Abstract
Acetylsalicylic acid (aspirin) is commonly used for primary and secondary prevention of cardiovascular diseases. Aspirin use is associated with better outcomes among COVID‐19 positive patients. We hypothesized that the aspirin use for primary cardiovascular disease prevention might have a protective effect on COVID‐19 susceptibility and disease duration. We conducted a retrospective population‐based cross‐sectional study, utilizing data from the Leumit Health Services database. The proportion of patients treated with aspirin was significantly lower among the COVID‐19‐positive group, as compared to the COVID‐19‐negative group [73 (11.03%) vs. 1548 (15.77%); P = 0.001]. Aspirin use was associated with lower likelihood of COVID‐19 infection, as compared to nonusers (adjusted OR 0.71 (95% CI, 0.52 to 0.99; P = 0.041). Aspirin users were older (68.06 ± 12.79 vs. 56.63 ± 12.28 years of age; P < 0.001), presented a lower BMI (28.77 ± 5.4 vs. 30.37 ± 4.55; P < 0.0189), and showed higher prevalence of hypertension (56, 76.71%), diabetes (47, 64.38%), and COPD (11, 15.07%) than the aspirin nonusers (151, 25.64%, P < 0.001; 130, 22.07%, P < 0.001; and 43, 7.3%, P = 0.023, respectively). Moreover, COVID‐19 disease duration (considered as the time between the first positive and second negative COVID‐19 RT–PCR test results) among aspirin users was significantly shorter, as compared to aspirin nonusers (19.8 ± 7.8 vs. 21.9 ± 7.9 P = 0.045). Among hospitalized COVID‐positive patients, a higher proportion of surviving subjects were treated with aspirin (20, 19.05%), as opposed to 1 dead subject (14.29%), although this difference was not significant (P = 0.449). In conclusion, we observed an inverse association between the likelihood of COVID‐19 infection, disease duration and mortality, and aspirin use for primary prevention.
Keywords: aspirin, COVID‐19, disease likelihood, Israeli cohort
Acetylsalicylic acid (aspirin) is used for the prevention of cardiovascular diseases and might be associated with better outcomes among COVID‐19 patients. We hypothesized that aspirin use for primary cardiovascular disease prevention might have effects on COVID‐19 susceptibility. We conducted a retrospective population‐based study, utilizing data from the Leumit Health Services database. We observed an inverse association between the likelihood of COVID‐19 infection, disease duration and mortality, and aspirin use for primary prevention.

Abbreviations
- COVID‐19
coronavirus SARS‐CoV‐2
- CVD
cardiovascular disease
- LHS
Leumit Health Services
- SES
socioeconomic status
- STING
stimulator of interferon genes
Introduction
Surveillance of the clinical characteristics of patients with coronavirus disease 2019 (COVID‐19) is important for clarifying the epidemiology of the disease. Several epidemiological studies have suggested that individuals with diabetes, cardiovascular disease (CVD), or chronic lung disease are at higher risk for COVID‐19 infection [1, 2]. While many reports on COVID‐19 have emphasized age‐, sex‐, and comorbid disease‐related differences in health outcomes, pharmacological treatment differences in infection susceptibility have yet to be described in‐depth.
Acetylsalicylic acid (aspirin) is among the most used medications in the world for the treatment of acute coronary syndromes and secondary prevention of CVD [3]. Recent trials have, however, revealed that low‐dose aspirin is not associated with significant differences in terms of primary CVD prevention [4, 5]. Therefore, the American College of Cardiology and the American Heart Association, who previously published guidelines for aspirin use in primary prevention, now discourages routine use of aspirin, particularly in patients with increased risk of bleeding [6]. Nevertheless, many individuals still receive low‐dose aspirin for primary prevention [7]. Aspirin has multiple effects on different components of innate and adaptive immunity and, therefore, can influence susceptibility to viral infections [8]. As such, we hypothesized that pre‐existing treatment with low‐dose aspirin might have a protective effect on COVID‐19 susceptibility and disease duration among COVID‐19‐infected subjects. We accordingly analyzed the prevalence of low‐dose aspirin therapy and clinical characteristics of that group in a large cohort of consecutive outpatients who tested positive in an RT–PCR assay designed to detect infection with COVID‐19.
Results
Data sources
We directed a retrospective population‐based cross‐sectional study utilizing data from the Leumit Health Services (LHS) database. LHS is a nation‐wide health maintenance organization in Israel, which provides services to some 725, 000 members. LHS maintains a wide‐ranging computerized database, continuously updated in terms of member hospitalizations and laboratory tests, demographics, medical diagnoses, and medical encounters. During each physician visit, the patient's diagnosis is entered or updated according to the International Classification of Diseases (9th revision, ICD‐9). At the same time, there is an ongoing process of validation by which the physicians are encouraged to report on patients, their diagnosis classification, medications, and services. As such, the validity of diagnoses entered into the registry is high for important medical diagnoses, in particular those based on the LHS laboratory data [9, 10, 11, 12].
The study period was from February 1, 2020, to June 30, 2020. The first COVID‐19 patient in Israel was diagnosed in February 2020, and on July 9, the criteria used for defining recovery from COVID‐19 were changed [13]. All LHS enrollees who had been tested for COVID‐19 during the study period were included in the present study. Testing for COVID‐19 infection was performed upon physician referral, according to Israel Ministry of Health criteria for COVID‐19 testing, which includes direct exposure to a confirmed COVID‐19 patient and/or presentation of symptoms suggesting COVID‐19 (essentially, a cough, shortness of breath, or any other respiratory symptom, with fever). Nasopharyngeal swabs were taken and examined for COVID‐19 by real‐time RT–PCR performed with internal positive and negative controls, according to World Health Organization guidelines. The Allplex 2019‐nCoV assay (Seegene, Seoul, Korea) was used until March 10, 2020, after which time the COBAS SARS‐CoV‐2 6800/8800 assay (Roche Pharmaceuticals, Basel, Switzerland) was employed. The study protocol was approved by the statutory clinical ethics committees of LHS and the Shamir Medical Center Institutional Review Board (Helsinki Committee Approval #0129‐20‐LEU, Shamir Medical Center) on human research.
Study subjects
Data on demographics, laboratory results, and ICD‐9 codes were derived from the LHS electronic medical record (EMR) system. COVID‐19 RT–PCR testing of samples derived from nasopharyngeal swabs was performed by experienced personnel in a single centralized laboratory according to international guidelines [14]. All consecutive patients aged ≥ 40 years who had been tested for COVID‐19 during the study period were included in the study. The EMR of each subject was reviewed, and those patients receiving low‐dose aspirin treatment 4 weeks prior to COVID‐19 RT–PCR testing were identified. Individuals who had been diagnosed with coronary artery disease, cerebrovascular disease, and/or peripheral vascular disease were classified as taking aspirin for secondary prevention and excluded from the study. Such exclusion was performed to reduce the risk of bias due to differences in social activity and hence differences in potential COVID‐19 exposure between healthy and more socially active populations and those patients suffering from cardiovascular diseases and, accordingly, less socially active.
Whole sample analysis from COVID‐19‐positive vs. COVID‐19‐negative patients
A total of 10 477 subjects were identified who had been tested for COVID‐19 by RT–PCR and who had received and purchased at least three prescriptions for aspirin for primary prevention. Comparison of various demographic and clinical characteristics in these subjects, both positive and negative for COVID‐19, is presented in Table 1. As compared to the COVID‐19‐negative group, the COVID‐19‐positive group was slightly younger (57.89 ± 13.83 years vs. 59.11 ± 14.53 years; P = 0.047) and included higher proportions of males [375 (56.65%) vs. 4307 (43.88%); P < 0.001] and persons from low‐medium socioeconomic status (SES) [506 (76.44%) vs. 5828 (59.38%); P < 0.001] presented a higher BMI (28.96 ± 5.29 kg·m−2 vs. 28.35 ± 5.61 kg·m−2; P = 0.012) and contained a significantly lower proportion of smokers [37 (7.3%) vs. 1806 (23.35%); P < 0.001]. The prevalence of arterial hypertension [207 (31.27%)] and COPD [1170 (11.92%)] was higher in the COVID‐19‐negative group than in the COVID‐19‐positive group [3719 (37.89%), P < 0.001, and 54 (8.16%), P = 0.003, respectively]. In contrast, the prevalence of obesity was higher in the COVID‐19‐positive group [232 (40.56%) vs. 2871 (34.3%); P = 0.002]. The prevalence of diabetes was similar in both groups. The proportion of patients treated with aspirin or statins was significantly lower among the COVID‐19‐positive group, as compared to the COVID‐19‐negative group [73 (11.03%) vs. 1548 (15.77%), P = 0.001, and 108 (16.31%) vs. 2198 (22.43%), P = 0.003, respectively]. Those subjects who had purchased at least three prescriptions for aspirin and statins were less associated with the likelihood of COVID‐19 infection than were those who did not [adjusted OR: 0.71 (95% CI, 0.52 to 0.99; P = 0.041) and adjusted OR 0.70 (95% CI, 0.53–0.92; P = 0.012)] (Table 1). Moreover, there were no differences between COVID‐19‐positive and COVID‐19‐negative subjects in terms of the proportion of patients treated with ACE inhibitors [65 (9.82%) vs. 1162 (11.86%)] and angiotensin II receptor blockers (ARBs) [28 (4.23%) vs. 540 (5.51%)] (Table 1).
Table 1.
Demographic and clinical characteristics of patients tested for COVID‐19.
|
COVID‐19‐positive n = 662 |
COVID‐19‐negative n = 9815 |
P‐value |
Multiple logistic regression model adjusted for sex and age OR (95% CI) |
Multiple logistic regression model adjusted for sex, age, smoking, medication use, and comorbidities* OR (95% CI) |
VIF when all covariates are in the model | |
|---|---|---|---|---|---|---|
| Age, years (mean ± SD) | 57.89 ± 12.83 | 59.11 ± 14.53 | 0.037 | 0.99 (0.99–1.00); P = 0.778 | 1.01 (0.99–1.02); P = 0.063 | 1.01 |
| Sex (male), n (%) | 375 (56.65%) | 4307 (43.88%) | < 0.001 | 1.70 (1.45–1.99); P < 0.001 | 2.06 (1.57–2.69); P < 0.001 | 1.02 |
| Low–medium SES, n (%) | 506 (76.44%) | 5828 (59.38%) | < 0.001 | 2.16 (1.79–2.60); P < 0.001 | 1.80 (1.41–2.31); P < 0.001 | 1.08 |
| BMI, kg·m−2 (mean ± SD) | 28.96 ± 5.29 | 28.35 ± 5.61 | 0.012 | 1.02 (1.01–1.03); P = 0.005 | 1.01 (0.99–1.03); P = 0.136 | 1.03 |
| Current smoking, n (%) | 37 (7.3%) | 1806 (23.35%) | < 0.001 | 0.23 (0.16–0.32); P < 0.001 | 0.24 (0.16–0.37); P < 0.001 | 1.73 |
| Comorbidity | ||||||
| Hypertension, n (%) | 207 (31.27%) | 3719 (37.89%) | 0.001 | 0.75 (0.62–0.91); P = 0.004 | 0.77 (0.59–0.99); P = 0.048 | 1.91 |
| Diabetes mellitus, n (%) | 177 (26.74%) | 2752 (28.04%) | 0.468 | 0.97 (0.80–1.17); P = 0.739 | 0.81 (0.60–1.07); P = 0.128 | 1.73 |
| COPD, n (%) | 54 (8.16%) | 1170 (11.92%) | 0.003 | 0.65 (0.49–0.87); P = 0.004 | 0.66 (0.45–0.96); P = 0.030 | 1.69 |
| Obesity, n (%) | 232 (35.04%) | 2871 (28.33%) | 0.002 | 1.27 (0.99–1.64); P = 0.079 | 1.21 (0.98–1.51); P = 0.067 | 1.85 |
| Laboratory data | ||||||
| HgbA1C % (mean ± SD) | 5.71 ± 1.52 | 5.59 ± 1.42 | 0.005 | 1.08 (1.02–1.15): P = 0.012 | 1.10 (1.00–1.21); P = 0.049 | 1.07 |
| Total cholesterol (mean ± SD) | 192.91 ± 40.93 | 197.14 ± 44.83 | 0.021 | 0.99 (0.99–1.00); P = 0.101 | 0.99 (0.98–1.00); P = 0.506 | 2.67 |
| LDL cholesterol (mean ± SD) | 117.71 ± 36.68 | 120.27 ± 39.54 | 0.114 | 0.99 (0.99–1.00); P = 0.259 | 1.00 (0.99–1.01); P = 0.659 | 2.21 |
| HDL cholesterol (mean ± SD) | 47.29 ± 12.11 | 48.83 ± 13.42 | 0.005 | 0.99 (0.99–1.00); P = 0.648 | 1.01 (0.99–1.02); P = 0.312 | 1.01 |
| Triglycerols (mean ± SD) | 136.38 ± 84.92 | 134.71 ± 85.97 | 0.649 | 0.99 (0.99–1.00); P = 0.669 | 0.99 (0.99–1.00); P = 0.853 | 1.03 |
| Medications | ||||||
| Aspirin, n (%) | 73 (11.03%) | 1548 (15.77%) | 0.001 | 0.63 (0.46–0.86); P = 0.004 | 0.71 (0.51; 0.99); P = 0.041 | 3.82 |
| ACE inhibitors | 65 (9.82%) | 1162 (11.86%) | 0.161 | 0.81 (0.62–1.07); P = 0.134 | 1.04 (0.75–1.46); P = 0.809 | 2.03 |
| ARB's | 28 (4.23%) | 540 (5.51%) | 0.183 | 0.78 (0.53–1.15); P = 0.214 | 0.89 (0.56–1.39); P = 0.610 | 2.44 |
| Statins | 108 (16.31%) | 2198 (22.43%) | 0.003 | 0.67 (0.54–0.83); P < 0.001 | 0.70 (0.53–0.92); P = 0.012 | 3.56 |
* Comorbidities ‐ hypertension, diabetes mellitus, COPD, obesity, allergic diseases, systemic and organ‐specific autoimmune diseases
# Not applicable
Demographic and clinical characteristics of COVID‐19‐positive subjects receiving aspirin treatment (aspirin users) vs. COVID‐19‐positive subjects not receiving aspirin treatment (aspirin nonusers) are presented in Table 2. Aspirin users were older (68.06 ± 12.79 vs. 56.63 ± 12.28 years of age; P < 0.001), presented a lower BMI (28.77 ± 5.4 vs. 30.37 ± 4.55; P < 0.0189), and showed higher prevalence of hypertension [56 (76.71%)], diabetes [47 (64.38%)], and COPD [11 (15.07%)] vs. aspirin nonusers [151 (25.64%), P < 0.001; 130 (22.07%), P < 0.001; and 43 (7.3%), P = 0.023, respectively]. Moreover, among aspirin users, a significantly higher proportion of subjects were treated with ACE inhibitors [25 (34.25%)], ARBs [10 (13.70%)], and statins [52 (71.23%)], relative to aspirin nonusers [40 (6.79%), P < 0.001; 18 (3.06%), P < 0.001; and 56 (9.51%), P < 0.001, respectively]. The variance inflation factor (VIF) was used to account for the problem of multicollinearity among independent variables used. All VIFs were less than 5, indicative of a degree of multicollinearity in our data, although one not sufficiently severe to warrant further corrective measures (see Table 1 and Fig. 1).
Table 2.
Demographic and clinical characteristics of COVID‐19‐positive patients with and without aspirin.
|
COVID‐19‐positive subjects with aspirin n = 73 |
COVID‐19‐positive subjects without aspirin n = 589 |
P‐value | |
|---|---|---|---|
| Age, years (mean ± SD) | 68.06 ± 12.79 | 56.63 ± 12.28 | < 0.001 |
| Sex (male), n (%) | 43 (58.90%) | 332 (56.37%) | 0.848 |
| Low–medium SES, n (%) | 50 (68.49%) | 456 (77.42%) | 0.511 |
| BMI, kg·m−2 (mean ± SD) | 28.77 ± 5.4 | 30.37 ± 4.55 | 0.0189 |
| Current smoking, n (%) | 2 (2.74%) | 35 (5.94%) | 0.278 |
| Comorbidity | |||
| Hypertension, n (%) | 56 (76.71%) | 151 (25.64%) | < 0.001 |
| Diabetes mellitus, n (%) | 47 (64.38%) | 130 (22.07%) | < 0.001 |
| COPD, n (%) | 11 (15.07%) | 43 (7.30%) | 0.023 |
| Obesity, n (%) | 34 (46.58%) | 198 (33.62%) | 0.059 |
| Laboratory data | |||
| HgbA1C % (mean ± SD) | 6.64 ± 1.62 | 5.63 ± 1.46 | < 0.001 |
| Total cholesterol (mean ± SD) | 176.89 ± 46.31 | 195.04 ± 39.73 | 0.0004 |
| LDL cholesterol (mean ± SD) | 102.91 ± 40.27 | 119.69 ± 35.76 | < 0.001 |
| HDL cholesterol (mean ± SD) | 44.49 ± 11.07 | 47.66 ± 12.19 | 0.035 |
| Triglycerols (mean ± SD) | 146.34 ± 70.21 | 135.06 ± 86.67 | 0.286 |
| Medications | |||
| ACE inhibitors | 25 (34.25%) | 40 (6.79%) | < 0.001 |
| ARBs | 10 (13.70%) | 18 (3.06%) | < 0.001 |
| Statins | 52 (71.23%) | 56 (9.51%) | < 0.001 |
| Time from 1st positive SARS‐CoV‐2 RT–PCR test result to 1st negative SARS‐CoV‐2 RT–PCR test result, days (mean ± SD) | 15.66 ± 7.15 | 18.38 ± 7.71 | 0.0052 |
| Time from 1st positive SARS‐CoV‐2 RT–PCR test result to 2nd negative SARS‐CoV‐2 RT–PCR test result, days (mean ± SD) | 19.81 ± 7.77 | 21.91 ± 7.88 | 0.045 |
Fig. 1.

Three peak age‐groups at high risk of contracting COVID‐19: 45–50, 50–60, and 60–75 years old (red bars). These three age‐groups were included in the subsets aspirin (A) and aspirin and statins (B) (the area highlighted in light blue). In the subsets of persons not treated with drugs, the age range of 40–45 years peaked (shown in the area highlighted in light pink). The delta density of the drugs was calculated by the formulae described in Methods.
Analysis of hospitalized vs. community‐treated COVID‐19‐positive patients
Demographic and clinical characteristics of hospital‐treated COVID‐19‐positive patients (n = 112) vs. community‐treated COVID‐19‐positive patients (n = 550) are presented in Table 3. Hospitalized patients were significantly older (62.89 ± 12.19 vs. 56.91 ± 12.19 years of age; P < 0.001) and showed higher prevalence of hypertension [46 (41.07%)], diabetes [47 (41.96%)], and COPD [15 (13.39)] than did community‐treated patients (161 (29.27)), P = 0.014; (130 (22.07%)), P < 0.001; and (39 (7.09)), P = 0.026, respectively). Hospitalized patients also had significantly higher hemoglobin A1C levels than did community‐treated patients (6.26% ± 1.85 and 5.70% ± 1.4, P = 0.002, respectively). Older age and higher ages of comorbidities were noted among hospital‐treated patients, as compared to community‐treated patients. This may be explained by the significantly higher proportion of subjects treated with aspirin [21 (18.75%) vs. 51 (9.27%), P = 0.003]. After adjustment for age, sex, and comorbidities, this difference disappeared [adjusted OR: 1.00 (95% CI, 0.47–2.57; P = 0.826)] (Table 3).
Table 3.
Demographic and clinical characteristics of hospital‐treated vs. COVID‐19‐positive patients.
|
Hospital‐treated COVID‐19‐positive patients n = 112 |
Community‐treated COVID‐19‐positive patients n = 550 |
P‐value |
Multiple logistic regression model adjusted for sex and age OR (95% CI) |
Multiple logistic regression model adjusted for sex, age, smoking status, medication use, and comorbidities* OR (95% CI) |
|
|---|---|---|---|---|---|
| Age, years (mean ± SD) | 62.89 ± 14.67 | 56.91 ± 12.18 | < 0.001 | 1.03 (1.01–1.04); P < 0.001 | 1.02 (1.00–1.05); P = 0.016 |
| Sex (male), n (%) | 62 (55.36%) | 313 (56.9%) | 0.762 | 0. 92 (0.61–1.40); P = 0.73 | 0.70 (0.38–1.05); P = 0.263 |
| Low–medium SES, n (%) | 80 (71.43%) | 426 (77.45%) | 0.170 | 0.77 (0.48–1.23); P = 0.278 | 0.69 (0.37–1.29); P = 0.243 |
| BMI, kg·m−2 (mean ± SD) | 29.05 ± 6.16 | 28.94 ± 5.1 | 0.860 | 1.00 (0.96–1.04); P = 0.84 | 0.92 (0.84–1.01); P = 0.088 |
| Currently smoking, n (%) | 6 (5.36%) | 31 (5.64%) | 0.884 | 1.04 (0.41–2.64); P = 0.92 | 1.07 (0.37–3.14); P = 0.888 |
| Comorbidity | |||||
| Hypertension, n (%) | 46 (41.07%) | 161 (29.27%) | 0.014 | 1.27 (0.81–1.99); P = 0.278 | 1.25 (0.65–2.40); P = 0.486 |
| Diabetes mellitus, n (%) | 47 (41.9%6) | 130 (23.64%) | < 0.001 | 1.98 (1.27–3.06); P = 0.002 | 1.27 (0.63–2.55); P = 0.491 |
| COPD, n (%) | 15 (13.39%) | 39 (7.09%) | 0.026 | 1.79 (0.94–3.44); P = 0.075 | 1.80 (0.80–4.08); P = 0.154 |
| Obesity, n (%) | 44 (39.29%) | 188 (34.18%) | 0.336 | 1.31 (0.83–2.04); P = 0.235 | 2.06 (0.83–5.11); P = 0.117 |
| Laboratory data | |||||
| HgbA1C % (mean ± SD) | 6.26 ± 1.85 | 5.70 ± 1.4 | < 0.001 | 1.24 (1.08–1.43); P = 0.002 | 1.30 (1.06–1.59); P = 0.009 |
| Total cholesterol (mean ± SD) | 195.76 ± 42.82 | 192.33 ± 40.55 | 0.431 | 1.00 (0.99–1.00); P = 0.453 | 1.00 (0.97–1.03); P = 0.842 |
| LDL cholesterol (mean ± SD) | 122.06 ± 34.39 | 116.82 ± 37.1 | 0.182 | 1.00 (0.99–1.00); P = 0.166 | 1.00 (0.97–1.03); P = 0.631 |
| HDL cholesterol (mean ± SD) | 46.51 ± 11.00 | 47.45 ± 12.31 | 0.470 | 0.98 (0.96–1.00); P = 0.222 | 0.96 (0.92–1.00); P = 0.116 |
| Triglycerols (mean ± SD) | 136.60 ± 67.65 | 136.34 ± 88.10 | 0.976 | 1.00 (0.99–1.00); P = 0.924 | 0.99 (0.98–1.00); P = 0.521 |
| Medications | |||||
| Aspirin, n (%) | 21 (18.75%) | 51 (9.27%) | 0.003 | 1.62 (0.90–2.92); P = 0.103 | 1.00 (0.47–2.57); P = 0.826 |
| ACE inhibitors, n (%) | 14 (12.50%) | 51 (9.27%) | 0.295 | 1.23 (0.65–2.33); P = 0.521 | 0.92 (0.34–1.97); P = 0.672 |
| ARBs, n (%) | 4 (3.57%) | 24 (4.36%) | 0.704 | 0.60 (0.20–1.83); P = 0.377 | 0.62 (0.15–1.82); P = 0.312 |
| Statins, n (%) | 24 (21.43%) | 84 (15.27%) | 0.108 | 1.22 (0.72–2.06); P = 0.457 | 0.98 (0.45–2.14); P = 0.976 |
* Comorbidities ‐ hypertension, diabetes mellitus, COPD, obesity, allergic diseases, systemic and organ‐specific autoimmune diseases
# Not applicable
Seven subjects of the 112 hospitalized COVID‐19‐positive patients died during the study period. Their demographic and clinical characteristics are presented in Table 4. Dead subjects were significantly older (80.71 ± 20.51 vs. 61.72 ± 13.50 years of age; P < 0.0007). After adjustment for other variables, only age, as a continuous variable, appeared to be a significant risk factor for mortality, introducing an additional 9% for every year [adjusted OR: 1.09 (95% CI, 1.02–1.17; P = 0.009)] (Table 4). Among surviving hospitalized patients, a higher proportion of subjects were treated with aspirin [20 (19.05%) vs. 1 (14.29%), adjusted OR: 0.36 (95% CI, 0.02–6.85)] and statins [23 (21.90) vs. 1 (14.29), OR: 0.31 (95% CI, 0.01–6.57)], although this difference was not significant (P = 0.449 and P = 0.514, respectively) (Table 4).
Table 4.
Demographic and clinical characteristics of dead vs. surviving hospital‐treated COVID‐19‐positive patients.
|
Dead n = 7 |
Survived n = 105 |
P‐value |
Multiple logistic regression model adjusted for sex and age OR (95% CI) |
Multiple logistic regression model adjusted for sex, age, smoking status, medication use, and comorbidities* OR (95% CI) |
|
|---|---|---|---|---|---|
| Age, years (mean ± SD) | 80.71 ± 20.51 | 61.72 ± 13.50 | 0.0007 | 1.09 (1.03–1.16); P = 0.005 | 1.09 (1.02–1.17); P = 0.009 |
| Sex (male), n (%) | 3 (42.86%) | 59 (56.19%) | 0.49 | 1.03 (0.19–5.54); P = 0.964 | 0.60 (0.06–5.67); P = 0.657 |
| Low–medium SES, n (%) | 4 (57.14%) | 76 (72.38%) | 0.39 | 0.66 (0.12–3.61); P = 0.635 | 0.76 (0.10–5.63); P = 0.791 |
| BMI, kg·m−2 (mean ± SD) | 30.76 ± 6.71 | 28.99 ± 6.17 | 0.62 | 1.06 (0.85–1.32); P = 0.564 | 1.09 (0.86–1.38); P = 0.439 |
| Currently smoking, n (%) | 0 | 6 (5.71) | 0.68 | # | # |
| Comorbidity | |||||
| Hypertension, n (%) | 3 (42.86%) | 43 (40.95%) | 0.92 | 0.51 (0.81–3.13); P = 0.465 | 1.31 (0.13–12.63); P = 0.337 |
| Diabetes mellitus, n (%) | 4 (57.14%) | 44 (41.90%) | 0.43 | 1.65 (0.29–9.34); P = 0.571 | 3.19 (0.29–35.03); P = 0.816 |
| COPD, n (%) | 1 (14.29%) | 14 (13.33%) | 0.94 | 0.68 (0.65–7.09); P = 0.749 | 0.56 (0.02–16.06); P = 0.343 |
| Obesity, n (%) | 1 (14.29%) | 43 (40.95%) | 0.68 | 0.72 (0.55–9.39); P = 0.802 | 0.75 (0.04; 12.49); P = 0.739 |
| Laboratory data | |||||
| HgbA1C % (mean ± SD) | 7.11 ± 2.01 | 6.37 ± 1.75 | 0.29 | 1.23 (0.79–1.92); P = 0.352 | 1.31 (0.75–2.32); P = 0.276 |
| Total cholesterol (mean ± SD) | 162.48 ± 42.96 | 197.79 ± 42.73 | 0.0028 | 0.97 (0.95–0.99); P = 0.046 | 0.91 (0.65–1.07); P = 0.058 |
| LDL cholesterol (mean ± SD) | 100.33 ± 20.93 | 123.38 ± 34.67 | 0.11 | 0.97 (0.95–1.00); P = 0.098 | 1.05 (0.76–1.47); P = 0.762 |
| HDL cholesterol (mean ± SD) | 42.66 ± 6.37 | 46.75 ± 11.21 | 0.38 | 0.92 (0.83–1.03); P = 0.166 | 1.09 (0.75–1.57); P = 0.645 |
| Triglycerides (mean ± SD) | 115.03 ± 79.43 | 137.90 ± 67.12 | 0.42 | 1.00 (0.98–1.01); P = 0.816 | 0.97 (0.89–1.05); P = 0.441 |
| Medications | |||||
| Aspirin, n (%) | 1 (14.29%) | 20 (19.05%) | 0.75 | 0.38 (0.04–3.59); P = 0.399 | 0.36 (0.02–6.85); P = 0.514 |
| ACE inhibitors, n (%) | 0 | 14 (13.33) | 0.302 | # | # |
| ARBs, n (%) | 0 | 4 (3.81) | 0.591 | # | # |
| Statins, n (%) | 1 (14.29) | 23 (21.90) | 0.449 | 0.35 (0.35–3.57); P = 0.381 | 0.31 (0.01–6.57); P = 0.449 |
* Comorbidities ‐ hypertension, diabetes mellitus, COPD, obesity, allergic diseases, systemic and organ‐specific autoimmune diseases
# Not applicable
Conversion time of SARS‐CoV‐2 PCR test results from positive to negative among COVID‐positive patients
To strengthen the hypothesis that aspirin pharmacotherapy for primary prevention has a possible protective effect against COVID‐19 infection, a separate analysis of the conversion time of a SARS‐CoV‐2 RT–PCR test result from positive to negative among COVID‐19‐positive subjects was performed. Demographic and clinical characteristics of COVID‐19‐positive patients with and without aspirin treatment are presented in Table 2. The time between the first positive SARS‐CoV‐2 RT–PCR test result and the first and second negative SARS‐CoV‐2 RT–PCR test results among aspirin users was significantly shorter, as compared to that time measured for aspirin nonusers [(15.66 ± 7.15 vs. 18.38 ± 7.71 days, P = 0.0052) and (19.81 ± 7.77 vs. 21.91 ± 7.88, P = 0.045), respectively] (Table 2). Figure 2 presents the effect of aspirin on conversion time among COVID‐19‐positive patients, using a statistical R function Welch two‐sample t‐test. This means that a significant difference exists in all tests, with a significance threshold of P‐value < 0.05. These results indicate a reduction in the duration of illness by an average of about 1.2 times in the case of those taking aspirin.
Fig. 2.

Effects of aspirin on conversion time of SARS‐CoV‐2 RT–PCR test results among COVID‐19‐positive patients. The P‐values have been calculated by the standard t‐test.
Discussion
A large, nation‐wide study revealed that the use of aspirin is associated with a decreased likelihood of a positive COVID‐19 test result. Indeed, aspirin gained remarkable popularity during the 1918 Spanish influenza pandemic, several decades prior to confirmation of its action on several components of innate immunity [8] and the in vitro efficacy against RNA viruses of the respiratory tract [15]. The basic mechanism of the antiviral activity of aspirin against RNA viruses relies on several biochemical and immunological pathways. Host response and clearance of viral infections heavily depend on the expression of type I interferon (IFN), which modulates cell responses and reprograms cells into an "antiviral state" [16]. RNA viruses, such as SARS‐CoV and MERS‐CoV, can escape immune system recognition via suppression of type I IFN signaling through an inhibition of STAT family transcription factor phosphorylation [17]. Another specific mechanism used by RNA viruses to evade host antiviral responses involves upregulation of prostaglandin E2 (PGE2) levels, which leads to an inhibition of type I IFN production and apoptosis in macrophages, thereby causing increased viral replication [18]. As low‐dose aspirin inhibits PGE2 biosynthesis [19], this mechanism might enhance antiviral immunity via induction of type I INF [20].
COVID‐19 uses the transmembrane angiotensin‐converting enzyme 2 (ACE2) as the host transmembrane cellular receptor for cell infection [21]. Higher ACE2 expression may be of benefit in preventing COVID‐19 infection, as COVID‐19 virus particles may compete with angiotensin‐2 protein for cell surface binding sites and cellular uptake [22]. In addition, the stimulator of interferon genes (STING) induces type I INF production when cells are infected with DNA and RNA viruses [23]. Furthermore, polymorphisms in the STING pathway contribute to the pathogenesis of COVID‐19 infection [24]. Interestingly, aspirin has been found to directly affect an acetylate cyclic GMP‐AMP synthase (cGAS) that activates a type I INF response via the STING pathway [25]. In COVID‐19, excessive angiotensin II signaling due to poor ACE2‐mediated conversion of angiotensin II at the cell surface could activate the STING pathway [26]. Accordingly, Chow et al. suggested that COVID severity would be reduced among aspirin users. In their recent retrospective on an observational cohort study of adult patients admitted with COVID‐19 to multiple hospitals in the United States, these authors found that aspirin use was associated with decreased rates of mechanical ventilation, ICU admission, and in‐hospital mortality [27].
The present study sought to better understand the potential favorable effects of aspirin in aiding the human immune system battle against COVID‐19. At the same time, several studies have revealed that platelets can associate with SARS‐CoV‐2 RNA and are activated in response to COVID‐19 infection [28, 29, 30]. Recently, Zhang et al. [31] demonstrated the expression of ACE2 and TMPRSS2 (transmembrane serine protease 2 protein), which facilitates entry of viruses into host cells by proteolytically cleaving and activating viral envelope glycoproteins. SARS‐CoV‐2 can bind CD147 and CD26 in an ACE2‐independent manner to interact with platelets [32, 33]. Further studies are, however, needed to determine whether aspirin can disturb interactions of SARS‐CoV‐2 with targets on platelets, thereby delaying viral infection and propagation.
The present study, to our knowledge, is the first to examine the relationship between low‐dose aspirin treatment and the likelihood of COVID‐19 infection among a large cohort of outpatients listed in a nation‐wide database. A major limitation in our observational study is the lack of control over treatments given to the study population. Unlike random assignment in clinical trials, where groups differ only in terms of treatment intervention, the treatment groups in observational studies, such as the present study, are likely to differ with respect to treatment intervention and other variables that can independently affect outcome. To avoid bias due to comparison of a healthy, more socially active population with patients suffering from severe cardiovascular comorbidity, we excluded all subjects taking aspirin for secondary prevention. We also applied multivariate logistic regression analysis adjusted for sex, age, smoking status, socioeconomic status (SES), chronic medication administration, laboratory data, and comorbidities and used the variance inflation factor (VIF) [34] to check for multicollinearity among independent variables. Another serious limitation is related to the observed shortening of PCR positivity among aspirin users, given how PCR testing was not performed on a daily basis in the community setting. However, two consecutive negative SARS‐CoV‐2 RT–PCR test results were recommended as a criterion for discharge and termination of social isolation by health authorities in different countries and were used for making a clinical decision regarding COVID‐19 management [35]. The shortening of the time needed for conversion of SARS‐CoV‐2 PCR test results from positive to negative observed here could reflect a resolution of symptoms [36] and have significant impact on COVID‐19 patients. Another limitation is related to the different COVID‐19 testing methods used in our cohort, given how the true sensitivity of testing remains unknown. Such limitation is typical of most similar epidemiological studies of COVID‐19 infection.
Fig. 3.
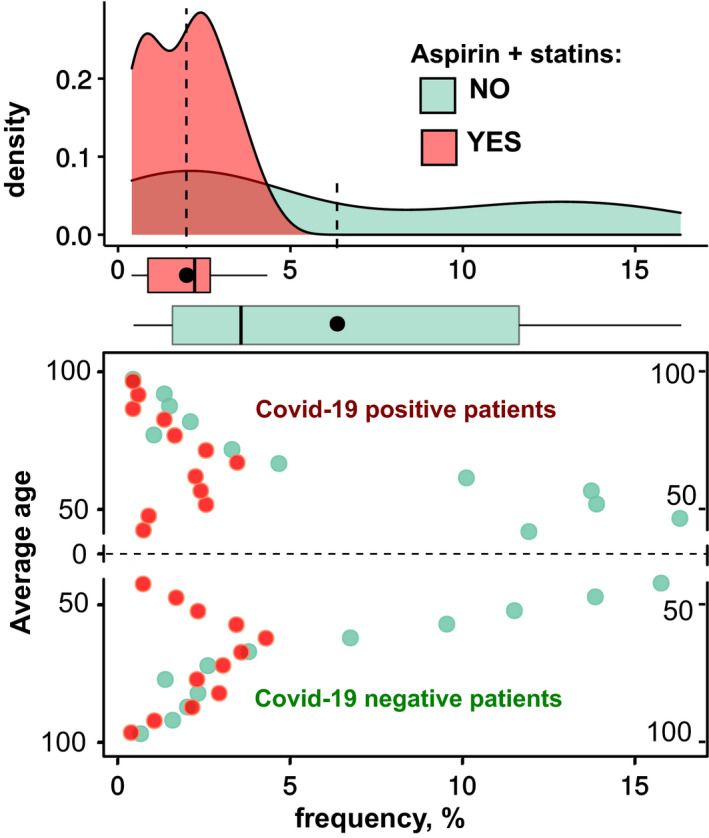
The frequency distributions (12 age‐groups ranging from 40 to 95+ years, in 5‐year steps) of COVID‐19‐positive patients (top area) and corresponding negative patients (bottom area), who took aspirin and statins (highlighted in red) and who did not take these drugs (highlighted in light green).
Currently, aspirin is proposed for use as an antithrombotic drug for treating COVID‐19 in patients with established hypercoagulability [35]. The RECOVERY II (Randomized Evaluation of COVID‐19 Therapy II) trial, a randomized clinical trial, is now being planned to test the effectiveness of low‐dose aspirin as an anti‐inflammatory and antithrombotic treatment in COVID‐19 patients [37, 38].
In conclusion, we observed an inverse association between the likelihood of COVID‐19 infection and aspirin use for primary prevention. Our data on the possible use of low doses of aspirin for the prevention of COVID‐19 infection are preliminary, yet intriguing. We thus need prompt clinical consideration of this safe, low‐cost drug with the potential to favorably alter COVID‐19 infection outcome. Such an effect would also provide immediate socioeconomical relief by reduction in COVID‐19 susceptibility. Therefore, our observations justify efforts to repeat this study using larger samples, including patients from other institutions.
Methods
Definitions
Medication use was deemed if the patient had been prescribed and had purchased at least three prescriptions of certain groups of medications, defined according to their Anatomical Therapeutic Chemical (ATC) codes, namely aspirin (acetylsalicylic acid; ATC code B01AC06), angiotensin‐converting enzyme inhibitors (ACEI; ATC codes C09A and C09B), angiotensin II receptor blockers (ARB; ATC codes C09C and C09D), and lipid‐lowering medications (statins; ATC code C10A), during the past twelve months. The period of disease duration was defined as the number of days between conversion of the first positive SARS‐CoV‐2 RT–PCR test result to the first two consecutive negative SARS‐CoV‐2 RT–PCR results (sampled at least 24 h apart), according to Israel Ministry of Health guidelines [13, 39].
SES data were organized according to the Israel Central Bureau of Statistics classification system that includes 20 subgroups, delineated according to home address. Classifications 1–9 are considered low–medium SES, while classifications 10–20 are considered upper medium–high SES. Smoking status was defined based upon last electronic medical record (EMR) documentation made by the family physician. Rates of missing data were generally similar for COVID‐19‐positive and COVID‐19‐negative subjects. Levels of missing BMI and laboratory data were less than 10%. The greatest amount of missing data addressed smoking status (23% of adherent and 25% of nonadherent subjects).
Statistical analysis
Differences in demographic and clinical characteristics between subjects with negative and positive COVID‐19 RT–PCR test results were analyzed using Student's t‐test and Fisher's exact χ2 test for continuous and categorical variables, respectively, based on normal distribution and variable characteristics. Categorical data are presented as counts and percentages. Data on continuous variables with normal distribution are presented as means and standard deviation (SD). We applied multiple imputations for missing data under the assumption that data were missing at random, conditional on the observed data. Any problem of multicollinearity among variables in the models was tested by calculating the VIF [34]. Multiple regression analyses adjusted for sex, age, smoking status, comorbidity, and chronic medication use served to estimate the odds ratios (OR) and 95% confidence interval (CI) for the independent association between aspirin treatment and COVID‐19 RT–PCR test results. All statistical analyses were conducted using stata 12 statistical package software (StataCorp, College Station, TX, USA). r functions (Shapiro–Wilk and Welch's two‐sample t‐test) were used for statistical assessment of disease duration figures. Visualizations relied on open‐source programs (Perl, R, GIMP, Inkscape). The ggplot2 R package (Hadley Wickham, Garrett Grolemund R for Data Science; O'Reilly Media, 2017) was used to produce figures, including scatter plots, area charts, bar charts, and 3D charts. Plots were prepared using in‐house scripts written by the Frenkel–Morgenstern group in R and Perl.
Formulae for calculating the density of aspirin and statins
is a set of the patients;
is a set of the aspirin levels;
is a set of the statin levels;
is a set of the aspirin and statin levels;
is a set of age levels (bins);
is a set of positive and negative COVID‐19 patients;
is a set of drugs used;
Conflict of interest
The authors declare no conflict of interest.
Author contributions
EM and ElM have designed the study, EM, IG, SV, AGC, MF‐M, and ElM have analyzed the data, AG and MF‐M have produced figures, EM has supervised the study, and all authors have written the manuscript.
Peer Review
The peer review history for this article is available at https://publons.com/publon/10.1111/febs.15784.
Acknowledgements
We thank the Leumit Health System for providing all the ethical applications for this study. MF‐M is supported by the Israel Innovation Authority (Kamin Grant #66824, 2019–2020) and COVID‐19 Data Science Institute (DSI) Grant, Bar‐Ilan University (#247017, 2020).
Eugene Merzon and Ilan Green contributed equally to this article
References
- 1. Grasselli G, Zangrillo A, Zanella A, Antonelli M, Cabrini L, Castelli A, Cereda D, Coluccello A, Foti G, Fumagalli R et al. (2020) Baseline characteristics and outcomes of 1591 patients infected with SARS‐CoV‐2 admitted to ICUs of the Lombardy Region, Italy. JAMA 323, 1574–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu YI, Zhang LI, Fan G, Xu J, Gu X et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desborough MJR & Keeling DM (2017) The aspirin story – from willow to wonder drug. Br J Haematol 177, 674–683. [DOI] [PubMed] [Google Scholar]
- 4. Gaziano JM, Brotons C, Coppolecchia R, Cricelli C, Darius H, Gorelick PB, Howard G, Pearson TA, Rothwell PM, Ruilope LM et al. (2018) Use of aspirin to reduce risk of initial vascular events in patients at moderate risk of cardiovascular disease (ARRIVE): a randomised, double‐blind, placebo‐controlled trial. Lancet 392, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McNeil JJ, Nelson MR, Woods RL, Lockery JE, Wolfe R, Reid CM, Kirpach B, Shah RC, Ives DG, Storey E et al. (2018) Effect of aspirin on all‐cause mortality in the healthy elderly. N Engl J Med 379, 1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, Himmelfarb CD, Khera A, Lloyd‐Jones D, McEvoy JW et al. (2019) 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines. J Am Coll Cardiol 74, e177–e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Patrono C & Baigent C (2019) Role of aspirin in primary prevention of cardiovascular disease. Nat Rev Cardiol 16, 675–686. [DOI] [PubMed] [Google Scholar]
- 8. Hussain M, Javeed A, Ashraf M, Zhao Y, Mukhtar MM & Rehman MU (2012) Aspirin and immune system. Int Immunopharmacol 12, 10–20. [DOI] [PubMed] [Google Scholar]
- 9. Pringle M, Ward P & Chilvers C (1995) Assessment of the completeness and accuracy of computer medical records in four practices committed to recording data on computer. Br J Gen Pract 45, 537–541. [PMC free article] [PubMed] [Google Scholar]
- 10. Rennert G & Peterburg Y (2001) Prevalence of selected chronic diseases in Israel. Isr Med Assoc J 3, 404–408. [PubMed] [Google Scholar]
- 11. Rennert G & Peterburg Y (2001) Prevalence of selected chronic diseases in Israel. Isr Med Assoc J 3, 404–408. [PubMed] [Google Scholar]
- 12. Chodick G, Heymann AD, Shalev V & Kookia E (2003) The epidemiology of diabetes in a large Israeli HMO. Eur J Epidemiol 18, 1143–1146. [DOI] [PubMed] [Google Scholar]
- 13. https://www.health.gov.il/Subjects/disease/corona/Documents/bz‐259818720.pdf
- 14. World Health Organization (WHO) (2020) Laboratory testing for 2019 novel coronavirus (2019‐nCoV) in suspected human cases. Interim guidance. https://apps.who.int/iris/bitstream/handle/10665/331501/WHO‐COVID‐19‐laboratory‐2020.5‐eng.pdf?sequence¼1&isAllowed¼y
- 15. Glatthaar‐Saalmüller B, Mair KH & Saalmüller A (2017) Antiviral activity of aspirin against RNA viruses of the respiratory tract‐an in vitro study. Influenza Other Respir Viruses 11, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lazear HM, Schoggins JW & Diamond MS (2019) Shared and distinct functions of type I and type III interferons. Immunity 50, 907–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Wit E, van Doremalen N, Falzarano D & Munster VJ (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C et al. (2014) Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 40, 554–568. [DOI] [PubMed] [Google Scholar]
- 19. Boutaud O, Sosa IR, Amin T, Oram D, Adler D, Hwang HS, Crews BC, Milne G, Harris BK, Hoeksema M et al. (2016) Inhibition of the biosynthesis of prostaglandin E2 by low‐dose aspirin: implications for adenocarcinoma metastasis. Cancer Prev Res (Phila) 9, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coulombe F, Jaworska J, Verway M, Tzelepis F, Massoud A, Gillard J, Wong G, Kobinger G, Xing Z, Couture C et al. (2014) Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I interferon and apoptosis in macrophages. Immunity 40, 554–568. [DOI] [PubMed] [Google Scholar]
- 21. Wu F, Zhao SU, Yu B, Chen Y‐M, Wang W, Song Z‐G, Hu YI, Tao Z‐W, Tian J‐H, Pei Y‐Y et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subir R, Jagat JM & Kalyan KG (2020) Pros and cons for use of statins in people with coronavirus disease‐19 (COVID‐19). Diabetes Metab Syndr 14, 1225–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakhaei P, Hiscott J & Lin R (2010) STING‐ing the antiviral pathway. J Mol Cell Biol 2, 110–112. [DOI] [PubMed] [Google Scholar]
- 24. Berthelot JM & Lioté F (2020) COVID‐19 as a STING disorder with delayed over‐secretion of interferon‐beta. EBioMedicine 56, 102801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berthelot JM, Drouet L & Lioté F (2020) Kawasaki‐like diseases and thrombotic coagulopathy in COVID‐19: delayed over‐activation of the STING pathway? Emerg Microbes Infect 9, 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sriram K & Insel PA (2020) A hypothesis for pathobiology and treatment of COVID‐19: the centrality of ACE1/ACE2 imbalance. Br J Pharmacol 177, 4825–4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chow JH, Khanna AK, Kethireddy S, Yamane D, Levine A, Jackson AM, McCurdy MT, Tabatabai A, Kumar G, Park P et al. (2020) Aspirin use is associated with decreased mechanical ventilation, ICU admission, and in‐hospital mortality in hospitalized patients with COVID‐19. Anesth Analg. 10.1213/ANE.0000000000005292. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28. Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, Petrey AC, Tolley ND, Guo LI, Cody M et al. (2020) Platelet gene expression and function in patients with COVID‐19. Blood 136, 1317–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zaid Y, Puhm F, Allaeys I, Naya A, Oudghiri M, Khalki L, Limami Y, Zaid N, Sadki K, Ben El Haj R et al. (2020) Platelets can associate with SARS‐Cov‐2 RNA and are hyperactivated in COVID‐19. Circ Res 127, 1404–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shen BO, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L et al. (2020) Proteomic and metabolomic characterization of COVID‐19 patient sera. Cell 182, 59–72, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang SI, Liu Y, Wang X, Yang LI, Li H, Wang Y, Liu M, Zhao X, Xie Y, Yang Y et al. (2020) SARS‐CoV‐2 binds platelet ACE2 to enhance thrombosis in COVID‐19. J Hematol Oncol 13, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seyedpour S, Khodaei B, Loghman AH, Seyedpour N, Kisomi MF, Balibegloo M, Nezamabadi SS, Gholami B, Saghazadeh A & Rezaei N (2021) Targeted therapy strategies against SARS‐CoV‐2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J Cell Physiol 236, 2364–2392. [DOI] [PubMed] [Google Scholar]
- 33. Vankadari N & Wilce JA (2020) Emerging WuHan (COVID‐19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect 9, 601–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim JH (2019) Multicollinearity and misleading statistical results. Korean J Anesthesiol 72, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guidance for discharge and ending of isolation of people with COVID‐19. https://www.ecdc.europa.eu/sites/default/files/documents/Guidance‐for‐discharge‐and‐ending‐of‐isolation‐of‐people‐with‐COVID‐19.pdf
- 36. Chang D, Mo G, Yuan X, Tao Y, Peng X, Wang FS, Xie L, Sharma L, Dela Cruz CS & Qin E (2020) Time kinetics of viral clearance and resolution of symptoms in novel coronavirus infection. Am J Respir Crit Care Med 201, 1150–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Violi F, Pastori D, Cangemi R, Pignatelli P & Loffredo L (2020) Hypercoagulation and antithrombotic treatment in coronavirus 2019: a new challenge. Thromb Haemost 120, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. https://clinicaltrials.gov/ct2/show/results/NCT04381936
- 39. https://govextra.gov.il/media/17976/coronavirus_med_guidelines.pdf
- 40. Wickham H & Grolemund G (2016) R for Data Science: Import, Tidy, Transform, Visualize, and Model Data, O'Reilly Media.


