Abstract
We compared the performance of two 96-well multiplex immunoassay platforms in assessing plasma cytokine concentrations in patients with glioblastoma (GBM; n = 27), individuals with melanoma, breast or lung cancer metastases to the brain (n = 17), and healthy volunteers (n = 11). Assays included a bead-based fluorescence MILLIPLEX® assay/Luminex (LMX) platform and 4 planar electrochemiluminescence kits from Meso Scale Discovery (MSD). The LMX kit evaluated 21 cytokines and the 3 MSD kits evaluated 20 cytokines in total, with 19 overlapping human cytokines between platforms (GM-CSF, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, IL-17A, IL-21, IL-23, MIP-1α, MIP-1β, MIP-3α, TNFα). The MSD platform had lower LLoQs (lower limits of quantification) than LMX for 17/19 cytokines, and higher LLoQs for IFN-γ and IL-21. The ULoQs were higher in LMX versus MSD assays for 17/19 shared analytes, but lower than MSD for IL-17A and IL-21. With LMX, all 19 shared analytes were quantifiable in each of 55 samples. Although MSD recombinant protein standard curves indicated lower LLoQs than LMX for most cytokines, MSD detected 7/19 (37%) native analytes in <75% of samples, including 0% detection for IL-21 and 8% for IL-23. The LMX platform categorized identical samples at greater concentrations than the MSD system for most analytes (MIP-1β the sole exception), sometimes by orders of magnitude. This mismatched quantification paradigm was supported by Bland-Altman analysis. LMX identified significantly elevated levels of 10 of 19 circulating cytokines in GBM: GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-17A, IL-21, IL-23, MIP-1α, and MIP-3α, consistent with prior findings and confirming the utility of applying appropriate multiplex immunoassay technologies toward developing a cytokine signature profile for GBM.
Keywords: Chemokine, cytokine, glioblastoma multiforme, glioma, brain metastases, fluorescence, electrochemiluminescence, multiplex immunoassay, in vitro assay, performance, precision, detection limit, lower limit of quantification, dynamic range
Introduction
Gliomas are the most frequently diagnosed primary intracranial, intraparenchymal neoplasm.1 Glioblastoma (GBM), the most common glioma histopathology (≈45% of all gliomas), is an aggressive malignancy with a 5-year relative survival of only ≈5%, despite extensive ongoing research into surgical, chemotherapy, and radiological treatments. However, brain metastases from other primary sites are the most common intracranial malignancies in adults, with an incidence of more than 170 000 per year in the US alone.2 By some estimates, more than 40% of cancer patients develop brain metastases, including up to 50% of patients with lung cancer, >25% of patients with breast cancer, and 20% of patients with melanoma.2
Cytokines and chemokines constitute a growing group of small (<40 kDa) secreted bioactive proteins that perform diverse interactive roles in molecular communication between cells and tissues.3,4 The balanced release of various cytokines is important for maintaining normal homeostasis, and is also an essential component of a well-regulated immune response that changes in diverse disease states including cancers such as GBM.5-7 Developing profiles of multiple interacting biomarkers in human plasma may have important utility in differentially diagnosing and staging both primary and secondary intracranial tumors, tracking disease progression, estimating prognosis, and selecting optimal management strategies and following treatment responses.8-10
Multiplex immunoassays employ a variety of technologies, including planar chemiluminescence and bead-based immunocapture suspension array platforms, to simultaneously quantify circulating levels of many cytokines and other biomarkers.11 These approaches are useful for deciphering the complex underlying biochemical mechanisms and interactions that occur during many disease states, have great possibilities for accelerating epidemiological research, and are emerging as valuable clinical diagnostic and prognostic health appraisal tools. This novel functionality is underscored by a progressive increase in FDA approvals of multiplex proteomic assays for clinical use.12 Multiplex immunoassays provide several advantages over singleplex immunoassays for obtaining the same cumulative information, including enhanced efficiency and reduced cost when concurrently measuring multiple analytes from single samples, increased throughput, and the mapping of complex biochemical networks that may enable developing personalized medical interventions.11,12 The relative benefits of multiplex protein analysis over singleplex immunoassays are most obvious with scarce and valuable samples that are restricted in volume or accessibility, and when streamlining budget, workload, and time expenditures are critical concerns.
Multiplex immunoassay platforms have distinct analytical performance capabilities, strengths, shortcomings, and instrumentation and operator requirements. The current study compared the analytical performance characteristics of 2 different commonly used multiplex platforms—a bead-based fluorescence assay and a planar chemiluminescence assay—to quantify circulating cytokine concentrations in human plasma samples and to assess operator time requirements for assay completion. Both methods employed 96-well microtiter plate formats and measured 19 common cytokine analytes in identical samples. Additionally, we performed a time-and-motion assessment of both operator-attentive and total time required for assay completion with each method. Our group is interested in identifying and characterizing blood tumor biomarkers in brain cancer, and the evaluated plasma samples were obtained from patients upon diagnosis of either primary glioblastoma or of brain metastases of various cancers originating in other organs. Plasma samples from ostensibly healthy subjects were assessed as controls. The purpose of our study was to interrogate multiplex methodologies to find the best platform to comprehensively characterize circulating cytokine profiles for oncological research, and to investigate their potential for identifying molecular signatures specific for GBM and for brain metastases from other primary sites.
Materials and Methods
Multiplex assay kits and instrumentation
This study compared 2 multiplex immunoassay detection systems. The bead-based fluorescence assay was the MILLIPLEX® MAP Human High Sensitivity T Cell Magnetic Bead Panel (Product #HSTCMAG28SMPX21; Merck EMD Millipore, Billerica, MA), that simultaneously evaluates 21 analytes (fractalkine, GM-CSF, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p70, IL-13, IL-17A, IL-21, IL-23, ITAC, MIP-1α, MIP-1β, MIP-3α, and TNFα) in suspension.13 This assay was run on the Luminex® FLEXMAP 3D® detection instrument operated with xPONENT Software V4.2 (both from Luminex Corp., Austin, TX). This system has improved sensitivity, broader dynamic range, and higher throughput than the predecessor Luminex 100/200™ technology used in earlier studies.11
The second platform was the electrochemiluminescence-based Meso Scale Discovery solid-matrix assay (MSD V-Plex® kits; Meso Scale Discovery, LLC, Rockville, MD). Because the MSD 96-well electrochemiluminescence immunoassay kits that we used were limited to assessing 10 cytokines per assay, we evaluated identical samples using 4 different kits, 3 customized, to achieve reasonable overlap with the 21-plex LMX assay. The MSD Proinflammatory Panel 1 (Product #K15049G) measures IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL 8, IL-10, IL-12 (p70), IL-13 and TNF-α). We also purchased 3 customized kits that measured 3 to 4 relevant molecules each: the TH17 V-Plex® (#K15085D) measured IL-21, IL-23, and MIP-3α; the Cytokine Panel 1 (#K15050D) measured GM-CSF, IL-5, IL-7, and IL-17A; and the Chemokine Panel 1 (#K15047D) measured IL-8 HA, MIP-1α, and MIP-1β. The Quickplex SQ120 detection instrument was operated using MSD Discovery workbench software v.3.0/4.0 (both from Meso Scale Discovery).14 The Luminex/MILLIPLEX and Meso Scale Discovery systems and associated results are hereafter abbreviated as “LMX” and “MSD,” respectively.
Together, the LMX and MSD assays measured 19 common cytokine analytes: GM-CSF, IFNγ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (p70), IL-13, IL-17A, IL-21, IL-23, MIP-1α, MIP-1β, MIP-3α, and TNFα. Kits were used before their stated expiration dates, and all assay steps were performed in accordance with manufacturer instructions.13,14 With both platforms, samples and accompanying kit standards were manually pipetted into assay plates. All samples were assayed in duplicate in all kits. The LMX assay used 25 µL of sample per duplicate well. The MSD kits used either 12.5 or 25 µL of sample per duplicate well, depending on the specific kit. Thus, the LMX kit required 50 µL of each sample and the MSD kits together required 150 µL of each sample to provide desired analyte overlap. One key difference is the LMX assay requires an overnight sample incubation time whereas the MSD offers a standard 2-hour or alternative overnight sample incubation. This experimental series used the manufacturer-preferred 2-hour incubation period for the MSD kits. Washing steps for the LMX assay were performed using an automated programmable washer-dispenser (BioTek MultiFlow FX reagent dispenser and BioTek 405 TS Microplate Washer (both from BioTek, Winooski, VT). Reagents dispensation and plate washing for the MSD assays were manually performed using a multichannel pipettor.
Samples
Plasma samples were centrifugally separated from venous blood samples drawn into EDTA-anticoagulant collection tubes and stored at −80°C until analysis by both assays, having undergone only a single freeze-thaw event. Demographic information on sample donors is provided in Table 1. Subjects included 27 persons diagnosed with glioblastoma (GBM) and 17 persons with brain metastases (BrMet) from primary lung, breast, or melanoma primary tumors. Plasma samples from 11 anonymous healthy control (HC) individuals were purchased from Discovery Life Sciences (Los Osos, CA). This study was performed under the approval and oversight of the Washington University Institutional Review Board. All volunteers provided written informed consent prior to enrollment and sample provision. Study performance complied with the tenets of the Declaration of Helsinki.
Table 1.
Demographics of plasma donors.
| Parameter | GBM | BrMet | HC |
|---|---|---|---|
| n | 27 | 17 | 11 |
| Gender, male/female, n | 14/13 | 4/13 | 1/10 |
| Age, y, mean ± SD | 56.7 ± 12.7 | 59.0 ± 13.2 | 35.1 ± 7.8 |
| Age, y, range | 25-82 | 31-81 | 26-47 |
Experimental overview
Prior to analysis, all samples were thawed on ice and centrifuged at 15 000 × g for 10 minute at 4°C to remove potential particulates. In all instances, samples were run in duplicate and each assay plate contained recombinant standards for each analyte and quality control (QC) samples provided with the kits. Calibration curves were established according to the user manuals using serial 4-fold dilutions of each kit’s stock analyte standards. Blank values were established using the sample diluent supplied with each immunoassay kit. All samples were diluted 1:2 or 1:4 using the sample diluent provided in each kit.
Assay performance characteristics
We assessed intra-assay precision, the lower and upper limits of quantification (LLoQ and ULoQ, respectively), and the resulting dynamic ranges of cytokine detection for both assays.15,16 The mean of 8 blank values measured on LMX and MSD plates were used to calculate the LLoQ, defined as the mean blank signal + 2.5 × SD. The ULoQ was defined as the highest mean standard curve value for each analyte in each kit.
Comparative evaluation of cytokine levels in human plasma
We measured the circulating concentrations of 19 cytokines in diverse plasma samples from the 3 subject groups (GBM, BrMet, HC) using both platforms, and evaluated whether values for any analyte were outside of the assays’ dynamic range of quantification. Bland-Altman correlation plots were generated to assess cytokine measurement agreement between LMX and MSD.17
Time and motion study
We evaluated and compared the precise time allocations required for labor (operator hands-on and/or mandatory observation) and total times including incubations to evaluate a similar number of analytes by both assay methods. All assays were performed by the same technician. Observation and timing data collection were conducted by Nexus (Plano, TX), an independent third-party healthcare consulting firm.
Data analysis
Data were analyzed using Prism v.8 graphing and statistical analysis software (GraphPad Software, Inc., San Diego, CA), and Excel v.16 (Microsoft Inc. Redmond). Analyte concentrations were determined by comparing sample readings to standard curves generated using a 4-parameter logistical curve fit algorithm (Belysa v.1 software from Millipore/Sigma for LMX, and MSD Discovery v.3.0/4.0 for MSD. Analyte concentrations are presented as pg/mL ± SD, range, or as number (% of samples), as appropriate. Means were compared by one-way ANOVA, followed by Tukey test for multiple comparisons. Proportions of samples within the dynamic range were compared for each analyte using the Fisher Exact test. Bland-Altman correlation plots show the mean ratios of MSD/LMX concentrations detected for each analyte within each sample, and include 95% limit-of-agreement representing mean values ±1.96 × SD. Any mean slope deviation from 1 was indicative of proportional (concentration-dependent) bias in 1 or both assays for that analyte.
Results
Analytical limits of detection and quantification, and dynamic range
The dynamic range was defined as the spread between the calculated LLoQ and the mean measured value of the highest standard curve point (ULoQ), and was determined for all 22 analytes coordinately and individually measured by the LMX and MSD systems. The dynamic ranges of quantification for all 22 combined cytokine analytes that both assay platforms measured are tabulated in Table 2, with the ranges of the 19 shared analytes shown graphically in Figure 1. Dynamic ranges for all analytes in both assays were within the expected ranges stated in assay lot-specific documentation. The MSD platform had lower analytical LLoQs than LMX for 17 of 19 (89%) shared cytokines. For IFN-γ, the LLoQ with LMX was 66% of the MSD value, and for both IL-8 and IL-21, the LMX values were 50% of LLoQs obtained using the MSD assay. The LMX assay had higher ULoQs for 17 of 19 (89%) analytes, ranging from 6% higher for MIP-1α to a maximal 3810% higher for IL-4. In 2 analytes, IL-17A and IL-21, the ULoQs determined by LMX were 55% and 66% of the ULoQ measurements obtained with the MSD assay, respectively.
Table 2.
Limits of quantitation and dynamic range of analytes measured by the bead-based fluorescence (LMX) and planar electrochemiluminescence (MSD) multiplex assays. Dynamic range spans the lower limit of quantitation (LLoQ) and upper limit of quantitation.
| 19 shared analytes | LMX | MSD | Range size ratio LMX/MSD | ||
| Dynamic range (pg/mL) | Dynamic range size (fold of LLoQ) | Dynamic range (pg/mL) | Dynamic range size (fold of LLoQ) | ||
| GM-CSF | 0.90-18 629 | 20 699 | 0.05-4393 | 87 860 | 0.23 |
| IFN-γ | 0.19-9639 | 50 731 | 0.29-4745 | 16 362 | 3.10 |
| IL-1β | 0.21-7315 | 34 833 | 0.03-1791 | 59 700 | 0.58 |
| IL-2 | 0.20-7633 | 38 165 | 0.07-5401 | 77 157 | 0.49 |
| IL-4 | 1.49-28 992 | 19 458 | 0.01-761 | 76 100 | 0.26 |
| IL-5 | 0.20-7581 | 37 905 | 0.08-3465 | 43 312 | 0.87 |
| IL-6 | 0.66-2912 | 4412 | 0.04-2524 | 63 100 | 0.07 |
| IL-7 | 3.62-5717 | 1579 | 0.06-3397 | 56 617 | 0.03 |
| IL-8 | 0.10-4795 | 47 950 | 0.02-2277 | 113 850 | 0.42 |
| IL-10 | 1.21-23 291 | 19 249 | 0.08-1806 | 22 575 | 0.85 |
| IL-12 p70 | 0.27-7909 | 29 293 | 0.03-2217 | 73 900 | 0.40 |
| IL-13 | 0.20-3913 | 19 565 | 0.11-1574 | 14 309 | 1.37 |
| IL-17A | 1.78-11 391 | 6399 | 0.23-20 872 | 90 748 | 0.07 |
| IL-21 | 0.11-3818 | 34 709 | 0.22-5786 | 26 300 | 1.32 |
| IL-23 | 12.15-124 034 | 10 208 | 0.22-20 833 | 94 695 | 0.11 |
| MIP-1α | 5.58-4604 | 825 | 1.17-4345 | 3714 | 0.22 |
| MIP-1β | 0.57-14 808 | 25 979 | 0.30-3528 | 11 760 | 2.21 |
| MIP-3α | 0.60-9400 | 15 667 | 0.08-1806 | 22 575 | 0.69 |
| TNF-α | 0.08-6529 | 81 612 | 0.07-1362 | 19 457 | 4.19 |
| Unshared analytes | Dynamic range (pg/mL) | Dynamic range (pg/mL) | |||
| Fractalkine | 19.38-285 596 | 14 737 | n/a | n/a | n/a |
| ITAC | 1.58-22 425 | 14 193 | n/a | n/a | n/a |
| IL-8 HA | n/a | n/a | 109-239 574 | 2198 | ‒ |
“n/a” indicates “not applicable” because that particular analyte was not included in the platform.
Figure 1.
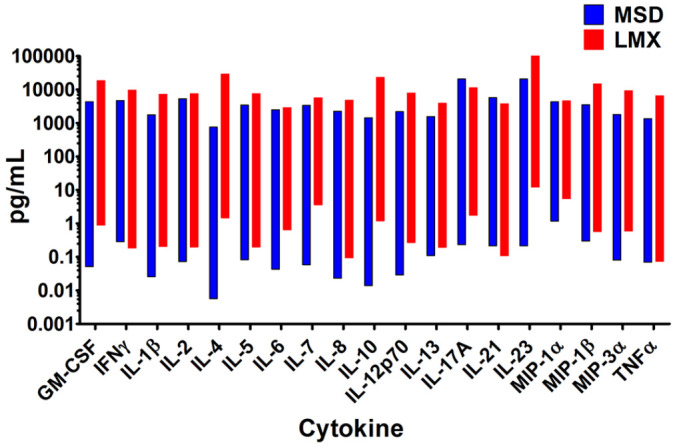
Dynamic ranges of the Luminex bead-based fluorescence (LMX) and Meso Scale Discovery electrochemiluminescence (MSD) multiplex cytokine immunoassay kits. Ranges of quantification were determined on standard calibration curves. With 3 exceptions (ie, IFN-γ, IL-8, and IL-21), the low-end of the remaining 16/19 shared analytes’ dynamic ranges was lower with the MSD platform compared to the LMX assay. Conversely, and with 2 exceptions (ie, IL-17A and IL-21), the high-end of the dynamic range was greater with the LMX assay versus the MSD assay. In 2 instances (IL-4 and IL-10), the differences in both upper and lower quantification levels differed by at least an order of magnitude between the 2 assays.
For 14 of 19 (74%) shared analytes, the MSD dynamic range was larger than that of the LMX platforms; in the remaining 5 instances (IFN-γ, IL-13, IL-21, MIP-1β, and TNF-α), the LMX range breadth was between 32% and 419% larger than the MSD range size (Table 2). Only in 2 analytes (IL-5 and IL-10) were the LMX and MSD dynamic range magnitudes within 20% of each other. In 2 instances (IL-4 and IL-10), the differences in both upper and lower quantification levels differed by at least an order of magnitude between the 2 assays.
Proportion of samples within assay quantification range
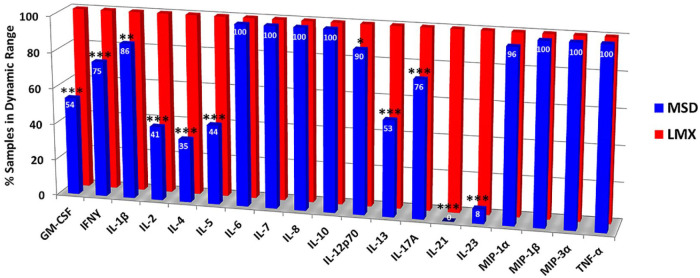
A total of 55 plasma samples from all 3 experimental groups were evaluated in duplicate wells using both platforms (Figure 2). With the LMX system, all of the 19 shared cytokines were quantifiable in 100% of samples tested. Despite having a lower LLoQ for a plurality of common analytes, in many instances the MSD chemiluminescence assay was unable to detect these cytokines in human plasma samples. All instances where experimental values fell outside of an analyte’s dynamic range were with readings below the LLoQ; no value ever exceeded the ULoQ for any analyte. The MSD platform detected and quantified 7 of 19 analytes (37%) in 100% of samples (ie, IL-6, IL-7, IL-8, IL-10, MIP-1β, MIP-3α, and TNF-α). The MSD assay detection/quantification rate for MIP-1α, IL-12p70, IL-1β, IL-17A, and IFN-γ in samples was 96%, 90%, 86%, 76%, and 75%, respectively. The MSD detection rate was <75% for the remaining 7 of 19 (37%) analytes, including a 0% detection rate for IL-21 and a 8% detection rate for IL-23. By contrast, the LMX system detected and quantified valid concentrations of IL-21 (range 1.8-18.9 pg/mL) and IL-23 (range 86.8-1922.2 pg/mL) in all samples. Using Fisher Exact test, 11/19 analytes were missed at a significantly greater frequency in samples using MSD versus LMX (Figure 2).
Figure 2.
Proportion of donor plasma samples inside the assay quantification range for the Luminex (LMX) and Meso Scale Discovery (MSD) multiplex cytokine immunoassay kits. With the LMX system, all of the 19 shared cytokines were quantifiable in 100% of samples tested. Although the MSD assay had a LLoQ below that of the LMX assay for 17/19 (89) common analytes in earlier standard curve performance evaluations, this sensitivity did not directly translate to evaluating cytokines in human plasma samples. The MSD platform detected and quantified 7 of 19 analytes (37%) in 100% of plasma samples. The MSD assay detection/quantification rate for MIP-1α, IL-12p70, IL-1β, IL-17A, and IFN-γ in samples was 96%, 90%, 86%, 76%, and 75%, respectively. The MSD detection rate was <75% in the remaining 7 of 19 (37%) analytes, including a 0% detection rate for IL-21 and a 8% detection rate for IL-23.
Asterisks indicate significant differences for 11/19 analytes between MSD and LMX by Fisher Exact test, with “*” indicating P < .05, “**” indicating P < .01, and “***” indicating P < .001. No asterisk indicates mathematically similar detection frequencies between platforms for those cytokines.
Analyte concentrations according to assay platform
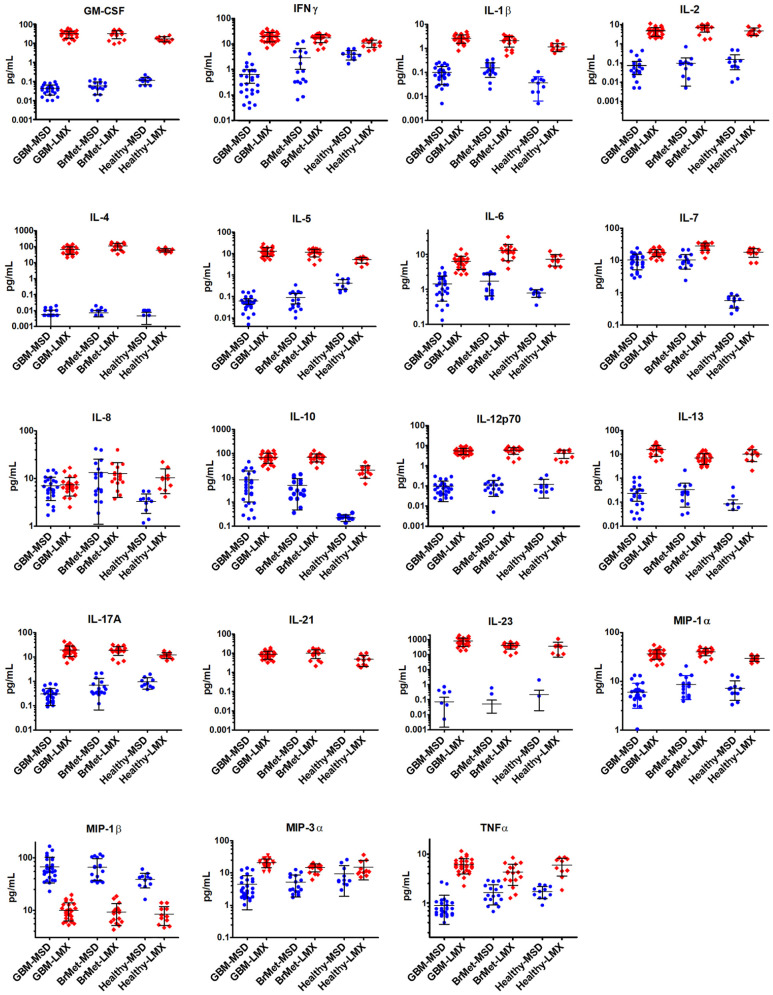
All 19 shared cytokine analytes were evaluated in 55 human plasma samples comprising 27 glioma subjects, 17 subjects with secondary brain metastases, and 11 controls (Figure 3 and Table 3). The LMX platform categorized identical samples at significantly greater concentrations than did the MSD system for most analytes. Clear differences occurred with IL-4, whereby LMX categorized cytokine concentrations at 4 orders of magnitude higher than did MSD, and GM-CSF at 2 to 3 orders of magnitude greater than MSD values. Notably, when the MSD platform evaluated IL-21, although the analyte quantification limit on standard curves was 0.22 pg/mL, no cytokine was detectable in any sample; the LMX platform, however, returned values in all of the identical samples, ranging from 1.8 to 18.9 pg/mL. Another prominent discrepancy between assay detection performance occurred with IL-23, which displayed a LLoQ that was 55-fold more sensitive with MSD (0.22 pg/mL) than LMX (12.15 pg/mL). While IL-23 was quantifiable by LMX in 100% of samples, the MSD assay detected the analyte in only 8% of the same samples, and categorized them at a much lower concentration (range 0.35-2.03 pg/mL) than did LMX (range 86.8-1922.2 pg/mL).
Figure 3.
Cytokine concentrations in human plasma samples. All 19 shared cytokine analytes were evaluated in 55 human plasma samples comprising 27 glioma subjects, 17 subjects with secondary brain metastases (BrMet), and 11 healthy controls (HC). Values shown are mean concentration ± SD. The Luminex (LMX) platform categorized identical samples at greater concentrations than the Meso Scale Discovery (MSD) system for most analytes. With IL-4, wherein LMX categorized cytokine concentrations at 4 orders of magnitude higher than MSD, and GM-CSF values at 2-3 orders of magnitude greater than MSD values. Notably, with IL-21, although the MSD lower quantification limit was 0.22 pg/mL, none was detectable by MSD in any sample. The LMX platform identified IL-21 in all samples (range 1.8-18.9 pg/mL). With IL-23, the LLoQ was 55-fold more sensitive using MSD (0.22 pg/mL) than LMX (12.15 pg/mL). While IL-23 was quantifiable by LMX in all samples, MSD detected the analyte in only 8% of samples, and categorized them at lower concentrations (range 0.24-2.03 pg/mL) than LMX (range 86.8-1922.2 pg/mL). The MSD platform quantified 7 cytokines in 100% of samples (IL-6, IL-7, IL-8, IL-10, MIP-1β, MIP-3α, and TNF-α), and expression trends of each of these across the 3 experimental groups were generally similar between MSD and LMX. With MSD, IL-7 levels appeared lower in HC samples versus GBM and BrMet, which was not observed with LMX assessment. While IL-17A was detected in 100% of samples by LMX and 76% by MSD, there a contradictory trend appeared in which IL-17 was lower in HC versus the 2 neoplasm groups with LMX, but higher with MSD. Only MIP-1β levels were categorized as greater using MSD (range 16.06-166.88 pg/mL) versus LMX (range 4.26-19.65 pg/mL).
Table 3.
Cytokine levels measured by a bead-based fluorescence multiplex assay in plasma samples from healthy controls (HC; n = 11), subjects with secondary brain metastases from other organs (BrMet; n = 17), and individuals with glioblastoma (GBM; n = 27).
| Cytokine | HC Conc. pg/mL ± SD | BrMet Conc. pg/mL ± SD | % of HC | P-value vs HC | GBM Conc. pg/mL ± SD | % of HC | P-value vs HC |
|---|---|---|---|---|---|---|---|
| GM-CSF | 16.8 ± 5.1 | 31.6 ± 13.9 | 188 | .011 | 31.2 ± 13.0 | 186 | .008 |
| IFN-γ | 10.8 ± 3.5 | 17.5 ± 6.2 | 162 | .037 | 20.0 ± 8.0 | 185 | .0012 |
| IL-1β | 1.1 ± 0.4 | 2.1 ± 0.9 | 191 | .026 | 2.6 ± 1.0 | 236 | .0001 |
| IL-2 | 4.7 ± 2.0 | 6.8 ± 2.8 | 145 | ns | 4.8 ± 2.3 | 102 | ns |
| IL-4 | 61.7 ± 16.2 | 111.1 ± 47.9 | 180 | .003 | 66.9 ± 32.1 | 108 | ns |
| IL-5 | 5.2 ± 1.6 | 11.5 ± 4.5 | 221 | .005 | 13.0 ± 5.6 | 250 | .0002 |
| IL-6 | 7.3 ± 2.6 | 12.9 ± 6.3 | 178 | .004 | 6.3 ± 2.6 | 86 | ns |
| IL-7 | 18.0 ± 5.5 | 27.8 ± 6.8 | 154 | <.001 | 17.6 ± 4.6 | 98 | ns |
| IL-8 | 10.4 ± 5.6 | 12.7 ± 8.8 | 122 | ns | 7.3 ± 3.6 | 70 | ns |
| IL-10 | 20.4 ± 10.8 | 69.7 ± 25.4 | 293 | <.0001 | 71.2 ± 31.1 | 286 | <.0001 |
| IL-12 p70 | 4.1 ± 1.8 | 5.9 ± 2.2 | 143 | ns | 5.5 ± 1.9 | 134 | ns |
| IL-13 | 10.3 ± 5.3 | 15.6 ± 7.4 | 151 | .039 | 7.1 ± 3.3 | 69 | ns |
| IL-17A | 12.1 ± 3.3 | 19.0 ± 7.4 | 157 | ns | 19.3 ± 8.8 | 159 | .028 |
| IL-21 | 4.9 ± 2.9 | 10.1 ± 4.9 | 206 | .005 | 8.8 ± 3.9 | 180 | .029 |
| IL-23 | 356 ± 290 | 394 ± 171 | 111 | ns | 792 ± 435 | 222 | .004 |
| MIP-1α | 29.4 ± 3.9 | 40.2 ± 6.9 | 137 | .0008 | 36.1 ± 7.7 | 123 | .030 |
| MIP-1β | 8.4 ± 3.3 | 9.2 ± 4.1 | 109 | ns | 10.0 ± 3.7 | 119 | ns |
| MIP-3α | 15.2 ± 9.1 | 14.9 ± 4.1 | 98 | ns | 20.8 ± 6.3 | 137 | .045 |
| TNF-α | 5.9 ± 2.3 | 4.2 ± 1.9 | 72 | ns | 6.0 ± 2.1 | 102 | ns |
Concentrations are shown as mean pg/mL ± SD. P-values were determined by one-way ANOVA adjusted for multiple comparisons using Tukey test. Bolded values indicate statistically significant differences from HC. Differences from HC that are not significant are labeled “ns”.
Of the 7 analytes in which the MSD platform detected quantifiable cytokine in 100% of samples (ie, IL-6, IL-7, IL-8, IL-10, MIP-1β, MIP-3α, and TNF-α), the trends in relative expression levels of these cytokines across the 3 experimental groups were similar between MSD and LMX in most instances. With MSD, values for IL-7 appeared lower in the HC samples versus GBM and BrMet samples, which was not observed with LMX assessment. With IL-17A, which was detected in 100% of samples by LMX and 76% of samples by MSD, there appeared to be a contradictory trend in which IL-17A was lower in HC plasma versus the 2 neoplasm samples with LMX, but higher with MSD. A similarly contrary relationship regarding IL-5 concentration in controls versus neoplasm groups was observed.
In a single instance, MIP-1β, cytokine levels quantified by MSD (range 16.06-166.88 pg/mL) were greater than concentrations determined by LMX (range 4.26-19.65 pg/mL), and the relative expression level relationships across the 3 experimental groups appeared similar with both MSD and LMX.
Cytokine profiles in healthy, GBM, and BrMet plasma
Because of the high frequency of non-quantifiable target cytokines using the MSD platform, we limited our analysis of patient versus HC to LMX findings, with which all analytes were detectable in all samples, although MSD findings are provided in Supplemental Table 1. Concentrations of multiple cytokines including GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-17A, IL-21, IL-23, MIP-1α, and MIP-3α were significantly elevated in GBM plasma compared to HC (Table 3). Of these analytes, measured values were at least double in GBM versus HC for 4, comprising IL-1β, IL-5, IL-10, and IL-23; levels for GM-CSF (186%), IFN-γ (185%), and IL-21 (180%) were very close to double in GBM versus HC plasma. Values of cytokines measured in BrMet samples are shown in Figure 3 and Table 3. Cytokines that were commonly elevated in GBM and BrMet plasma versus controls included GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-21, and MIP-1α. Analytes that were uniquely upregulated in GBM but not BrMet plasma included IL-17. IL-23 and MIP-3α, all of which have inflammatory functions and suspected roles in tumorigenesis.18-20 Analytes that were uniquely elevated in BrMet but not GBM samples compared to controls were the classical Th2 cytokines IL-4, IL-6 and IL-13,21and IL-7, a promoter of both T- and B-cell development.22 Interpretation of the significance of the various analytes measured in BrMet plasma is confounded because patients had received some type of chemotherapy regimen for their non-brain primary tumor that likely affected cytokine expression.
Correlation analysis
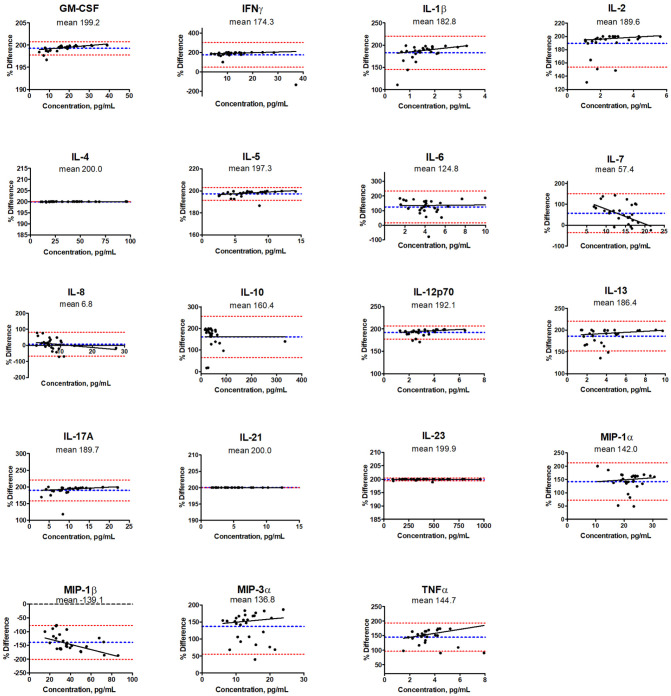
Correlations between LMX and MSD assay categorization of levels of the 19 shared analytes measured in human plasma were graphed on Bland-Altman plots (Figure 4).17 Log2 values of the mean MSD/LMX analyte concentration ratios shown on the y-axis estimates systematic bias. Good correlation is assumed between assays if the mean value approximates zero deviation. The 95% limits of agreement lines represent ±1.96 SDs from the mean. For most shared analytes, the mean concentration ratio was shifted from zero to a positive value, indicating that LMX classified the target cytokine in the same plasma samples at a higher concentration than MSD. The sole exception was with MIP-1β, where MSD characterized samples as containing markedly higher concentrations of cytokine than LMX. Analyte-concentration-dependent effects (ie, proportional bias) were suggested by the slope of the mean line, and were particularly notable for IL-7 and MIP-1β. The only native cytokine whose concentration was characterized nearly identically by both assays was IL-8. Interpretation of these data becomes less certain when SDs are large and 95% limits of agreement are spread further apart, reflective of widely distributed data points away from the mean. The maximum mean differences were restricted to −200 or +200 due to software limitations, but can theoretically be larger. In the case of IL-21, no cytokine was identified within the MSD assay’s dynamic range of quantitation in any sample; hence, the true mean difference is not 200, but infinity.
Figure 4.
Correlation analysis of concentrations of the 19 cytokines measured in 27 human plasma samples from patients with glioblastoma by the Luminex (LMX) and Meso Scale Discovery (MSD) multiplex assays. Log2 values of MSD/LMX concentration ratios are shown on the y-axis, and estimate systematic measurement differences between methods when the mean value deviates from zero. A negative shift of the blue mean indicates that the MSD assay classifies the analyte at concentrations lower than LMX, and vice versa. The red ±1.96 SD lines represent 95% limits of agreement, and when SDs are large or n-values are low, then interpretation can be ambiguous. For 17 of 19 shared analytes, the mean concentration ratio was shifted to a positive value, indicating that LMX classified the target cytokine in identical plasma samples at a higher concentration than MSD. The sole exception was with MIP-1β, where MSD characterized samples as containing markedly higher concentrations of cytokine than LMX. Analyte-concentration-dependent effects were suggested by the slope of the mean line, and were notable for IL-7 and MIP-1β. The only cytokine similarly characterized by both assays was IL-8.
Time in motion study
Hands-on labor time was 1 hour 38 minutes for LMX and 2 hours 42 minutes for the MSD assays, reflecting the increased number of MSD plates required. The total time required for performing the single Luminex (LMX) 21-plex and 4 Meso Scale Discovery (MSD) 3- to 10-plex assays was approximately 21.1 hours and 6.2 hours, respectively. This included a protocol-specified overnight primary antibody/sample incubation for LMX, and a 2-hour primary antibody/sample incubation for MSD (the alternative manufacturer-allowed overnight incubation was not used).
Discussion
Cytokines are critical mediators of diverse disease states, and have multiple pleotropic actions in different organs.3,4 Earlier categorization of inflammatory and anti-inflammatory cytokine categories has been revised with the appreciation that cytokines function in complex interacting pathways, often regulate the expression of other cytokines, and their inflammatory or anti-inflammatory roles depending upon their environmental context.23,24 Understanding the function of complex cytokine networks in pathogenesis requires comparison of expression profiles of multiple cytokines in health and disease states.25 Cytokines can serve as molecular biomarkers that provide insight into the development, progression, and prognosis of both acute health threats such as sepsis26 and trauma,27 and of chronic disorders including, for example, autoimmune,28 cardiopulmonary,29,30 and neoplastic diseases.5
Glioblastoma and brain metastases are particularly lethal neoplastic disorders with limited treatment options,1,2 whose pathogenesis may be modulated by multiple cytokines.6-10 Cytokines play important roles in immune cell infiltration and function. Although the brain is generally an immunologically specialized site, immune cell infiltration occurs in GBM tissue, with macrophages/microglial cells comprising a significant proportion of the tumor mass.31,32 We hypothesized that cytokines released from the GBM or brain metastatic tissues, and from immune cells in the tumor microenvironment, can be non-invasively identified in the circulation. These unique molecular profiles might provide information on disease existence, type, responsiveness to treatment, and prognosis. We evaluated these hypothesized biomarker signatures using multiplex proteomic immunoassays.
Multiplex assays have advantages over classical singleplex colorimetric ELISAs in that they simultaneously quantify numerous bioactive molecules within a single small sample volume to give a more complete overview of circulating analytes.12,33,34 Multiplex immunoanalysis streamlines cytokine profiling with improved productivity, while decreasing financial, time, and personnel expenditures needed to evaluate many analytes individually.12,15,16 These benefits are particularly valuable when evaluating diverse cytokines in rare or size-limited samples.
To identify potential circulating GBM-associated biomarkers, we compared concentrations of 19 cytokines in plasma from newly-diagnosed GBM patients and healthy individuals, using 2 different multiplex immunoassay platforms.13,14 The dynamic range of fluorescence and electrochemiluminescence multiplex immunoassays is generally several orders of magnitude greater than that of singleplex ELISAs.12,33,34 In our study, the MSD platform calibration curves suggested superior sensitivity than the LMX assay, with lower LLoQs for 89% of analytes, having 12 of 19 analytes’ LLoQ registering below 100 fg/mL. With native proteins in plasma samples, however, the MSD assay was surprisingly unable, in our hands, to detect or quantify many cytokines that LMX assessed in the 1 to 100 pg/mL range. This incongruous analyte classification was confirmed by correlation analyses. An example of this disparity was IL-21, which LMX quantified in all 55 of 55 samples (range 1.8-18.9 pg/mL) but was not detected by MSD in any sample even though MSD calibrator curves indicated a LLoQ of 0.22 pg/mL. Although many parameters can affect multiplex immunoassay function, we speculate that, while the MSD assay is excellent at quantifying the kit-specific recombinant cytokine fragments used for calibration, it may be less efficient in detecting native proteins, at least in the sample types/collection media we tested. While this manuscript was in preparation, another publication reported similar findings in which MSD missed detection of considerably more overlapping cytokine analytes in 62 (similarly EDTA-anticoagulated) human plasma samples than simultaneously performed LMX assays.35 Hands-on time required for LMX assessment was only 60% of vigilant time required with MSD because we needed to use 4 MSD plates to obtain desired analyte overlap with a single LMX 21-plex kit. Although assay time determinations were only performed once per platform in this study, our findings concur well with another report in which hands-on labor time to assay 16 overlapping cytokines in human plasma was reduced by 36% using LMX (1 hour 37 minutes) compared to the MSD platform (2 hour 33 minutes).35 Given the frequency of undetected analytes in samples with the MSD platform, we focused on assessing results obtained using the LMX assay.
Multiple cytokines were upregulated in the plasma of GBM and/or BrMet subjects compared to circulating levels in healthy volunteers. In GBM plasma, we identified significant elevation of 10 of 19 evaluated cytokines versus control samples, comprising GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-17A, IL-21, IL-23, MIP-1α, and MIP-3α. Of these analytes, measured values were at least double in GBM versus HC for 4, comprising IL-1β, IL-5, IL-10, and IL-23; concentrations were nearly double in GBM samples for GM-CSF, IFN-γ, and IL-21. Cytokines that were commonly elevated in GBM and BrMet plasma versus healthy control samples were GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-21, and MIP-1α. Analytes uniquely upregulated in GBM but not BrMet plasma included IL-17A, IL-23, and MIP-3α, all of which have inflammatory activity and are suspected to play proliferative and/or metastatic roles in solid tumors including glioblastoma.18-20 Analytes uniquely elevated in BrMet but not GBM plasma versus controls were IL-4, IL-6, IL-7, IL-10, IL-13, and MIP-3α. Besides IL-5, classical Th2 cytokines (ie, IL-4, IL-10, and IL 13) were notably elevated in BrMet but not GBM samples compared to healthy control plasma. Cytokines that were commonly elevated in GBM and BrMet plasma versus controls included GM-CSF, IFN-γ, IL-1β, IL-5, IL-10, IL-21, and MIP-1α. Analytes that were uniquely upregulated in GBM but not BrMet plasma included IL-17A and IL-23, both associated with glioblastoma progression,18,19 and MIP-3α (chemokine CCL20), which is an inflammatory chemokine with known roles in solid tumor progression and metastasis.20 MIP-3α contributes to the progression of liver, colon, breast, pancreatic, and gastric cancers.36 In glioblastoma, MIP-3α released from neighboring astrocytes may enhance malignancy by inducing hypoxia-induced factor-1 (HIF-1) expression in tumor cells, thereby promoting their survival and functionality in the hypoxic tumor environment.37
Analytes that were uniquely elevated in BrMet but not GBM samples compared to controls were the classical Th2 cytokines IL-4, IL-6, and IL-13,21 and IL-7, a promoter of both T- and B-cell development22 that is also a prognostic biomarker of improved survival in malignant glioblastoma.38 However, BrMet samples came from individuals who had variable histories of receiving chemotherapy for their primary non-brain tumors, which can induce cytokine expression,39 thereby confounding meaningful comparison with GBM findings. All GBM samples were obtained at the time of initial diagnosis from individuals who were treatment naïve, so cytokines differentially expressed in GBM plasma might be part of a valid GBM-specific biomarker profile.
A similar study used bead-based immunoassays to profile 48 circulating cytokine levels, and identified an 18-cytokine signature that discriminated 26 healthy subjects from 148 GBM patients with a diagnostic accuracy of 95.40%.40 Of the 18 markers detected by those researchers, 5 shared cytokines were similarly upregulated in GBM in our study: GM-CSF, IFN-γ, IL-12, IL-17A, and MIP-1α. When we queried the US National Cancer Institute’s Cancer Genome Atlas (TCGA) microarray database (available at https://portal.gdc.cancer.gov/projects/TCGA-GBM), both GM-CSF and IFN-γ showed increased transcript levels in GBM versus normal brain tissues, suggesting the tumor tissue itself as a primary source of these 2 circulating analytes. Conversely, IL-17A was upregulated in GBM plasma by our protein analysis compared to control plasma, although transcript levels of IL-17A were decreased in GBM tissue versus normal brain tissue in the TCGA database. This suggests that cells extraneous to the primary tumor mass are responsible for the increased circulating IL-17A that we detected in GBM plasma, but that these biomarkers, together with others, might provide a cytokine profile specific to GBM.
By univariate and multivariate cox regression survival analyses, the earlier group determined that out of their identified 18-cytokine GBM signature, only IL-17A and IL-4 had good prognostic value.40 We identified a 1.7-fold increase in plasma IL-17A levels in GBM versus healthy subjects, but did not identify a difference in circulating IL-4 concentrations. Greater numbers of T helper type 17 (Th17) cells in gliomas are associated with higher number of myeloid (CD11b) cells as well as the expression of TGF-β1 and IL-6,41 though we saw no change in IL-6 and did not measure TGF-β1. IL-1β and IL-23, both of which we observed to be elevated in GBM plasma, are critical in inducing the Th17 phenotype in humans.42,43 Glioma-associated Th17 cells are potentially non-cytotoxic and may contribute to immune suppression,41 and thereby disease progression. Ultimately, the collective pattern of expression of these molecules might serve as a useful clinical biomarker signature for detecting GBM and tracking progression. There may be similar utility for cytokine signatures in patients with brain metastases, although it is not surprising that the composition of cytokines detected may differ if systemic disease burden is variable.
Our findings support the discoveries of other researchers toward identifying a GBM-specific circulating cytokine profile. Differences between our findings and those from a previous large cytokine profiling study40 may have resulted from sampling differences, whereby we tested plasma from blood collected in EDTA-anticoagulant solution and immediately centrifuged then frozen, while the prior study collected sera from whole blood specimens that were separated after allowing sample coagulation overnight at 4°C before freezer storage. Future studies will ascertain whether tightly-coordinated sampling and assay parameter selection can produce more congruity in establishing a definitive and prognostic GBM cytokine biomarker signature. Additional work will be needed to determine how these cytokines change through the course of a patient’s disease and whether cytokine expression in cerebrospinal fluid may also yield important insights. Future experiments should evaluate additional molecules with known or suspected prognostic value in assessing GBM. It will be important to establish cytokine baselines at diagnosis, as reported here, and then follow longitudinally how expression may change throughout treatment, particularly with the increasing interest in immunotherapy. Limitations of this study include the relatively low number of GBM samples available for testing and the limited number of cytokines evaluated. We recognize that the complexities of multiplex proteomic immunoassay procedures will require extensive validation before being acceptable for clinical use. Ideally, all findings would be repeatable with the same samples using simplex immunoassays employing the exact same reference cytokines used in the multiplex platform. Nonetheless, this study demonstrated that appropriate multiplex platforms have important utility for dissecting the complex molecular mechanisms that drive the pathology of diverse health disorders, including GBM and metastatic cancers.
Supplemental Material
Supplemental material, sj-pdf-1-bmi-10.1177_11772719211006666 for Cytokine Profiling in Plasma from Patients with Brain Tumors Versus Healthy Individuals using 2 Different Multiplex Immunoassay Platforms by Diane Elizabeth Bender, Maximilian O Schaettler, Kathleen CF Sheehan, Tanner M Johanns and Gavin P Dunn in Biomarker Insights
Acknowledgments
The authors thank Matthew Silverman PhD (Biomedical Publishing Solutions, Shrewsbury MA) for expert assistance in data analysis and manuscript development; Dr. Silverman’s fee was paid by Luminex Corporation (Austin, TX).
Footnotes
Author Contributions: All authors were involved in the study conception, design, performance, analysis, and manuscript generation and revisions. All authors approved the final version for publication.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by Luminex, and some assay kits were provided by Merck EMD Millipore (Billerica, MA).
Declaration Of Conflicting Interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: GD reports being cofounder of Immunovalent Therapeutics Incorporated (St. Louis, MO), a company that develops immunologic cancer therapies, and is a member of the Scientific Advisory Board of Ziopharm Oncology (Boston, MA). No other author has any potential conflicts of interest to disclose.
ORCID iDs: Maximilian O Schaettler  https://orcid.org/0000-0001-7545-6342
https://orcid.org/0000-0001-7545-6342
Kathleen CF Sheehan  https://orcid.org/0000-0002-3357-8285
https://orcid.org/0000-0002-3357-8285
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Enrique GV, Irving SR, Ricardo BI, et al. Diagnosis and management of brain metastases: an updated review from a radiation oncology perspective. J Cancer Metastasis Treat. 2019;5:54. [Google Scholar]
- 3. Kany S, Vollrath JT, Relja B. Cytokines in inflammatory disease. Int J Mol Sci. 2019;20: E6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285:2944-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218-e228. [DOI] [PubMed] [Google Scholar]
- 6. Feng Y, Wang J, Tan D, Cheng P, Wu A. Relationship between circulating inflammatory factors and glioma risk and prognosis: a meta-analysis. Cancer Med. 2019;8:7454-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christofides A, Kosmopoulos M, Piperi C. Pathophysiological mechanisms regulated by cytokines in gliomas. Cytokine. 2015;71:377-384. [DOI] [PubMed] [Google Scholar]
- 8. Brown NF, Carter TJ, Ottaviani D, Mulholl P. Harnessing the immune system in glioblastoma. Br J Cancer. 2018;119:1171-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lynes JP, Nwankwo AK, Sur HP, et al. Biomarkers for immunotherapy for treatment of glioblastoma. J Immunother Cancer. 2020;8:e000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sims JS, Ung TH, Neira JA, Canoll P, Bruce JN. Biomarkers for glioma immunotherapy: the next generation. J Neurooncol. 2015;123:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKay HS, Margolick JB, Martínez-Maza O, et al. Multiplex assay reliability and long-term intra-individual variation of serologic inflammatory biomarkers. Cytokine. 2017;90:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 2015;9:406-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luminex Inc. Human T-cell 21 multiplex assay protocol and user manual and Flexmap 3D with xPONENT 4.2 overview. 2021. Accessed February 1, 2021. https://www.emdmillipore.com/US/en/product/MILLIPLEX-MAP-Human-High-Sensitivity-T-Cell-Panel-Premixed-21-plex-Immunology-Multiplex-Assay,MM_NF-HSTCMAG28SPMX21#anchor_PR, and https://www.rndsystems.com/products/flexmap-3d-with-xponent-42_flexmap-3d-ruo
- 14. Meso Scale Diagnostics LLC. V-Plex® protocols and user manuals. 2021. Accessed February 1, 2021. https://www.mesoscale.com/en/products_and_services/assay_kits/v-plex
- 15. Crabb-Breen E, Reynolds SM, Cox C, et al. Multisite comparison of high-sensitivity multiplex cytokine assays. Clin Vaccine Immunol. 2011;18:1229-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem 2010;56:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altman DG, Bland JM. Assessing agreement between methods of measurement. Clin Chem. 2017;63:1653-1654. [DOI] [PubMed] [Google Scholar]
- 18. Wang B, Zhao CH, Sun G, et al. IL-17 induces the proliferation and migration of glioma cells through the activation of PI3K/Akt1/NF-κB-p65. Cancer Lett. 2019;447:93-104. [DOI] [PubMed] [Google Scholar]
- 19. Yan J, Smyth MJ, Teng MWL. Interleukin (IL)-12 and IL-23 and their conflicting roles in cancer. Cold Spring Harb Perspect Biol. 2018;10:a028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elhousiny M, Miller K, Ariyawadana A, Nimmo A. Identification of inflammatory mediators associated with metastasis of oral squamous cell carcinoma in experimental and clinical studies: systematic review. Clin Exp Metastasis. 2019;36:481-492. [DOI] [PubMed] [Google Scholar]
- 21. Becker Y. Molecular immunological approaches to biotherapy of human cancers–a review, hypothesis and implications. Anticancer Res. 2006;26:1113-1134. [PubMed] [Google Scholar]
- 22. Barata JT, Durum SK, Seddon B. Flip the coin: IL-7 and IL-7R in health and disease. Nat Immunol. 2019;20:1584-1593. [DOI] [PubMed] [Google Scholar]
- 23. Cavaillon JM. Pro- versus anti-inflammatory cytokines: myth or reality. Cell Mol Biol (Noisy-le-grand). 2001;47:695-702. [PubMed] [Google Scholar]
- 24. Schmitz ML, Weber A, Roxlau T, Gaestel M, Kracht M. Signal integration, crosstalk mechanisms and networks in the function of inflammatory cytokines. Biochim Biophys Acta. 2011;1813(12):2165-2175. [DOI] [PubMed] [Google Scholar]
- 25. Morel PA, Lee REC, Faeder JR. Demystifying the cytokine network: mathematical models point the way. Cytokine. 2017;98:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumoto H, Ogura H, Shimizu K, et al. The clinical importance of a cytokine network in the acute phase of sepsis. Sci Rep. 2018;8:13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muire PJ, Mangum LH, Wenke JC. Time course of immune response and immunomodulation during normal and delayed healing of musculoskeletal wounds. Front Immunol. 2020;11:1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martinez-Quiles N, Goldbach-Mansky R. Updates on autoinflammatory diseases. Curr Opin Immunol. 2018;55:97-105. [DOI] [PubMed] [Google Scholar]
- 29. Bartekova M, Radosinska J, Jelemensky M, Dhalla NS. Role of cytokines and inflammation in heart function during health and disease. Heart Fail Rev. 2018;23:733-758. [DOI] [PubMed] [Google Scholar]
- 30. Barnes PJ. Targeting cytokines to treat asthma and chronic obstructive pulmonary disease. Nat Rev Immunol. 2018;18:454-466. [DOI] [PubMed] [Google Scholar]
- 31. Yang I, Han SJ, Kaur G, Crane C, Parsa AT. The role of microglia in central nervous system immunity and glioma immunology. J Clin Neurosci. 2010;17:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Badie B, Schartner J. Role of microglia in glioma biology. Microsc Res Tech. 2001;54:106-113. [DOI] [PubMed] [Google Scholar]
- 33. Shah K, Maghsoudlou P. Enzyme-linked immunosorbent assay (ELISA): the basics. Br J Hosp Med (Lond). 2016;77:C98-101. [DOI] [PubMed] [Google Scholar]
- 34. Chen Z, Dodig-Crnković T, Schwenk JM, Tao S. Current applications of antibody microarrays. Clin Proteomics. 2018;15:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Günther A, Becker M, Göpfert J, Joos T, Schneiderhan-Marra N. Comparison of bead based fluorescence versus planar electrochemiluminescence multiplex immunoassays for measuring cytokines in human plasma. Front Immunol. 2020;11:572634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen W, Qin Y, Liu S. CCL20 signaling in the tumor microenvironment. Adv Exp Med Biol. 2020;1231:53-65. [DOI] [PubMed] [Google Scholar]
- 37. Jin P, Shin SH, Chun YS, et al. Astrocyte-derived CCL20 reinforces HIF-1-mediated hypoxic responses in glioblastoma by stimulating the CCR6-NF-κB signaling pathway. Oncogene. 2018;37:3070-3087. [DOI] [PubMed] [Google Scholar]
- 38. Lin Y, Zhang G, Zhang J, et al. A panel of four cytokines predicts the prognosis of patients with malignant gliomas. J Neurooncol. 2013;114:199-208. [DOI] [PubMed] [Google Scholar]
- 39. Edwardson DW, Parissenti AM, Kovala AT. Chemotherapy and inflammatory cytokine signalling in cancer cells and the tumour microenvironment. Adv Exp Med Biol. 2019;1152:173-215. [DOI] [PubMed] [Google Scholar]
- 40. Nijaguna MB, Patil V, Hegde AS, et al. An eighteen serum cytokine signature for discriminating glioma from normal healthy individuals. PLoS One. 2015;10:e0137524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paladugu M, Thakur A, Lum LG, Mittal S, Parajuli P. Generation and immunologic functions of Th17 cells in malignant gliomas. Cancer Immunol Immunother. 2013;62:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aliahmadi E, Gramlich R, Grutzkau A, et al. TLR2-activated human langerhans cells promote Th17 polarization via IL-1beta, TGF-beta and IL-23. Eur J Immunol. 2009;39:1221-1230. [DOI] [PubMed] [Google Scholar]
- 43. Yu RY, Gallagher G. A naturally occurring, soluble antagonist of human IL-23 inhibits the development and in vitro function of human Th17 cells. J Immunol. 2010;185:7302-7308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-bmi-10.1177_11772719211006666 for Cytokine Profiling in Plasma from Patients with Brain Tumors Versus Healthy Individuals using 2 Different Multiplex Immunoassay Platforms by Diane Elizabeth Bender, Maximilian O Schaettler, Kathleen CF Sheehan, Tanner M Johanns and Gavin P Dunn in Biomarker Insights