Abstract
Background
The ongoing coronavirus disease 2019 (Covid‐19) pandemic has been rapidly spreading throughout the world with confirmed case numbers already exceeding 75 million. Although nasopharyngeal swabs are the most commonly utilized samples for based severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) RNA detection, collecting these specimens requires healthcare workers and necessitates the use of personal protective equipment as it presents a nosocomial transmission risk. We aimed to assess the diagnostic value of saliva samples in mildly symptomatic and asymptomatic patients with confirmed Covid‐19.
Methods
We performed a cohort study to validate the use of saliva for SARS‐CoV‐2 detection in mildly symptomatic and asymptomatic patients with a confirmed diagnosis of Covid‐19. Saliva samples of the patients were analyzed by reverse‐transcriptase polymerase chain reaction (RT‐PCR).
Results
In May 2020, 28 asymptomatic and 25 mildly symptomatic patients were enrolled in the study. The median age was 37 years (range 4–70). None of the patients had a fever on presentation. Among 53 patients with SARS‐CoV‐2 detected in the nasopharyngeal sample, the real‐time RT‐PCR was positive in the saliva specimens in 48 (90.56%) patients. The mean cycle threshold (CT) values for nasopharyngeal and saliva specimens (27.80 ± 3.44 and 30.64 ± 2.83, respectively) were significantly correlated between the two sample types (p = .016). The mean CT values of nasopharyngeal and saliva samples in mildly symptomatic and asymptomatic patients (27.18 ± 3.53 and 30.24 ± 3.29 vs. 28.36 ± 3.31 and 30.98 ± 2.39, respectively) were not significantly different (p = .236 and p = .733, respectively).
Conclusions
Saliva specimens can be considered as a reliable and less resource‐intensive alternative to nasopharyngeal specimens for screening asymptomatic SARS‐CoV‐2 infections.
Keywords: Covid‐19, diagnosis, RT‐PCR, saliva, SARS‐CoV‐2
1. INTRODUCTION
A cluster of patients with pneumonia of unknown cause was reported in Wuhan, China in December 2019. Shortly after, a new coronavirus, currently known as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), was identified as the etiologic agent. 1 , 2 The infection has spread rapidly worldwide due to the high transmissibility of SARS‐CoV‐2. 3 On March 11, 2020, the WHO declared coronavirus disease 2019 (Covid‐19) a global pandemic. As of the end of June 2020, more than 10 million confirmed cases of Covid‐19 have been reported worldwide with over 500,000 deaths. 4
High attack rate and transmission from even asymptomatic carriers to others require timely diagnosis of suspected cases and strict infection control measures and public health interventions to contain the outbreak. 3 , 5
Real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) based SARS‐CoV‐2 RNA detection in respiratory samples is the gold standard for Covid‐19 diagnosis. 6 For initial diagnostic testing of SARS‐CoV‐2, Centers for Disease Control recommends collecting and testing an upper respiratory specimen. A nasopharyngeal swab and/or an oropharyngeal swab are often recommended for screening or diagnosis of early infection but nasal mid‐turbinate or anterior nares samples are also acceptable. 7
On the other hand, it has been demonstrated that saliva has a high concordance rate with nasopharyngeal specimens for RT‐PCR detection of respiratory pathogens, including two seasonal human coronaviruses. 8 , 9
To date, there are a few studies regarding the possible role of saliva in the detection of SARS‐CoV‐2 in hospitalized severe Covid‐19 patients. The purpose of this study was to assess saliva samples' diagnostic value in mildly symptomatic and asymptomatic patients with confirmed Covid‐19 disease.
2. METHODS
2.1. Patient recruitment/ethical approval
Consecutive patients admitted to Kafkas University Faculty of Medicine Research hospital, Kars, Turkey, in May 2020 who tested positive by RT‐PCR for SARS‐CoV‐2 in nasopharyngeal specimens were enrolled in the study. All included patients were tested on the basis of clinical indications and/or contact tracing.
Saliva samples were collected within 12 h after the diagnosis of Covid‐19 provided by RT‐PCR on nasopharyngeal swabs by simply asking the patient to spit 3–4 ml of saliva into a sterile container. Patients were instructed and supervised by registered nurses. All saliva samples were transported to the laboratory in 30 min upon collection and stored at +4°C until RNA extraction. RNA extracts were stored at −20°C until RT‐PCR was performed.
Age, sex, presenting symptoms, and comorbidities were recorded for each patient. Laboratory tests including complete blood count, coagulation parameters, biochemical analysis were obtained on admission, and chest computed tomography was performed if clinically indicated.
This study was approved by both the Republic of Turkey Ministry of Health COVID‐19 Scientific Research Evaluation Commission (Approval date: 02/05/2020; number: 2020‐05‐02T16‐07‐46) and the Local Ethics Committee of Kafkas University Faculty of Medicine (Approval date: 06/05/2020 number: 80576354‐050‐99/131). The informed consent form had been obtained from each patient before the saliva specimens were collected.
2.2. Nucleic acid extraction and real time RT‐PCR
Saliva specimens were resuspended in 2 ml of PBS, 150 µl was subjected to RNA extraction by a Patho Gene‐spin Viral DNA/RNA Extraction Kit (Intron Biotechnology Inc.). RNA extraction was performed according to the manufacturer's instructions. The SARS‐CoV‐2 virus in nasopharyngeal swab and saliva specimens was detected by the real‐time RT‐PCR technique via using a Roche Lightcycler‐96 device (Roche Diagnostic Systems). RT‐PCR was performed using a SARS‐CoV‐2 (2019‐nCoV) qPCR Detection Kit (Bioeksen R&D) that is targeting a SARS‐CoV‐2‐specific RNA dependent RNA polymerase (RdRp) gene fragment. The final PCR concentration was 20‐µl (10‐µl qPCR master mix, 5‐µl primer/probe set, and 5‐µl template). The nucleic acid amplification was performed with the following PCR steps: Reverse transcription stage (45°C, 15 min, and 1 cycle), initial activation stage (95°C, 3 min, and 1 cycle), and amplification stage (Denaturation: 95°C, 5 s, annealing, and extension: 55°C, 35 s, and 45 cycles). All samples were run in two replicates, together with a SARS‐Cov‐2 positive control, and negative control. For data analysis, the method was used and a cycle threshold (CT) value less than 40 is interpreted as positive for SARS‐CoV‐2 RNA.
2.3. Statistical methods
Statistical analysis was performed with the Statistical Package for the Social Sciences version 22.0 (SPSS; IBM). The “number,” “percentage,” “mean,” and “SD” were used for the descriptive statistics of the continuous variables. The Pearson correlation analysis was used to evaluate two numerical data (CT values of two different samples). The independent samples t‐test or Mann–Whitney U test were used to compare two independent groups. The Pearson χ 2 or Fisher's exact tests were used to analyze categorical data. The results were evaluated with a confidence interval of 95%, and the level of significance, p, was set at 0.05.
3. RESULTS
The cohort consisted of 53 patients with confirmed Covid‐19 infection, of whom 21 (39.6%) were males. The median age of patients was 37 years, with a range from 4 to 70 years.
None of the patients had a fever on presentation. The most common symptom was dry cough in 15 (28.3%) patients, followed by fatigue in 10 (18.9%) and sore throat in eight (15.1%) patients, and two (3.8%) patients presented with anosmia. Twenty‐eight patients (52.8%) were considered to have an asymptomatic course and the remaining 25 (47.2%) patients were mildly symptomatic.
Seven (13.2%) patients had chronic medical illnesses and the most common underlying disease was hypertension in six (11.3%) patients and one (1.9%) patient had diabetes.
Nine (%17) patients had patchy ground‐glass opacities with mainly subpleural distribution on chest tomography. Two of these patients had hypertension and the remaining seven had no underlying conditions.
No significant hematological or biochemical abnormal results were observed in the study group. Demographic and laboratory findings of asymptomatic and mildly symptomatic patients are shown in Table 1. There were no significant differences between the two groups, except asymptomatic patients were younger than those with mild symptoms (p = .026).
Table 1.
Demographic and laboratory findings of mildly symptomatic and asymptomatic patients
| Mildly symptomatic (n = 25) | Asymptomatic (n = 28) | ||
|---|---|---|---|
| Mean ± SD | Mean ± SD | p | |
| Age | 44.08 ± 16.66 | 31.85 ± 18.9 | 0.026 |
| Gender, male, n (%) | 10 (40%) | 11 (39.3%) | 0.596 |
| Laboratory findings | |||
| Leukocyte count (×109/L, normal range 3.7–10.4) | 4.79 ± 1.26 | 5.61 ± 1.52 | 0.064 |
| Lymphocyte count (×109/L, normal range 0.9–3.7) | 1.36 ± 0.58 | 1.74 ± 0.53 | 0.183 |
| Hemoglobin (g/dl, normal range 10.8–15.1) | 14.1 ± 1.9 | 14.4 ± 2.1 | 0.288 |
| Platelet count (×109/L, normal range 149–371) | 222 ± 55 | 235 ± 66 | 0.362 |
| Alanine aminotransferase (U/L, normal range 0–40) | 22.5 ± 14.3 | 31.5 ± 29.6 | 0.176 |
| Aspartate aminotransferase (U/L, normal range 0–41) | 24.8 ± 9.5 | 29.1 ± 12.6 | 0.052 |
| Urea (mg/dL, normal range 10–50) | 30.5 ± 14.0 | 31.4 ± 9.1 | 0.652 |
| Creatinine (mg/dl, normal range 0.7–1.2) | 0.89 ± 0.28 | 0.81 ± 0.36 | 0.189 |
| Lactate dehydrogenase (U/L, normal range 135–225) | 214 ± 53 | 200 ± 41 | 0.534 |
| Cycle threshold | |||
| Nasopharyngeal swab | 27.18 ± 3.53 | 28.36 ± 3.31 | 0.236 |
| Saliva | 30.24 ± 3.29 | 30.98 ± 2.39 | 0.733 |
Only 10 patients (18.9%) required supplemental nasal oxygen for brief periods at the beginning of their hospitalization, none of the patients required intubation or admitted to the intensive care unit. None of the patients died in the study period.
Among 53 patients with SARS‐CoV‐2 detected in the nasopharyngeal sample, the real‐time RT‐PCR for the same virus was positive in the saliva specimens in 48 (90.56%) patients. The mean CT values for nasopharyngeal and saliva specimens were 27.80 ± 3.44 and 30.64 ± 2.83, respectively. The mean CT values were significantly correlated between the two sample types (p = .016) (Figure 1).
Figure 1.
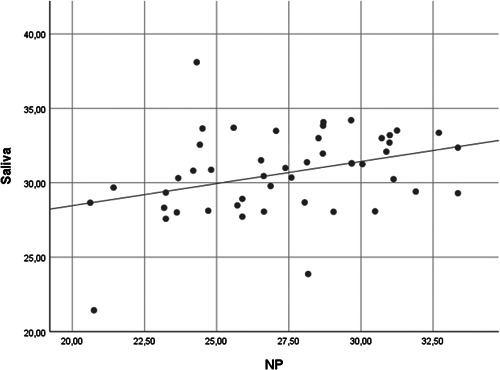
Scatter graph for correlation between the mean cycle threshold values for NP and saliva specimens. NP, nasopharyngeal
The mean CT values of nasopharyngeal and saliva samples in mildly symptomatic and asymptomatic patient groups were 27.18 ± 3.53 and 30.24 ± 3.29 versus 28.36 ± 3.31 and 30.98 ± 2.39, respectively. There were no statistically significant differences between the two groups (p = .236 and p = .733, respectively).
We also evaluated the mean CT values for patients with or without evidence of pneumonia on tomography. The mean CT values for nasopharyngeal and saliva specimens in patients with pneumonia were lower than those without pneumonia (26.23 ± 3.56 and 29.15 ± 3.90 vs. 28.12 ± 3.36 and 30.94 ± 2.52, respectively) although the difference was not significant (p = .077 and p = .154, respectively).
4. DISCUSSION
Early and accurate diagnosis of patients with SARS‐CoV‐2 infection is the cornerstone to containment of the ongoing Covid‐19 pandemic. Without the availability of effective vaccines or treatments, the only way to reduce SARS‐CoV‐2 transmission is to identify and isolate persons who are contagious. 10
The current gold standard for Covid‐19 diagnosis is real‐time RT‐PCR based SARS‐CoV‐2 detection in respiratory samples. Although other upper and lower respiratory tract samples such as oropharyngeal, nasal swabs, or sputum are acceptable, the most utilized specimen type for diagnosis is nasopharyngeal samples. 7
However, the nasopharyngeal sample's sensitivity is variable, partly due to deficiency in sampling technique and requiring experienced healthcare personnel. Furthermore, the collection of nasopharyngeal specimens causes discomfort to patients due to the procedure's invasiveness. 11 Moreover, nasopharyngeal sampling requires close contact between the patient and the healthcare worker and presents a considerable transmission risk to healthcare workers because it can induce patients to sneeze or cough, generating aerosols. 12 , 13 Therefore, this procedure requires using personal protective equipment including an N95 respirator, eye protection, gloves, and a gown when collecting specimens. 7
As the pandemic is evolving, global shortages of key equipment needed to care for patients such as personal protective equipment and adequate swabs or viral transport medium for sampling procedures becomes a growing problem, especially in resource‐limited settings. 14 These challenges are expected to be further exacerbated as the Covid‐19 pandemic intensifies in low‐income countries. 13 These limitations related to the nasopharyngeal sampling necessitates the validation of new diagnostic approaches.
Saliva sampling is an appealing alternative to nasopharyngeal swabs, as collecting saliva is noninvasive, easy to self‐administer, and diminishes the need for personal protective equipment use of healthcare workers. 13
Previous reports indicated that SARS‐CoV‐2 can be detected from saliva samples of Covid‐19 patients. To et al. demonstrated that SARS‐CoV‐2 could be detected in the saliva specimens of 11 of the 12 patients studied. 15 Patient characteristics were not identified in this study. Azzi et al. reported 100% concordance between saliva and nasopharyngeal samples for RT‐PCR detection of SARS‐CoV‐2 in severe or very severe Covid‐19 patients. 11 Similarly, Wyllie et al. compared SARS‐CoV‐2 detection from patient‐matched nasopharyngeal and saliva samples in patients with severe Covid‐19 infection and suggested that saliva may be an appropriate alternative to nasopharyngeal swabs for SARS‐CoV‐2 testing. 13 A recent meta‐analysis reported that the sensitivity of RT‐PCR in saliva samples among Covid‐19 infected patients was 91%, ranging from 78% to 100%. 16
The aforementioned studies were largely carried out in hospitalized patients with severe Covid‐19 infection to assess the utility of saliva samples for RT‐PCR diagnosis. Here, we demonstrated the feasibility and acceptability of using saliva as a diagnostic specimen for the detection of SARS‐CoV‐2 in mildly symptomatic and asymptomatic patients.
We have found that RT‐PCR positivity was 90.56% in saliva samples of mildly symptomatic and asymptomatic confirmed Covid‐19 patients. The mean CT values were correlated in both sample types (p = .016). The mean CT values in nasopharyngeal and saliva samples of both asymptomatic and mildly symptomatic patients were not significantly different.
We also have shown that saliva samples were comparably positive with nasopharyngeal samples in both patient groups with or without radiologic evidence of lung involvement, although the mean CT values in patients with lung involvement were lower. As CT values can be considered as a semiquantitative indicator of viral load, the higher CT values in both saliva and nasopharyngeal samples of patients without radiologic evidence of lung involvement suggests lower viral loads in asymptomatic patients. This finding is in line with previous reports suggesting higher viral loads in severe cases. 17
The findings of our study indicate that consistent RT‐PCR positivity in saliva samples of asymptomatic Covid‐19 patients should draw attention to the transmission potential of this patient group. To et al. confirmed the presence of live SARS‐CoV‐2 in saliva by positive cultures. Consequently, SARS‐CoV‐2 may be transmitted through saliva directly or indirectly even from patients without having any respiratory symptoms. 15 Indeed, it has been assumed that the rapid spread of the coronavirus pandemic results from high transmissibility of SARS‐CoV‐2 from undocumented infections. 18 Therefore recognition of asymptomatic or mildly symptomatic patients is crucial in regard to optimizing infection control and isolation practices and implementing effective public health measures.
Nasopharyngeal sampling is not an ideal screening procedure to be adopted for massive screening practices. Given the high sensitivity and concordance with nasopharyngeal samples, saliva specimens may be considered as a reliable and suitable alternative to nasopharyngeal specimens, especially in settings where large numbers of tests should be carried out.
Saliva samples can easily be provided by patients themselves without necessitating healthcare workers and is a resource sparing sample collection method reducing the need for both personal protective equipment and consumables such as swabs and viral transport media. Accordingly, the use of saliva specimens will reduce the risk of health‐care‐associated SARS‐CoV‐2 transmission.
We have several limitations in our study. First, we could not measure the viral load in our patients due to the absence of a reliable positive control in our laboratory. Additionally, although we have demonstrated RT‐PCR positivity in saliva samples, we have not conducted viral cultures. Also, we included patients with a confirmed diagnosis of Covid‐19 by means of nasopharyngeal sampling. Thus, we cannot provide data regarding the specificity of saliva specimens for the Covid‐19 diagnosis. Further studies should include patients prospectively to assess the sensitivity and specificity of using saliva specimens in larger cohorts.
Previous reports suggested the use of saliva as an alternative specimen type for diagnosis in severe or very severe cases of Covid‐19. The population analyzed in our study was homogenous, and mostly comprised of asymptomatic individuals identified through contact tracing. We have shown that the sensitivity of SARS‐CoV‐2 detection from saliva is comparable to nasopharyngeal swabs in mildly symptomatic and asymptomatic patients.
In conclusion, while there are efforts for validation of new diagnostic approaches such as rapid antigen tests, our results indicate that saliva may be considered as an appropriate and reliable alternative to nasopharyngeal samples for screening asymptomatic SARS‐CoV‐2 infections.
AUTHOR CONTRIBUTIONS
Study design: Emin Ediz Tutuncu. Literature search: Emin Ediz Tutuncu, Didem Ozgur, Murat Karamese. Sample collection: Emin Ediz Tutuncu. Experimental studies: Didem Ozgur, Murat Karamese. Data analysis and interpretation: Emin Ediz Tutuncu, Didem Ozgur, Murat Karamese. Statistical analysis: Murat Karamese. Manuscript preparation and editing: Emin Ediz Tutuncu, Didem Ozgur. Manuscript review: Emin Ediz Tutuncu, Didem Ozgur, Murat Karamese
ACKNOWLEDGMENT
This study was supported by the donation of a qPCR Detection Kit from Bioeksen (Bioeksen R&D, Istanbul, Turkey).
Tutuncu EE, Ozgur D, Karamese M. Saliva samples for detection of SARS‐CoV‐2 in mildly symptomatic and asymptomatic patients. J Med Virol. 2021;93:2932–2937. 10.1002/jmv.26821
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Tan W, Zhao X, Ma X, et al. A novel coronavirus genome identified in a cluster of pneumonia cases—Wuhan. China 2019. China CDC Weekly. 2020;2(4):61‐62. [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization . Coronavirus disease (COVID‐19) situation report. https://www.who.int/docs/default-source/coronaviruse/20200630-covid-19-sitrep-162.pdf?sfvrsn=e00a5466_2 (accessed December 20, 2020).
- 5. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Pevention . Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons for Coronavirus Disease 2019 (COVID‐19). www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html (accessed June 29, 2020).
- 8. Kim Y, Yun SG, Kim MY, et al. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription‐PCR. J Clin Microbiol. 2016;55(1):226‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. To KK, Lu L, Yip CC, et al. Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect. 2017;6(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome–related coronavirus‐2: a narrative review. Ann Intern Med. 2020;172(11):726‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Azzi L, Carcano G, Gianfagna F, et al. Saliva is a reliable tool to detect SARS‐CoV‐2. J Infect. 2020;81(1):e45‐e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. To KKW, Tsang OTY, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wyllie AL, Fournier J, Casanovas‐Massana A, et al. Saliva is more sensitive for SARS‐CoV‐2 detection in COVID‐19 patients than nasopharyngeal swabs. medRxiv. 2020. 10.1101/2020.04.16.20067835 [DOI] [Google Scholar]
- 14. Ranney ML, Griffeth V, Jha AK. Critical supply shortages—the need for ventilators and personal protective equipment during the Covid‐19 pandemic. N Engl J Med. 2020;382(18):e41. [DOI] [PubMed] [Google Scholar]
- 15. KKW To, OTY Tsang, CCY Yip, Chan KH, Wu TC, JMC Chan. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Czumbel LM, Kiss S, Farkas N, et al. Saliva as a candidate for COVID‐19 diagnostic testing: a meta‐analysis. medRxiv. 2020. 10.1101/2020.05.26.20112565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect Dis. 2020;20(6):656‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS‐CoV‐2). Science. 2020;368(6490):489‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


