. Data points (solid circles) represent mean ±SD or single concentration values of ivermectin that inhibited viral activity by 50% (EC50). The solid vertical line and the shaded area represent the average of concentration of ivermectin that caused cytotoxicity in 50% of utilized mammalian cells and 95% CI, respectively. High selectivity is indicated by EC50 values outside and to the left of the 95% CI. BHK‐21, Baby hamster kidney cells; Huh, Human liver cells; Vero, Monkey kidney epithelial cells; BoHV, Bovine herpesvirus; CHKV, Chikungunya virus; DENV, Dengue virus; PRRS, Betaarterivirus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus; SuHV, Suid herpesvirus; WNV, West Nile virus; YFV, Yellow fever virus; and Zika, Zika virus.
3.2. Qualitative and quantitative antiviral effects of ivermectin based on in vivo studies in animals
In animals, antiviral effects of ivermectin have been examined in different arthropod models of infection including mosquitoes, biting midges and crayfish, and mammalian hosts including mice and pigs (Table 3). At a wide range of nanomolar to micromolar concentrations that had no effect on arthropods, ivermectin significantly inhibited infection and/or replication of Dengue virus in Aedes albopictus [21], Bluetongue virus in Culicoides sonorensis [13] and parvovirus of crayfish [22]. However, ivermectin had no effect on infection and/or dissemination of Zika virus in Aedes aegypti [23], West Nile virus in Culex tarsalis [24] and Epizootic haemorrhagic disease virus in Culicoides sonorensis (Figure 3 ). In mammalian hosts, administration of 0.2 mg/kg of ivermectin for 2–6 days inhibited the replication of Porcine circovirus in visceral organs including the brain, liver, heart, kidneys, spleen and lymph nodes over 21 days in piglets [25]. A single ivermectin dose of 0.2 mg/kg did not prevent infection but it inhibited replication of Suid herpesvirus in the brain and kidneys, and mortality at day 7 post‐infection in mice [11]. One study showed that administration of 4 mg/kg of ivermectin for 2 days before infection and at days 1, 2 and 4 after infection did not prevent the infection or mortalities caused by Zika virus in mice [26]. There was significant heterogeneity in all animal studies examined herein (I 2 = 98%; p < 0.00001; Figure 3 ). While these data necessitate scrutiny and qualification of each study individually, pooled meta‐analyses showed that the antiviral effects of ivermectin outlined above were statistically significant. Standardized mean differences and 95% confidence intervals for ivermectin versus respective controls were −3.95 (−5.60, −2.30) (Z = 4.69; p < 0.00001; Figure 3 ) for tests in arthropods and −5.71 (−8.91, −2.51) (Z = 3.50; p < 0.0005; Figure 3 ) for tests in mammals.
Figure 3.
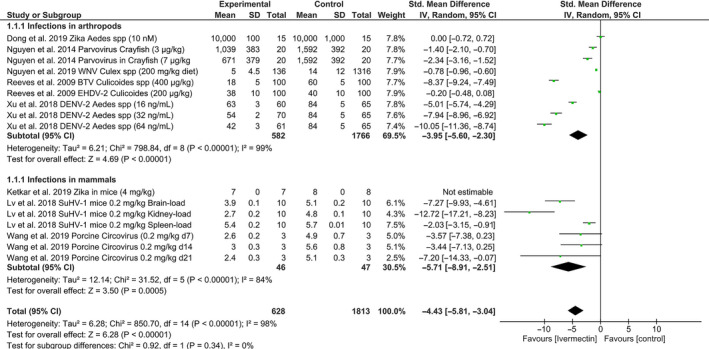
A forest plot showing effects of ivermectin on infection and transmission of different viruses in arthropod and mammalian models. Meta‐analyses included 4 studies for infections in arthropods and 3 studies for mammalian hosts. Data include all comparators over time, dose range and assessments of viral replication in multiple organs where indicated. Comparisons were made using standard mean differences and a random effects model. BTV, Bluetongue virus; DENV, Dengue virus; EHD, Epizootic haemorrhagic disease virus; SuHV, Suid herpesvirus; WNV, West Nile virus; and Zika, Zika virus.
3.3. Identified nuclear location signals in genomes of studied viruses
Since the mode of action of ivermectin was shown to be interfering with the action of importin α (see Introduction) which aids the transit of viral proteins through the nuclear pore complex into the nucleus, then the role of nuclear location signals (NLSs) necessary for this transit of viral proteins was examined. Of the studies of ivermectin against viruses in cell cultures, 16.6% were viruses with a DNA genome, while the remainder, surprisingly, were RNA viruses (84.4%) dominated by the family Flaviviridae (59%), in particular Dengue viruses (Table 2). Perhaps this should not have been surprising as Pryor et al. [27] demonstrated multiple NLS in Dengue virus, particularly in protein N5, while Wagstaff et al. [5] showed that ivermectin could clearly block Dengue virus replication. Our analysis of the proteins of Dengue virus 2 found four of the ten major proteins had possible NLS that would allow transit into the nucleus (Table 4). This might explain the dominance of flaviviruses in our analysis.
Table 4.
The possible nuclear location signals in Dengue and SARS‐CoV‐2 viruses.
| Virus | Proteins 5′–3′ Direction | Possible Nuclear Location Signal |
|---|---|---|
| Dengue Virus 2 | Anchored capsid protein | 4QRKK A K;67 KRWGTI KKSK;73 KKSKAINVLRGFRKEIGRMLNILNRRRRS |
| NC_001474.2 | Membrane glycoprotein precursor | 199EHRREKRS |
| Envelope protein | None | |
| Nonstructural protein NS1 | None | |
| Nonstructural protein NS2A | [1399SRTSKKR\S] | |
| RNA helicase NS3 | 1616 D KKGK;1659 RKRR;1931QRRGR;2062ERKKLK | |
| Nonstructural protein NS4A | None | |
| Protein 2 K | None | |
| Nonstructural protein NS4B | None | |
| RNA‐dependent RNA polymerase NS5 | 2862 KKLMKITAEWLWKELGKKK;2948 KREKK;3378 KRFRR | |
| SARS‐CoV‐2 | ORF1ab; (nsp8) | 3148 KK V K;3688 KK I K;3952 KK‐21aa‐KKCK;4156 RK‐15aa‐QRK Y K; |
| NC_045512.2 | 4960 KK W K;11932 KK L KK;11995 RK‐20aa‐D KR A K; | |
| gene S | None | |
| ORF3a | None | |
| gene E | None | |
| gene M | None | |
| ORF6 | None; bait sequence | |
| ORF7a | None | |
| ORF7b | None | |
| ORF8 | None | |
| gene N | 742 KKSAAEASKKPRQKR;1104PKK D KKKK; [1147PQRQKK] | |
| ORF10 | None |
Basic amino acids are bolded; possibly disrupting bulky and hydrophobic amino acids are red; possible near NLS are in [brackets]. The amino acid codes are in single letter format for efficient space utilization.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The results of Caly et al. [28] demonstrated marginal selective activity of ivermectin against SARS‐CoV‐2 (Figure 2 ). This led them to hypothesize that there might be NLS in the proteins of SARS‐CoV‐2 and a similar mechanism might be at work. An extremely thorough investigation of open reading frame 6 (ORF6) (ORF7 in the NCBI entry) of severe acute respiratory syndrome coronavirus (SARS‐CoV‐1) demonstrated that its 3′ mock NLS was a sequestrating bait for importin β, while the 5′ end was transmembrane‐anchored into the membranes of the rough endoplasmic reticulum/Golgi apparatus [29]. This led to sequestration of importin β, downregulating the STAT1 signalling function and preventing cells from producing interferon γ via the interferon regulatory factor (IRF) genes. This prevents the cell from entering a virus‐resistant state. ORF6 of SARS‐CoV‐2 is identical to SARS for 42 of 61 amino acids (aa); similar (functional, non‐homologous replacement) for a further 12 of 61 aa (Figure 4 ). By substituting hydrophobic Ala at the 3′ end, Frieman et al. [29] demonstrated the critical motif was aa(49−53), but not aa(54−58) or aa(59−63). Examination of this area shows a possible bipartite NLS spanning Lys+Lys46 to unconventional Tyr+Pro63 motif (see Materials and methods). The experimental hydrophobic series of five Ala is right behind the leading basic duo Lys‐Lys, thus disrupting the binding in this area and functionality [29]. SARS‐CoV‐2 area is identical to SARS at 9/16 aa and similar at 5/9 aa, predicting an almost similar bait/mock NLS activity for importin β (Figure 4 , Table 4). An anomaly is that SARS‐CoV‐2 ORF is two aa shorter missing the terminal Tyr‐Pro. A check (Aug 2020) of all the SARS‐CoV‐2 ORF6 sequences in NCBI GenBank all terminated here demonstrating it is real, not a strain artefact and not a sequencing error. However, a few factors suggest this is not at all acting as an NLS but more as a mock NLS with its positive charged tail (sequestering bait) from Arg/Lys38 onwards, trapping and effectively downregulating importin β. These factors include the non‐conventional nature of the possible NLS sequence, the changes in SARS‐Cov‐2 from SARS‐CoV‐1 where the leading Lys‐Lys46 are changed to Glu‐Asn46 and, the loss of unconventional Tyr‐Pro63 trailing bipartite signal.
Figure 4.
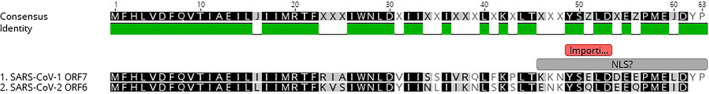
An illustration of NLS in the open reading frame 6 (ORF6) (ORF7 in the NCBI entry) of severe acute respiratory syndrome coronavirus (SARS‐CoV‐1) and SARS‐CoV‐2. ORF6 of SARS‐CoV‐2 was identical to SARS‐CoV‐1 for 42 of 61 amino acids (aa); similar (functional non‐homologous replacement) for a further 12 of 61 aa. SARS‐CoV‐2 NLS was identical to SARS‐CoV‐1 at 9/16 aa and similar at 5/9 aa, predicting an almost similar bait activity for importin β. SARS‐CoV‐2 ORF was two aa shorter missing the terminal Tyr‐Pro. The small red box shows the area experimentally disrupted by substituting bulky Ala [27].
The apparent critical role of this short protein in coronaviruses evading the innate immune response would lead to it being a major target for small interfering RNA (siRNA) degradation delivered in liposomes via a nasal spray after swabbing for testing for SARS‐CoV‐2 (see review of La Fauce and Owens) [30]. Formiga et al. [3] outlined other possible delivery systems for micro‐ and nanoparticles. If a longer lasting therapeutic was needed, then short hairpin RNA (shRNA) delivered in a plasmid could be substituted instead [30]. Indeed, Shi et al. [31] have shown siRNA was effective against structural proteins of SARS‐CoV in Vero cells with a 70% reduction. A quick analysis of the ORF6 sequence for siRNA targeting using siDirect version 2.0,[32] identified multiple candidates including one at 136–158 bp with reduced off‐target effects which would cleave and then degrade the 3′ bait/mock NLS in the area that was identified by Frieman et al. [29] as critical for activity.
A recent review paper on antivirals against coronaviruses towards controlling SARS‐CoV‐2 did not identify any drug targeting ORF6 [33], so given the above information, we suggest this might be a fruitful target for an antiviral. In ORF1ab, seven potential NLSs were identified (Table 4) notably all of them Chelsky style NLS (Table 1), either simple or bipartite. Interestingly, all but the bipartite one at 3952(Lys‐Lys‐21aa‐Lys‐Lys‐Cys‐Lys) have a disrupting bulky or hydrophobic aa in the Chelsky signal that would likely prevent them operating as a NLS. It appears as if evolutionary pressure has silenced these NLSs so the viral proteins are not sequestered into the nucleus. On the other hand, we can see no logical reason why 3952NLS would not be functional, so the translated and cleaved protein, nsp8, would be moved sometimes into the nucleus by importin α. Thus, ivermectin should have a role in slowing the translocation of nsp8 into the nucleus. However, nsp8 role is to act with nsp7 as a co‐factor to nsp12 which is the highly conserved viral RNA‐dependent RNA polymerase [34] necessary for viral replication. Nsp8 has several DNA and RNA‐binding residues [34], which may suggest an undetected role with the DNA in the nucleus, the major source of DNA in a cell. It is speculated that this role might be to mine nucleotides for viral replication.
Gene N (Table 4), encoding for the nucleocapsid protein, has a large area of basic aa (742aa onwards) in which there are many strong potential NLSs, both Chelsky style and hexapeptide strings of basic Lys with a Glu after the first two Lys. However, using confocal microscopy of intact N protein there was no evidence of SARS‐CoV‐1 coronavirus nucleocapsid proteins being found in the nucleus or nucleoli despite the identification of the same or similar NLS found by ourselves and Rowland et al. [35] On the other hand, Timani et al. [36] also using confocal microscopy on experimental fragments of the N protein of SARS‐CoV‐1 demonstrated accumulation of these fragments in the nucleus and nucleolus, suggesting functional NLSs. In a brilliant paper using electron microscopy by Wolff et al., [37] the nucleocapsid proteins are in the cytosol capturing the viral RNA as it leaves the double membraned viral replication organelle. Taken all together, it is most likely, the positive charged, mock NLSs capture and coat the negative charged phosphate backbone of the viral RNA to start the formation of the virions in the cytosol. Therefore, these mock NLSs in intact N protein [35] do not have the opportunity to function as NLS as they are immediately sequestered by viral RNA in the cytosol.
4. DISCUSSION
This study presents an integrative review with a critical appraisal of the qualitative and quantitative antiviral properties of ivermectin. For in vitro studies, susceptibility to ivermectin based on established EC50 values seemed to depend on the virus strain in some cases (Table 1 and Figure 2 ). For instance, the Kunjin strain of West Nile virus was more susceptible than the NY99 strain with a fivefold difference in EC50 values for viral replication. Similarly, evaluations of relative potency and safety margins in different mammalian cells revealed that ivermectin exerted no selective antiviral activity against Betaarterivirus, Venezuelan equine encephalitis virus, Equine herpesvirus 1 and Bovine herpes virus 1. While there are no clear and apparent reasons for these species or strain differences in susceptibility to ivermectin, this may be attributable, at least in part, to specific differences in the molecular targets of ivermectin.
Another important consideration relates to potency, relative selectivity and toxicity of ivermectin. Our evaluation of in vitro studies showed that ivermectin exerts selectivity for some viruses in ex vivo mammalian cell infection models utilizing Vero, Huh and BHK cells. However, the micromolar concentration range required to inhibit replication by 50% for most viruses may be a cause for concern. Clinically approved formulations of ivermectin can be administered orally, subcutaneously, intramuscularly or topically with a recommended dose range of 150–200 µg/kg in humans and 6–500 µg/kg in animals depending on species and formulation, and the indicated clinical applications. With this dose range, pharmacokinetic characterizations have shown that attainable peak plasma concentrations increase with dose and may range from 3–48 ng/mL in dogs, 21–82 ng/mL in horses, 7–40 ng/mL in pigs, 9–60 ng/mL in sheep, 12–133 ng/mL in cattle and 20–81 ng/mL in humans [38, 39, 40]. A study on safety and tolerability of escalating doses of ivermectin in healthy humans showed that a single dose (120 mg) that is 10‐fold bigger than the clinically recommended dose (200 µg/kg) was well tolerated and it yielded a peak plasma concentration equivalent to 248 ng/mL with an elimination half‐life of 19 hr [41]. Similarly, population‐based pharmacokinetic modelling revealed that ivermectin administered orally for three days at 600 µg/kg would yield maximal median plasma concentrations of 105–119 ng/mL (0.12–0.14 µM) and an elimination half‐life of 3–5 hr [42]. These data indicate that even with extremely high doses of ivermectin, attainable peak plasma concentrations would remain markedly lower than established EC50 concentrations for most viruses in vitro, albeit significantly higher than 0.5–1 ng/mL that is optimal for the anthelmintic activity. The use of extremely high doses of ivermectin would increase the prospect of adverse drug‐drug interactions in patients requiring polypharmacy, as is often the case in viral infections [2, 43]. It is uncertain, therefore, that the utilization of immortalized neoplastic cell lines in vitro will effectively determine the selectivity of ivermectin and represent its potential clinical efficacy against viral infections in vivo. Thus, if ivermectin is to be repurposed as an antiviral agent, established antiviral properties based in vitro experiments should be critically evaluated in validated models of infection in animals in vivo. We show that a limited number of studies have examined antiviral effects of ivermectin in multicellular organisms. Most tests have been done in arthropod models of infection with only three experimental studies in mammals, and several registered but yet inconclusive human trials against SARS‐Cov‐2 [2] and Dengue virus at ClinicalTrials.gov. Collective efficacy data are promising given that pooled meta‐analyses demonstrate a significant antiviral effect overall (Table 2 and Figure 3 ). However, caution should be exercised in interpreting these data as the applicability will depend on broader questions such as which viral and animal species should be targeted, what would be the optimal dosing regimen and what costs or benefits would ensue. All these questions notwithstanding, the merits and potential applications of some individual studies are worth noting. In a study on Cherax quadricarinatus crayfish with pre‐existing gill parvovirus, for example, non‐toxic, well‐tolerated intramuscular doses of ivermectin (3–7 µg/kg) significantly reduced lesions associated with this infection [22]. Since this viral infection is an important cause of economic loss in farmed crustaceans such as prawns in aquaculture, this selective antiviral activity may offer an additional tool to control infections. For prawns and crayfish particularly, application trials would need to consider formulations and dosing regimen of ivermectin that are commercially viable, suitable for large‐scale administration and, with minimal public health concerns.
Another promising prospect is in the application of ivermectin to target infection, replication and transmission of arboviruses within arthropod vectors. It is shown that nanomolar concentrations of ivermectin that were not arthropodicidal significantly reduced infection and dissemination of Bluetongue virus in Culicoides sonorensis and Dengue virus in Aedes albopictus mosquitoes (Table 3, Figure 3 ). In contrast, a similar strategy of feeding ivermectin‐treated blood showed that nanomolar concentrations of ivermectin were arthropodicidal against Culex tarsalis, Aedes aegypti and Culicoides sonorensis, but with no significant effect on intra‐vector infection rates and replication of West Nile virus, Zika virus and Epizootic haemorrhagic disease virus, respectively (Table 3). Ivermectin already has approved label indications for the control of ticks, mites, flies and lice on livestock in agriculture and, scabies and filariasis in humans and companion animals. In humans, the recommended dose (150 mg/kg) of ivermectin used to treat filarial infections yielded plasma concentrations that significantly reduced the survival of Anopheles mosquitoes and the transmission of malaria [44, 45, 46]. Given the demonstrated antiviral effects of ivermectin in arthropods, a parallel application could be treating humans and livestock with clinically approved doses of ivermectin to reduce arthropod vector abundance and lower infection rates as well as transmission of arboviruses such as Bluetongue virus for which susceptibility has been demonstrated. Interestingly, after a single therapeutic dose of ivermectin in humans, the attained peak plasma concentration (40–45 ng/mL) closely matches the concentration range (16–64 ng/mL) that was effective in reducing the replication of Dengue virus in Aedes albopictus [21]. While there is potential for this particular application to control disease in mammals, using ivermectin as a strategic tool in controlling arboviruses and associated diseases can only be effective if it is preceded with a critical evaluation of the direct health, social and economic benefits. Such detailed epidemiological and economic evaluations are beyond the scope of the current review but are necessary given the significant economic losses associated with arboviruses. Annual global livestock economic losses attributable to reduced milk production, loss of body condition, veterinary treatments and diagnostics, and mortalities due to infections with Bluetongue virus have been estimated at 3.0 billion US dollars [47]. In humans, there is an estimated annual total of 58.4 million symptomatic dengue virus infections with a total global cost of 9 billion US dollars annually [48].
In vertebrate, mammalian models of viral infection, effects of ivermectin as an antiviral agent have been tested and reported in only 3 peer‐reviewed studies to date [11, 25, 26]. Pooled data from these 3 studies show a strong general antiviral effect but results from individual studies are equivocal thus far. At much higher dose (4 mg/kg) one day before infection and for 3 days after infection with a Senegalese strain of Zika virus in 5‐week‐old Ifnar1−/− mice, ivermectin did not inhibit infections or prevent mortalities [26]. Based on pharmacokinetic evaluations and the need for a much higher dose of ivermectin to match micromolar concentrations that were effective against Zika virus in vitro, this result is not surprising and may seem to discourage any follow‐up pre‐clinical trials against this virus in laboratory animals. It is worth noting, however, that this particular study had a number of limitations and ranked poorly on the qualitative evaluation of individual studies. The test was conducted in relatively young mice with a homozygous interferon alpha/beta receptor subunit gene knockout (see Introduction). Inherently, these mice are highly susceptible to viral infections and this may have contributed to marked mortality in the small number of mice tested, despite administering ivermectin. In addition, this study did not evaluate tissue viral loads in response to treatment with ivermectin or confirm that mortalities were indeed due to infection with Zika virus. These arguments seem to be supported at least in part, by contrasting results from two other in vivo tests utilizing different viruses and animal models of infection. In 6‐ to 8‐week‐old BALB/c mice and 30‐day‐old piglets, ivermectin (0.2 mg/kg) caused a significant reduction in the replication and viral DNA copies in visceral organs following infection with Suid herpesvirus and Porcine circovirus, respectively [11, 25]. Interestingly, these data are discordant with results from in vitro tests where micromolar concentrations of ivermectin were required to inhibit viral replication of Porcine circovirus in PK‐15 cells and Suid herpesvirus in BHK‐21 cells. This suggests that for some viral infections, ivermectin at currently recommended therapeutic doses may exert efficacy in vivo even if effective concentrations in vitro are not attainable without causing considerable toxicity. This argument has also been advanced previously [6, 8], and it seems plausible because the proposed antiviral mechanism targets mammalian cell proteins that are important for intracellular transport. These critical functions are then hijacked by viruses to enhance viral replication. The fact that ivermectin may serve as a mammalian host‐directed antiviral agent implies that reducing viral load by even a modest amount at a low dose could be supplementary in enhancing the immune system in fighting viral infections [49]. Indeed, immune stimulatory effects of ivermectin have been documented [50], with treatment at 0.2 mg/kg significantly enhancing antibody production against sheep red blood cells as well as helper T lymphocyte and macrophage‐dependent responses in the CD‐1 strain of mice. Together, these observations may negate the need for comprehensive toxicological re‐evaluation of higher ivermectin doses for putative use as a broad‐spectrum antiviral agent, but carefully designed pre‐clinical and clinical trials to confirm these effects are still needed.
With regard to coronaviruses and the possible mode of action of ivermectin by blocking the NLS of importin α, we can only find one efficiently functional NLS in the co‐factor protein nsp8 that is not logically rapidly sequestered for other viral functions. Three proteins of SARS‐CoV‐2 had apparent or mock NLS; nsp8, ORF6 and the nucleocapsid protein N. Logically, mostly these proteins will be utilized quickly for other functions outside the nucleus and only minor leakage into the nucleus would occur except for possibly nsp8 that does not have an essential role, just a co‐factor role outside the nucleus and this needs closer scrutiny. It appears the application of ivermectin should have a clinical effect that is poorly understood at the moment. Of considerable interest is the paper of Giri et al. on the intrinsically disordered protein regions of the SARS‐like coronaviruses examined by a computational approach [34]. Completely independently, using vastly different methods, they identified only the nucleocapsid N, nsp8 and ORF 6 as proteins of high intrinsic disorder. We were not aware of this publication when conducting our review, so we find it incongruous that the same proteins were identified by total independent scientific approaches and different goals. This intriguing co‐incidence deserves further scrutiny.
5. CONCLUSION
This review provides a critical assessment of the potential for repurposing ivermectin (a clinically approved drug for parasitic infections) as a broad‐spectrum antiviral drug. Molecular studies have identified the inhibition of nuclear translocation of viral proteins, facilitated by mammalian host processes as the main target. Other off‐target effects such as stimulation of immune responses against viral infections are possible but have not been directly investigated. The bulk of current knowledge in this field comes from in vitro studies done by infecting cell cultures. Testing of the antiviral effects of ivermectin in in vivo animal infection models is very limited but the available data are promising, and this may be particularly true for infections with arboviruses. Given that viral infections remain one of the major causes of economic losses in medicine and agriculture, the potential to develop ivermectin as an additional antiviral agent should be pursued with an emphasis of pre‐clinical trials in validated models of infection. However, given the coronavirus attack on importin β, the use of ivermectin to further block importin α seems counter‐intuitive to being a high priority treatment in the clinical arena without further pharmacological investigation of ivermectin. Nevertheless, there does appear to be a functional NLS in ORF1ab encoding the cleaved protein nsp8 which would be hampered by ivermectin and this should be examined further as a priority.
6. AUTHORS’ CONTRIBUTION
Both authors (RTK and LO) contributed equally to the conception of research ideas, literature searches and appraisal, data extraction and validation, and writing of the manuscript.
7. ETHICAL APPROVAL
This research work did not require direct experimentation and the use of animals or humans to generate data. No ethics approval was required.
DECLARATION OF INTERESTS
None to declare.
Funding information
This research did not receive any funding from agencies in the public, commercial or not‐for‐profit organizations. The study was carried out as part of our routine work.
REFERENCES
- 1. Heidary F, Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID‐19 complementary regimen. J. Antibiot. (Tokyo). 2020;73:593‐602. 10.1038/s41429-020-0336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jans DA, Wagstaff KM. Ivermectin as a broad‐spectrum host‐directed antiviral: the real deal? Cells. 2020;9(9):2100‐ 10.3390/cells9092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Formiga FR, Leblanc R, Reboucas JDS et al. Ivermectin: an award‐winning drug with expected antiviral activity against COVID‐19. J. Control Release. 2020; 329: 758–761. 10.1016/j.jconrel.2020.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mastrangelo E, Pezzullo M, Burghgraeve T et al. Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 2012;67:1884‐1894. 10.1093/jac/dks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin as specific inhibitor of importin α/β‐mediated nuclear import able to inhibit replication of HIV‐1 and dengue virus. Biochem. J. 2012;443:851‐856. 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tay MYF, Fraser JE, Chan WKK et al. Nuclear localization of dengue virus (DENV) 1–4 non‐structural protein 5; protection against all 4 DENV serotypes by the inhibitor ivermectin. Antiviral Res. 2013;99:301‐306. 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 7. Croci R, Bottaro E, Chan KWK et al. Liposomal systems as nanocarriers for the antiviral agent ivermectin. Int. J. Biomater. 2016;2016:1‐15. 10.1155/2016/8043983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang SNY, Atkinson SC, Wang C et al. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 2020;177:104760. 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 9. Lundberg L, Pinkham C, Baer A et al. Nuclear import and export inhibitors alter capsid protein distribution in mammalian cells and reduce Venezuelan Equine Encephalitis Virus replication. Antiviral Res. 2013;100:662‐672. 10.1016/j.antiviral.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 10. Wagstaff KM, Rawlinson SM, Hearps AC, Jans DA. An alphaScreen®‐based assay for high‐throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen. 2011;16:192‐200. 10.1177/1087057110390360. [DOI] [PubMed] [Google Scholar]
- 11. Lv C, Liu W, Wang B et al. Ivermectin inhibits DNA polymerase UL42 of pseudorabies virus entrance into the nucleus and proliferation of the virus in vitro and vivo. Antiviral Res. 2018;159:55‐62. 10.1016/j.antiviral.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 12. Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group . Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. J. Clin. Epidemiol. 2009;62:1006‐1012. 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 13. Reeves WK, Nol P, Miller MM, Jones GZ. Effects of ivermectin on the susceptibility of Culicoides sonorensis (Diptera: Ceratopogonidae) to bluetongue and epizootic hemorrhagic disease viruses. J. Vector Ecol. 2009;34:161‐163. 10.1111/j.1948-7134.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 14. Hooijmans CR, Rovers MM, de Vries RB. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014;14:43. 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? BioEssays. 2000;22:532‐544. . [DOI] [PubMed] [Google Scholar]
- 16. Boulikas T. Putative localization signals (NLS) in protein transcription factors. J. Cell Biochem. 1994;55:32‐58. 10.1002/jcb.240550106. [DOI] [PubMed] [Google Scholar]
- 17. Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411‐415. 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou W, Zhang J, Yang B, Zhou L, Hu Y. The nuclear localization signal of the NS1 protein is essential for Periplaneta fuliginosa densovirus infection. Virus Res. 2009;145:134‐140. 10.1016/j.virusres.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 19. Lee YJ, Lee C. Ivermectin inhibits porcine reproductive and respiratory syndrome virus in cultured porcine alveolar macrophages. Arch. Virol. 2016;161:257‐268. 10.1007/s00705-015-2653-2. [DOI] [PubMed] [Google Scholar]
- 20. Raza S, Shahin F, Zhai W et al. Ivermectin inhibits bovine herpesvirus 1 DNA polymerase nuclear import and interferes with viral replication. Microorganisms. 2020;8:409. 10.3390/microorganisms8030409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu T, Han Y, Liu W et al. Antivirus effectiveness of ivermectin on dengue virus type 2 in Aedes albopictus. PLoS Negl. Trop. Dis. 2018;12:e0006934. 10.1371/journal.pntd.0006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen KY, Sakuna K, Kinobe R, Owens L. Ivermectin blocks the nuclear location signal of parvoviruses in crayfish, Cherax quadricarinatus. Aquaculture. 2014;420–421:288‐294. 10.1016/j.aquaculture.2013.11.022. [DOI] [Google Scholar]
- 23. Dong S, Kang S, Dimopoulos G. Identification of anti‐flaviviral drugs with mosquitocidal and anti‐Zika virus activity in Aedes aegypti. PLoS Negl. Trop. Dis. 2019;13:e0007681. 10.1371/journal.pntd.0007681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nguyen C, Gray M, Burton TA et al. Evaluation of a novel West Nile virus transmission control strategy that targets Culex tarsalis with endectocide‐containing blood meals. PLoS Negl. Trop. Dis. 2019;13:e0007210. 10.1371/journal.pntd.0007210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X, Lv C, Ji X et al. Ivermectin treatment inhibits the replication of Porcine circovirus 2 (PCV2) in vitro and mitigates the impact of viral infection in piglets. Virus Res. 2019;263:80‐86. 10.1016/j.virusres.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 26. Ketkar H, Yang L, Wormser GP, Wang P. Lack of efficacy of ivermectin for prevention of a lethal Zika virus infection in a murine system. Diagn. Microbiol. Infect. Dis. 2019;95:38‐40. 10.1016/j.diagmicrobio.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pryor MJ, Rawlinson SM, Butcher RE et al. Nuclear localization of dengue virus nonstructural protein 5 through its importin α/β‐recognized nuclear localization sequences is integral to viral infection. Traffic. 2007;8:795‐807. 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 28. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA‐approved drug ivermectin inhibits the replication of SARS‐CoV‐2 in vitro. Antiviral Res. 2020;178:104787. 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frieman M, Yount B, Heise M et al. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007;81:9812‐9824. 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. La Fauce K, Owens L. RNA interference with special reference to combating viruses of crustacean. Indian J. Virol. 2012;23:226‐243. 10.1007/s13337-012-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi Y, Yang DH, Xiong J et al. Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res. 2005;15:193‐200. 10.1038/sj.cr.7290286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naito Y, Ui‐Tei K. siRNA design software for a target gene‐specific RNA interference. Front. Genet. 2012;3:102. 10.3389/fgene.2012.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santos IA, Grosche VR, Bergamini FRG, et al. Antivirals against coronaviruses: candidate drugs for SARS‐CoV‐2 treatment? Front. Microbiol. 2020;11:1818. 10.3389/fmicb.2020.01818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giri R, Bhardwaj T, Shegane M et al. Understanding COVID‐19 via comparative analysis of dark proteomes of SARS‐CoV‐2, human SARS and bat SARS‐like coronaviruses. Cell Mol. Life. Sci. 2020;25:1‐34. 10.1007/s00018-020-03603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rowland RRR, Chauhan V, Fang Y et al. Intracellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein: absence of nucleolar accumulation during infection and after expression as a recombinant protein in vero cells. J. Virol. 2005;79:11507‐11512. 10.1128/JVI.79.17.11507-11512.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Timani KA, Liao Q, Ye L et al. Nuclear/nucleolar localization properties of C‐terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23‐34. 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wolff G, Limpens RWAL, Zevenhoven‐Dobbe JC et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395‐1398. 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Canga AG, Prieto AMS, Liebana MJD et al. The pharmacokinetics and interactions of ivermectin in humans—a mini‐review. AAPS J. 2008;10:42‐46. 10.1208/s12248-007-9000-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Canga AG, Prieto AMS, Liebana MJD et al. The pharmacokinetics and metabolism of ivermectin in domestic animal species. Vet. J. 2009;179:25‐37. 10.1016/j.tvjl.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 40. Navarro M, Camprubi D, Requena‐Mendez A et al. Safety of high dose ivermectin: a systematic review and meta‐analysis. J. Antimicrob. Chemother. 2020;75:827‐834. 10.1093/jac/dkz524. [DOI] [PubMed] [Google Scholar]
- 41. Guzzo CA, Furtek CI, Porras AG et al. Safety, tolerability, and pharmacokinetics of escalating high doses of ivermectin in healthy adult subjects. J. Clin. Pharmacol. 2002;42:1122‐1133. 10.1177/009127002401382731. [DOI] [PubMed] [Google Scholar]
- 42. Smit MR, Ochomo EO, Waterhouse D et al. Pharmacokinetics‐pharmacodynamics of high‐dose ivermectin with dihydroartemisinin‐piperaquine on mosquitocidal activity and QT‐prolongation (IVERMAL). Clin. Pharmacol. Ther. 2019;105:388‐401. 10.1002/cpt.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lemaitre F, Solas C, Gregoire M et al. Potential drug‐drug interactions associated with drugs currently proposed for COVID‐19 treatment in patients receiving other treatments. Fundam. Clin. Pharmacol. 2020;34:530‐547. 10.1111/fcp.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobylinski KC, Deus KM, Butters MP et al. The effect of oral anthelmintics on the survivorship and re‐feeding frequency of anthropophilic mosquito disease vectors. Acta Trop. 2010;116:119‐126. 10.1016/j.actatropica.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Butters MP, Kobylinski KC, Deus KM et al. Comparative evaluation of systemic drugs for their effects against Anopheles gambiae. Acta Trop. 2012;121:34‐43. 10.1016/j.actatropica.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alout H, Krajacich BJ, Meyers JI et al. Evaluation of ivermectin mass drug administration for malaria transmission control across different West African environments. Malar. J. 2014;13:417. 10.1186/1475-2875-13-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rushton J, Lyons N. Economic impact of Bluetongue: a review of the effects on production. Vet. Ital. 2015;51:401‐406. 10.12834/VetIt.646.3183.1. [DOI] [PubMed] [Google Scholar]
- 48. Shepard DS, Undurraga EA, Halasa YA, Stanaway JD. The global economic burden of dengue: a systematic analysis. Lancet Infect. Dis. 2016;16:935‐941. 10.1016/S1473-3099(16)00146-8. [DOI] [PubMed] [Google Scholar]
- 49. Bray M, Rayner C, Noel F, Jans D, Wagstaff K. Ivermectin and COVID‐19: A report in Antiviral Research, widespread interest, an FDA warning, two letters to the editor and the authors responses. Antiviral Res. 2020;178:104805. 10.1016/j.antiviral. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blakley BR, Rousseaux CG. Effect of ivermectin on the immune response in mice. Am. J. Vet. Res. 1991;52:593‐595. [PubMed] [Google Scholar]
- 51. Varghese FS, Kaukinen P, Glasker S et al. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antiviral Res. 2016;126:117‐124. 10.1016/j.antiviral.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 52. Saudi M, Zmurko J, Kaptein S et al. In search of Flavivirus inhibitors part 2: tritylated, diphenylmethylated and other alkylated nucleoside analogues. Eur. J. Med. Chem. 2014;76:98‐109. 10.1016/j.ejmech.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 53. Pattnaik A, Palermo N, Sahoo BR et al. Discovery of a non‐nucleoside RNA polymerase inhibitor for blocking Zika virus replication through in silico screening. Antiviral Res. 2018;151:78‐86. 10.1016/j.antiviral.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 54. Atkinson SC, Audsley MD, Lieu KG et al. Recognition by host nuclear transport proteins drives disorder‐to‐order transition in Hendra virus V. Sci. Rep. 2018;8:358. 10.1038/s41598-017-18742-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bennett SM, Zhao L, Bosard C, Imperiale MJ. Role of a nuclear localization signal on the minor capsid proteins VP2 and VP3 in BKPyV nuclear entry. Virology. 2015;474:110‐116. 10.1016/j.virol.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shechter S, Thomas DR, Lundberg L et al. Novel inhibitors targeting Venezuelan equine encephalitis virus capsid protein identified using In Silico Structure‐Based‐Drug‐Design. Sci. Rep. 2017;7:17705. 10.1038/s41598-017-17672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Slonska A, Cymerys J, Skwarska J, Golke A, Banbura MW. Influence of importin alpha/beta and exportin 1 on equine herpesvirus type 1 (EHV‐1) replication in primary murine neurons. Pol. J. Vet. Sci. 2013;16:749‐751. 10.2478/pjvs-2013-0106. [DOI] [PubMed] [Google Scholar]
- 58. Azeem S, Ashraf M, Rasheed MA, Anjum AA, Hameed R. Evaluation of cytotoxicity and antiviral activity of ivermectin against Newcastle disease virus. Pak. J. Pharm. Sci. 2015;28:597‐602. [PubMed] [Google Scholar]


