Abstract
Motile cilia are molecular machines used by a myriad of eukaryotic cells to swim through fluid environments. However, available molecular structures represent only a handful of cell types, limiting our understanding of how cilia are modified to support motility in diverse media. Here, we use cryo‐focused ion beam milling‐enabled cryo‐electron tomography to image sperm flagella from three mammalian species. We resolve in‐cell structures of centrioles, axonemal doublets, central pair apparatus, and endpiece singlets, revealing novel protofilament‐bridging microtubule inner proteins throughout the flagellum. We present native structures of the flagellar base, which is crucial for shaping the flagellar beat. We show that outer dense fibers are directly coupled to microtubule doublets in the principal piece but not in the midpiece. Thus, mammalian sperm flagella are ornamented across scales, from protofilament‐bracing structures reinforcing microtubules at the nano‐scale to accessory structures that impose micron‐scale asymmetries on the entire assembly. Our structures provide vital foundations for linking molecular structure to ciliary motility and evolution.
Keywords: centrioles, cryo‐electron tomography, cryo‐FIB milling, motile cilia, sperm
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton
In situ cryo‐electron tomography uncovers structural specialisations of sperm flagella in several mammalian species.

Introduction
Cilia, also called flagella, are evolutionarily ancient organelles used by diverse eukaryotic cell types and organisms to propel themselves through fluid environments or to move fluid across their surfaces (Mitchell, 2017; Wan, 2018). These intricate molecular machines are paragons of self‐organization built from an extensive array of active and passive structural elements that, together, are able to spontaneously generate oscillatory wave‐like motion (Gaffney et al, 2011). The basic architecture of motile cilia is conserved across broad swaths of the eukaryotic tree, providing information on the minimal structures needed for spontaneous undulation (Brokaw, 2009). However, because they operate in a wide range of environments, cilia from different cell types generate different waveforms (Khan & Scholey, 2018) that are modulated by fluid viscosity (Smith et al, 2009).
The motile cilium is a continuous assembly of compound microtubules (Ishikawa, 2017). The base of the cilium is the centriole or basal body, which is typically a cylinder of triplet microtubules. The centriole transitions into the axoneme, which consists of nine doublet microtubules arrayed around a central pair of singlet microtubules. Axonemal microtubules anchor hundreds of dynein motors and accessory proteins to power and regulate movement. Axoneme structure has been studied extensively by cryo‐electron tomography (cryo‐ET) in Chlamydomonas, Tetrahymena, and sea urchin sperm (Nicastro et al, 2006, 2011; Pigino et al, 2012; Owa et al, 2019). Recent studies have begun to shed light on species‐ and cell type‐specific specializations (Lin et al, 2014a; Yamaguchi et al, 2018; Imhof et al, 2019; Greenan et al, 2020), motivating efforts to expand the pantheon of organisms and cell types used in axoneme research.
Perhaps the most striking example of ciliary diversity across species is in sperm, which are highly specialized for a defined function—to find and fuse with the egg. Sperm consist of a head, which contains the genetic payload, and a tail, which is a modified motile cilium. Despite their streamlined structure, sperm are simultaneously the most diverse eukaryotic cell type (Gage, 2012; Lüpold & Pitnick, 2018), reflecting the sheer range of reproductive modes and fertilization arenas, from watery media for marine invertebrates and freshwater species to the viscous fluids of the female reproductive tract for mammals. Because motility is crucial to sperm function, the natural variation of sperm form presents a unique opportunity to understand the structural diversification of motile cilia.
Mammalian sperm flagella are characterized by accessory structures that surround and dwarf the axoneme (Fawcett, 1975), unlike marine invertebrates whose sperm tails consist essentially of the axoneme (Fawcett, 1970). In mammalian sperm, axonemal doublets are associated with filamentous cytoskeletal elements called outer dense fibers (ODFs) for most of their lengths. The ODFs are further surrounded by a sheath of mitochondria in the midpiece and by a reticular structure called the fibrous sheath in the principal piece. These accessory structures are proposed to stabilize beating of the long flagella of mammalian sperm (Lindemann, 1996; Lindemann & Lesich, 2016). The accessory structures also facilitate movement through the viscous fluids of the female reproductive tract by suppressing buckling instabilities that would otherwise cause sperm to swim in circles (Gadêlha & Gaffney, 2019). Indeed, many cases of male infertility are linked to defects in these accessory elements (Serres et al, 1986; Haidl et al, 1991; Zhao et al, 2018). However, we still do not fully understand how these accessory structures modulate the flagellar beat since there is very little structural information on how they interact with the axoneme proper.
Another distinguishing feature of mammalian sperm flagella is that they are not anchored by a basal body (Avidor‐Reiss, 2018). Instead, the base of the mammalian sperm flagellum is surrounded by a large cytoskeletal scaffold called the connecting piece. The isolated bovine sperm connecting piece was characterized by cryo‐ET, revealing a complex asymmetric assembly (Ounjai et al, 2012). However, the purification process resulted in loss of the centrioles. Thus, there is still a paucity of structural information on the flagellar base in intact cells and on how it varies across species that often have very different head shapes.
Sperm have two centrioles that are located in the neck, where the nucleus attaches to the flagellum. The centriole closer to the nucleus is referred to as the proximal centriole (PC) and the one at the base of the flagellum the distal centriole (DC). During spermiogenesis in mammals, the DC is remodeled to the point that it no longer resembles a canonical centriole. This was thought to represent a process of degeneration (Manandhar et al, 2000), but recent work showed that the DC is in fact a functional centriole that participates in orchestrating the first zygotic division (Fishman et al, 2018). Such drastic deviations from canonical centriole structure have not been investigated in detail.
Here, we combine cryo‐focused ion beam (cryo‐FIB) milling‐enabled cryo‐ET (Marko et al, 2007; Rigort et al, 2012) with subtomogram averaging to image mature sperm from three mammalian species—the pig (Sus scrofa), the horse (Equus caballus), and the mouse (Mus musculus)—that differ in terms of gross morphology, motility, and metabolism (Fig 1A–C). We leverage the uniquely multi‐scale capabilities of cryo‐ET to define the molecular architecture of microtubule‐based assemblies and their critical interactions with accessory structures. We take advantage of the highly streamlined shape of sperm in order to define how these structures and relationships change throughout the flagellum.
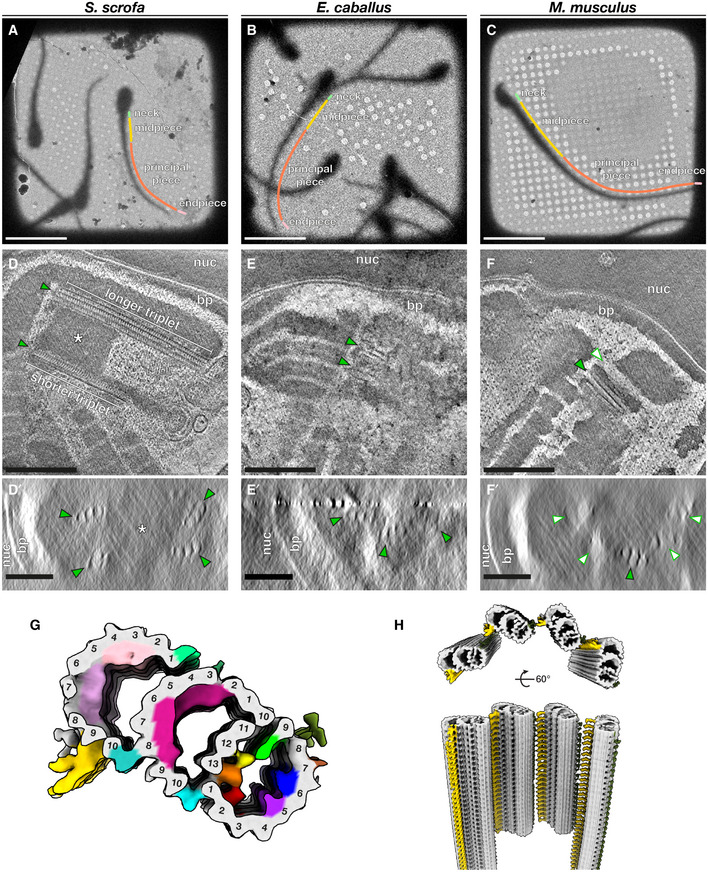
Figure 1. The proximal centriole (PC) in mammalian sperm is asymmetric and contains novel microtubule inner proteins.
-
A–CLow‐magnification cryo‐EM projection images of pig (A), horse (B), and mouse (C) sperm. Different regions of the flagellum discussed in this paper are annotated as follows: green—neck, yellow—midpiece, coral—principal piece, pink—endpiece.
-
D–FTomographic slices through cryo‐FIB‐milled lamellae of pig (D), horse (E), and mouse (F) sperm. Transverse slices (D’–F’) show complete triplets in the pig (D’) and the horse (E’), but not in the mouse (F’). Complete triplets are indicated by green arrowheads with black outlines, while degenerated triplets are indicated by white arrowheads with green outlines. Note the electron‐dense material in the lumen of the pig sperm proximal centriole that is continuous with the connecting piece (asterisks in D and D’).
-
GIn situ structure of triplet microtubules from the proximal region of the pig sperm PC with the tubulin backbone in gray and microtubule inner protein densities colored individually. A‐tubule MIPs are colored: MIP1—green, MIP2—yellow, MIP3—orange, MIP4—red, MIP5—purple, MIP6—blue. B‐tubule MIPs are colored: MIP7—magenta. C‐tubule MIPs are colored: MIP8—light pink, MIP9—pink. The A‐C linker is colored gold and the putative A‐link is colored olive green.
-
HReconstruction of the proximal region of the pig sperm PC generated by plotting the average back into the original particle positions and orientations in the tomogram. This plotback only contains four triplets as only part of the centriole was captured in the lamella. The A–C linker is colored in gold and the putative A‐link in olive green.
Data information: Labels: nuc—nucleus, bp—baseplate. Scale bars: (A–C) 20 µm; (D–F) 250 nm; (D’–F’) 100 nm.
We define the architecture of the flagellar base and show that the ODFs are anchored through an intricate structure that forms a large, asymmetric chamber around the centrioles. We show that the ODFs are directly coupled to axonemal microtubules in the principal piece, but not in the midpiece. We find that mammalian sperm microtubules are additionally decorated throughout by large, protofilament‐bridging microtubule inner protein densities. Thus, mammalian sperm flagella are modified across scales—from large accessory structures that increase the effective size and rigidity of the entire assembly to extensive microtubule inner proteins that likely reinforce the microtubules themselves. We further discuss the implications of these accessory structures to ciliary motility.
Results
The base of the flagellum is anchored through a large, asymmetric chamber around the centrioles
The neck region containing the PC and DC is too thick (~600–700 nm) for direct imaging by cryo‐ET, so in order to image sperm centrioles in their native subcellular milieu, we used cryo‐FIB milling to generate thin lamellae suitable for high‐resolution imaging (Fig 1). Cryo‐ET of lamellae containing the PC confirmed that it is indeed composed of triplet microtubules in pig and in horse sperm (Fig 1D–F). Unexpectedly, we found that triplets of the pig sperm PC are not all the same length (Figs 1, EV1). Shorter triplets are grouped on one side of the centriole, giving the PC a striking dorsoventral asymmetry (Fig EV1A). Consistent with previous reports that the PC degenerates in rodents (Woolley & Fawcett, 1973; Manandhar et al, 1998), the PC was not prominent in mouse sperm. However, cryo‐ET showed unequivocally that some centriolar microtubules remain (Fig 1F), demonstrating that degeneration is incomplete. We observed complete triplets as well as triplets in various stages of degeneration, including triplets in which only the B‐tubule had degraded (Fig 1F’).
Figure EV1. Structural features of the pig sperm proximal centriole (PC).

- PC triplets have unequal lengths (top panel), and shorter triplets are grouped on one side, giving the PC dorsoventral asymmetry (bottom panel).
- Many of the microtubule inner proteins (MIPs) in the pig sperm PC are not found in other mammalian centriole structures.
- Details of the MIP densities in the pig sperm PC.
We determined the in situ structure of the pig sperm PC by subtomogram averaging (Fig 1G). Because only parts of the PC were captured in cryo‐FIB‐milled lamellae, our average includes only particles from the proximal ~400 nm of the centriole. While the overall structure of the PC triplet is similar to other centriole structures (Li et al, 2012; Guichard et al, 2013; Greenan et al, 2018, 2020; Le Guennec et al, 2020), it differs in terms of the microtubule inner protein densities (MIPs; Fig EV1B). We observed nine MIPs, six in the A‐tubule, one in the B‐tubule, and two in the C‐tubule (Fig 1G and EV1C). In the A‐tubule, most of the MIPs are unique, including MIP2 (yellow) that binds to protofilament A12, MIP3 (orange) bridging protofilaments A13 and A1, MIP4 (red) that binds to A2, MIP5 (purple) that binds to A5, and MIP6 (blue) bridging A6 and A7. MIP1 (green), a prominent MIP associated with protofilament A9, was also reported in centrioles isolated from CHO cells (Greenan et al, 2018), Trichonympha (Guichard et al, 2013), Chlamydomonas, and Paramecium (Le Guennec et al, 2020), and in basal bodies from bovine respiratory epithelia (Greenan et al, 2020). The seam is located between protofilaments A9 and A10 (Ichikawa et al, 2017; Ma et al, 2019), which suggests that MIP1 is a highly conserved seam‐stabilizing or seam‐recognizing structure.
In the B‐tubule, we observed a large helical MIP7 (magenta) bridging protofilaments B3‐9. We observed two groups of unique MIPs in the C‐tubule, MIP8 (light pink) associated with C2‐C4 and MIP9 (pink) with C5–C7. The inner junctions between A‐ and B‐tubules (cyan) and between B‐ and C‐tubules (turquoise) are non‐tubulin proteins that repeat every 8 nm and are staggered relative to each other when viewed from the luminal side of the triplet (Fig EV1C). We resolved density for the A–C linker (gold), which is associated with protofilaments C9 and C10, and possibly for the A‐link (olive green), associated with protofilament A8/A9 (Fig 1G and H).
The B‐ and C‐tubule MIPs we observed are not present in other centriole structures. However, helical MIPs have been observed in the transition zone of bovine respiratory cilia (Greenan et al, 2020). Unlike other mammalian centriole structures, we do not observe MIPs that bridge B1‐B2 or C1‐C2 (Fig EV1B). It is difficult to tell whether these differences in MIP patterns are due to differences in cell type or species. As this is the first in situ structure of any mammalian centriole, these differences may also be because previous structures were of isolated centrioles. Nonetheless, it is clear that there is great diversity in how core centriolar microtubules are accessorized, which raises questions about the functions of these MIPs.
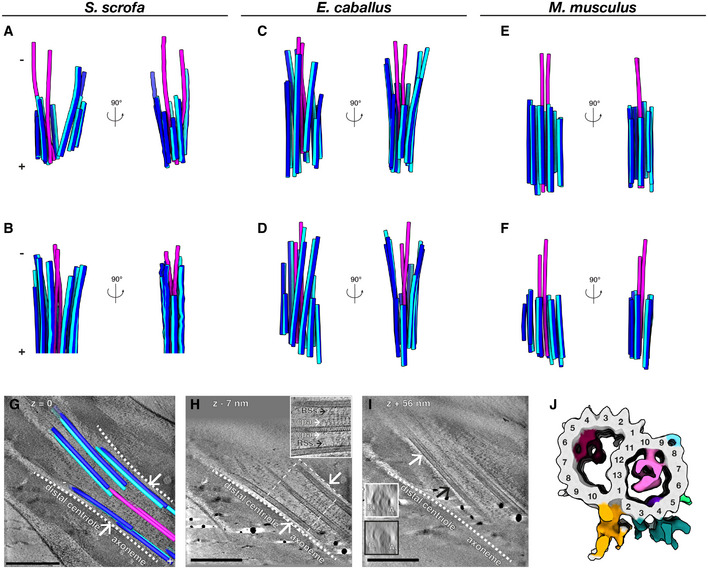
We next determined the organization of the atypical DC by tracing microtubules through Volta phase plate (VPP; Danev et al, 2014) cryo‐tomograms of whole sperm (Fig 2). The DC consists of doublet microtubules, with a pair of singlets extending through the lumen (Fig 2A–F). In pig and in horse sperm (Fig 2A–D), doublets extend almost as far proximally as the central pair. In a further departure from canonical centriole structure, DC doublets are splayed open and arranged asymmetrically around the singlets. The central singlets themselves are spaced inconsistently, suggesting that they lack the projections characteristic of the central pair apparatus (CPA). Mouse sperm lack the splayed doublets, but they also have a pair of singlets extending beyond the axoneme (Fig 2E and F).
Figure 2. The distal centriole (DC) in mammalian sperm is composed of doublet microtubules arrayed asymmetrically around a pair of singlet microtubules.
-
A–FMicrotubules in the DC of pig (A, B), horse (C, D), and mouse (E, F) sperm traced from Volta phase plate cryo‐tomograms of intact sperm. Doublets are colored blue (A‐tubule in light blue, B‐tubule in dark blue), while singlets are pink.
-
G–ITomographic slices through cryo‐FIB‐milled lamellae of the DC‐to‐axoneme transition in pig sperm show how the change in geometry (G, white arrows and white dashed lines) coincides with the appearance of axoneme accessory structures (H, white arrows) and with density in the A‐tubule (I, compare insets in white and black boxes). In (I), the white and black arrows indicate where the cross‐sections in white and black boxes were taken from.
-
JIn situ structure of the pig sperm DC microtubule doublet with the tubulin backbone in gray and microtubule inner protein densities colored individually. The gold and turquoise densities on the luminal side of the doublet are consistent with the positions of parts of the inner scaffold. This structure represents the DC doublets closest to the axoneme (the area shown in (G–I)).
Data information: Labels: RSs—radial spokes, cpa—central pair apparatus, At—A‐tubule, Bt—B‐tubule. Scale bars: 250 nm.
To more precisely define the DC‐to‐axoneme transition, we imaged cryo‐FIB‐milled sperm (Fig 2G–J). We directly observed this transition in situ in pig sperm, defined by the appearance of axonemal accessory proteins such as the radial spokes and the projections of the CPA (Fig 2H). The onset of the axoneme coincides with a change in microtubule geometry (Fig 2G), suggesting that the splayed‐open doublets are indeed characteristic of the DC. The transition zone also coincides with an increase in density in the A‐tubule (Fig 2I), suggesting that binding of axonemal accessory structures is related to the regulated binding of A‐tubule MIPs. We then determined the structure of doublet microtubules from the region of the DC closest to the axoneme by subtomogram averaging, revealing the presence of MIPs distinct from those in the PC (Fig 2J). We resolved some density for structures on the luminal side of the doublet (gold and turquoise), which may correspond to parts of the inner scaffold (Li et al, 2012; Le Guennec et al, 2020).
The flagellar waveform depends greatly on the properties of the base (Riedel‐Kruse & Hilfinger, 2007), but there is very little information on how this region is organized in three dimensions in any cell type. In order to capture the full three‐dimensional complexity of the flagellar base, we took advantage of enhanced contrast provided by the VPP, which allowed us to trace microtubules while retaining the context of the surrounding connecting piece (Fig 3A–C, Movies [Link], [Link], [Link]). Semi‐automated neural network‐based segmentation (Chen et al, 2017) revealed that the connecting piece forms a large chamber enclosing the sperm centrioles. Although precise dimensions and shapes of the connecting piece differ across species (Fig 3D–F), its general architecture appears to be conserved across mammalian species.
Figure 3. The connecting piece forms a large, asymmetric chamber around the sperm centrioles.
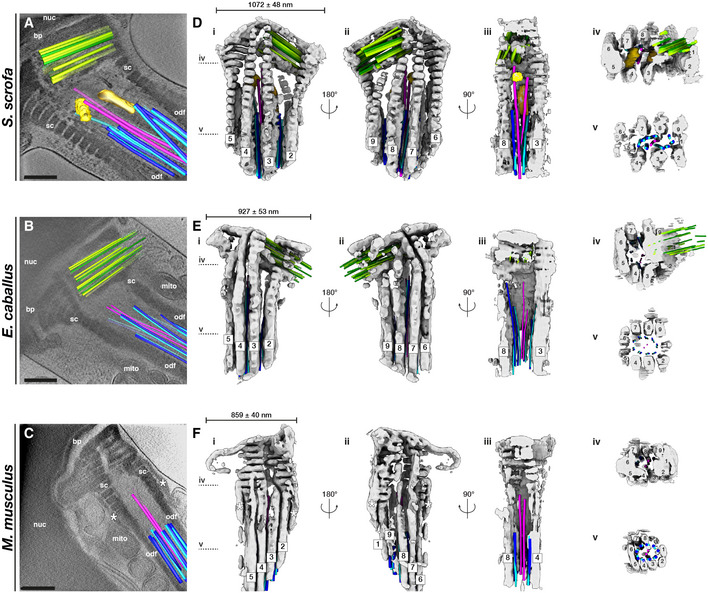
-
A–CSlices through Volta phase plate cryo‐tomograms of the neck region in intact pig (A), horse (B), and mouse (C) sperm. Proximal centriole triplets are shown in green, distal centriole doublets in blue (A‐tubule in light blue, B‐tubule in dark blue) and singlets in pink, and electron‐dense bars in yellow. Note the electron‐dense structures flanking the connecting piece in the mouse (asterisks in C, F).
-
D–FThree‐dimensional architecture of the flagellar base, with the connecting piece in gray, the proximal centriole in green, distal centriole doublets in blue (A‐tubule in light blue, B‐tubule in dark blue) and singlets in pink, and electron‐dense bars in yellow. The connecting piece was segmented semi‐automatically with a neural network, while microtubules were traced manually.
Data information: Labels: nuc—nucleus, bp—baseplate, sc—striated columns, odf—outer dense fibers, mito—mitochondria. Scale bars: 250 nm.
The proximal region of the connecting piece consists of striated columns (SCs), called such because of their banded appearance. Following the numbering scheme laid out in (Ounjai et al, 2012), we found that the SCs follow a conserved pattern of grouping and splitting. The proximal connecting piece can be grossly divided into left and right regions. The right region forms the proximal centriolar vault where columns 8, 9, 1, 2, and 3 merge, whereas the left region comprises columns 4, 5, 6, and 7 (Fig 3D–F, panels iv). The columns gradually separate, eventually splitting into nine separate columns that fuse distally with the ODFs (Fig 3D–F, panels v).
The connecting piece displays both marked left–right asymmetry and dorsoventral asymmetry in all three species. The PC is embedded within the proximal region of the connecting piece, and always on the same side. In pig sperm, one side of the proximal centriolar vault is formed by the Y‐shaped SC 9, which also gives the entire connecting piece dorsoventral asymmetry (Fig 3D, panel ii). The material of the connecting piece extends through the interstices of the PC triplets (Fig 1D–F) and is continuous with electron‐dense material within the proximal lumen of the PC (asterisks in Fig 1D and D’). Intriguingly, the dorsoventral asymmetry of the pig sperm PC is defined relative to the connecting piece, with the side of the shorter triplets always facing the Y‐shaped segmented column 9 (Fig 3D).
The pig sperm connecting piece also has two electron‐dense bars associated with the central singlets of the DC (Fig 3A and D, yellow and goldenrod), which resemble the bars observed in the bovine sperm connecting piece (Ounjai et al, 2012). These bars are conspicuously absent from horse and from mouse sperm. Instead, mouse sperm have two electron‐dense structures flanking the SCs (asterisks in Fig 3C and F), an arrangement reminiscent of the distribution pattern of the centrosomal protein speriolin (Goto et al, 2010; Ito et al, 2019).
The mammalian sperm axoneme anchors unique accessory structures and species‐specific microtubule inner proteins
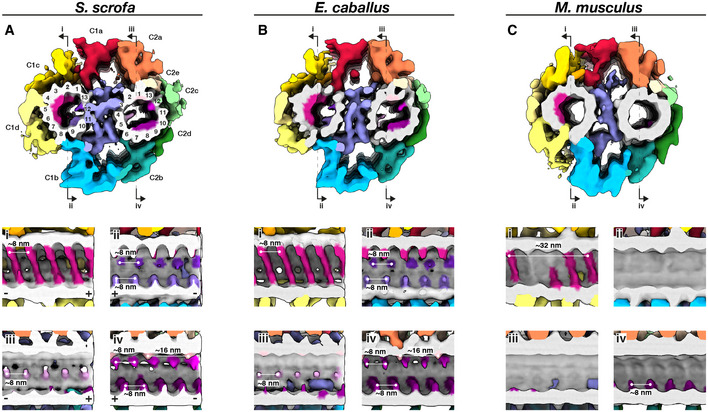
To gain insight into the molecular architecture of the axoneme, we determined in situ structures of the central pair apparatus (Fig 4) and of the 96‐nm axonemal doublet repeat (Fig 5). Our structures of the CPA are the first from any mammalian system, and our structures of the doublets are the first from any mammalian sperm, thus filling crucial gaps in the gallery of axoneme structures. The overall architecture of the mammalian CPA projection network is similar across the three species we examined (Fig EV2) and resembles that of the CPA from Chlamydomonas and from sea urchin sperm (Carbajal‐González et al, 2013; Fu et al, 2019). Indeed, mutations in hydin, a component of the C2b projection, impair ciliary motility in both Chlamydomonas and mice (Lechtreck et al, 2008). However, the mammalian sperm CPA lacks the C1f projection found between the C1b and C1d projections in Chlamydomonas and sea urchin.
Figure 4. The mammalian sperm central pair apparatus (CPA) has a conserved projection network but species‐specific microtubule inner proteins.
-
A–CWhole‐population in situ structures of the 32‐nm CPA repeat from pig (A), horse (B), and mouse (C) sperm. Panels (i), (ii), (iii), and (iv) are cut‐through views of the lumens of the C1 (i and ii) and C2 (iii and iv) microtubules. Microtubules are shown in gray and microtubule protein densities are colored individually. The projection network is colored according to (Carbajal‐González et al, 2013).
Figure 5. The mammalian sperm axoneme anchors unique accessory structures and species‐specific microtubule inner proteins.
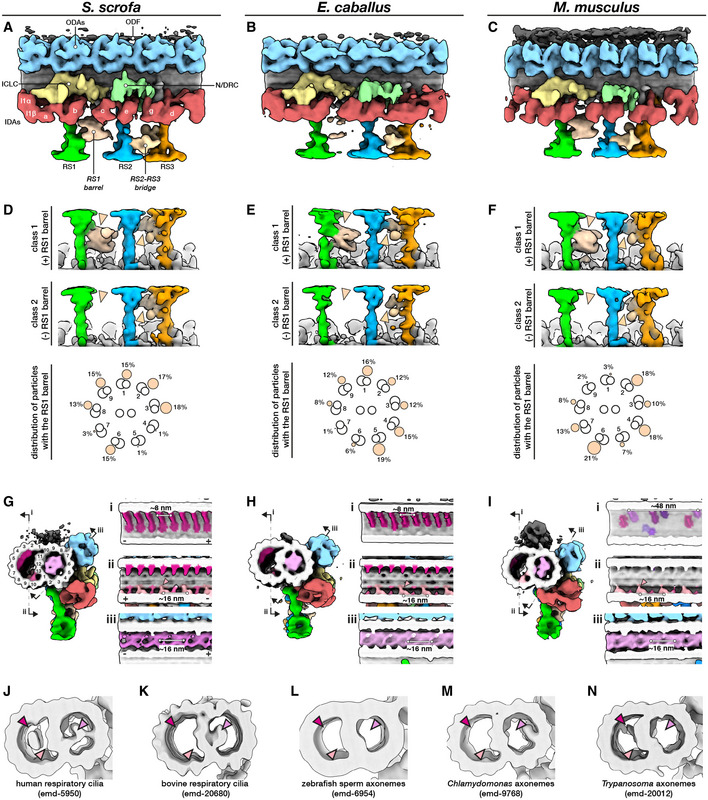
-
A–CIn situ structures of the 96‐nm repeat from pig (A), horse (B), and mouse (C) sperm principal pieces.
-
D–FClassification focused on the RS1 barrel revealed two distinct classes of particles, one with (top panels) and one without (middle panels) the structure. Particles with the RS1 barrel are distributed asymmetrically around the axoneme (bottom panels).
-
G–IMicrotubule inner proteins in axonemes from pig (G), horse (H), and mouse (I) sperm.
-
J–NMicrotubule inner proteins in axonemes from other cell types and organisms. Arrowheads correspond to locations of MIPs that are prominent in mammalian sperm: magenta—helical MIP in the B‐tubule; pink—large A‐tubule MIP; light pink—additional density associated with MIP3a.
Data information: Labels: ODAs—outer dynein arms, IDAs—inner dynein arms, ICLC—intermediate chain/light chain of the I1 dynein, N/DRC—nexin/dynein regulatory complex, ODF—outer dense fiber, RS1‐3—radial spokes 1–3.
Figure EV2. Structural details of the central pair apparatus projection network in mammalian sperm.
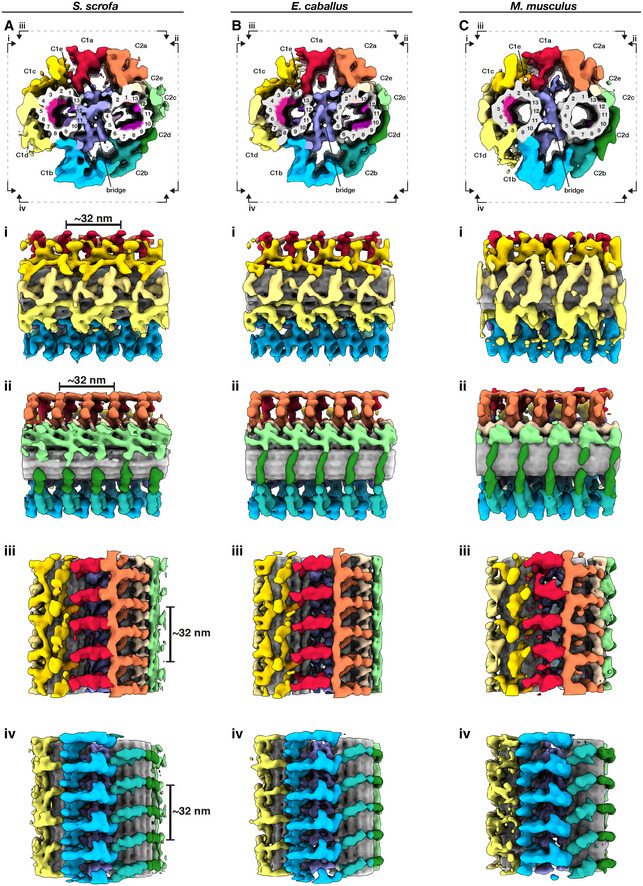
-
A–CWhole‐population in situ structures of the 32‐nm CPA repeat from pig (A), horse (B), and mouse (C) sperm. Individual rotated views show details of C1 projections (i), C2 projections (ii), the interface between C1a and C2a projections (iii), and the interface between C1b and C2b projections (iv).
Mammalian CPAs have several MIPs (Fig 4) that are absent from Chlamydomonas and from sea urchin sperm, which have only small MIPs or no MIPs, respectively. Similar to the PC (Fig 1g), pig and horse CPAs have large helical MIPs in the C1 microtubule that bridge protofilaments 1‐7 and repeat every 8 nm (Fig 4A and B, panels i). The corresponding MIPs in the mouse CPA appear to bridge fewer protofilaments and to repeat with a longer period, but this will need to be confirmed unequivocally with better‐resolved structures (Fig 4C, panel i). Pig and horse also have smaller C1 MIPs on the side facing the bridge, with one bridging protofilaments 9 and 10 and the other jutting out from between protofilaments 12 and 13 (Fig 4A and B, panels ii). Both of these MIPs repeat every 8 nm, and both are absent from the mouse (Fig 4C, panel ii). In the C2 microtubule, the pig and the horse have several other MIPs that are also absent from the mouse. These include a MIP that protrudes out from between protofilaments 4 and 5 and repeats every 8 nm (Fig 4A and B, panel iii), a MIP that binds between protofilaments 1 and 13 and repeats every 16 nm, and a MIP that extends from protofilament 12 and repeats every 8 nm (Fig 4A and B, panel iv). A fourth B‐tubule MIP bridges protofilaments 7–9 in the pig and the horse, but this MIP is smaller and only bridges protofilaments 8 and 9 in the mouse (Fig 4A–C, panels iv).
Our in situ structures of the 96‐nm axonemal doublet repeat from mammalian sperm revealed density for attachment to the outer dense fibers (ODFs) as well as novel structural features associated with the radial spokes (RSs; Figs 5 and EV3). In particular, we observed a barrel‐shaped structure associated with RS1 (the RS1 barrel) and an extensive bridge linking the stalks of RS2 and RS3 (the RS2‐RS3 bridge). These structures are absent from all other axoneme structures reported so far (Fig EV3A–G).
Figure EV3. Comparing the mammalian sperm axoneme with other structures.
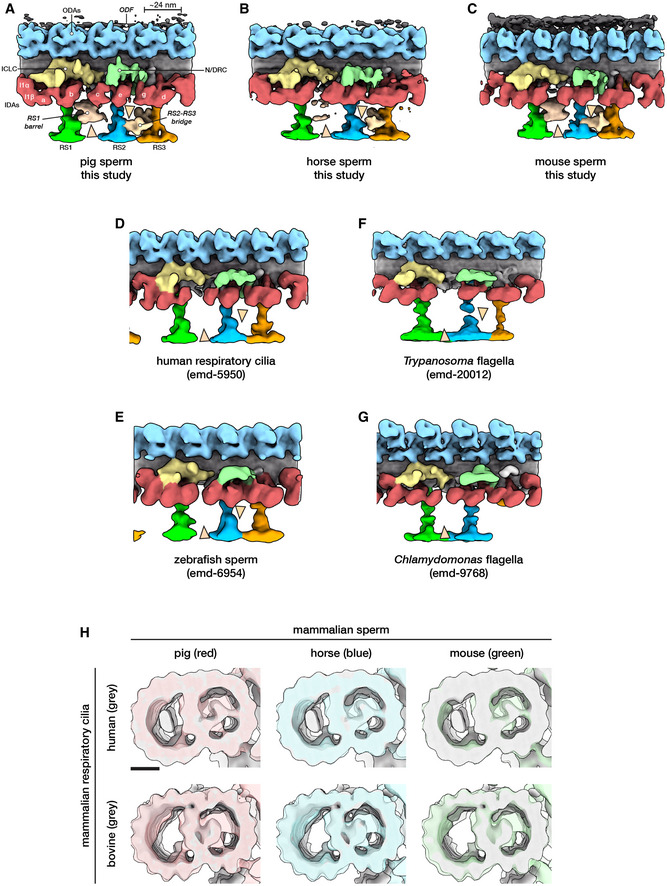
-
A–GStructures of the 96‐nm axonemal repeat from various species in cell types. Arrowheads mark the novel RS1 barrel and RS2‐RS3 bridge so far seen only in mammalian sperm.
-
HA‐tubule MIPs are present in mammalian respiratory cilia, but the A‐MIP in mammalian sperm is larger and makes more extensive connections with the A‐tubule itself. Scale bar: 10 nm.
Data information: Labels: ODAs—outer dynein arms, IDAs—inner dynein arms, ICLC—intermediate chain/light chain of the I1 dynein, N/DRC—nexin/dynein regulatory complex, ODF—outer dense fiber, RS1‐3—radial spokes 1–3.
By focused classification, we resolved two distinct classes of particles, one with and the other without density for the RS1 barrel (Fig 5D–F). By separating out only particles with the barrel, classification also allowed us to improve the density for this structure. The RS1 barrel is ~18 nm long and ~16 nm wide and makes two major contacts with RS1, one at the base of the head and one at the middle of the stalk. Interestingly, the RS1 barrel is radially distributed asymmetrically around the axoneme. Although the RS1 barrel is not particularly enriched in any individual doublet position, it seems to occur less frequently in specific positions. In the pig, only 1% of the particles were found in each of doublets 4 and 5, while only 3% were in doublet 7. In the horse, only 1% were found in doublet 7. In the mouse, only 2% were found in doublet 9 and 3% in doublet 1.
Comparing mammalian sperm axonemes to those from other species reveals large variations in MIP densities (Fig 5G–N). Mammalian sperm have a large MIP that fills almost the entire lumen of the A‐tubule (the A‐MIP, Fig 5G–I, bottom panels), which explains why the A‐tubule appears dark in cross‐section. The A‐MIP makes extensive contacts with the A‐tubule, including protofilaments A1‐A3, A5‐A6, and A8‐A13, and has an overall periodicity of ~16 nm. Because the sperm A‐MIP makes contacts with nearly all protofilaments of the A‐tubule, it seems plausible that it would affect the mechanics of the doublet. The A‐MIP also makes contacts with the same protofilament 9 to which the ODFs attach, which suggests that it may also functionalize the outer surface of the A‐tubule for ODF docking. A‐tubule MIPs are present in axonemes from human (Fig 5J; Lin et al, 2014a) and bovine (Fig 5K) respiratory cilia (Greenan et al, 2020), but these are not as extensive as the A‐MIP in mammalian sperm (Figs 5G–I and EV3H). Zebrafish (Fig 5L) (Yamaguchi et al, 2018) and sea urchin sperm (Nicastro et al, 2011; Lin et al, 2014b) do not have large MIPs in the A‐tubule, nor do Chlamydomonas (Fig 5M; Nicastro et al, 2011; Ma et al, 2019; Owa et al, 2019) or Trypanosoma (Fig 5N; Imhof et al, 2019).
In all axoneme structures reported so far, B‐tubules contain MIP3a and MIP3b, which bind to protofilaments B9 and B10 with staggered ~16‐nm repeats (Fig 5G–I). However, in mammalian sperm, MIP3a has an additional density that links it to protofilament A13 (Fig 5G–I, panels ii, pink arrowheads; Fig EV3H). In pig and horse sperm, a helical MIP with an ~8‐nm repeat bridges protofilaments B2‐B7 (Fig 5G–I, panels i). Mouse sperm do not have this large MIP and instead have smaller MIPs that bind with an overall apparent periodicity of ~48‐nm (Fig 5I, panel i). Large B‐tubule MIPs have so far only been seen in human respiratory cilia (Fig 5J; Lin et al, 2014a) and in Trypanosoma (the ponticulus, Fig 5N; Imhof et al, 2019), but the morphometry of these MIPs differs from the helical MIPs in mammalian sperm.
A crucial comparison comes from the structure of axonemes from mouse respiratory cilia (Ueno et al, 2012). Mouse respiratory cilia lack the large A‐MIP that is so prominent in mouse sperm, which points to cell type‐specific differences in the MIP repertoire. Similarly, the RS1 barrel and the RS2‐RS3 bridge (Fig 5A–F) are present in sperm, but not in respiratory cilia (Fig EV3A–E). Because the radial spokes are key regulators of flagellar motility, as evidenced by the fact that radial spoke defects cause a number of ciliopathies (Lin et al, 2014a), the RS1 barrel and the RS2‐RS3 bridge are likely to play a significant role in modulating sperm motility. As with many of the structures we report, defining why exactly they are needed in mammalian sperm would help us better understand the intricacies of flagellar organization.
Outer dense fibers are directly coupled to axonemal doublets in the principal piece but not in the midpiece
To resolve how the ODFs associate with the microtubule doublets of the axoneme, we aligned and averaged particles from the principal piece, then classified them with a mask on the ODF‐doublet attachment. Our structures reveal that, for doublets associated with ODFs, the ODFs are directly coupled to protofilament 9 of the A‐tubule (Fig 6A–C). The ODF‐doublet attachment consists of a pair of linkers spaced ~8 nm apart, with each pair spaced ~16 nm from the next, yielding an apparent overall periodicity of ~24 nm that is consistent across species. Direct ODF‐microtubule coupling provides the elusive structural mechanism by which forces from axoneme bending can be transmitted through the ODFs to the base of the flagellum. As such, in the principal piece, the ODFs should be considered a part of the axoneme proper. This increases the effective diameter of the axoneme and also translates to an increase in bending moments (Lindemann, 1996; Lindemann & Lesich, 2016). However, the ODFs themselves are not of the same size, taper along the flagellum and terminate at different points, which leads to an anisotropic bending stiffness along the tail (Lindemann, 1996; Gadêlha & Gaffney, 2019).
Figure 6. The attachment of outer dense fibers to axonemal doublets varies along the length of the sperm flagellum.

-
A–CIn situ structures of the 96‐nm axonemal repeat from the principal piece of pig (A), horse (B), and mouse (C) sperm after classification focused on the ODF attachment.
-
D–LAxoneme structures with particles from tomograms from (D, G, J) only the midpiece, (E, H, K) only the proximal principal piece, and (F, I, L) only the distal principal piece. Left panels (D–L) show transverse slices through sperm flagella, while right panels (D’–L’) show subtomogram averages of the 96‐nm axoneme repeat from the corresponding regions of the tail. In the left panels, one doublet‐ODF pair is traced to emphasize the gap between the ODF and the doublet in the midpiece. The ODF is traced in gray with a black outline, and the doublet is traced in blue (light blue for the A‐tubule, dark blue for B‐tubule). Doublets 3 and 8 are numbered to aid visualization.
Data information: Labels: ODF—outer dense fiber, mito—mitochondria, fs—fibrous sheath. Scale bars: 250 nm.
To determine how ODF‐doublet association changes along the flagellum, we then separately averaged the 96‐nm repeat from the midpiece, proximal principal piece, and distal principal piece (Fig 6D–L). Structures from the distal principal piece, after the ODFs had terminated, did not show density for the ODFs (Fig 6F’, I’, L’). In the proximal principal piece, the ODFs are directly attached to the axoneme via the A‐tubule as described above (Fig 6E’, H’, K’). In the midpiece, the ODFs are at their largest but, surprisingly, are not directly connected to the microtubule doublets (Fig 6D’, G’, J’). This inhomogeneous pattern of association seems to be a general feature of mammalian sperm, as we observed it in all three species we examined. Our data thus reveal that the organization of accessory structures in mammalian sperm flagella is more complex than previously thought.
Singlet microtubules in the endpiece are capped by a conserved plug but contain species‐specific microtubule inner proteins
We then determined how the doublets of the axoneme transition into singlets of the endpiece (Fig 7A–C). We found that doublets can transition into singlets by two possible arrangements, either by a doublet splitting into two independent singlets (Fig 7D–F, left panels) or by the B‐tubule terminating abruptly (Fig 7D–F, right panels). Similar patterns have been observed in human sperm (Zabeo et al, 2019).
Figure 7. Singlet microtubules in the endpiece are capped by a conserved plug but contain species‐specific microtubule inner proteins.
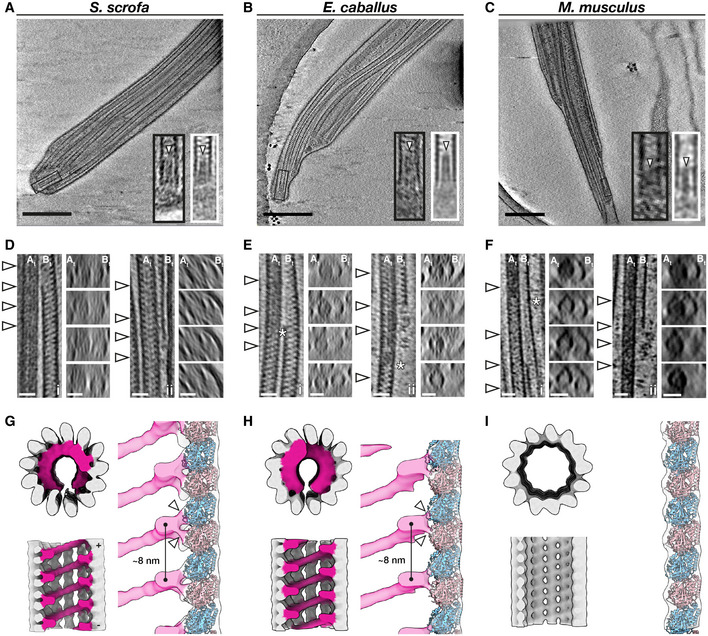
-
A–CSlices through defocus cryo‐tomograms of the endpiece in pig (A), horse (B), and mouse (C) sperm. Insets show digital zooms (black boxes) and subtomogram averages (white boxes) of microtubule tips. Note the presence of a plug (arrowhead) extending into the lumen.
-
D–FRepresentative examples of the mechanisms by which doublets can transition into singlets. Doublets can split into two complete singlets (panels i), or the B‐tubule can terminate abruptly with the A‐tubule extending as a singlet (panels ii). White arrowheads in the left panels indicate approximate locations of transverse slices shown in the right panels. Asterisks mark approximate locations of the change in A‐tubule luminal density.
-
G–IIn situ structures of singlet microtubules from pig (G), horse (H), and mouse (I) sperm endpieces. Note how the MIP binds to two tubulin monomers (white arrowheads).
Data information: Labels: At—A‐tubule, Bt—B‐tubule. Scale bars: (A–C) 250 nm, (D–F) 25 nm.
We further observed that the doublet‐to‐singlet transition is also associated with loss of density in the A‐tubule (Fig 7D–F, asterisks), although the precise location of this change relative to the splitting event varies. After the splitting event, 8‐nm striations previously seen only in the B‐tubule were visible in both singlets. The A‐MIP thus seems to be a marker of the axoneme proper: the A‐tubule lumen transitions from “empty” to “full” at the DC‐to‐axoneme transition (Fig 2I), whereas it goes from “full” to “empty” at the doublet‐to‐singlet transition.
We averaged endpiece singlets from the three species and found that they are consistently comprised of 13 protofilaments (Fig 7G–I). In the pig and the horse, singlets contain a helical MIP that follows the microtubule lattice, similar to the MIP previously described for human sperm endpieces (Zabeo et al, 2018). Our higher‐resolution averages now reveal that this helical MIP makes independent contacts with both tubulin monomers (Fig 7G and H, right panels). This helical MIP is very similar to MIP7 we observed in the B‐tubule of the PC (Fig 1G), to the helical MIP on the outer wall of the B‐tubule of the axonemal doublets (Fig 5G–I), and to the helical MIPs in the CPA (Fig 4A and B). Intriguingly, this MIP is absent from endpiece singlets in the mouse (Fig 7I), where axonemal B‐tubules and CPA microtubules also lack the large multi‐protofilament‐spanning MIPs present in the pig and horse (Figs 4C and 5I).
We further observed that microtubule termini are capped in all three species (Fig 7A–C, black boxes). Averaging confirmed the presence of a plug extending ∼30 nm into the microtubule lumen (Fig 7A–C, white boxes). Normally, ciliary length control and turnover of axonemal components is mediated by the intraflagellar transport (IFT) system. However, IFT is absent in mature spermatozoa (San Agustin et al, 2015), which raises the question of how microtubule length and stability are maintained in these cells. These capping structures may stabilize free microtubule ends and prevent them from depolymerizing.
Discussion
Accessory structures are integral parts of mammalian sperm flagella and impose large mechanical asymmetries and anisotropies on the core axoneme
The presence of accessory structures in mammalian sperm flagella has long been recognized, but details of their three‐dimensional organization and interactions with the core axoneme have remained elusive. Our comparative structural analysis revealed that the accessory elements impose striking multi‐scale asymmetries and anisotropies on the sperm flagellum. Of particular relevance to wave generation, we found prominent asymmetry in the connecting piece at the base of the flagellum (Fig 3). This large‐scale asymmetry could bias basal sliding to one side relative to the head, consequently polarizing inter‐doublet sliding moments transmitted to the rest of the axoneme. Indeed, in order to swim forward, mouse sperm flagella must balance intrinsic asymmetry of the waveform with episodic switching of side‐to‐side asymmetric bends (Babcock et al, 2014). Asymmetric counter‐bends are also observed in rat sperm flagella, showing that shear developed in the distal flagellum is not identical when the flagellum is bent in the two opposing directions relative to the head (Lindemann et al, 2005). Polarized waveforms have also been reported for human and bovine sperm (Friedrich et al, 2010; Saggiorato et al, 2017; Gadêlha et al, 2020). Mathematical models incorporating basal sliding have confirmed the critical role of this phenomenon on waveform generation, but no modeling framework has attempted to study the effects of basal asymmetry that are now evident in our structures.
We also found that, in the principal piece, the ODFs are directly coupled to a defined protofilament on the axonemal doublets (Figs 5 and 6). Such linkages provide a structural mechanism by which forces from axoneme bending can be transmitted through the ODFs to the base of the flagellum. Because the ODFs are anchored through the connecting piece at the base, these linkages also provide a mechanism by which changes in the base can be transmitted to the axoneme further down the tail. The ODFs also impose asymmetry and anisotropy on the flagellum; they gradually taper along the flagellum, but each ODF does so at a different rate, in contrast to the assumptions made in mathematical models incorporating the ODFs (Riedel‐Kruse & Hilfinger, 2007; Gadêlha & Gaffney, 2019).
Beyond the gradual tapering and staggered termination of the ODFs, we show that a further source of proximal‐distal asymmetry is the association of the ODFs with the axonemal doublets themselves (Fig 6). Intriguingly, this arrangement has actually been suggested previously based on measurements of ODF‐doublet spacing in thin‐section TEM of bull sperm (Lesich et al, 2014), and we now show that it holds true at the molecular level in three other mammalian species. This configuration would allow the ODFs to slide relative to the axoneme while being restrained by the mitochondrial sheath, lending flexibility to the midpiece. This is proposed to support formation of the extreme bends in the midpiece seen during hyperactivation (Lindemann & Lesich, 2016). Midpiece flexibility is crucial for sperm motility and hyperactivation, and mice lacking the catalytic subunit of a sperm‐specific calcineurin isoform are infertile because they have more rigid midpieces (Miyata et al, 2015). Midpiece stiffness decreases as sperm transit through the epididymis (Jeulin et al, 1996; Miyata et al, 2015), which suggests that ODFs start out directly linked to the axoneme along their entire lengths and later locally detach in the midpiece.
Our comparative approach revealed that aforementioned details of how accessory structures relate to the microtubule core are conserved across species, suggesting that this architecture is fundamental for mammalian sperm motility. However, we also observe substantial inter‐species variation, such as in the shape of the connecting piece (Fig 3) and in the dimensions of ODFs (Fig 6). Comparative motility studies suggest that sperm flagella with larger ODFs form arcs with larger radii of curvature (Phillips, 1972). In contrast, how differences in the flagellar base and in head shape relate to differences in motility remains largely unexplored. We hope our structures will motivate and inform further theoretical and empirical work on the role of the base in shaping the flagellar beat.
Mammalian sperm are characterized by large, helical, protofilament‐bridging microtubule inner proteins that may affect microtubule bending stiffness
Our structures show that ciliary MIPs are highly diverse, both within a single cilium and across species (Figs 1, 2, 4, 5 and 7). By comparing several ciliary assemblies across species, we show that there are fundamental differences in the MIP repertoire across lineages. Most notably, large helical protofilament‐bridging MIPs are present in essentially all microtubule‐based assemblies throughout the sperm flagellum in pig and in horse, whereas they are reduced in corresponding structures in mouse sperm (Figs 4, 5 and 7). These MIPs seem to be characteristic of mammalian sperm flagella as they are absent from axonemes of zebrafish sperm, sea urchin sperm, and mammalian respiratory cilia (Fig EV3). Similar MIPs are present in human sperm endpiece singlets and in bovine respiratory epithelia, although in the latter they are restricted to the transition zone (Greenan et al, 2020). It is plausible that the helical MIPs are formed by variations of the same core protein complex, but direct confirmation awaits higher‐resolution structures (Ichikawa et al, 2019; Ma et al, 2019; Khalifa et al, 2020), genetic perturbation experiments, and direct labeling. If this were the case, however, this complex would have to adapt to the subtle differences in curvature between the walls of the centriolar/axonemal B‐tubules and the 13‐protofilament singlets. We did not observe large differences in MIP densities along the proximodistal axis of the flagellum (Fig 6), but more systematic and exhaustive sampling may yet uncover subtle molecular asymmetries.
The intimate and extensive contacts that the helical MIPs make with the tubulin lattice suggest that such a MIP would affect the mechanical properties of the microtubule itself. Microtubules are characterized by a paradoxical length‐dependent bending stiffness attributed to the low shear modulus between adjacent protofilaments (Kurachi et al, 1995; Pampaloni et al, 2006; Taute et al, 2008). The protofilament‐bridging MIPs observed here may decrease inter‐protofilament shearing, increasing the shear modulus and the resulting bending stiffness of sperm microtubules. This adaptation might be necessary to withstand the large forces involved in moving the long flagellum of mammalian sperm, potentially reducing the length dependency of bending stiffness in MIP‐reinforced microtubules. These MIPs may also act as structural braces that suppress internal buckling within the axoneme under large loading in high viscosity fluids.
Concluding Remarks
This study exemplifies the need for comparative studies of cilia and flagella, both from different species and from different cell types of the same species. Our study motivates future efforts to define how species‐specific features of flagellar architecture affect the hydrodynamics of sperm motility. Such endeavors will likely involve the synthesis of structural cell biology, motility analysis, and mathematical modeling. The structures we present here provide crucial resources for understanding how the ancient and conserved ciliary microtubule core is ornamented to support motility through diverse fluid environments.
Materials and Methods
Sperm collection and preparation
Pig sperm samples were purchased from an artificial insemination company (Varkens KI Nederland), stored at 18°C, and prepared for imaging within 1 day of delivery. Sperm were layered onto a discontinuous gradient consisting of 4 ml of 35% Percoll® (GE Healthcare) underlaid with 2 ml of 70% Percoll®, both in HEPES‐buffered saline (HBS: 20 mM HEPES, 137 mM NaCl, 10 mM glucose, 2.5 mM KCl, 0.1% kanamycin, pH 7.6) and centrifuged at 750 g for 15 min at RT. Pelleted cells were washed once in phosphate‐buffered saline (PBS: 137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4), resuspended in PBS, and counted.
Horse semen was collected from mature Warmblood stallions using a Hanover artificial vagina in the presence of a teaser mare. After collection, semen was filtered through gauze to remove gel fraction and large debris, then transported to the laboratory at 37°C and kept at room temperature until further processing. Semen was diluted in INRA96 (IMV Technologies) to obtain a sperm concentration of 30 × 106 cells/ml. After this, sperm were centrifuged through a discontinuous Percoll gradient for 10 min at 300 g followed by 10 min at 750 g. The remaining pellet was resuspended in 1 ml of PBS and centrifuged again for 5 min at 750 g.
Mouse sperm were collected from the cauda epididymis of adult male C57BL/6 mice as described (Hutcheon et al, 2017). Briefly, male mice were culled as described (Mederacke et al, 2015) and the cauda epididymides were dissected with the vas deferens attached and placed in a 500 µl droplet of modified Biggers, Whitten, and Whittingham media (BWW: 20 mM HEPES, 91.5 mM NaCl, 4.6 mM KCl, 1.7 mM D‐glucose, 0.27 mM sodium pyruvate, 44 mM sodium lactate, 5 U/ml penicillin, and 5 µg/ml streptomycin, adjusted to pH 7.4 and an osmolarity of 300 mOsm/kg). To retrieve the mature cauda spermatozoa from the epididymides, forceps were used to first gently push the stored sperm from the vas deferens, after which two incisions were made with a razor blade in the cauda. Spermatozoa were allowed to swim out of the cauda into the BWW over a period of 15 min at 37°C, after which the tissue was removed and sperm were loaded onto a 27% Percoll density gradient and washed by centrifugation at 400 g for 15 min. The pellet consisting of an enriched sperm population was resuspended in BWW and again centrifuged at 400 g for 2 min to remove excess Percoll.
Cryo‐EM grid preparation
Typically, 3 µl of a suspension containing either 2–3 × 106 cells/ml (for whole cell tomography) or 20–30 × 106 cells/ml (for cryo‐FIB milling) was pipetted onto a glow‐discharged Quantifoil R 2/1 200 mesh holey carbon grid. One µl of a suspension of BSA‐conjugated gold beads (Aurion) was added, and the grids then blotted manually from the back (opposite the side of cell deposition) for ∼3 s (for whole cell tomography) or for ∼5‐6 s (for cryo‐FIB milling) using a manual plunge‐freezer (MPI Martinsried). Grids were immediately plunged into a liquid ethane‐propane mix (37% ethane) (Tivol et al, 2008) cooled to liquid nitrogen temperature. Grids were stored under liquid N2 until imaging.
Cryo‐focused ion beam milling
Grids were mounted into modified Autogrids (ThermoFisher) for mechanical support. Clipped grids were loaded into an Aquilos (ThermoFisher) dual‐beam cryo‐focused ion beam/scanning electron microscope (cryo‐FIB/SEM). All SEM imaging was performed at 2 kV and 13 pA, whereas FIB imaging for targeting was performed at 30 kV and 10 pA. Milling was typically performed with a stage tilt of 18°, so lamellae were inclined 11° relative to the grid. Each lamella was milled in four stages: an initial rough mill at 1 nA beam current, an intermediate mill at 300 pA, a fine mill at 100 pA, and a polishing step at 30 pA. Lamellae were milled with the wedge pre‐milling technique (Schaffer et al, 2017) and with expansion segments (Wolff et al, 2019).
Tilt series acquisition
Tilt series were acquired on either a Talos Arctica (ThermoFisher) operating at 200 kV or a Titan Krios (Thermo Fisher) operating at 300 kV, both equipped with a post‐column energy filter (Gatan) in zero‐loss imaging mode with a 20‐eV energy‐selecting slit. All images were recorded on a K2 Summit direct electron detector (Gatan) in either counting or super‐resolution mode with dose‐fractionation. Tilt series were collected using SerialEM (Mastronarde, 2005) at a target defocus of between −4 and −6 µm (conventional defocus‐contrast) or between −0.5 and −1.5 µm (for tilt series acquired with the Volta phase plate). Tilt series were typically recorded using either strict or grouped dose‐symmetric schemes, either spanning ± 56° in 2° increments or ± 54° in 3° increments, with total dose limited to ∼100 e‐/Å2.
Tomogram reconstruction
Frames were aligned either post‐acquisition using Motioncor2 1.2.1 (Zheng et al, 2017) or on‐the‐fly using Warp (Tegunov & Cramer, 2019). Frames were usually collected in counting mode, but when appropriate super‐resolution frames were binned 2X during motion correction. Tomograms were reconstructed in IMOD (Kremer et al, 1996) using weighted back‐projection, with a SIRT‐like filter (Zeng, 2012) applied for visualization and segmentation. Defocus‐contrast tomograms were CTF‐corrected in IMOD using ctfphaseflip while VPP tomograms were left uncorrected.
Tomogram segmentation
Segmentation was generally performed semi‐automatically using the neural network‐based workflow implemented in the TomoSeg package in EMAN 2.21 (Chen et al, 2017) Microtubules, however, were traced manually in IMOD. Segmentation was then manually refined in Chimera 1.12 (Pettersen et al, 2004) or in ChimeraX (Goddard et al, 2018). Visualization was performed in ChimeraX.
For the pig sperm connecting piece, 7 tomograms were analyzed, each from a different cell, from three different animals. For the horse connecting piece, 13 tomograms were analyzed, each from a different cell, from two different animals. For the mouse connecting piece, 14 tomograms were analyzed, each from a different cell, from two different animals. Four additional tomograms of mouse sperm connecting pieces from FIB‐milled lamellae were analyzed, each from a different cell, from two different animals.
Subtomogram averaging
Subtomogram averaging with missing wedge compensation was performed using PEET 1.13.0 (Nicastro et al, 2006; Heumann et al, 2011). Resolution was estimated using the Fourier shell correlation (FSC) at a cut‐off of 0.5 (Nicastro et al, 2006). Alignments were generally performed first on binned data, after which aligned positions and orientations were transferred to less‐binned data using scripts generously provided by Dr. Daven Vasishtan. After alignment, classification was performed in order to assess heterogeneity and to identify cases of misalignment. Missing wedge‐corrected classification was performed by first running a principal components analysis using the pca function, followed by k‐means clustering using the clusterPca function (Heumann et al, 2011). The resulting class averages were manually inspected, and similar classes were combined. Specific averaging strategies are described below. Details of acquisition parameters and particle numbers are summarized in Table 1.
Table 1.
Averaging summary.
| Parameter | Species | ||
|---|---|---|---|
| Sus scrofa | Equus caballus | Mus musculus | |
| Proximal centriole | |||
| Sample type | lamellae | – | – |
| No. of animals/cells/tomograms | 1/3/3 | ||
| Microscope (accelerating voltage) | Arctica (200 kV) | – | – |
| Pixel size (Å) | 8.68 | – | – |
| Symmetry | C1 | – | – |
| Number of particles | 202 | – | – |
| Estimated resolution (Å) | 30 | – | – |
| Distal centriole | |||
| Sample type | lamellae | – | – |
| No. of animals/cells/tomograms | 1/2/2 | ||
| Microscope (accelerating voltage) | Arctica (200 kV) | – | – |
| Pixel size (Å) | 8.68 | – | – |
| Symmetry | C1 | – | – |
| Number of particles | 120 | – | – |
| Estimated resolution (Å) | 30 | – | – |
| 96‐nm axonemal repeat | |||
| Sample type | Whole cells | Whole cells & lamellae | Whole cells |
| No. of animals/cells/tomograms | 2/12/23 | 2/15/16 | 2/13/14 |
| Microscope (accelerating voltage) | Arctica (200 kV) | Arctica (200 kV) | Krios (300 kV) |
| Pixel size (Å) | 11.32 | 11.32 | 14.16 |
| Symmetry | C1 | C1 | C1 |
| Number of particles | 1,438 | 634 | 1,112 |
| Estimated resolution (Å) | 37 | 40 | 40 |
| Central pair complex | |||
| Sample type | Whole cells | Whole cells & lamellae | Whole cells & lamellae |
| No. of animals/cells/tomograms | 2/16/30 | 2/16/18 | 2/18/19 |
| Microscope (accelerating voltage) | Arctica (200 kV) | Arctica (200 kV) | Arctica (200 kV) |
| Pixel size (Å) | 11.32 | 11.32 | 11.32 |
| Symmetry | C1 | C1 | C1 |
| Number of particles | 978 | 461 | 281 |
| Estimated resolution (Å) | 36 | 34 | 49 |
| Endpiece singlets | |||
| Sample type | Whole cells | Whole cells | Whole cells |
| No. of animals/cells/tomograms | 1/6/6 | 1/6/6 | 2/4/4 |
| Microscope (accelerating voltage) | Arctica (200 kV) | Arctica (200 kV) | Krios (300 kV) |
| Pixel size (Å) | 3.50 | 5.66 | 7.08 |
| Symmetry | C1 | C1 | C1 |
| Number of particles | 5,799 | 9,136 | 1,581 |
| Estimated resolution (Å) | 19 | 19 | 30 |
Proximal centriole triplets, axonemal doublets, central pair apparatus
Microtubule‐based structures were manually traced in IMOD, and model points were added every 8 nm (for triplets, (Greenan et al, 2018)), 32 nm (for the CPA, (Carbajal‐González et al, 2013)), or 96 nm (for doublets, (Nicastro et al, 2006)) using addModPts. Subtomograms of approximately 70 nm × 42 nm × 70 nm (for centriole triplets), 100 nm × 100 nm × 100 nm (for the CPA), and 100 nm × 100 × 100 nm (for doublets) were computationally aligned and averaged.
For averaging triplets and doublets, particles with similar orientations (e.g., positions 9, 1, and 2) were first averaged. Sub‐averages were then manually rotated along the y‐axis using modifyMotiveList to align them with a common reference, followed by an alignment with a restricted search around the y‐axis. If necessary, sub‐averages were flipped to the right orientation using modifyMotiveList in order to generate grand averages.
The plotback of the proximal centriole was generated in IMOD by first running createAlignedModel to generate model files reflecting updated particle positions and orientations after alignment. The subtomogram average was then thresholded and saved as an isosurface model, which was then placed back into the tomograms using clonemodel.
Endpiece singlets
Endpiece singlets were manually traced in IMOD, and model points were added every 8 nm using addModPts. Subtomograms of approximately 35 nm × 35 nm × 35 nm were computationally aligned and averaged. An initial alignment was performed to align protofilaments, after which a mask was used to focus alignment on the helical MIP. The mask was then expanded to include the microtubule, and a final restricted alignment was performed.
Microtubule plugs
Particles were picked manually, and their initial orientations were defined using stalkInit. Alignments allowed for large search ranges around the particle long axis (defined as the y‐axis), with limited search ranges around the x‐ and z‐axes.
Author contributions
PM, MZ, HH, EGB, and BMG provided sperm samples. MRL, MCR, and RTR prepared samples for cryo‐EM. MRL performed cryo‐FIB milling. MRL, MCR, RTR, SCH, and TZ‐B‐M collected cryo‐ET data. MRL and MCR processed data. MRL, MCR, and TZ‐B‐M analyzed data. MRL, HB, and TZ‐B‐M wrote the manuscript, and all authors contributed to revisions.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Acknowledgements
The authors thank Dr. M. Vanevic for enabling this project with superb computational support; Dr. D. Vasishtan for providing scripts and advice that greatly facilitated subtomogram averaging; Ingr. C. T. W. M. Schneijdenberg and J. D. Meeldijk for management and maintenance of the Utrecht University EM Square facility; Stal Schep (Tull en het Waal, The Netherlands) for providing horse semen; M. W. Haaker and M. Houweling for providing mouse reproductive tracts; Prof. F. Förster and Prof. A. Akhmanova for critical reading of the manuscript; and Prof. E. Y. Jones for insightful discussions. This work benefitted from access to the Netherlands Center for Electron Nanoscopy (NeCEN) with support from operators Dr. R. S. Dillard and Dr. C. Diebolder and IT support from B. Alewijnse. This work was funded by NWO Start‐Up Grant 740.018.007 to T. Z., and M.R.L. is supported by a Clarendon Fund‐Nuffield Department of Medicine Prize Studentship.
The EMBO Journal (2021) 40: e107410.
Data availability
Subtomogram averages have been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) with the following accession numbers:
EMD‐12067 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12067).
EMD‐12068 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12068).
EMD‐12069 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12069).
EMD‐12070 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12070).
EMD‐12071 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12071).
EMD‐12072 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12072).
EMD‐12076 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12076).
EMD‐12077 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12077).
EMD‐12078 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12078).
EMD‐12079 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12079).
EMD‐12131 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12131).
EMD‐12132 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12132).
EMD‐12133 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12133).
EMD‐12134 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12134).
EMD‐12135 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12135).
EMD‐12136 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12136).
References
- Avidor‐Reiss T (2018) Rapid evolution of sperm produces diverse centriole structures that reveal the most rudimentary structure needed for function. Cells 7: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock DF, Wandernoth PM, Wennemuth G (2014) Episodic rolling and transient attachments create diversity in sperm swimming behavior. BMC Biol. 12: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ (2009) Thinking about flagellar oscillation. Cell Motil Cytoskeleton 66: 425–436 [DOI] [PubMed] [Google Scholar]
- Carbajal‐González BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D (2013) Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton 70: 101–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Dai W, Sun SY, Jonasch D, He CY, Schmid MF, Chiu W, Ludtke SJ (2017) Convolutional neural networks for automated annotation of cellular cryo‐electron tomograms. Nat Methods 14: 983–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W (2014) Volta potential phase plate for in‐focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA 111: 15635–15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW (1970) A comparative view of sperm ultrastructure. Biol Reprod Suppl 2: 90–127 [PubMed] [Google Scholar]
- Fawcett DW (1975) The mammalian spermatozoon. Dev Biol 44: 394–436 [DOI] [PubMed] [Google Scholar]
- Fishman EL, Jo K, Nguyen QPH, Kong D, Royfman R, Cekic AR, Khanal S, Miller AL, Simerly C, Schatten G et al (2018) A novel atypical sperm centriole is functional during human fertilization. Nat Commun 9: 2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich BM, Riedel‐Kruse IH, Howard J, Jülicher F (2010) High‐precision tracking of sperm swimming fine structure provides strong test of resistive force theory. J Exp Biol 213: 1226–1234 [DOI] [PubMed] [Google Scholar]
- Fu G, Zhao L, Dymek E, Hou Y, Song K, Phan N, Shang Z, Smith EF, Witman GB, Nicastro D (2019) Structural organization of the C1a‐e‐c supercomplex within the ciliary central apparatus. J Cell Biol 218: 4236–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadêlha H, Gaffney EA (2019) Flagellar ultrastructure suppresses buckling instabilities and enables mammalian sperm navigation in high‐viscosity media. J R Soc Interface 16: 20180668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadêlha H, Hernández‐Herrera P, Montoya F, Darszon A, Corkidi G (2020) Human sperm uses asymmetric and anisotropic flagellar controls to regulate swimming symmetry and cell steering. Sci Adv 6: eaba5168 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gaffney EA, Gadêlha H, Smith DJ, Blake JR, Kirkman‐Brown JC (2011) Mammalian Sperm Motility: Observation and Theory. Annu Rev Fluid Mech 43: 501–528 [Google Scholar]
- Gage MJG (2012) Complex sperm evolution. Proc Natl Acad Sci 109: 4341–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE (2018) UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, O’Brien DA, Eddy EM (2010) Speriolin is a novel human and mouse sperm centrosome protein. Hum Reprod 25: 1884–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan GA, Keszthelyi B, Vale RD, Agard DA (2018) Insights into centriole geometry revealed by cryotomography of doublet and triplet centrioles. Elife 7: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan GA, Vale RD, Agard DA (2020) Electron cryotomography of intact motile cilia defines the basal body to axoneme transition. J Cell Biol 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Hachet V, Majubu N, Neves A, Demurtas D, Olieric N, Fluckiger I, Yamada A, Kihara K, Nishida Y et al (2013) Native architecture of the centriole proximal region reveals features underlying its 9‐fold radial symmetry. Curr Biol 23: 1620–1628 [DOI] [PubMed] [Google Scholar]
- Haidl G, Becker A, Henkel R (1991) Poor development of outer dense fibres as a major cause of tail abnormalities in the spermatozoa of asthenoteratozoospermic men. Hum Reprod 6: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Heumann JM, Hoenger A, Mastronarde DN (2011) Clustering and variance maps for cryo‐electron tomography using wedge‐masked differences. J Struct Biol 175: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheon K, McLaughlin EA, Stanger SJ, Bernstein IR, Dun MD, Eamens AL, Nixon B (2017) Analysis of the small non‐protein‐coding RNA profile of mouse spermatozoa reveals specific enrichment of piRNAs within mature spermatozoa. RNA Biol 14: 1776–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Khalifa AAZ, Kubo S, Dai D, Basu K, Maghrebi MAF, Vargas J, Bui KH (2019) Tubulin lattice in cilia is in a stressed form regulated by microtubule inner proteins. Proc Natl Acad Sci 116: 19930–19938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, Bui KH (2017) Subnanometre‐resolution structure of the doublet microtubule reveals new classes of microtubuleassociated proteins. Nat Commun 8: 15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof S, Zhang J, Wang H, Bui KH, Nguyen H, Atanasov I, Hui WH, Yang SK, Zhou ZH, Hill KL (2019) Cryo electron tomography with Volta phase plate reveals novel structural foundations of the 96‐nm axonemal repeat in the pathogen Trypanosoma brucei . Elife 8: 1–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T (2017) Axoneme structure from motile cilia. Cold Spring Harb Perspect Biol 9: a028076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito C, Akutsu H, Yao R, Yoshida K, Yamatoya K, Mutoh T, Makino T, Aoyama K, Ishikawa H, Kunimoto K et al (2019) Odf2 haploinsufficiency causes a new type of decapitated and decaudated spermatozoa, Odf2‐DDS, in mice. Sci Rep 9: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeulin C, Lewin LM, Chevrier C, Schoevaert‐Brossault D (1996) Changes in flagellar movement of rat spermatozoa along the length of the epididymis: manual and computer‐aided image analysis. Cell Motil Cytoskeleton 35: 147–161 [DOI] [PubMed] [Google Scholar]
- Khalifa A, Ichikawa M, Dai D, Kubo S, Black C, Peri K, McAlear TS, Veyron S, Yang SK, Vargas J et al (2020) The inner junction complex of the cilia is an interaction hub that involves tubulin post‐translational modifications. Elife 9: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Scholey JM (2018) Assembly, functions and evolution of archaella, flagella and cilia. Curr Biol 28: R278–R292 [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Kurachi M, Hoshi M, Tashiro H (1995) Buckling of a single microtubule by optical trapping forces: direct measurement of microtubule rigidity. Cell Motil Cytoskeleton 30: 221–228 [DOI] [PubMed] [Google Scholar]
- Le Guennec M, Klena N, Gambarotto D, Laporte MH, Tassin A, van den Hoek H, Erdmann PS, Schaffer M, Kovacik L, Borgers S et al (2020) A helical inner scaffold provides a structural basis for centriole cohesion. Sci Adv 6: eaaz4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB (2008) Mutations in Hydin impair ciliary motility in mice. J Cell Biol 180: 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesich KA, de Pinho TG, Dang L, Lindemann CB (2014) Ultrastructural evidence that motility changes caused by variations in ATP, Mg2+, and ADP correlate to conformational changes in reactivated bull sperm axonemes. Cytoskeleton 71: 649–661 [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez JJ, Marshall WF, Agard DA (2012) Three‐dimensional structure of basal body triplet revealed by electron cryo‐tomography. EMBO J 31: 552–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D (2014b) Structural mechanism of the dynein power stroke. Nat Cell Biol 16: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, Nicastro D (2014a) Cryo‐electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat Commun 5: 5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB (1996) Functional significance of the outer dense fibers of mammalian sperm examined by computer simulations with the geometric clutch model. Cell Motil Cytoskeleton 34: 258–270 [DOI] [PubMed] [Google Scholar]
- Lindemann CB, Lesich KA (2016) Functional anatomy of the mammalian sperm flagellum. Cytoskeleton 73: 652–669 [DOI] [PubMed] [Google Scholar]
- Lindemann CB, Macauley LJ, Lesich KA (2005) The counterbend phenomenon in dynein‐disabled rat sperm flagella and what it reveals about the interdoublet elasticity. Biophys J 89: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüpold S, Pitnick S (2018) Sperm form and function: what do we know about the role of sexual selection? Reproduction 155: R229–243 [DOI] [PubMed] [Google Scholar]
- Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, Zhang R (2019) Structure of the decorated Ciliary doublet microtubule. Cell 179: 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar G, Simerly C, Schatten G (2000) Highly degenerated distal centrioles in rhesus and human spermatozoa. Hum Reprod 15: 256–263 [DOI] [PubMed] [Google Scholar]
- Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G (1998) Centrosome reduction during mouse spermiogenesis. Dev Biol. 203: 424–434 [DOI] [PubMed] [Google Scholar]
- Marko M, Hsieh C, Schalek R, Frank J, Mannella C (2007) Focused‐ion‐beam thinning of frozen‐hydrated biological specimens for cryo‐electron microscopy. Nat Methods 4: 215–217 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005) Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152: 36–51 [DOI] [PubMed] [Google Scholar]
- Mederacke I, Dapito DH, Affò S, Uchinami H, Schwabe RF (2015) High‐yield and high‐purity isolation of hepatic stellate cells from normal and fibrotic mouse livers. Nat Protoc 10: 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR (2017) Evolution of Cilia. Cold Spring Harb Perspect Biol 9: a028290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata H, Satouh Y, Mashiko D, Muto M, Nozawa K, Shiba K, Fujihara Y, Isotani A, Inaba K, Ikawa M (2015) Sperm calcineurin inhibition prevents mouse fertility with implications for male contraceptive. Science 350: 442–445 [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW (2011) Cryo‐electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci 108: E845–E853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948 [DOI] [PubMed] [Google Scholar]
- Ounjai P, Kim KD, Lishko PV, Downing KH (2012) Three‐dimensional structure of the bovine sperm connecting piece revealed by electron cryotomography. Biol Reprod 87: 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owa M, Uchihashi T, Yanagisawa H, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi K, Ando T, Kikkawa M (2019) Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun 10: 1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampaloni F, Lattanzi G, Jonáš A, Surrey T, Frey E, Florin EL (2006) Thermal fluctuations of grafted microtubules provide evidence of a length‐dependent persistence length. Proc. Natl Acad Sci USA 103: 10248–10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Phillips DM (1972) Comparative analysis of mammalian sperm motility. J Cell Biol 53: 561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T (2012) Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol 178: 199–206 [DOI] [PubMed] [Google Scholar]
- Riedel‐Kruse IH, Hilfinger A (2007) How molecular motors shape the flagellar beat. HFSP J 1: 192–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigort A, Bauerlein FJB, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM (2012) Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci 109: 4449–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggiorato G, Alvarez L, Jikeli JF, Kaupp UB, Gompper G, Elgeti J (2017) Human sperm steer with second harmonics of the flagellar beat. Nat Commun 8: 1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin JT, Pazour GJ, Witman GB (2015) Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell 26: 4358–4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer M, Mahamid J, Engel BD, Laugks T, Baumeister W, Plitzko JM (2017) Optimized cryo‐focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J Struct Biol 197: 73–82 [DOI] [PubMed] [Google Scholar]
- Serres C, Feneux D, Jouannet P (1986) Abnormal distribution of the periaxonemal structures in a human sperm flagellar dyskinesia. Cell Motil Cytoskeleton 6: 68–76 [DOI] [PubMed] [Google Scholar]
- Smith DJ, Gaffney EA, Gadelha H, Kapur N, Kirkman‐Brown JC (2009) Bend propagation in the flagella of migrating human sperm, and Its modulation by viscosity. Cell Motil Cytoskeleton 66: 220–236 [DOI] [PubMed] [Google Scholar]
- Taute KM, Pampaloni F, Frey E, Florin EL (2008) Microtubule dynamics depart from the wormlike chain model. Phys Rev Lett 100: 1–4 [DOI] [PubMed] [Google Scholar]
- Tegunov D, Cramer P (2019) Real‐time cryo‐electron microscopy data preprocessing with Warp. Nat Methods 16: 1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ (2008) An improved cryogen for plunge freezing. Microsc Microanal 14: 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Ishikawa T, Bui KH, Gonda K, Ishikawa T, Yamaguchi T (2012) Mouse respiratory cilia with the asymmetric axonemal structure on sparsely distributed ciliary cells can generate overall directional flow. Nanomedicine Nanotechnology. Biol Med 8: 1081–1087 [DOI] [PubMed] [Google Scholar]
- Wan KY (2018) Coordination of eukaryotic cilia and flagella. Essays Biochem. 62: 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Limpens RWAL, Zheng S, Snijder EJ, Agard DA, Koster AJ, Bárcena M (2019) Mind the gap: Micro‐expansion joints drastically decrease the bending of FIB‐milled cryo‐lamellae. J Struct Biol 208: 107389 [DOI] [PubMed] [Google Scholar]
- Woolley DM, Fawcett DW (1973) The degeneration and disappearance of the centrioles during the development of the rat spermatozoon. Anat Rec 177: 289–301 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Oda T, Kikkawa M, Takeda H (2018) Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly. Elife 7: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeo D, Croft JT, Höög JL (2019) Axonemal doublet microtubules can split into two complete singlets in human sperm flagellum tips. FEBS Lett 593: 892–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabeo D, Heumann JM, Schwartz CL, Suzuki‐Shinjo A, Morgan G, Widlund PO, Höög JL (2018) A lumenal interrupted helix in human sperm tail microtubules. Sci Rep 8: 2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng GL (2012) A filtered backprojection algorithm with characteristics of the iterative landweber algorithm. Med Phys 39: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Z, Ping P, Wang G, Yuan X, Sun F (2018) Outer dense fibers stabilize the axoneme to maintain sperm motility. J Cell Mol Med 22: 1755–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: Anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Review Process File
Data Availability Statement
Subtomogram averages have been deposited in the Electron Microscopy Data Bank (https://www.ebi.ac.uk/pdbe/emdb/) with the following accession numbers:
EMD‐12067 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12067).
EMD‐12068 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12068).
EMD‐12069 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12069).
EMD‐12070 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12070).
EMD‐12071 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12071).
EMD‐12072 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12072).
EMD‐12076 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12076).
EMD‐12077 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12077).
EMD‐12078 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12078).
EMD‐12079 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12079).
EMD‐12131 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12131).
EMD‐12132 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12132).
EMD‐12133 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12133).
EMD‐12134 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12134).
EMD‐12135 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12135).
EMD‐12136 (https://www.ebi.ac.uk/pdbe/entry/emdb/EMD‐12136).


