Summary
Viruses have evolved to manipulate host lipid metabolism to benefit their replication cycle. Enveloped viruses, including coronaviruses, use host lipids in various stages of the viral life cycle, particularly in the formation of replication compartments and envelopes. Host lipids are utilised by the virus in receptor binding, viral fusion and entry, as well as viral replication. Association of dyslipidaemia with the pathological development of Covid‐19 raises the possibility that exploitation of host lipid metabolism might have therapeutic benefit against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). In this review, promising host lipid targets are discussed along with potential inhibitors. In addition, specific host lipids are involved in the inflammatory responses due to viral infection, so lipid supplementation represents another potential strategy to counteract the severity of viral infection. Furthermore, switching the lipid metabolism through a ketogenic diet is another potential way of limiting the effects of viral infection. Taken together, restricting the access of host lipids to the virus, either by using lipid inhibitors or supplementation with exogenous lipids, might significantly limit SARS‐CoV‐2 infection and/or severity.
Keywords: Covid‐19, inflammation, lipids inhibitors, lipids supplementation, SARS‐CoV‐2
Abbreviations
- AA
arachidonic acid
- ACAT
Acyl‐CoA cholesterol acyltransferase
- ACC
acetyl CoA carboxylase
- ACE2
angiotensin‐converting enzyme 2
- AM
alveolar macrophage
- ATII
alveolar type II
- CerS
ceramide synthase
- CERT
ceramide transfer protein
- cPLA2α
cytosolic phospholipase A2α enzyme
- DAG
diacylglycerol
- DGAT
diacylglycerol acyltransferase
- DHA
docosahexaenoic acid
- DHET
dihydroxyeicosatrienoic acid
- DMV
double membrane vesicle
- DPPC
dipalmitoyl‐phosphatidylcholine
- EET
epoxyeicosatrienoic acid
- EPA
eicosapentaenoic acid
- FASN
fatty acid synthetase enzyme
- FFA
free fatty acid
- HDL
high‐density lipoprotein
- HE
hemagglutinin‐esterase
- HMGR
3‐hydroxy‐3‐methyl‐glutaryl‐Coenzyme A reductase
- IL‐6
interleukin‐6
- LA
linoleic acid
- LD
lipid droplet
- LDL
low‐density lipoprotein
- LE/L
late endosomes/ lysosomes
- LPA
lysophosphatidic acid
- LPC
lysophosphatidylcholine
- LPCAT
lysophosphatidylcholine acyl transferase
- LPE
lysophosphatidylethanolamine
- LPLs
lysophospholipids
- LPS
lysophosphatidylserine
- LTPs
lipid transfer proteins
- MBCD
methyl‐Β‐cyclodextrin
- MCTs
medium chain triglycerides
- MUFA
monounsaturated fatty acid
- NF‐KB
nuclear factor kappa‐light‐chain‐enhancer of activated B cells
- NPC
Niemann‐Pick C
- OA
oleic acid
- OEA
oleoylethanolamide
- OSBP
oxysterol binding protein
- PA
palmitic acid
- PC
phosphatidyl choline
- PCSK9
proprotein convertase subtilisin/kexin type 9
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PI3P
phosphatidylinositol‐3‐phopshate
- PI4P
phosphatidylinositol‐4‐phopshate
- Pls
phospholipids
- PS
phosphatidylserine
- PUFA
polyunsaturated fatty acid
- S1P1
sphingosine‐1‐phosphate receptor 1
- SCD1
stearoyl‐CoA desaturase 1
- SP
surfactant protein
- SPT
serine palmitoyl transferase
- SREBP
sterol regulatory element‐binding protein
- TG
triglycerides
- TLR
Toll like receptor
- TNF‐α
tumour necrosis factor alpha
- VLDL
very low‐density lipoprotein
1. INTRODUCTION
Coronaviruses including severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) are family of enveloped viruses 1 (Figure 1). The virion production involves major changes in the host cellular lipidome. 2 , 3 Since coronaviruses lack the basic metabolic processes, 4 , 5 they manipulate the host lipid metabolism in various stages of the life cycle. 6 Viral infections modify the host lipid synthesis, transportation and metabolism for its replication and to get the lipids required for the formation of their envelopes and double membrane vesicles (DMV). 5 The lipids are critically required for virus invasion, attachment, fusion and replication. 7 Lipids play crucial role as source of energy and signalling in SARS‐CoV‐2 life cycle. 8 Therefore, we aimed to study the importance of host lipids in SARS‐CoV‐2 infection in order to explore novel therapeutic host lipid targets for the development of effective, broad spectrum and safe antiviral therapies and to avoid the evolved drug resistance by the virus.
FIGURE 1.
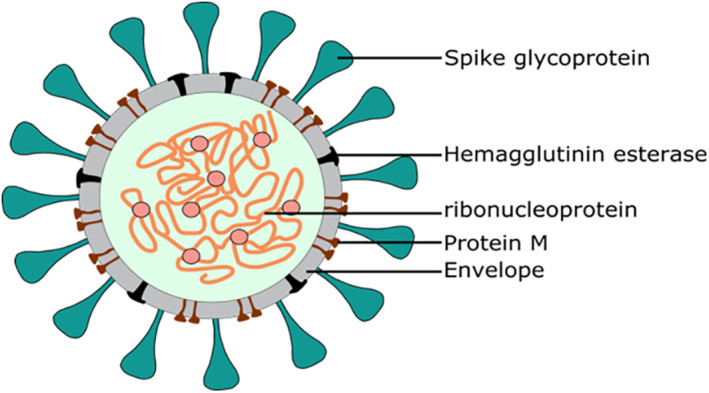
Structural features SARS‐CoV‐2
2. SARS‐CoV‐2 INFECTION ALTERED THE HOST LIPID METABOLISM
A significant elevated lipogenesis has been observed as a sign of SARS‐CoV‐2‐infection. 2 Lipidomics changes associated with Covid‐19 disease give mechanistic insight about the optimal lipid microenvironment required during the viral infection. Shen et al. 9 reported a significant alteration in the serum lipid levels of SARS‐CoV‐2‐infected patients when compared to healthy individuals (Figure 2). They reported a significant decrease in over 100 lipids including sphingolipids, glycerophospholipids and fatty acids in the serum of SARS‐CoV‐2‐infected patients. 9 Similarly, Hu et al. 10 showed that total cholesterol (TC), high‐density lipoprotein (HDL) cholesterol and low‐density lipoprotein (LDL) cholesterol levels were sharply decreased in the serum of infected patient when compared to healthy control. On the other hand, lipidomics analysis of virus‐infected alveolar cells showed significant increase in multiple lipid pathways (Figure 2) and lipid modifying enzymes. 11
FIGURE 2.
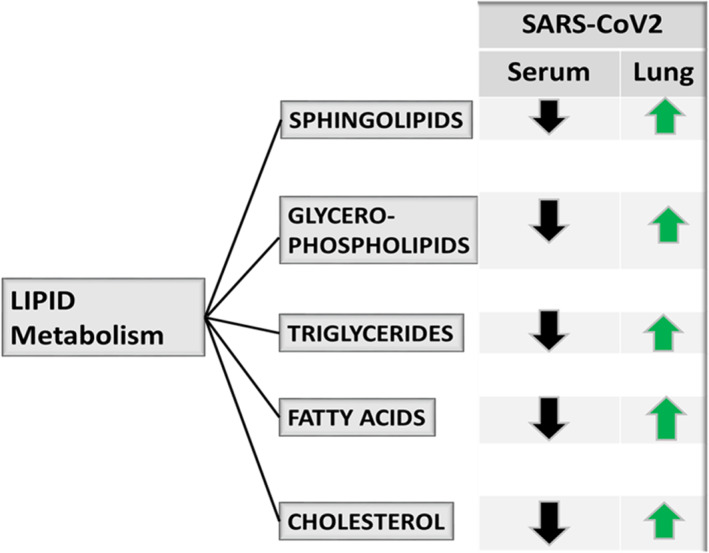
SARS‐CoV‐2 infection altered the host lipid metabolism
3. LIPIDS MIGRATE FROM SERUM TO ALVEOLAR SPACE TO SUPPORT SARS‐CoV‐2 INFECTION
A significant reduction in the serum lipid levels of Covid‐19 patients was observed, which is dramatically associated with the severity of the disease symptoms. 12 The progression in SARS‐CoV2 infection is associated with the leaking of plasma lipids and cholesterol into the alveolar space. 12 However, the signal leading to the transport of lipids from blood to alveolar space is not yet identified. The occupancy of angiotensin‐converting enzyme 2 (ACE2) receptor by the viral glycoprotein may be a major signal, particularly that the increase in lung cholesterol can result in increasing the ACE2 trafficking to the infection site. 13 Although the intra‐alveolar microenvironment is separated from the systemic circulation, it is believed that neutral fat, cholesterol (free and esterified forms) and phospholipids (PLs) are transported in the circulation combined with one another and with lipoproteins as macromolecular complexes. 14 Alveolar type II (ATII) cells have long been known to bind and take up lipoproteins, including HDL, LDL and very LDL (VLDL), and to resecrete PLs and cholesterol, 15 , 16 , 17 which are mostly used up to support the viral infection at the alveolar cells.
The increase in lipid metabolism following viral infection indicated that the virus highjacks the host cells to utilize the host lipids for their own propagation. The aforementioned data indicated the importance of host lipids and their relocalisation to the site of viral infection. Further, viral binding may signal the transportation of lipids from the circulation to the alveolar space. Here, we are proposing that viral binding, entry and replication disrupt the lipids balance at the alveolar space, site of infection, leading to transportation of lipids from the circulation to the alveolar space. In order to explain this mechanism, the importance of host lipids to the virus is highlighted.
4. HOST LIPIDS MANIPULATION DURING VIRUS INVASION
4.1. At the surfactant
Surfactant is the major component within the lung defence system that has unique properties displaying both lung stabilizing and antimicrobial properties. 18 Surfactant is synthesised within the ATII epithelial cells. 19 Surfactant is actively being secreted and its materials are constantly being exchanged and recycled into the ATII cells to maintain constant surfactant pool size, 20 , 21 while some materials can be degraded by alveolar macrophages (AM) to lipids that can support the viral infection (Figure 3). These lipids include palmitic acid (PA), phosphatidyl choline (PC) and cholesterol.
FIGURE 3.
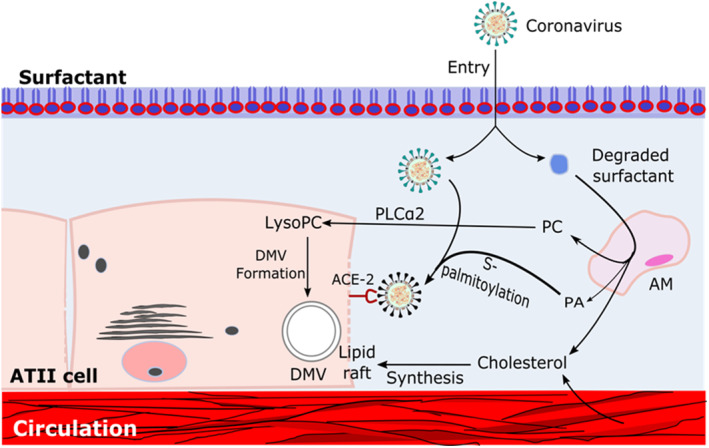
Role of host lipids in viral entry and invasion. ACE‐2; Angiotensin‐converting enzyme 2, AM; Alveolar macrophage, ATII; Alveolar type II cells, DMV; Double membrane vesicle, PLCa2; Cellular phospholipase, PA; Palmitic acid, PC; Phosphatidyl choline
Pulmonary surfactant is a complex mixture of lipids and proteins that forms a film lines the alveolar air‐surface interface and protect the lungs from pathogen invasion. 21 Disruption of surfactant lipids can disturb the surface tension lowering ability and allow the pathogens entry. 22 However, the process by which the virus penetrates the surfactant and reaches the ATII cells is not known. Several studies demonstrate that human viruses including SARS‐CoV‐2 can significantly alter surfactant phospholipids composition and causing loss of surfactant and hence promote the viral penetration. 23 On the other hand, binding of surfactant proteins (SP), which importantly contribute to the surfactant behaviour as a defence system, 24 , 25 to the virus occurs by recognition of hemagglutinin and neuraminidase glycans on the surface of the virus, thereby hindering the ability of the virus to enter the cell. 26 However, the hemagglutinins found on SARS‐CoV‐2 exhibit antigenic variations that resulted in reduced binding, leading to greater virulence and subsequent high mortality and morbidity in patients. 26 SARS‐CoV‐2 hemagglutinin‐esterase (HE) provides classical glycan‐binding lectin activity, while exhibiting hemagglutination and destruction of the surfactant proteins 27 (Figure 1).
4.2. At the alveolar cell
4.2.1. Host lipids potentiate the viral attachment and entry to ATII cells
Viral attachment to the host cell receptor is lipid‐dependent. 28 The entry of SARS‐CoV‐2 is mediated by the binding of viral spike (S) protein to ACE2 receptor, which is localised in cholesterol‐rich microdomains within the lipid rafts 29 (Figure 3). Lipid rafts are regions of the plasma membrane characterised by high concentrations of sphingolipids (sphingomyelin and sphingoglycolipids) and cholesterol. 30 Therefore, cholesterol increases the expression of ACE2 receptor and hence facilitates the interaction between the S protein and ACE2 receptor. 28 S‐palmitoylation is a unique protein lipidation process essential for viral invasion. 31 S‐palmitoylation of SARS‐CoV‐2 S protein has been reported to facilitate its anchor and fusion with the host cellular membrane receptor (Figure 3). Furthermore, it has been reported that the HDL scavenger receptor B type 1 (SR‐B1) facilitates ACE2‐dependent uptake of SARS‐CoV‐2 by augmenting the virus attachment to ATII cells. 32 Viral internalisation process occurs by the fusion of the envelope lipid with the plasma membrane, which is mediated by endocytosis. Inhibition of viral endocytosis by cholesterol depletion indicates the importance of raft‐mediated endocytosis for the viral entry. 33
4.2.2. Virus hijacks the host lipids for viral replication and envelope formation
4.2.2.1. Lipids manipulation for the formation of DMVs and envelopes
Coronaviruses hijack the host cells for the formation of replication and transcription complex, the double membrane vesicles (DMVs). 34 , 35 Formation of DMVs is a key factor for viral replication that provides a favourable barrier to protect the viral replication compartments from the host innate immune responses. 36 DMVs formation requires specific lipid compositions, the unsaturated lysophospholipids (Figure 3). 37 Coronaviruses manipulate the host cellular pathways particularly the lipid metabolism and lipid trafficking to ensure the availability of required lipids for DMVs formation (Figure 3). 38
DMVs biogenesis is a complex process that critically requires modification of lipid composition either via lipid transfer proteins sterol regulatory element‐binding proteins (SREBP) or lipid biosynthesis via cellular cytosolic phospholipase A2α enzyme (cPLA2α). 39 SREBPs play essential role in the formation of DMVs, viral protein palmitoylation and viral replication. 40 cPLA2α is a crucial lipid processing enzyme that plays a critical role in the formation of DMVs (Figure 3). 41 Inhibition of SREBPs and cPLA2α can be employed as potential strategy to eradicate coronavirus. Furthermore, viruses stimulate the synthesis of lipid components including in particular lysophospholipids (LPLs) to support the rapid DMVs biogenesis. Therefore, inhibition of LPL synthesis resulted in loss of DMVs and dramatic reduction in viral replication (Figures 3 and 4). 42
FIGURE 4.
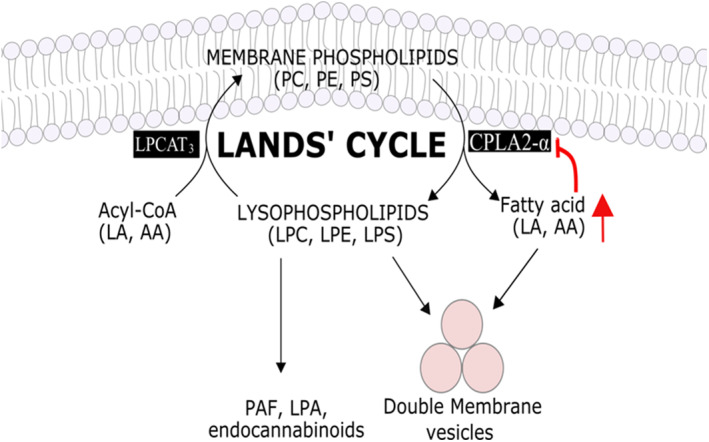
Land’s cycle showing the importance of cPLA2‐α, LA and AA in the formation of lysophospholipids, critically important in virus DMV and envelope formation. Red arrows indicated the negative feedback mechanism following exogenous supplementation of AA
Viruses access the energy required for their replication and assembly via lipid droplets. Lipid droplet serve as storage of neutral lipid including triglycerides and cholesterol esters, which can be employed for energy production, signalling and cell immune response. 43 Monocytes from SARS‐CoV‐2‐infected patients showed accumulated level of triglycerides and cholesterol ester with increased level of diacylglycerol acyltransferase (DGAT) enzyme required for TGs synthesis. 44 This indicates the utilisation of lipid stores from the lipid droplets during viral infection. Utilisation of lipid stores serve to provide energy and substrates for the virus replication and to dampen the host antiviral responses. 45
Viral envelopes are made of lipid bilayer, which can supply the required protection from environmental stresses including the effect of host defensive systems. SARS‐CoV‐2 lipid bilayer is rich in fatty acids such as linoleic acid (LA), arachidonic acid (AA), PA and oleic acid (OA), while LA and AA play protective role during the virus entry. 46 Furthermore, several studies confirmed that enveloped viruses including coronaviruses are developed or bud from the lipid rafts. 30 , 47
4.2.2.2. Virus enhances the intracellular transportation of host lipids
A quick process for the intracellular lipids transportation is required to support the process of viral replication, which is achieved by nonvesicular lipid transfer proteins (LTPs). Viruses hijack the LPTs to mobilize the lipids required for the formation of replication organelles. Cholesterol is transported by oxysterol binding protein (OSBP), 48 while ceramide transportation occurs by ceramide transfer protein (CERT). 49 Both are significantly required in viral replication. Furthermore, Niemann‐pick C (NPC) is required to carry and transfer the free and recycled cholesterol to the site of replication.
4.2.2.3. Virus manipulates the intracellular host lipid biosynthesis
Biosynthesis of fatty acids is initiated by the carboxylation of citrate with acetyl CoA carboxylase to malonyl‐CoA, which is then converted to PA by FASN (Figure 5 ). PA is then elongated and converted to diacylglycerols and then TAGs by DGAT1. PA serves as precursor for the biosynthesis of phospholipids and sphingolipids. 45 Furthermore, PA plays important role in the palmitoylation of viral protein (Figure 3). Cholesterol biosynthesis is catalysed by HMGR from acetyl CoA. 50 Cholesterol is then esterified by acyl‐CoA cholesterol acyltransferase and employed in lipid droplets formation. Viral infection induced the lipids hydrolysis to yield necessary lipid metabolites such as lysophospholipid and FFAs that are required for viral infection (Figure 3, 4, 5 ).
FIGURE 5.
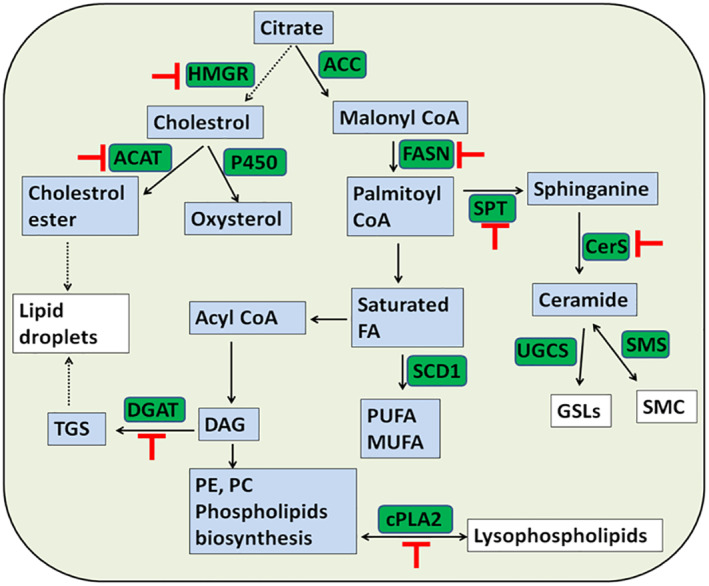
Intracellular host lipids metabolism. Important lipids to the virus are indicated in white color, critical biosynthetic enzymes are indicated in green color, and their potential inhibition is indicated in red color
5. HOST LIPIDS AS THERAPEUTIC TARGETS FOR THE DEVELOPMENT OF ANTIVIRAL DRUGS
The significant changes of host lipidomics due to viral infection and the importance of host lipids to the viral pathogenicity indicated that targeting the host lipids can offer excellent potential to counteract the SARS‐CoV‐2 infection. The aforementioned data indicated that host lipid biosynthesis, metabolism, transfer and lipid modifying enzymes can serve as excellent targets for the development of anti‐SARS‐CoV‐2 drugs. Host‐targeting strategies can avoid the impact of antiviral drug resistance when compared to viral‐targeting strategies. Careful selection of a target with less side effects on the host, while providing broad‐spectrum antiviral activity can serve as excellent opportunity to overcome this pandemic life‐threatening disease. Here, we have identified most promising host lipid targets and their potential inhibitors (Figures 6 and 7 and Tables 1 and 2).
FIGURE 6.
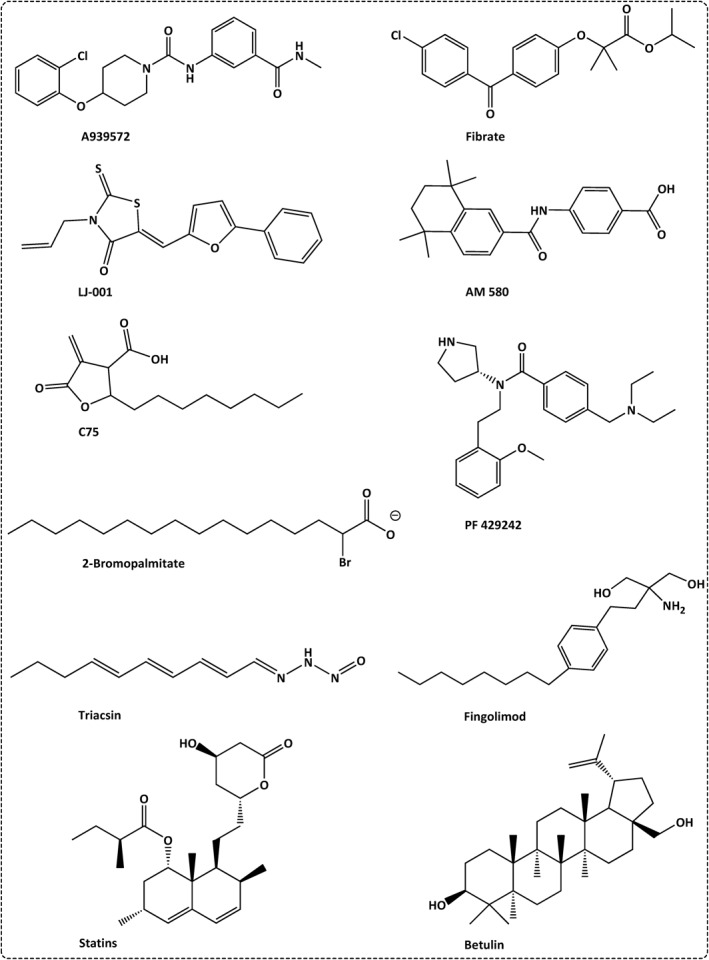
Identified inhibitors that can be employed to target the host lipids, utilized in the invasion and pathogenesis of SARS‐CoV‐2
FIGURE 7.
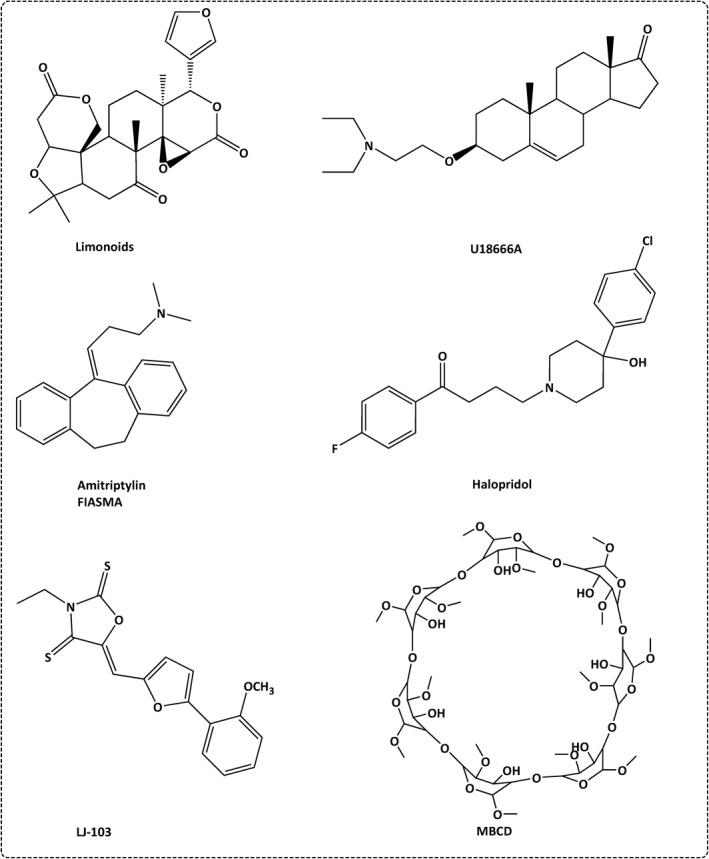
Lipids inhibitors that interfere with SARS‐CoV‐2 entry
TABLE 1.
Host lipids as targets to inhibit SARS‐CoV‐2 infection
| Host lipid | Effect of viral infection | Role in viral pathogenesis | Strategy of lipid target therapy | Inhibitor | FDA‐approved drugs |
|---|---|---|---|---|---|
| Lipid raft (cholesterol and sphingolipids) | Increased to mediate the viral
|
|
|
|
|
| Sphingomyelin |
|
|
|
|
|
| Cholesterol regulator |
|
|
|
|
|
| Palmitic acid (S‐palmitoylation) |
|
|
|
|
|
| LPLs |
|
|
|
|
|
| PI3P |
|
|
|
|
|
| Fatty acid |
|
|
|
|
|
| TGs |
|
|
|
|
|
| Unsaturated fatty acid |
|
|
|
|
|
| Lipid droplet |
|
|
|
|
|
| Sterol |
|
|
|
|
|
| Sphingolipid |
|
|
|
|
|
| Lipid transfer |
|
|
|
|
|
TABLE 2.
Supplementation of bioactive lipids to inhibit SARS‐CoV‐2 infection
| Lipid | Role in Covid‐19 | Mechanism | Example |
|---|---|---|---|
| Phytosterol |
|
|
|
| Elovanoids |
|
|
|
| Bioactive lipids (PUFAs) including LA, AA, DHA and EPA |
|
|
|
5.1. Targeting the host lipids, employed during viral invasion
5.1.1. Targeting the host lipid raft and viral fusion
Cholesterol‐lowering agents can inhibit the optimum lipid microenvironment required for viral infection (Figure 6 and Table 1). Methyl‐β‐cyclodextrin interacts with the lipid raft via its lipophilic core, and hence competes with the viral binding site 51 (Figure 7 and Table 1). On the other hand, statins inhibit HMGR enzyme, the rate‐limiting enzyme in cholesterol biosynthesis and hence reduce the available cholesterol. 28 , 52 Consequently, it lowers the expression of membrane ACE2 receptors and blocks the viral entry. 53
Plant phytosterols such as betulinic acid are lipophilic compounds with cholesterol‐like structures, which can interact with the lipid rafts, decrease the membrane cholesterol and hence can inhibit the viral attachment to the host cell 51 , 54 (Table 2). Interestingly, phytosterols are synthesised naturally with high levels in oilseed plants such as rapeseed, corn and wheat, and nuts such as pine nuts and pistachios. 55
5.1.2. Lipid hinders the viral entry
Elovanoids are polyunsaturated fatty acids (PUFA) with prohomeostatic lipid mediator activity (Figure 6 and Table 2). Elovanoids downregulate the expression of ACE2 and enhance the expression of a set of protective proteins including acid sphingomyelinase that hinder the viral binding to ACE2 receptor. 56 Viral entry is associated with the activation of acid sphingomyelinase and creation of ceramide‐rich patches on the plasma membrane. Therefore, inhibition of acid sphingomyelinase can block SARS‐CoV‐2 infection. 57
5.1.3. Targeting the S‐palmitoylation process
Since ZDHHC5 S‐palmitoyltransferase is important for viral S protein attachment. It can be a potential therapeutic target for the inhibition of SARS‐CoV‐2 infection. 31 2‐Bromopalmitate was confirmed as a candidate compound to inhibit S‐palmitoylation 58 (Figure 6 and Table 1).
5.1.4. Targeting viral endocytosis
Endocytosis is a pH‐dependent process, which is facilitated by cysteine proteases, such as cathepsin B or L, allowing the release of viral nucleocapsid into the cytoplasm. Chloroquine neutralizes the endosome‐lysosomal acidic pH and thus blocks the protease activity and viral internalisation. 50 On the other hand, PVS34 perturbs the structure of viral membrane (Figure 6 and Table 1). PVS34 inhibits the phosphoinositide (PI) 3‐kinase centres, which functions in autophagy, and endocytosis. VPS34 inhibits the required membranes components needed for the formation of SARS‐CoV‐2 particles. 59
5.1.5. Targeting the lipid droplets
Triacsin C blocks the lipid droplet formation through the inhibition of triglycerides‐modulating enzymes particularly the long chain fatty acyl CoA synthetase. 60 A922500 is a potent DGAT inhibitor that can inhibit the lipid droplet formation and hence can stop the production of infectious progeny from SARS‐CoV‐2‐infected cells 60 (Figure 6 and Table 1).
5.1.6. Targeting the viral DMVs formation
AM580, a stable retinobenzoic acid derivative, exhibits potent antiviral activity by blocking the activation of SREBP pathway and hence inhibits DMVs formation 4 (Figure 6 and Table 1). Betulin inhibits SREBP cleavage and maturation. 61 cPLA2α inhibitor such as pyrrolidine‐2 significantly decreases the formation of DMVs 41 (Figure 6 and Table 1). Phosphatidylinositol‐4‐phopshate is a potent membrane modifiers that can interfere with the formation of DMVs. 42
5.1.7. Targeting the intracellular lipid transportation
OSBP inhibitors such as TTP‐8307, OSW‐1 and itraconazole are potent antiviral drugs. 48 HPA‐12 and limonoids are CERT 62 and sphingomyelin synthesis inhibitors, 63 respectively (Figure 7 and Table 1). Inhibition or loss of NPC can block cholesterol transport and hence impairs the infectivity of SARS‐CoV‐2. U18666A is NPC inhibitor that blocks the movement of cholesterol out of lysosomes and hence impairs the viral pathogenicity. 29 Haloperidol is an anti‐SARS‐CoV‐2 candidate, since it hinders the viral entry and replication by inhibiting cholesterol trafficking from the late endosomes/lysosomes (LE/L) 29 (Figure 7 and Table 1).
5.2. Targeting the host lipid biosynthetic pathways
Lipidomics profile of coronavirus‐infected patients showed significant increase in fatty acid synthesis with the upregulation of AA, LA (the metabolic precursor of AA), PA and OA metabolism, in addition to significant elevated levels of lysophospholipids (LPC, LPE). Both, LA and AA play important role in the regulation of cPLA2α, and hence the level of lysophospholipids that are required in viral envelope and DMV formation (Figure 4). Therefore, exogenous supplementation of LA and AA can suppress viral infection via negative feedback inhibition mechanism of cPLA2α, thus reducing the production of lysophospholipids (Figure 4). 64 Coronavirus infection is associated with the selective upregulation of key lipid‐modifying enzyme, cPLA2α and the high production of glycerophospholipids, LPL, fatty acids 41 , 64 and the long‐chain PUFA. 65 Lysophospholipids is required by SARS‐CoV for the optimal formation of replicative organelles (Figure 4). 66 LPL is a crucial component required in the formation of DMVs. 41 cPLA2 inhibition can significantly suppress the formation of virus progeny. 41 Plant‐based natural supplements could be a good source of LA, which accounts for more than 50% of the lipid content in plant seed oil such as nuts. 67
SARS‐CoV‐2‐infection is associated with an increase in the levels of lipid‐modifying enzymes including the fatty acid synthetase enzyme (FASN), 45 and HMGR, the rate‐limiting enzyme in cholesterol synthesis and the activation of transcriptional factor SERBP, which regulates the synthesis of cholesterol. 68 SARS‐CoV‐2 infection also caused an increase in the production of inositol (PI), ceramide sphingolipid, phosphatidyl choline, lysophosphatidyl inositol and lysophatidylcholine. 69 , 70 Collectively, disruption of the aforementioned lipid microenvironments can significantly inhibit SARS‐CoV‐2 infection.
Lipid‐lowering agents such as statins can inhibit HMGR, and hence inhibit cholesterol biosynthesis, leading to disturbance in lipid raft formation, inhibition of viral replication and immunomodulation activity. 71 Fibrates are triglyceride‐lowering agents that target the fatty acid synthesis and showed potent antiviral activity (Figure 6 and Table 1). Annexin A2 is a natural inhibitor of PCSK9, a cholesterol homeostasis regulator, which was recommended with statin in the treatment of Covid‐19 patients. 72
Viral nsp3 activates fatty acid synthesis. 73 Therefore, the use of FASN inhibitors such as the natural product cerulenin and the synthetic inhibitor C75 can be employed as potential anti‐SARS‐CoV‐2 drugs (Figure 6 and Table 1). 74 Stearoyl‐CoA desaturase1 (SCD1) is the rate‐limiting step in MUFA and PUFA biosynthesis. The piperazine derivative A939572 is SCD1 inhibitor, suggesting its potential antiviral activity (Figure 6 and Table 1). 75 Fingolimod (Figure 6) is a sphingomimetics drug employed in clinical trial for the treatment of Covid‐19 due to its selective inhibition activity of S1P1 (sphingosine‐1‐phosphate‐1) and its immunosuppressive activity in the late severe infection via the inhibition of TLR‐mediated immune response (Table 1). 50
5.3. Targeting the viral lipid to inhibit SARS‐CoV‐2 fusion and invasion
LJ001 is a membrane‐binding compound that is selectively targets the viral unsaturated phospholipids and inhibits viral entry. 50 Unfortunately, LJ001 shows poor physiological stability, and hence used as lead compound for the development of more effective and stable antiviral drugs such as LJ103 (Figure 7).
6. LIPIDS SUPPLEMENTATION AS INHIBITORY MECHANISM TO SARS‐CoV‐2
PUFAs including docosahexaenoic acids (AA, DHA, omega‐3 fatty acid), and eicosapentaenoic acid were consistently upregulated in cells infected with the virus. 79 It has been shown that PUFAs can inactivate SARS‐CoV‐2 by blocking the viral proliferation and by inducing the leakage and lysis of viral envelope. 80 Therefore, PUFAs supplementation can help in reducing the susceptibility of SARS‐CoV‐2 infection. Eicosanoids are proinflammatory mediators and signalling molecules that elevated in SARS‐CoV‐2 infected cells (Table 2). 81 Further, AA is an endogenous antiviral compound released by the immune cells in response to viral infection in order to inactivate SARS‐CoV2. 82 Therefore, its exogenous supplementation can provide an inhibition activity to SARS‐CoV‐2 (Table 2).
It has been shown that LA can tightly bind with the three composite pockets present on S protein, forming a complex that interrupts the binding with ACE2 receptor. 84 Therefore, LA intake, alone or more likely when synergised with remdesivir, can lead to efficient suppression of SARS‐CoV‐2 replication. 84 LA is an essential “omega‐6” fatty acid that must be obtained from the diet, since it cannot be synthesised in the human body (Table 2). Therefore, the proper intake of these PUFAs can result in significant reduction in the viral loads and hence can decrease the morbidity and mortality associated with SARS‐CoV‐2 infection. 82
Glycerophospholipids are essential structural and functional components of the cellular membrane. Cellular phospholipase, cPLA2α, releases lysophospholipids and free fatty acids from glycerophospholipids‐based membranes. 41 The released small bioactive lipid molecules (LA and AA) are required together with the produced lysophospholipids to form the specialised DMVs, required in viral replication (Figure 4). 41 It has been reported that inhibition of cPLA2α resulted in a significant reduction in lysophospholipids, DMV formation and viral replication in infected cells. 41
Yan et al. 64 reported that lysophospholipids and FAs downstream of cPLA2 activation are upregulated following the HCoV‐229E infection. Therefore, the upregulation of these lipid species including LA and AA were believed to promote the efficient coronavirus replication. 64 However, they have noticed a significant reduction in virus replication when they exogenously supplemented to HCoV‐229E‐ or MERS‐CoV‐infected cells 64 (Figure 4). This indicated that exogenous supplementation of these FAs can disrupt the equilibrium between membrane phospholipids and lysophospholipids and hence interfere with the optimal replication of the virus. In addition, supplementation of LA and AA can disturb the LA–AA metabolism and can result in feedback reversion of lysophospholipids to phospholipids through Lands' cycle (Figure 4). 85
6.1. Plant sources of essential fatty acids (LA and AA)
LA (C18:3) and AA (C20:4) are among the essential fatty acids that must be provided in human's diets. 86 Plants have been recommended as source of these essential fatty acids. Higher concentration of AA was reported in Sonchus oleraceus (Sow‐thistle), Chenopodium album (goosefoot) and Parietaria diffusa (pellitory‐of‐the‐wall). 87 They all are edible wild plants that have been proposed for human consumption in the Mediterranean region. 87 , 88 On the other hand, plants rich in LA can include Nigella sativa, 89 Artemisia species 90 and Brassica napus (Rapeseed). However, the fatty acids contents in plants can be affected by environmental conditions including drought, 91 temperature 92 and salinity. 92 Therefore, it is important to assess the precise amounts of fatty acids from different regions in order to administer the right dose at the right condition.
6.2. Switching the host metabolism by lipids supplementation
Switching the host metabolism from carbohydrate‐dependent glycolytic state to fat‐dependent ketogenic state can significantly affect the viral replication. 93 Supplementation of medium‐chain triglycerides such as lauric acid can result in significant reduction in viral envelope formation. Therefore, it can be employed as prophylactic strategy for normal people and adjunct therapy in case of infected individuals. 93
6.3. Surfactant‐based therapeutics supplementation
SARS‐CoV‐2 infection is associated with depleted surfactant. Therefore, it has been suggested that supplementation with synthetic surfactant, KL4 (a mix of DPPC, palmitoyl‐oleoyl phosphatidylglycerol, PA and 21‐amino acid synthetic peptide) can play a protective role in Covid‐19 pandemic. 94
7. CRITICAL ROLE OF LIPID SUPPLEMENTATION IN RESOLVING THE INFLAMMATION CAUSED BY SARS‐CoV‐2
The pathogenicity of SARS‐CoV‐2 is associated with excessive inflammation, oxidative stress and release of cytokines. Imbalance between proinflammatory, anti‐inflammatory eicosanoids is initiated in Covid‐19 disease. 95
7.1. Autacoids (eicosanoids)
AA‐derived autacoids or eicosanoids are inflammatory lipid mediators. 81 AA is metabolised by epoxygenase to epoxyeicosatrienoic acids (EET). EET is inflammatory regulator that reduces the stress‐based cytokines release. 81 However, EET is mainly converted to dihydroxyeicosatrienoic acids by the soluble epoxide hydrolase (sEH), and hence increases the release of cytokines. Therefore, inhibition of sEH can increase the EET level, and hence can suppress the release of IL‐6 and activation of NF‐kB 81 (Table 3). EET can shift the AA metabolism to proresolving lipid mediators such as lipoxin, resolvin, protectins and meostasis. Proresolving lipids can restore the normal balance and protect the lung. Lipoxin controls the inflammation without imparting immunosuppression activity. 96 Omega‐3 fatty acids are rich in EET that possess anti‐inflammatory activity (Table 3). 81
TABLE 3.
Role of lipids in inflammation due to SARS‐CoV‐2 infection and their anti‐inflammatory activity
| Lipid | Function | Treatment strategy |
|---|---|---|
| Epoxyeicosatrienoic acid (EET) | Shifts the AA metabolism towards the formation of protective pro‐solving lipid mediators. | Inhibition of sEH will increase the ETT level and subsequently decreases the IL‐6 level and NF‐Kb activation |
| Oleoylethanolamide (OEA) |
|
Exogenous administration of OEA |
7.2. Cannabinoids
Oleoylethanolamide (OEA) is a cannabinoid derived from oleic acid. It has potential removal activity of respiratory pathogen and can attenuate the inflammatory responses due to SARS‐CoV‐2 infection (Table 3). 97 Further, OEA can decrease TLR expression; thus reducing IL‐6 and TNF‐α during SARS‐CoV‐2 infection. TLR can initiate the proinflammatory cytokines release.
8. CONCLUSION
In summary, SARS‐CoV‐2 infection is associated with host lipids migration from the circulation to the alveolar space in order to support the viral invasion and pathogenicity. In response, host lipids biosynthesis and release are enhanced to cover the viral needs. Furthermore, lipids such as AA and LA are released to limit the inflammation at the initiation of infection. Therefore, restricting the viral access to the host lipids either by applying specific inhibitors or supplementation with specific lipids can be of great importance to limit the viral infection and severity of disease.
CONFLICT OF INTERESTS
The authors declared that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Eman Humaid Alketbi helped in writing the lipid supplementation and data interpretation. Rania Hamdy helped in collecting the data, designing and writing the first draft in addition to data interpretation. Abdalla El‐Kabalawy helped in drawings and analysis of the data. Viktorija Juric and Marc Pignitter helped in collecting data regarding host lipid inhibitors. Kareem Mosa helped in writing and analysing the lipid supplementation as therapy. Ahmed M. Almehdi helped in supervising and interpreting the data. Ali A. El‐Keblawy helped in writing the lipid supplementation, environmental factors and analysing the data. Sameh S. M. Soliman developed the idea, collected the data, designed the manuscript, wrote the first and final drafts, supervised the process of writing and analysis and analysed and interpreted the manuscript.
ACKNOWLEDGEMENTS
The authors acknowledge the generous support from Sandooq Al‐Watan and University of Sharjah to Sameh S. M. Soliman.
Alketbi EH, Hamdy R, El‐Kabalawy A, et al. Lipid‐based therapies against SARS‐CoV‐2 infection. Rev Med Virol. 2021;31(5):e2214. doi: 10.1002/rmv.2214
Eman Humaid Alketbi and Rania Hamdy contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available within this article.
REFERENCES
- 1. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet, 2020;395:470‐473. 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gualdoni GA, Mayer KA, Kapsch A‐M, et al. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc Natl Acad Sci USA. 2018;115:E7158‐E7165. 10.1073/pnas.1800525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagy PD, Strating JRPM, van Kuppeveld FJM. Building viral replication organelles: close encounters of the membrane types. PLoS Pathog. 2016;12:e1005912. 10.1371/journal.ppat.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yuan S, Chu H, Chan JF‐W, et al. SREBP‐dependent lipidomic reprogramming as a broad‐spectrum antiviral target. Nat Commun. 2019;10:120. 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murillo A, Vera‐Estrella R, Barkla BJ, Méndez E, Arias CF. Identification of host cell factors associated with astrovirus replication in caco‐2 cells. J Virol. 2015;89:10359‐10370. 10.1128/JVI.01225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ketter E, Randall G. Virus impact on lipids and membranes. Ann Rev Virol. 2019;6:319‐340. 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- 7. Lorizate M, Kräusslich H‐G. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011;3:a004820. 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernández‐Oliva A, Ortega‐González P, Risco C. Targeting host lipid flows: exploring new antiviral and antibiotic strategies. Cell Microbiol. 2019;21:e12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID‐19 patient sera. Cell. 2020;182:59‐72. 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu X, Chen D, Wu L, He G, Ye W. Low serum cholesterol level among patients with COVID‐19 infection in Wenzhou. China. Lancet. 2020. 10.2139/ssrn.3544826. [DOI] [Google Scholar]
- 11. Nguyen A, Guedán A, Mousnier A, et al. Host lipidome analysis during rhinovirus replication in HBECs identifies potential therapeutic targets. J Lip Res. 2018;59:1671‐1684. 10.1194/jlr.M085910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei X, Zeng W, Su J, et al. Hypolipidemia is associated with the severity of COVID‐19. J Clin Lipidol. 2020;14:297‐304. 10.1016/j.jacl.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. 10.1016/S0140-6736(20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyd GS, Oliver MF. The circulating lipids and lipoproteins in coronary artery disease. Postgraduate Med J, 1957;32:2‐26. 10.1136/pgmj.33.375.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guthmann F, Harrach‐Ruprecht B, Looman AC, Stevens PA, Robenek H, Rüstow B. Interaction of lipoproteins with type II pneumocytes in vitro: morphological studies, uptake kinetics and secretion rate of cholesterol. Eur J Cell Biol, 1997;74:197‐207. [PubMed] [Google Scholar]
- 16. Mallampalli RK, Salome RG, Bowen SL, Chappell DA. Very low density lipoproteins stimulate surfactant lipid synthesis in vitro. J Clin Investig, 1997;99:2020‐2029. 10.1172/jci119370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Voyno‐Yasenetskaya TA, Dobbs LG, Erickson SK, Hamilton RL. Low density lipoprotein‐ and high density lipoprotein‐mediated signal transduction and exocytosis in alveolar type II cells. Proc Natl Acad Sci USA, 1993;90:4256‐4260. 10.1073/pnas.90.9.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glasser JR, Mallampalli RK. Surfactant and its role in the pathobiology of pulmonary infection. Microbes Infect, 2012;14:17‐25. 10.1016/j.micinf.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barreira ER, Precioso AR, Bousso A. Pulmonary surfactant in respiratory syncytial virus bronchiolitis: the role in pathogenesis and clinical implications. Pediatr Pulmonol. 2011;46:415‐420. 10.1002/ppul.21395. [DOI] [PubMed] [Google Scholar]
- 20. Goerke J. Pulmonary surfactant: functions and molecular composition. Biochimica Biophysica Acta, 1998;1408:79‐89. 10.1016/S0925-4439(98. [DOI] [PubMed] [Google Scholar]
- 21. Rooney SA, Young SL, Mendelson C. Molecular and cellular processing of lung surfactant 1. FASEB J. 1994;8:957‐967. [DOI] [PubMed] [Google Scholar]
- 22. Markart P, Ruppert C, Wygrecka M, et al. Patients with ARDS show improvement but not normalisation of alveolar surface activity with surfactant treatment: putative role of neutral lipids. Thorax. 2007;62:588‐594. 10.1136/thx.2006.062398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miakotina OL, McCoy DM, Shi L, Look DC, Mallampalli RK. Human adenovirus modulates surfactant phospholipid trafficking. Traffic. 2007;8:1765‐1777. 10.1111/j.1600-0854.2007.00641.x. [DOI] [PubMed] [Google Scholar]
- 24. Mulugeta S, Beers M. Surfactant protein C: its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006;8(8):2317‐2323. [DOI] [PubMed] [Google Scholar]
- 25. Maina JN, West JB, Orgeig S, et al. Recent advances into understanding some aspects of the structure and function of mammalian and avian lungs. Pysiol Biochem Zool. 2010;83:792‐807. 10.1086/652244. [DOI] [PubMed] [Google Scholar]
- 26. Qi L, Kash JC, Dugan VG, et al. The ability of pandemic influenza virus hemagglutinins to induce lower respiratory pathology is associated with decreased surfactant protein D binding. Virology. 2011;412:426‐434. 10.1016/j.virol.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim C‐H. SARS‐CoV‐2 evolutionary adaptation toward host entry and recognition of receptor O‐acetyl sialylation in virus‐host interaction. Int J Mol Sci. 2020;21:4549. 10.3390/ijms21124549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo H, Huang M, Yuan Q, et al. The important role of lipid raft‐mediated attachment in the infection of cultured cells by coronavirus infectious bronchitis virus beaudette strain. PLoS One. 2017;12:e0170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sturley S, Rajakumar T, Hammond N, et al. Potential COVID‐19 therapeutics from a rare disease: weaponizing lipid dysregulation to combat viral infectivity. J Lipid Res. 2020;61(7):972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lenard J. Viral membranes. Encyclop Virol. 2008:308‐314. 10.1016/B978-012374410-4.00530-6. [DOI] [Google Scholar]
- 31. Santos‐Beneit F, Raškevičius V, Skeberdis VA, Bordel S. A metabolic modeling approach reveals promising therapeutic targets and antiviral drugs to combat COVID‐19. Research Square. 2020;1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wei C, Wan L, Yan Q, et al. HDL‐scavenger receptor B type 1 facilitates SARS‐CoV‐2 entry. Nature Metab. 2020;2:1391‐1400. 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 33. Yang N, Shen H‐M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID‐19. Int J Biol Sci. 2020;16:1724‐1731. 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Snijder EJ, Van Der Meer Y, Zevenhoven‐Dobbe J, et al. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927‐5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Al‐Mulla HM, Turrell L, Smith NM, et al. Competitive fitness in coronaviruses is not correlated with size or number of double‐membrane vesicles under reduced‐temperature growth conditions. MBio. 2014;5:e01107‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Den Boon JA, Ahlquist P. Organelle‐like membrane compartmentalization of positive‐strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241‐256. [DOI] [PubMed] [Google Scholar]
- 37. Strating JR, van Kuppeveld FJ. Viral rewiring of cellular lipid metabolism to create membranous replication compartments. Curr Opin Cell Biol. 2017;47:24‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Z, He G, Filipowicz NA, et al. Host lipids in positive‐strand RNA virus genome replication. Front Microbiol. 2019;10:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang J, Lan Y, Sanyal S. Membrane heist: coronavirus host membrane remodeling during replication. Biochimie. 2020;179:229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Investig. 2002;109:1125‐1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Müller C, Hardt M, Schwudke D, Neuman BW, Pleschka S, Ziebuhr J. Inhibition of cytosolic phospholipase A2α impairs an early step of coronavirus replication in cell culture. J Virol. 2018;92:e01463‐01417. 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wolff G, Melia CE, Snijder EJ, Bárcena M. Double‐membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28(12):1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Deevska GM, Nikolova‐Karakashian MN. The expanding role of sphingolipids in lipid droplet biogenesis. Biochimica Biophysica Acta. 2017;1862:1155‐1165. [DOI] [PubMed] [Google Scholar]
- 44. da Silva Gomes Dias S, Soares VC, Ferreira AC, et al. Lipid droplets fuel SARS‐CoV‐2 replication and production of inflammatory mediators. PLoS Pathog, 2020;16:e1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pagliari F, Marafioti MG, Genard G, et al. ssRNA virus and host lipid rearrangements: is there a role for lipid droplets in SARS‐CoV‐2 infection? Front Mol Biosci. 2020;7:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tizaoui C. Ozone: a potential oxidant for COVID‐19 virus (SARS‐CoV‐2). Ozone Sci Eng. 2020;42:378‐385. [Google Scholar]
- 47. Briggs JAG, Wilk T, Fuller SD. Do lipid rafts mediate virus assembly and pseudotyping? J general Virol. 2003;84:757‐768. 10.1099/vir.0.18779-0. [DOI] [PubMed] [Google Scholar]
- 48. Shahmohamadnejad S, Nabavi SF, Habtemariam S, et al. May we target double membrane vesicles and oxysterol‐binding protein to combat SARS‐CoV‐2 infection? Cell Biol Int. 2020;44(9):1770–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shulla A, Randall G. (+) RNA virus replication compartments: a safe home for (most) viral replication. Curr Opin Microbiol. 2016;32:82‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Abu‐Farha M, Thanaraj TA, Qaddoumi MG, Hashem A, Abubaker J, Al‐Mulla F. The role of lipid metabolism in COVID‐19 virus infection and as a drug target. Int J Mol Sci. 2020;21:3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baglivo M, Baronio M, Natalini G, et al. Natural small molecules as inhibitors of coronavirus lipid‐dependent attachment to host cells: a possible strategy for reducing SARS‐COV‐2 infectivity? Acta Bio Medica Atenei Parm. 2020;91:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fenyvesi É, Szemán J, Csabai K, Malanga M, Szente L. Methyl‐beta‐cyclodextrins: the role of number and types of substituents in solubilizing power. J Pharm Sci. 2014;103:1443‐1452. [DOI] [PubMed] [Google Scholar]
- 53. Cagno V, Tintori C, Civra A, et al. Novel broad spectrum virucidal molecules against enveloped viruses. PloS One. 2018:13:e0208333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Verma S. HIV: a raft‐targeting approach for prevention and therapy using plant‐derived compounds. Curr drug targets. 2009;10:51‐59. [DOI] [PubMed] [Google Scholar]
- 55. Moreau RA, Nyström L, Whitaker BD, et al. Phytosterols and their derivatives: structural diversity, distribution, metabolism, analysis, and health‐promoting uses. Prog Lipid Res. 2018;70:35‐61. 10.1016/j.plipres.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 56. Calandria J, Bhattacharjee S, Kautzmann M‐AI, et al. Elovanoids downregulate canonical SARS‐CoV‐2 cell‐entry mediators and enhance protective signaling in human alveolar cells. Research Square. 2020;1‐13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Carpinteiro A, Edwards MJ, Hoffmann M, et al. Inhibition of acid sphingomyelinase blocks infection with SARS‐CoV‐2. Cell Rep Med. 2020;1,100142:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jennings BC, Nadolski MJ, Ling Y, et al. 2‐Bromopalmitate and 2‐(2‐hydroxy‐5‐nitro‐benzylidene)‐benzo [b] thiophen‐3‐one inhibit DHHC‐mediated palmitoylation in vitro. J Lip Res. 2009;50:233‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silvas JA, Jureka AS, Nicolini AM, Chvatal SA, Basler CF. Inhibitors of VPS34 and lipid metabolism suppress SARS‐CoV‐2 replication. Biorxiv. 2020. 10.1101/2020.07.18.210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dias, SSDSG , Soares, VC , Ferreira, AC , et al . Lipid droplets fuel SARS‐CoV‐2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16(12):e1009127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang J‐J, Li J‐G, Qi W, et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metabol. 2011;13:44‐56. [DOI] [PubMed] [Google Scholar]
- 62. Hullin‐Matsuda F, Tomishige N, Sakai S, et al. Limonoid compounds inhibit sphingomyelin biosynthesis by preventing CERT protein‐dependent extraction of ceramides from the endoplasmic reticulum. J Biol Chem. 2012;287:24397‐24411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ďuriš A, Tomas W, Moravčíková D, et al. Expedient and practical synthesis of CERT‐dependent ceramide trafficking inhibitor HPA‐12 and its analogues. Org Lett. 2011;13:1642‐1645. [DOI] [PubMed] [Google Scholar]
- 64. Yan B, Chu H, Yang D, et al. Characterization of the lipidomic profile of human coronavirus‐infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11:73. 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thomas T, Stefanoni D, Reisz JA, et al. COVID‐19 infection alters kynurenine and fatty acid metabolism, correlating with IL‐6 levels and renal status. JCI Insight. 2020;5(14):e140327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fuller N, Rand R. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys J. 2001;81:243‐254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Marangoni F, Agostoni C, Borghi C, et al. Dietary linoleic acid and human health: focus on cardiovascular and cardiometabolic effects. Atherosclerosis. 2020;292:90‐98. 10.1016/j.atherosclerosis.2019.11.018. [DOI] [PubMed] [Google Scholar]
- 68. Deng Y, Almsherqi ZA, Ng MM, Kohlwein SD. Do viruses subvert cholesterol homeostasis to induce host cubic membranes? Trends Cell Biol. 2010;20:371‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma J, Ting N‐C, Kuo M‐L. LPS elevates ceramide levels through regulating ORMDL3 expression in macrophages and acute respiratory distress syndrome model. J Immunol, 2020;204(1 Supplement):144 [Google Scholar]
- 70. Li Y, Zhang Y, Dai M, et al. Changes in lipid metabolism in patients with severe COVID‐19. Research Square. 2020;1‐25. [Google Scholar]
- 71. Heaton N, Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368‐375. 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Katsiki N, Banach M, Mikhailidis DP. Lipid‐lowering therapy and renin‐angiotensin‐aldosterone system inhibitors in the era of the COVID‐19 pandemic. Arch Med Sci AMS. 2020;16:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Álvarez E, DeDiego ML, Nieto‐Torres JL, Jiménez‐Guardeño JM, Marcos‐Villar L, Enjuanes L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non‐structural protein 3 and is ubiquitinated. Virology. 2010;402:281‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martín‐Acebes MA, Vázquez‐Calvo Á, Saiz J‐C, Sobrino F. Lipid involvement in viral infections: present and future perspectives for the design of antiviral strategies. Lipid Metab. 2012:291‐322. [Google Scholar]
- 75. Rathi AK, Syed R, Shin H‐S, Patel RV. Piperazine derivatives for therapeutic use: a patent review (2010‐present). Expert Opin Ther Pat. 2016;26:777‐797. [DOI] [PubMed] [Google Scholar]
- 76. Sorokin AV, Karathanasis SK, Yang ZH, Freeman L, Kotani K, Remaley AT. COVID‐19—associated dyslipidemia: implications for mechanism of impaired resolution and novel therapeutic approaches. FASEB J. 2020;34:9843‐9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cottam EM, Maier HJ, Manifava M, et al. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335‐1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013‐1019. [DOI] [PubMed] [Google Scholar]
- 79. Yan B, Zou Z, Chu H, et al. Lipidomic profiling reveals significant perturbations of intracellular lipid homeostasis in enterovirus‐infected cells. Int J Mol Sci. 2019;20:5952. 10.3390/ijms20235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoxha M. What about COVID‐19 and arachidonic acid pathway? Eur J Clin Pharmacol. 2020;76:1501‐1504. 10.1007/s00228-020-02941-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hammock BD, Wang W, Gilligan MM, Panigrahy D. Eicosanoids: the overlooked storm in COVID‐19? Am J Pathol. 2020;190:1782‐1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Das UN. Can bioactive lipids inactivate coronavirus (COVID‐19)? Arch Med Res 2020;51:282‐286. 10.1016/j.arcmed.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Das UN. Bioactive lipids in COVID‐19‐further evidence. Arch Med Res. 2020;S0188‐4409:31263‐31267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Toelzer C, Gupta K, Yadav SKN, et al. Free fatty acid binding pocket in the locked structure of SARS‐CoV‐2 spike protein. Science. 2020;370:725‐730. 10.1126/science.abd3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang L, Shen W, Kazachkov M, et al. Metabolic interactions between the Lands cycle and the Kennedy pathway of Glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell. 2012;24:4652‐4669. 10.1105/tpc.112.104604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schulze MB, Minihane AM, Saleh RNM, Risérus U. Intake and metabolism of omega‐3 and omega‐6 polyunsaturated fatty acids: nutritional implications for cardiometabolic diseases. Lancet Diab Endocrinol. 2020;8:915‐930. 10.1016/S2213-8587(20)30148-0. [DOI] [PubMed] [Google Scholar]
- 87. Vardavas C, Majchrzak D, Wagner K‐H, Elmadfa I, Kafatos A. Lipid concentrations of wild edible Greens in crete. Food Chem. 2006;99:822‐834. 10.1016/j.foodchem.2005.08.058. [DOI] [Google Scholar]
- 88. Ceccanti C, Landi M, Benvenuti S, Pardossi A, Guidi L. Mediterranean wild edible plants: weeds or "new functional crops. Mol. 2018;23:2299. 10.3390/molecules23092299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kiani M, Alahdadi I, Soltani E, Boelt B, Benakashani F. Variation of seed oil content, oil yield, and fatty acids profile in Iranian Nigella sativa L. landraces. Ind Crops Prod. 2020;149:112367. 10.1016/j.indcrop.2020.112367. [DOI] [Google Scholar]
- 90. Kursat M, Emre İ, Yilmaz O, Civelek S, Demir E, Turkoglu I. Phytochemical contents of five Artemisia species. Not Sci Biol. 2015;7:495‐499. 10.15835/nsb749683. [DOI] [Google Scholar]
- 91. Elferjani R, Soolanayakanahally R. Canola responses to drought, heat, and combined stress: shared and specific effects on carbon assimilation, seed yield, and oil composition. Front Plant Sci. 2018;9:1224. 10.3389/fpls.2018.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hashem HA, Hassanein RA, Bekheta MA, El‐Kady FA. Protective role of selenium in Canola (Brassica napus L.) plant subjected to salt stress. Egypt J Exp Biol. 2013;9:199‐211. [Google Scholar]
- 93. Soliman S, Faris ME, Ratemi Z, Halwani R. Switching host metabolism as an approach to dampen SARS‐CoV‐2 infection. Ann Nutr Metab. 2020;76:297–303. 10.1159/000510508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Edser C. SURFACTANTS VERSUS COVID‐19. Focus Surfact. 2020;2020:1‐2. [Google Scholar]
- 95. Iddir M, Brito A, Dingeo G, et al. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: considerations during the COVID‐19 crisis. Nutrients. 2020;12:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Flock M, Rogers C, Prabhu K, et al. Immunometabolic role of long‐chain omega‐3 fatty acids in obesity‐induced inflammation. Diabetes Metab Res Rev. 2013;29. 10.1002/dmrr.2414. [DOI] [PubMed] [Google Scholar]
- 97. Ghaffari S, Roshanravan N, Tutunchi H, Ostadrahimi A, Pouraghaei M, Kafil B. Oleoylethanolamide, a bioactive lipid amide, as a promising treatment strategy for coronavirus/covid‐19. Arch Med Res. 2020;51(5):464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within this article.


