Abstract
To investigate the dynamic changes of Krebs von den Lungen‐6 (KL‐6) among patients with coronavirus disease 2019 (COVID‐19) and the role of KL‐6 as a noninvasive biomarker for predicting long‐term lung injury, the clinical information and laboratory tests of 166 COVID‐19 patients were collected, and a correlation analysis between KL‐6 and other parameters was conducted. There were 17 (10.2%, 17/166) severe/critical and 149 (89.8%, 149/166) mild COVID‐19 patients in our cohort. Serum KL‐6 was significantly higher in severe/critical COVID‐19 patients than in mild patients (median 898.0 vs. 451.2 U/ml, p < .001). KL‐6 was next confirmed to be a sensitive and specific biomarker for distinguishing mild and severe/critical patients and correlate to computed tomography lung lesions areas. Serum KL‐6 concentration during the follow‐up period (>100 days postonset) was well correlated to those concentrations within 10 days postonset (Pearson r = .867, p < .001), indicating the prognostic value of KL‐6 levels in predicting lung injury after discharge. Finally, elevated KL‐6 was found to be significantly correlated to coagulation disorders, and T cells subsets dysfunctions. In summary, serum KL‐6 is a biomarker for assessing COVID‐19 severity and predicting the prognosis of lung injury of discharged patients.
Keywords: COVID‐19, KL‐6, lung injury, prognosis
Highlights
KL‐6 is a biomarker for assessing COVID‐19 severity
KL‐6 predicts the prognosis of COVID‐19 patients
KL‐6 is associated with CT lung lesions areas.
1. INTRODUCTION
The coronavirus disease 2019 (COVID‐19), caused by the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has caused a global pandemic and claimed over 1 million lives by October 31. 1 The clinical spectrum of COVID‐19 ranges from asymptomatic, to mild, to moderate, to severe and critical. Severe and critical patients are characterized by respiratory failure and require oxygen supply and even invasive mechanical ventilation. Although SARS‐CoV‐2 infects could potentially infect various tissues and cells via its receptors angiotensin‐converting enzyme 2 (ACE2) and TMPRSS2, 2 the virus predominately replicates and causes mainly lung injury in clinical observations. Electron microscopy observed the SARS‐CoV‐2 replication foci in bronchial and lung type II pneumocytes and the direct viral‐induced cell lysis. 3 , 4 , 5 Viral invasion in the lung also alerts the innate immune cells per se, which subsequently sends danger signals and recruit other immune cells to the lung for combating the invader virus. Excessive accumulation and activation of tons of immune cells in the lung inevitably produce cytokines and chemical mediators, which evoke the “cytokine storm” affecting other organs especially in severe/critical patients. 6 Notably, coagulation abnormalities, such as elevated d‐dimer and lactate dehydrogenase (LDH), prolonged prothrombin time (PT) 7 are identified to be associated with poor prognosis and high mortality. 8 However, easy and affordable biomarkers indirectly reflecting lung injury are rarely employed to evaluate the long‐term outcome, and the relationship between lung injury and coagulation abnormalities in COVID‐19 patients remains uninvestigated.
Krebs von den Lungen‐6 (KL‐6) is a glycoprotein mainly produced by damaged or regenerating lung type II pneumocytes. Elevated serum KL‐6 concentrations have been utilized as a surrogate of interstitial lung diseases for more than 2 decades. 9 , 10 , 11 Recently KL‐6 was reported to be associated with COVID‐19 severity. 12 , 13 , 14 A significantly elevated level of KL‐6 was suggested to indicate a severe status and poor prognosis. However, their conclusions were only based on a small cohort of 22 patients and 2 severe patients, respectively. Further confirmation of the prognostic value of KL‐6 in a larger and longitudinal cohort with a comprehensive analysis is critical but unfortunately missing. Here, we profiled the longitudinal and dynamic changes of KL‐6 in 166 COVID‐19 patients from symptom onset to 6 months postdischarge, and also analyzed the association between KL‐6 and coagulation and immune parameters.
2. MATERIALS AND METHODS
2.1. Study population
One hundred sixty‐six COVID‐19 patients hospitalized at Guangzhou Eighth People's Hospital were included in this study. Patients were diagnosed as mild or severe/critical according to Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (seventh edition, General Office of National Health Commission). The patient clinic and laboratory tests were collected from the medical records of Guangzhou Eighth People's Hospital. The study was approved by the medical ethics committee of the Guangzhou Eighth People's Hospital (No. 202001134). Written consent was obtained from all patients.
2.2. KL‐6 assay
Serum KL‐6 was measured by a commercial Diagnostic Kit (Cat. 200309; Kangrun Biotech) based on a standard curve of serial dilutions of KL‐6 standard sample with Kaeser 1000 Automatic Chemiluminescence Immunoassay Analyzer provided by the Kangrun Biotech.
2.3. Flow cytometry analysis
Five‐hundred microliters of peripheral blood mononuclear cells were isolated from 10 ml of whole blood of patients and processed with flow cytometry with a panel of antibodies 12 including CD3‐Pacific‐Blue anti‐human antibody (B286012; Biolegend), CD4‐APC/cy7 anti‐human antibody (B299289; Biolegend), CD8‐BV510 anti‐human antibody (B303256; Biolegend), CD161‐FITC anti‐human antibody (B302548; Biolegend), CD25‐APC anti‐human antibody (B294860; Biolegend), CD127‐PE/cy7 anti‐human antibody (B286366; Biolegend), CXCR5‐Perp/cy5.5 anti‐human antibody (B295344; Biolegend), and PD‐1‐PE anti‐human antibody (B304891; Biolegend). Data were analyzed using FlowJo software (BD‐Biosciences).
2.4. Statistics analysis
Continuous variables were expressed as medians (interquartile) or mean ± SEM. Categorical variables were summarized as the counts and percentages in each category. Student's t tests were applied to continuous variables, the Mann–Whitney U test and χ 2 test were used for categorical variables as appropriate. Areas under the receiver operating characteristic (ROC) curves (area under the curve) of KL‐6, LDH, and d‐dimer were assessed for distinguishing mild and severe/critical patients. Pearson's or Spearman's rank correlation was used to explore the correlations between different parameters as appropriate. A linear regression model was established to predict the KL‐6 levels at greater than 100 days postsymptom onset based on the values of KL‐6 within 10 days postonset. p < .05 was considered statistically significant. Statistical analysis was performed with IBM SPSS Statistics 26. Graphic representations were performed with GraphPad Prism 8 software.
3. RESULTS
3.1. Serum KL‐6 was substantially elevated in severe/critical COVID‐19 patients
In total, 166 COVID‐19 patients were enrolled in this study (Table 1). A total of 149 (89.8%) patients were diagnosed as mild and 17 (10.2%) were as severe/critical. To get the base level of KL‐6 level in the whole population, we measured the samples from 59 healthy volunteers (n = 59) and found that the median value of serum KL‐6 was 180.9 U/ml (Figure 1 and Table S1). Patients with COVID‐19 had significantly higher levels of KL‐6 compare to healthy volunteers (p < .001, Figure 1). The severe/critical patients had even significantly higher levels of serum KL‐6 than the mild (median 898.0 vs. 452.1 U/ml, p < .001, Table 1 and Figure 1). We found that KL‐6 increased from symptom onset, reached a peak approximately within a month, and then gradually decreased (Figure S1). How long it requires to reach KL‐6 peak will be critical to reflect the extent and the speed of lung injury in patients with COVID‐19. Here, we defined the day with the highest KL‐6 among more than three detections within 1 month postonset as the peak day. We found a delayed peak day for severe/critical patients compared to the mild (21.8 ± 6.0 vs. 15.3 ± 7.0 [mean ± SEM] days, p = .015, Figure S2). Besides, we observed that it took 17.5 ± 1.9 (mean ± SEM) days for mild and 10.6 ± 1.8 (mean ± SEM) days for severe/critical patients to reach a higher level of KL‐6 (cutoff = 600 U/ml, p = .011, Figure S3). Altogether, our results indicated that lung injury deteriorated rapidly and that continuous and progressive lung injury among severe/critical COVID‐19 patients.
Table 1.
Demographic, clinical, and laboratory findings of patients with COVID‐19
| Parameters | Mild cases (n = 149) | Severe/critical cases (n = 17) | p |
|---|---|---|---|
| Age, years | 48.0 (34.5–62.0) | 55.0 (53.0–68.0) | <.001 |
| Sex | .464 | ||
| Male | 65 (43.6%) | 9 (52.9%) | – |
| Female | 84 (56.4%) | 8 (47.1%) | – |
| Outcome | .102 | ||
| Discharged | 149 (100%) | 16 (94.1%) | – |
| hospitalized | 0 (0%) | 1 (5.9%) | – |
| Hospital stay, days | 18.0 (13.5–26.0) | 23.0 (14.5–37.0) | .213 |
| KL‐6, U/ml | 452.1 (325.6–641.3) | 898.0 (567.7–1278.9) | <.001 |
| LDH, U/L | 180.0 (151.0–230.0) | 266.0 (229.0–398.5) | .001 |
| CK, U/L | 52 (36–77) | 59 (26–120) | .344 |
| PT, s | 13.6 (13.2–14.1) | 15.3 (14.4–16.0) | .754 |
| d‐dimer, μg/L | 1210 (800–1640) | 3380 (2368–10165) | .003 |
| PTA, % | 94.0 (87.0–100.0) | 74.0 (68.3–82.8) | <.001 |
| INR | 1.04 (1.00–1.09) | 1.20 (1.12–1.27) | <.001 |
| TT, s | 16.0 (15.5–16.7) | 18.0 (16.1–22.8) | .106 |
| APTT, s | 38.7 (36.1–42.5) | 54.4 (39.6–65.2) | .002 |
| Fib, g/L | 3.98 (3.17–4.88) | 5.05 (4.32–6.04) | <.001 |
| FDP, mg/L | 1.78 (1.30–2.82) | 6.95 (5.15–18.84) | .011 |
| PLT, 109/L | 210.5 (170.8–254.3) | 192.0 (117.5–272.5) | .766 |
| MPV, fL | 10.4 (9.7–11.0) | 11.7 (9.9–12.6) | <.001 |
| PLT‐PCT, % | 0.22 (0.18–0.27) | 0.23 (0.15–0.30) | .969 |
| PDW, % | 11.8 (10.4–13.4) | 14.3 (11.1–16.2) | <.001 |
| P‐LCR, % | 27.6 (22.1–32.8) | 38.0 (23.4–44.6) | <.001 |
| RBC, 1012/L | 4.26 (3.88–4.74) | 2.88 (2.55–3.82) | .216 |
| HGB, g/L | 130.5 (119.0–143.0) | 98.0 (75.5–117.0) | <.001 |
| Hct, % | 38.5 (35.5–41.7) | 28.2 (23.1–34.8) | <.001 |
| Urea, μmol/L | 3.99 (3.43–4.82) | 10.64 (4.64–18.29) | .025 |
| UA, mmol/L | 288.7 (235.0–346.8) | 189.3 (88.0–231.0) | .024 |
| Cr, μmol/L | 66.9 (55.1–79.6) | 83.8 (49.3–164.6) | .005 |
| Cys‐C, mg/L | 0.88 (0.77–0.99) | 2.21 (1.02–4.29) | <.001 |
| eGFR, ml/min/1.73 m2 | 129.1 (108.9–149.2) | 82.3 (35.8–162.1) | .005 |
| aCL IgG, CPLU/ml | 2.30 (1.57–3.56) | 3.28 (2.66–4.38) | .283 |
| CD3+ T, % | 58.4 (45.9–66.1) | 21.6 (11.6–44.8) | <.001 |
| CD3+CD4+ T, % | 56.3 (44.9–66.0) | 44.4 (29.6–57.5) | .030 |
| Tregs, % | 5.7 (4.7–7.7) | 9.3 (5.3–12.6) | .001 |
| CD4+CD161+ T, % | 12.6 (9.0–18.7) | 12.2 (9.2–18.7) | .869 |
| CD3+CD8+ T, % | 36.8 (28.6–48.3) | 44.0 (37.2–55.9) | .040 |
| CD8+CD161+ T, % | 9.4 (6.8–12.4) | 22.6 (15.0–35.3) | <.001 |
| CD4+/CD8+, % | 1.5 (0.9–2.3) | 1.0 (0.5–1.5) | .017 |
| CD4+PD‐1+ T, % | 10.4 (8.0–14.6) | 20.5 (11.8–30.8) | <.001 |
| CD4+CXCR5+ T, % | 15.2 (11.8–20.6) | 7.8 (3.8–16.6) | .005 |
Note: Data are median (IQR) or n (%). p values were calculated by Mann–Whitney U test, Student's t test, or χ 2 test, as appropriate.
Abbreviations: aCL IgG, anti‐cardiolipin immunoglobulin G; APTT, activated partial thromboplastin time; CD, cluster of differentiation; CK, creatine kinase; COVID‐19, coronavirus disease 2019; Cr, creatinine; CXCR5, chemokine receptor type 5; Cys‐C, cystatin C; eGFR, estimated glomerular filtration rate; FDP, fibrin degradation product; Fib, fibrinogen; Hct, hematocrit; HGB, hemoglobin; INR, international normalized ratio; KL‐6, Krebs von den Lungen‐6; LDH, lactate dehydrogenase; MPV, mean platelet volume; PD‐1, programmed death‐1; PDW, platelet distribution width; P‐LCR, platelet‐large cell ratio; PLT, platelet; PLT‐PCT, platelet crit; PT, prothrombin time; PTA, prothrombin time activity; RBC, red blood cells; Tregs, regulatory T cells; TT, thrombin time; UA, uric acid.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 1.
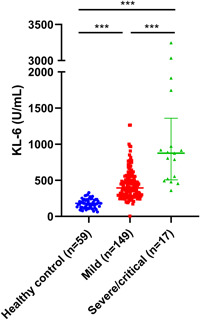
Serum Krebs von den Lungen‐6 (KL‐6) levels of healthy control (n = 59), mild (n = 149), and severe/critical coronavirus disease 2019 patients (n = 17). ***p < .001
Recently serum KL‐6 as well as other parameters, for example, d‐dimer and LDH, 15 , 16 , 17 , 18 were suggested as a biomarker of COVID‐19 severity, but the conclusion was drawn from a small cohort (n = 22 or 2). 12 , 13 In our cohort (n = 166), ROC analysis supported the use of serum KL‐6 distinguishing mild and severe/critical patients (Table S2 and Figure S4). The AUC of KL‐6 was 0.793 (95% confidence interval: 0.718–0.868. p < .001) and the best cutoff value was 642.3 U/ml (75.3% sensitivity and 73.3% specificity) or 788.2 U/ml (86.4% sensitivity and 62.2% specificity).
3.2. Early serum KL‐6 predicted the prognosis of lung injury of discharged COVID‐19 patients
For each patient with COVID‐19, the KL‐6 level at greater than 100 days postonset was linearly correlated to his KL‐6 level within 10 days postonset (Pearson r = .867, p < .001). A linear regression model predicted KL‐6 at greater than 100 days postonset = 87.729 + 0.563 × (KL‐6 within 10 days postonset) (r = .867, p < .001, Figure 2). For each COVID‐19 case with higher serum KL‐6, for example, ≥300 U/ml during hospitalization, a linear correlation of KL‐6 between the two intervals were also found (Pearson r = .794, p < .001), and a linear regression model predicted KL‐6 at greater than 100 days postonset = 106.8 + 0.539 × (KL‐6 within 10 days postonset) (r = .836, p < .001, Figure 2). Since the hospital stay of COVID‐19 was usually less than a month (Table 1), these data indicated that early serum KL‐6 could be a potent predictor for the prognosis of lung injury of discharged patients.
Figure 2.
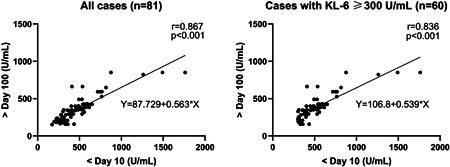
Linear regression models predict Krebs von den Lungen‐6 (KL‐6) at >100 days postonset among all coronavirus disease 2019 (COVID‐19) cases (left panel, n = 81) and COVID‐19 cases with KL‐6 ≥300 U/ml during hospitalization (right panel, n = 60)
3.3. Late serum KL‐6 was associated with computed tomography lung lesions areas
For those patients conducting computed tomography (CT) examination, we also explored whether there was an association between CT lung lesions areas and the KL‐6 values within the previous week (−7 days) and within the next week (+7 days). We found that KL‐6 values within the previous week had no significant correlations with lung lesions areas (p > .05, Figure 3A). In contrast, KL‐6 values within the next week were linearly correlated to CT left lung, right lung, and left plus right lung lesions areas (p < .001, Figure 3B). Therefore, the late serum KL‐6 was correlated to CT lung lesions areas based on our findings.
Figure 3.
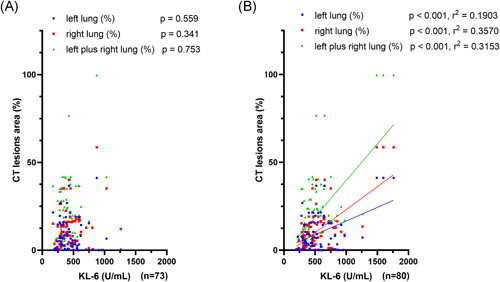
Linear correlations between computed tomography (CT) lung lesions areas and the Krebs von den Lungen‐6 (KL‐6) values within the previous week (A) (n = 73) and within the next week (B) (n = 80)
3.4. Elevated serum KL‐6 was correlated to coagulation dysfunctions
Coagulation dysfunctions have been reported in severe COVID‐19 patients. 8 , 15 , 19 , 20 In our cohort, compared to the mild, the coagulation dysfunctions, for example, significantly elevated d‐dimer (p = .003) and decreased prothrombin time activity (PTA, p < .001), were observed in severe/critical patients (Table 1). A Spearman's rank correlation analysis revealed that KL‐6 was moderately correlated to fibrin degradation product (FDP, r = .483, p < .001), and weakly correlated to d‐dimer (r = .343, p < .001), thrombin time (TT, r = .305, p < .001), and fibrinogen (Fib, r = .275, p < .001, Table S3). For mild patients, KL‐6 was weakly correlated to TT (r = .227, p = .008), d‐dimer (r = .200, p = .011), and Fib (r = .188, p = .029) (Table S3). Actually, for most mild patients, the coagulation indexes were normal during most of the hospital stay, as shown in Figure S5. For severe/critical patients, KL‐6 was strongly correlated to d‐dimer (r = .692, p < .001), FDP (r = .641, p = .001), and moderately correlated to the international normalized ratio (r = .517, p = .001), PT (r = .512, p = .001), PTA (r = −.512, p = .001), and TT (r = .423, p = .008) (Table S3). In contrast to mild patients, an elevated KL‐6 was usually accompanied by coagulation dysfunctions among severe/critical patients, as shown in Figure 4, which shows the dynamic profile of KL‐6 and coagulation indexes of a severe/critical case. In all, these data suggested a correlation between elevated KL‐6 and coagulation dysfunctions among patients with COVID‐19.
Figure 4.
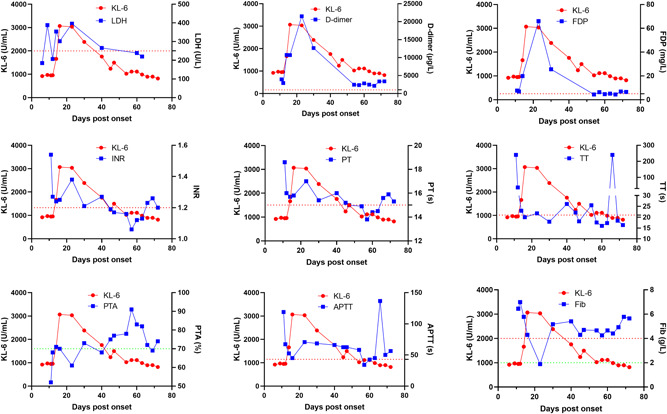
Dynamic profile of the coagulation indexes of a severe/critical patient. LDH is also shown as a biomarker for COVID‐19 severity. A dotted line in red or green represents the upper or lower normal limit of each index (right Y‐axis), respectively. APTT, activated partial thromboplastin time; COVID‐19, coronavirus disease 2019; FDP, fibrin degradation product; Fib, fibrinogen; INR, international normalized ratio; KL‐6, Krebs von den Lungen‐6; LDH, lactate dehydrogenase; PT, prothrombin time; PTA, prothrombin time activity; TT, thrombin time
3.5. Elevated serum KL‐6 was correlated to T cells subsets dysfunctions
For all patients with COVID‐19, the KL‐6 level was negatively correlated to CD3+ T cell counts (Table S3). Of note, the CD3+ T cell count was significantly lower in severe/critical patients than in the mild (median 21.6% vs. 58.4%, p < .001, Table 1), indicating lymphocytopenia in severe/critical patients. Serum KL‐6 was positively correlated to CD4+PD‐1+ T cells (r = .348, p < .001), a negative immune climate marker (Table S3). For mild patients, a weak correlation between KL‐6 and CD4+PD‐1+ T cells (r = .273, p = .003), and CD3+ T cells (r = −.325, p < .001) was found (Table S3). Excessive activation of the immune system and production of inflammatory cytokines, termed as “cytokine storm,” was considered to be one of the main causes of lung injury in severe COVID‐19 patients. 21 , 22 Here, we showed that among severe/critical patients, serum KL‐6 were positively correlated to CD4+CXCR5+ T cells (r = .535, p = .003), and CD4+/CD8+ ratio (r = .511, p = .005), and CD3+CD4+ T cells (r = .510, p = .006), as shown in Table S3. With an increase of serum KL‐6 among severe/critical patients, the proportion of CD3+CD4+ T helper cells increased, and CD4+CXCR5+ T cells were also upregulated, indicating the lung injury in severe/critical patients was accompanied by an enhanced inflammation response. In contrast, serum KL‐6 among severe/critical patients was negatively correlated to Tregs (r = −.516, p = .005), CD3+CD8+ T cells (r = −.475, p = .011), and CD8+CD161+ T cells (r = −.425, p = .034). The dynamic profile of KL‐6 and T cell clusters of a severe/critical patient was shown in Figures S6 and S7. These results indicated that among severe/critical COVID‐19 patients with elevated KL‐6, CD3+CD8+ cytotoxic T cells were excessively depleted, and Tregs was also downregulated, characterized by weakened immunosuppression and excessive activation of the T cell immunity, which may jointly lead to lung injury of severe/critical COVID‐19 patients. Since T cell subsets are usually not determined in clinical course, an elevated KL‐6 may be an alternative indicator of immune dysfunctions, especially among severe/critical COVID‐19 patients due to their significant correlations.
4. DISCUSSION
In this study, we showed a complete dynamic profile of serum KL‐6 among COVID‐19 patients from disease onset to postdischarge. In addition, we found that serum KL‐6 of discharged COVID‐19 patients could be predicted by its value at an early stage (Figure 2), rendering us an ideal tool to evaluate the prognosis of lung injury of COVID‐19. Undoubtedly, pulmonary function testings would provide a more comprehensive assessment of the recovery of discharged patients.
One of the most important questions in controlling COVID‐19 is to identify the risk factors of severe illness or death. 23 Here, we confirmed that serum KL‐6 was associated with COVID‐19 severity in a cohort including 17 severe/critical patients. In addition, we also found a linear correlation between serum KL‐6 and CT lung lesions areas, which links to COVID‐19 progress. 24 Therefore, COVID‐19 patients with an elevated KL‐6 level on admission deserve a close observation in case of progression.
Coagulation dysfunctions like high d‐dimer levels on admission have been proved to be associated with poor overall survival. 25 , 26 Innate immune response to SARS‐CoV‐2, dysfunctional ACE2, and inflammation activation may participate in the coagulation dysfunctions, 27 but the specific mechanism is unknown. Here, we demonstrated that KL‐6 was correlated to coagulation indexes, for example, among severe/critical patients, KL‐6 was strongly correlated to d‐dimer (r = .692, p < .001) and FDP (r = .641, p = .001, Table S3), both of which have been suggested as risk factors of COVID‐19 severity. 28 It is worth noting that for mild patients, KL‐6 was weakly correlated to some coagulation indexes (Table S3), which were normal during most of the hospital stay (Figure S5), and serum KL‐6 of some mild patients remained low from symptom onset (Figure S1). So the interpretation of these correlations in mild patients should be careful.
The role of host immunity in COVID‐19‐associated morbidity and mortality remains controversial. It is generally considered that overactivation of the immune system termed as “cytokine storm” mediates lung injury, 6 , 21 , 29 , 30 , 31 while another study hold that immunosuppression characterized COVID‐19 infections. 32 In this cohort, compared to mild COVID‐19, the severe/critical patients had significantly lower CD3+ T cells and CD3+CD4+ T cells, and significantly higher Tregs and CD4+PD‐1+ T cells, supporting immunosuppression in them. Because we did not determine the immunological indexes of healthy control here, whether immunosuppression also occurred in mild patients remains unclear. We found that KL‐6 was negatively correlated to CD3+ T cells (r = −.325, p < .001) and positively correlated to CD4+PD‐1+ T cells (r = .273, p = .003) among mild patients, indicating that lung injury was likely associated with immunosuppression in this population. However, this result should be interpreted with caution because the correlation coefficients are weak and as mentioned above, we did not determine whether immunosuppression indeed occurred in mild patients. The elevation of KL‐6 in severe/critical patients was correlated to overactivation of immunity, for example, KL‐6 was positively correlated to CD4+CXCR5+ T cells, and CD4+/CD8+ ratio, CD3+CD4+ T cells, and negatively correlated to Tregs, CD3+CD8+ T cells, and CD8+CD161+ T cells (Table S3). These findings may explain why dexamethasone, a kind of immunosuppressants, resulted in lower mortality among severe COVID‐19 patients. 33
The limitations of our study include that there were a small number of severe/critical patients (n = 17, 10.2% [17/166]) compared to the mild (n = 149, 89.8% [149/166]) included in the cohort (n = 166). The percentage of severe/critical patients here (10.2%) may represent the real proportion of it in natural SARS‐CoV‐2 infection in Guangzhou, as a previous study conducted by China Medical Treatment Expert Group for COVID‐19 reported that the proportion of severe cases in mainland China were 15.7% (173/1099), 34 which was similar to that in our study. By now the cumulative number of locally confirmed cases in Guangzhou was 377 according to National Health Commission Report. 35 Thus, collecting lots of severe/critical patients in this region seems difficult.
In summary, our findings support KL‐6 as a biomarker of COVID‐19 severity, and also a predictor of the prognosis of lung injury of discharged patients. The dynamic profile of KL‐6 was closely correlated to coagulation disorder and immune dysfunction, especially among severe/critical patients, highlighting the possibility that coagulation disorder and immune dysfunction may be a contributor to lung injury of COVID‐19, and the underlying mechanism requires further research.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Xiaoping Tang, Fengyu Hu, and Feng Li conceived the study and supervised all aspects of the study. Kai Deng, Qinghong Fan, Yanhong Yang, Xizi Deng, Ruiying He, Yizhou Tan, Yun Lan, Xilong Deng, Yuejun Pan, Yaping Wang, Yujuan Guan, Huiyuan Liu, Fengjuan Chen, Xiaoneng Mo, Xinghua Tan, Chun Luo, Xueliang Wen, Ying Liu, Jinxin Liu, and Lieguang Zhang participated in the experiment. Kai Deng, Qinghong Fan, Yanhong Yang, Yaping Wang, and Fengjuan Chen collected the data. Xiaoping Tang, Fengyu Hu, Feng Li, Kai Deng, and Qinghong Fan analyzed the data and prepared the manuscript.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGMENTS
This study was supported by Guangdong Provincial Department of Science and Technology Fund (No. 2020B1111330002). The authors would like to thank Guangzhou Kangrun Biotech Co. for their technical assistance.
Deng K, Fan Q, Yang Y, et al. Prognostic roles of KL‐6 in disease severity and lung injury in COVID‐19 patients: A longitudinal retrospective analysis. J Med Virol. 2021;93:2505–2512. 10.1002/jmv.26793
Kai Deng, Qinghong Fan, and Yanhong Yang contributed equally to this study.
Contributor Information
Xiaoping Tang, Email: tangxiaopinggz@163.com.
Fengyu Hu, Email: gz8hhfy@126.com.
Feng Li, Email: gz8h_lifeng@126.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed October 30, 2020.
- 2. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016‐1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yao XH, He ZC, Li TY, et al. Pathological evidence for residual SARS‐CoV‐2 in pulmonary tissues of a ready‐for‐discharge patient. Cell Res. 2020;30(6):541‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li H, Liu L, Zhang D, et al. SARS‐CoV‐2 and viral sepsis: observations and hypotheses. Lancet. 2020;395(10235):1517‐1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian Z, Travanty EA, Oko L, et al. Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome‐coronavirus. Am J Respir Cell Mol Biol. 2013;48(6):742‐748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu B, Huang S, Yin L. The cytokine storm and COVID‐19. J Med Virol. 2020;93:250‐256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H, Cai S, Li Y, et al. Prognostic factors for COVID‐19 pneumonia progression to severe symptoms based on earlier clinical features: a retrospective analysis. Front Med. 2020;7:557453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Levi M, Thachil J, Iba T, Levy JH. Coagulation abnormalities and thrombosis in patients with COVID‐19. Lancet Haematol. 2020;7(6):e438‐e440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakajima H, Harigai M, Hara M, et al. KL‐6 as a novel serum marker for interstitial pneumonia associated with collagen diseases. J Rheumatol. 2000;27(5):1164‐1170. [PubMed] [Google Scholar]
- 10. Kobayashi J, Kitamura S. KL‐6: a serum marker for interstitial pneumonia. Chest. 1995;108(2):311‐315. [DOI] [PubMed] [Google Scholar]
- 11. d′Alessandro M, Bergantini L, Cameli P, et al. Krebs von den Lungen‐6 as a biomarker for disease severity assessment in interstitial lung disease: a comprehensive review. Biomark Med. 2020;14(8):665‐674. [DOI] [PubMed] [Google Scholar]
- 12. d'Alessandro M, Cameli P, Refini RM, et al. Serum KL‐6 concentrations as a novel biomarker of severe COVID‐19. J Med Virol. 2020;92:2216‐2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura H, Miyagi K, Otsuki M, et al. Serum KL‐6 can distinguish between different phenotypes of severe COVID‐19. J Med Virol. 2020;93:158‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. d'Alessandro M, Bergantini L, Cameli P, et al. Peripheral biomarkers' panel for severe COVID‐19 patients [published online ahead of print October 2, 2020]. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323:1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kander T. Coagulation disorder in COVID‐19. Lancet Haematol. 2020;7:e630‐e632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh P, Schwartz RA. Disseminated Intravascular coagulation: a devastating systemic disorder of special concern with COVID‐19. Dermatol Ther. 2020;33:e14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID‐19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID‐19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lipsitch M, Swerdlow DL, Finelli L. Defining the epidemiology of Covid‐19—studies needed. N Engl J Med. 2020;382(13):1194‐1196. [DOI] [PubMed] [Google Scholar]
- 24. Li M, Lei P, Zeng B, et al. Coronavirus disease (COVID‐19): spectrum of CT findings and temporal progression of the disease. Acad Radiol. 2020;27(5):603‐608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fei Y, Tang N, Liu H, Cao W. Coagulation dysfunction: a hallmark in COVID‐19. Arch Pathol Lab Med. 2020;144:1223‐1229. [DOI] [PubMed] [Google Scholar]
- 26. Lin J, Yan H, Chen H, et al. COVID‐19 and coagulation dysfunction in adults: a systematic review and meta‐analysis. J Med Virol. 2021;93:934‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J, Saguner AM, An J, Ning Y, Yan Y, Li G. Dysfunctional coagulation in COVID‐19: from cell to bedside. Adv Ther. 2020;37(7):3033‐3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rod JE, Oviedo‐Trespalacios O, Cortes‐Ramirez J. A brief‐review of the risk factors for covid‐19 severity. Rev Saude Publica. 2020;54:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID‐19: an overview of the involvement of the chemokine/chemokine‐receptor system. Cytokine Growth Factor Rev. 2020;53:25‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the “Cytokine Storm” in COVID‐19. J Infect. 2020;80(6):607‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soy M, Keser G, Atagunduz P, Tabak F, Atagunduz I, Kayhan S. Cytokine storm in COVID‐19: pathogenesis and overview of anti‐inflammatory agents used in treatment. Clin Rheumatol. 2020;39(7):2085‐2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Remy KE, Mazer M, Striker DA, et al. Severe immunosuppression and not a cytokine storm characterize COVID‐19 infections. JCI Insight. 2020;5:e140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Group RC, Horby P, Lim WS, et al. Dexamethasone in hospitalized patients with Covid‐19—preliminary report [published online ahead of print July 17, 2020]. N Engl J Med. 2020. [Google Scholar]
- 34. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. http://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml. Accessed September 30, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


